Abstract
Components of multiprotein complexes are routinely determined by using proteomic approaches. However, this information lacks functional content except when new complex members are identified. To analyze quantitatively the abundance of proteins in human Mediator we used normalized spectral abundance factors generated from shotgun proteomics data sets. With this approach we define a common core of mammalian Mediator subunits shared by alternative forms that variably associate with the kinase module and RNA polymerase (pol) II. Although each version of affinity-purified Mediator contained some kinase module and RNA pol II, Mediator purified through F-Med26 contained the most RNA pol II and the least kinase module as demonstrated by the normalized spectral abundance factor approach. The distinct forms of Mediator were functionally characterized by using a transcriptional activity assay, where F-Med26 Mediator/RNA pol II was the most active. This method of protein complex visualization has important implications for the analysis of multiprotein complexes and assembly of protein interaction networks.
Keywords: multidimensional protein identification technology, proteomics, spectrum counting, mass spectrometry
Since its discovery in yeast (1, 2) and subsequent isolation and initial characterizations in human cells (3), the transcriptional coactivator complex Mediator has been the subject of numerous studies to characterize both its composition and function. Researchers have used a combination of immunoprecipitation (3–14), ion exchange/size-exclusion chromatography (8, 10, 15–19), glycerol gradient (6, 7, 12, 13, 19, 20), SDS/PAGE/silver stain/Western blot analysis (6, 7, 9, 10, 15, 18, 21), MS (9, 16, 19, 21, 22), and ChIP (21, 23) to determine the composition of Mediator complexes. Varying in size and subunit composition, the larger complexes (thyroid hormone receptor-associated protein complex, SRB/Med-containing cofactor complex, negative regulator of activated transcription, vitamin D receptor interacting protein complex, and activator recruited cofactor) ranged from 1 to 2 MDa and were composed of ≈30 subunits (3, 9–16, 21, 22, 24), whereas the smaller complexes (cofactor required for Sp1, positive cofactor 2, and positive cofactor 4) ranged from ≈500 to 700 kDa and were composed of ≈9–17 subunits (5–7, 17–20, 25, 26). The main shared difference between large and small complexes appears to be the presence or absence of the kinase module. Furthermore, different complexes possessed different levels of basal transcription, which could be accounted for by several possibilities, including purification method (high ionic strength may have washed away necessary proteins), the adding back of RNA polymerase (pol) II and basal transcription factors, and testing only for activated transcription.
Although these studies form the foundation for our present understanding of Mediator, the relationship between different forms of Mediator was unclear and the composition of each form was unclear. The first attempt at standardizing Mediator complex purifications was carried out by using FLAG-tagged subunits of Mediator analyzed by multidimensional protein identification technology (MudPIT) (27). This study definitively established a set of consensus Mediator subunits, determined that each of these subunits was present in almost every preparation, identified new subunits (Cdk8L and Med13L), and demonstrated that RNA pol II subunits were present in all preparations (27). Nevertheless, our previous study provided no quantitative information, and the relative subunit stoichiometries and abundances were not determined.
We therefore established a strategy to use MS as a means of quantifying the relative abundance of all proteins present in Mediator complexes. Although this study applies the same analytical means used to establish Mediator composition (27, 28), it implements methods of data analysis that account for protein size and variability between runs to normalize relative protein abundance between samples (termed NSAF values for normalized spectral abundance factor) (29). NSAF is based on spectral counting, which has been demonstrated to be an effective quantitative proteomics approach (30–35). In spectral counting, larger proteins would be expected to generate more peptides and therefore more spectral counts than smaller proteins. Consequently, the number of spectral counts for each protein must be divided by the mass (33) or protein length, which defines the spectral abundance factor (SAF) (31). However, to accurately account for run to run variation, individual SAF values must be normalized to one by dividing by the sum of all SAFs for proteins in the complex, resulting in the NSAF value (29). In a manner similar to microarray data analysis, NSAFs values are hence standardized across distinct preparations of Mediator to allow direct comparison between individual runs.
The current quantitative analysis points out significant and functionally relevant differences between Mediator preparations purified through different FLAG-tagged subunits: a common core of mammalian Mediator subunits is shared by alternative forms that variably associate with the kinase module and RNA pol II. Importantly, to substantiate this methodology as an accurate and reliable method, not only to determine relative protein abundance, but also to predict complex activity, the MS data were verified by several orthogonal methods. We carried out an AQUA experiment (36) using 15N Med9, and relative protein abundance estimated by NSAF was corroborated by Western blot analysis to multiple proteins in different Mediator modules and RNA pol II, as well as by in vitro transcription assays using a purified basal transcription system (37). As opposed to previous studies using spectral counting methods, in the current article we demonstrate and verify the ability of the NSAF approach to determine relative protein abundance in statistically supported structure/function analysis of multiprotein complexes.
Results
Mediator Core Subunits.
Based on a combination of biochemical, genetic, and electron microscopy data, largely performed with the Mediator complex from Saccharomyces cerevisiae, Mediator subunits have been assigned to several subassemblies, called the head, middle, tail, and kinase modules (38). Mediator interacts with RNA pol II to form a larger complex that has been referred to as the RNA pol II holoenzyme (1, 4, 39). Because Mediator proteins have been assigned to particular modules based largely on studies of the S. cerevisiae complex (38), and because metazoan Mediator includes additional subunits not found in yeast (27), there are six Mediator subunits that have not yet been localized to the head, middle, or tail regions.
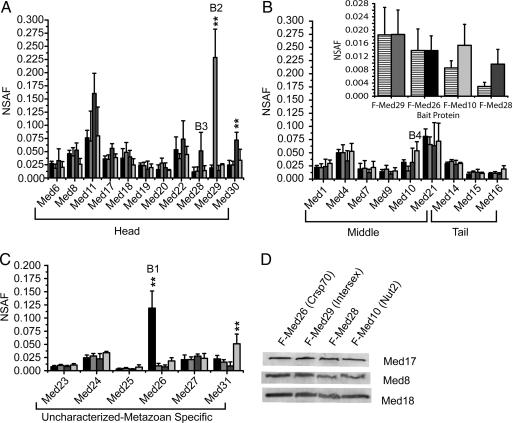
NSAF values measured for head, middle, tail, and unassigned subunits in each of the four Mediator preparations are plotted in Fig. 1 A–C. Regardless of the bait used to purify the complexes, each core mediator protein appears to be present at largely the same levels in each preparation. Statistically significant exceptions included the baits Med26 and Med29, which were recovered at much higher levels in their own pull-downs, and Med30 in the F-Med28 pull-downs and Med31 in the F-Med10 preparations [Fig. 1 and supporting information (SI)]. To validate the NSAF approach we carried out an AQUA experiment (36) by adding various absolute amounts of recombinant 15N Med9 purified from Escherichia coli into each of the four Mediator preparations analyzed (Fig. 1B Inset). NSAF values for 15N Med9 showed a linear response over an ≈6-fold range of abundance, whereas NSAF values for endogenous Med9 in each of the four Mediator preparations were not significantly different. To validate further the NSAF measurements we performed quantitative Western blotting using antibodies directed against Med17, Med8, and Med18. Both NSAF values (Fig. 1 A–C) and Western blotting (Fig. 1D) demonstrate that the abundance of each of these components does not vary even when different FLAG-tagged baits are used to purify Mediator. However, various levels are observed for different proteins. For example, Med21 is overall the most abundant protein component in the middle module, which suggests that it is present at more than one copy per Mediator complex or exists outside of Mediator, either on its own or as part of another protein complex present in the preparation. On the other end of the spectrum, Med25 is the least abundant of all core Mediator components regardless of the bait used to purify Mediator (Fig. 1B). This NSAF result is in excellent agreement with previous studies that demonstrated that Med25, which interacts with VP16, is easily dissociated from Mediator and is present at substoichiometric levels (11, 40).
Fig. 1.
NSAF analysis of human Mediator core components. NSAF values for Mediator components are plotted for each of the preparations purified through F-Med26, which is bait 1 (B1), F-Med29 (B2), F-Med28 (B3), and F-Med10 (B4). (A) Head components of Mediator. (B) Middle and tail components. (C) Unlocalized components. Each column is the average of four independent analyses, and error bars represent one SD of the data. NSAFs that were statistically different (P < 0.05) from NSAF values measured in the other three preparations (SI) are designated with a vertical ∗∗. (B Inset) The endogenous Med9 NSAF data (solid bars) compared with a 15N Med9 standard curve (striped bars) where the absolute amount of purified 15N Med9 added to F-Med29, F-Med26, F-Med10, and F-Med28 preparations was 0.128 μg, 0.113 μg, 0.042 μg, and 0.018 μg, respectively. (D) Quantitative Western blotting of the human Mediator components Med17, Med8, and Med18 from each of the affinity-purified Mediator preparations.
Mediator Association with Kinase Module and RNA Pol II.
The most striking differences between Mediator complexes purified through different baits were in the relative amounts of kinase module (Fig. 2) and RNA pol II (Fig. 3) present in each preparation. The kinase module components Cdk8L, Med12, Med13, and Med13L were all significantly decreased in Mediator purified through F-Med26 compared with Mediator purified through F-Med10, F-Med28, or F-Med29 (Fig. 2A and SI). Although NSAF values measured for the other two components of the kinase module, Cdk8 and Cyclin C, did not achieve rigorous statistical significance, quantitative Western blotting revealed that indeed Cdk8 was not detected and Cyclin C was only weakly detected in F-Med26 Mediator (Fig. 2B). On the other hand, the Mediator preparations purified through F-Med10, F-Med28, or F-Med29 all contained similar, and substantially higher, levels of kinase module components as determined both by the NSAF value (Fig. 2A), quantitative Western blotting (Fig. 2B), and statistical analysis (see SI). Next, Mediator purified through different FLAG-tagged subunits also contained various amounts of RNA pol II (Fig. 3). As shown in Fig. 3A, comparison of NSAF values for RNA pol II subunits indicates that similar amounts of RNA pol II were recovered in F-Med10 and F-Med26 Mediator, whereas Mediator purified through F-Med28 and F-Med29 contained statistically significantly less RNA pol II. Again, this was validated with quantitative Western blotting against the RNA pol II subunits Rpb1 and Rpb9, where F-Med26 Mediator had the most Rpb1 and Rpb9 followed by F-Med10, with F-Med29 and F-Med28 preparations having the least RNA pol II subunits (Fig. 3B).
Fig. 2.
NSAF analysis of the kinase module of human Mediator. (A) NSAF values for kinase module components are plotted for each of the baits used to affinity-purify Mediator. Listed on the x axis is the bait protein. Above each bait protein from left to right are Cdk8, Cdk8L, cyclin C, Med12, Med13, and Med13L. Each column is the average of four independent analyses, and error bars represent one SD of the data. Because of sequence homology NSAF values for Cdk8/Cdk8L are likely to be uniformly inflated because shared peptides were used to calculate NSAFs. Statistically significant (P < 0.05) average ± SD NSAF values for kinase module components in F-Med26 Mediator that were lower than NSAF values measured from the other three preparations are shown with vertical ∗∗ (see SI). (B) Quantitative Western blotting of the kinase module components Cdk8 and Cyclin C from each of the affinity-purified Mediator preparations. The Med17 Western blot analysis is the same as shown in Fig. 1 and is provided as a frame of reference.
Fig. 3.
NSAF analysis of RNA pol II components copurifying with human Mediator. (A) NSAF values for RNA pol II components are plotted for each of the baits used to purify Mediator. From left to right above each bait is the average NSAF value (with error bars) of Rpb1 to Rpb12 in numerical order. RNA pol II components in F-Med26 and F-Med10 purifications that were statistically different (P < 0.05) from all other preparations are represented by a vertical ∗∗, and RNA pol II components in F-Med26 and F-Med10 preparations that were only significantly different from F-Med28 and F-Med29 preparations are represented by ∗. Rpb7 in F-Med10 was significantly different from F-Med26 and F-Med29 but not F-Med28, and this is represented by + (see SI). (B) Quantitative Western blotting of the RNA pol II components Rpb1 and Rpb9 from each of the affinity-purified Mediator preparations. The Med17 Western blot analysis is the same as shown in Fig. 1 and is provided as a frame of reference. (C) Basal transcription performed on a naked DNA template with and without added RNA pol II. The amount of added eluate was normalized to Med17 subunit as determined from Western blot analysis. Transcription from the samples without added RNA pol II could only come from RNA pol II present in the Mediator preparations.
The kinase module is believed to function as an inhibitor of transcription while Mediator associated with RNA pol II is believed to be active. Based on NSAF values, statistical analysis, and quantitative Western blotting, F-Med26 and F-Med10 Mediator had enriched levels of RNA pol II, whereas F-Med26 Mediator preparations were depleted of kinase module compared with F-Med10 (Fig. 2). F-Med28 and F-Med29 Mediator had similar levels of kinase module compared with F-Med10, but diminished levels of RNA pol II components. These results predict that F-Med26 Mediator should have increased activity when compared with the other preparations. As an independent measurement of functional RNA pol II present in our preparations of Mediator we performed in vitro transcription assays using a highly purified, reconstituted transcription system. Under the reaction conditions used in this assay, none of the Mediator preparations affected basal transcription from the Adenovirus 2 major late promoter when reactions contained saturating amounts of TBP, TFIIB, TFIIE, TFIIF, TFIIH, and purified pol II (Fig. 3C, lanes 2–6). When purified pol II was omitted, however, transcription was highest in the presence of Mediator purified through F-Med26 followed by F-Med10 (Fig. 3C, lanes 7 and 8), consistent with NSAF data indicating that these preparations contained the highest amounts of RNA pol II, while very little transcription was observed in reactions with F-Med28 or F-Med29 Mediator. A possible explanation for the difference in activity between F-Med26 and F-Med10 Mediators is the F-Med26 population contains slightly more RNA pol components coupled with dramatically less kinase module compared with the F-Med10 preparations, as supported by the kinase module and RNA pol II NSAF analysis and quantitative Western blotting. Furthermore, of the five RNA pol II components statistically different between F-Med26 and F-Med10 preparations, four were increased in abundance in F-Med26 Mediator (Rpb1, 3, 7, and 9), while only Rpb5 was increased in abundance in F-Med10 preparations (Fig. 3 and SI).
Discussion
In this report we have demonstrated a statistically robust approach for structure function analyses of multiprotein complexes using label-free quantitative MudPIT and mammalian Mediator. Conclusions drawn from our quantitative analyses of MudPIT data using NSAFs were validated by an AQUA experiment (36) using 15N-labeled Med9, quantitative Western blotting, and transcription assays. Hence, with the use of NSAF, MudPIT can be a powerful tool not only for protein identification but also for obtaining information about the abundance of proteins present in purified multiprotein complexes. The most striking aspect of the data is that the subunit that is FLAG-tagged has a sizable impact on the makeup of bulk Mediator that is purified. Based on the results of our NSAF analyses we conclude that there is little significant difference in the relative stoichiometry of most Mediator subunit complexes purified through any of four different baits, Med26, Med10, Med28, and Med29. These findings define a common core of Mediator subunits that are likely present in all or most Mediator complexes found in cells.
We nevertheless find significant differences in the apparent compositions of complexes purified through different epitope-tagged subunits, arguing for the existence of multiple forms of Mediator complex that share a common core of subunits. Based on the MS data from this study, Mediator purified through these FLAG-tagged subunits falls into three main complexes (Fig. 4). On one end of the spectrum is the F-Med26 Mediator, which resembles the previously described cofactor required for Sp1 (5–7, 19, 20, 26) and the positive cofactor 2 (17, 18) purifications that have been reported to be highly active in support of activated transcription. This complex appears to be almost completely lacking in kinase module, with a high percentage of associated RNA pol II (Fig. 4A). It is important to note that a small amount of kinase module subunits were isolated in association with F-Med26; hence, although the majority of Mediator that contain Med26 lack kinase module, it is possible for both Med26 and the kinase module to interact simultaneously with the core complex.
Fig. 4.
Bait-dependent distribution of Mediator, kinase, and RNA pol modules. (A) The NSAF values for each protein in each of the three overall modules (Mediator, kinase, and RNA pol II) was summed as percentage of whole determined for each Mediator bait used. Filled squares, open circles, and filled circles represent the percent of the complex that was Mediator, RNA pol II, and kinase module, respectively. (B) A proposed model for the states of Mediator in a cellular environment where Mediator with the kinase module is transcriptionally inactive and Mediator with the RNA pol II module is transcriptionally active. An important question is whether the pol and kinase modules can interact with the same Mediator molecule or are mutually exclusive.
On the other end of the spectrum are the F-Med28/Med29 complexes that resemble the thyroid hormone receptor-associated protein complex (3, 14, 21, 22), SRB/Med-containing cofactor complex (9), negative regulator of activated transcription (16), vitamin D receptor interacting protein complex (12, 13), SRB (8, 15), and activator recruited cofactor-L (10, 11) purifications described previously and likely represent the majority of the transcriptionally repressed Mediator present in the cells. Based on NSAF analyses of MudPIT data and assays for RNA pol II activity we found that Mediator purified through F-Med28 and F-Med29 is apparently associated with less RNA pol II than Mediator purified through F-Med10 (Fig. 4A). Association of F-Med28 and F-Med29 with Mediator clearly does not prevent binding of RNA pol II (or vice versa), since Med28 and Med29 do not appear to be depleted in Mediator purified through F-Med26, which is associated with a large amount of RNA pol II. Association of the kinase module with Mediator has been proposed to block binding of RNA pol II; however, Mediator purified through F-Med28 or F-Med29 does not contain substantially more kinase module than Mediator purified through F-Med10.
These F-Med28/Med29 Mediator complexes incorporate a relatively high percentage of kinase module, with little associated RNA pol II (Fig. 4A). The F-Med10 preparation appears to offer a multifaceted picture, both in terms of complex composition and functionality. It is possible that this preparation either is composed of (i) one large complex incorporating Mediator with kinase module and RNA pol II or (ii) several smaller complexes, Mediator/pol II, Mediator/kinase module and Mediator alone as well as the larger complex described above (Fig. 4B). Diverse functionality can be hypothesized for the larger complex, which is not mutually exclusive and cannot be ruled out by the present literature. The larger complex (Mediator/kinase/pol II) can potentially represent both an active and repressive form of Mediator, governed by small changes in subunit composition that are promoter specific (21), or changes in overall three dimensional structure when bound to different activators (26). Another possibility is that this larger complex represents a transition from active to repressive Mediator (or vice versa). There is support in the literature for repressed Mediator, after stimuli from an activator, to lose the kinase module, incorporate Med26 and bind RNA pol II in a stepwise fashion, before transcription initiation (23). It is possible that a transition state exists that incorporates all of these proteins (4). Further fractionation of the F-Med10 preparation (along with MudPIT analysis and transcription assays) has the potential to clearly delineate overall Mediator composition and the functionality associated to each complex.
From an analytical standpoint, the method of protein complex characterization described in this article has important implications. MudPIT is routinely used to identify proteins in complexes (27), but we demonstrate a statistically robust approach for quantitative analysis of multiprotein complexes that yields structure/function insights. By replicate analysis of multiprotein Mediator complexes using distinct Mediator proteins as baits to analyze the complex, the NSAF approach revealed unique protein abundances and complex structures. Available antibodies validated these results and the functional significance was demonstrated with a transcription assay. The NSAF approach for relative abundance determination has excellent potential for statistically robust structure function analyses of multiprotein complexes where existing methods are unable to provide comprehensive information and a full suite of antibodies is not available.
Materials and Methods
Recombinant 15N Med9 Purification.
Recombinant Med9 cloned into a pET vector in E. coli strain BL21(DE3) (Novagen, Madison, WI) and expressed as a 6×His fusion protein (Novagen) was grown in Bioexpress cell growth media (U-15N, 98%) (Cambridge Isotope Laboratories, Andover, MA). Recombinant 15N Med9 was purified from E. coli extracts first by Ni-NTA agarose chromatography (Qiagen, Valencia, CA), then by ion exchange chromatography on a TSKgel DEAE-5PW column (Tosoh Biosciences, Tokyo, Japan) using a 25-min 0.005–0.25 M sodium chloride gradient generated by a Beckman Coulter System Gold HPLC (Beckman Coulter, Fullerton, CA). Protein concentration was determined by protein assay (Bio-Rad, Hercules, CA), and 5 μg of purified 15N Med9 was trypsin-digested (Roche, Mannheim, Germany) and analyzed on a LCQ Deca XP Plus mass spectrometer (Thermo Electron, Waltham, MA) to determine purity based on percentage of Med9 spectral counts (87%).
Mediator Preparations and MudPIT Analysis.
Nuclear extracts were prepared according to the method of Dignam et al. (41) from 6 × 109 HeLa cells stably expressing N-terminally FLAG-tagged Med26 (formerly Crsp70), Med29 (formerly Intersex), Med28, or Med10 (formerly Nut2) subunits. Nuclear extracts were subjected to anti-FLAG agarose immunoaffinity chromatography (Sigma, St. Louis, MO) as described in ref. 27 and eluted with FLAG peptide (Sigma). Four independent preparations of Mediator complex were generated from each FLAG-tagged cell line. Recombinant 15N Med9 was added to all four replicates of all four FLAG-tagged preparations in various amounts as follows: 0.113 μg added to F-Med26, 0.128 μg added to F-Med29, 0.018 μg added to F-Med28, and 0.042 μg added to F-Med10 Mediator.
Each sample was digested with trypsin (Roche) and analyzed by MudPIT as described previously (28, 42) on an LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). Variations to standard MudPIT approaches where mixtures were resolved by an eight-step chromatographic program consisting of 5% buffer C (500 mM ammonium acetate, 5% acetonitrile, and 0.1% formic acid), 15% buffer C, 30% buffer C, 50% buffer C, 50% buffer C, 70% buffer C, and two successive steps using 100% buffer C as the salt bumps within standard MudPIT chromatographic programs (42).
Data sets were searched by using SEQUEST against a database of 56,838 protein sequences, combining 28,242 Homo sapiens proteins (from the National Center for Biotechnology Information, February 17, 2005), 177 common contaminants like keratin and Igs, and their corresponding shuffled sequences (i.e., each H. sapiens and contaminant sequence was randomized maintaining the same amino acid composition and length). SEQUEST was run to search for methionine oxidation and with no enzyme specificity, an approach that has been demonstrated to be superior for minimizing false positive identifications (43). Tandem MS data sets were searched twice to match spectra to peptides generated from endogenous 14N Mediator protein and to 15N peptides derived from the spiked recombinant Med19 protein (29, 35). Protein lists were generated by using DTASelect (44) based on the following filtering criteria: each peptide had to be at least 7 aa long and fully tryptic, with a ΔCn of at least 0.08, and minimum Xcorr of 1.5 for +1, 1.9 for +2, and 2.6 for +3 charged precursor ions (45). Ambiguous peptides and redundant proteins were removed by using DTASelect (44). By using the parameters described in the methods, the average peptide false positive rates according to the equation used by Elias et al. (45) were 0.85% for Med10, 0.80% for Med26, 0.5% for Med28, and 1.3% for Med29. The peptide identifications from a representative run of each analysis are provided in the SI.
Spectral count normalization was applied to proteins belonging to the Mediator or RNA pol II complexes to estimate the relative levels of each subunit within the complexes. Here, for each protein k involved in Mediator/pol II, we calculated a SAF normalized against the whole protein complex (NSAF) as follows:
 |
in which the total number of tandem MS spectra matching peptides from protein k (SpC) was divided by the protein's length (L), then divided by the sum of SpC/L for all N proteins in Mediator (32 subunits) and pol II (12 subunits).
Western Blot Analyses.
To compare levels of individual subunits present in each sample, Western blot analyses were performed and normalized to the level of Med17, because MS data demonstrated that this subunit should be of similar abundance in each preparation. Once the amount of individual Mediator preparation needed to produce equal blotting of Med17 was determined, all Western blot analyses were produced by using the same volumes. Proteins were transferred onto poly(vinylidene difluoride) and blotted by using primary antibodies against Med17 and Med18 (Cocalico Biologicals, Reamstown, PA), Med8 (46), Cdk8 (Santa Cruz Biotechnology, Santa Cruz, CA), cyclin C (Neomarkers, Fremont, CA), Rpb1 and Rpb9 (Protein One, Bethesda MD), and secondary antibodies against mouse or rabbit IgG (GE Healthcare, Piscataway, NJ). Blots were developed with the ECL Plus Western Blotting Detection System (GE Healthcare) and imaged by using a Typhoon 9400 (GE Healthcare).
Transcription Assays.
Transcription reactions were performed as described (37) with the EcoR1 to NdeI fragment of pDN-AdML as template purified RNA pol II from calf thymus (a kind gift of Avi Gnatt, University of Maryland School of Medicine, Baltimore, MD) and TFIIH from rat liver, and recombinant TBP, TFIIB, TFIIE, and TFIIF. Template, RNA pol II, and transcription factors were incubated at 28°C for 30 min to allow formation, and transcription was initiated by addition of 50 μM ATP, UTP, and GTP, 6 μM CTP, 7 μCi of [α-32P]CTP (600 Ci/mmol; GE HealthCare), and 8 mM MgCl2. After 30 min at 28°C, reactions were stopped by addition of EDTA to 0.25 mM, NaCl to 0.15 M, 0.02 mg of Proteinase K, and 0.04 mg of tRNA. Reaction products were subjected to electrophoresis on a 6% polyacrylamide (19:1 acrylamide:bisacrylamide) gel containing 6 M urea and 0.5× TBE and imaged with Typhoon 8600 (Molecular Dynamics).
Supplementary Material
Acknowledgments
This work was supported by the Stowers Institute for Medical Research and by National Institutes of Health Grant R37GM41628 (to R.C.C.).
Abbreviations
- SAF
spectral abundance factor
- NSAF
normalized SAF
- MudPIT
multidimensional protein identification technology
- pol
polymerase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606379103/DC1.
References
- 1.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Bjorklund S, Jiang YW, Kim YJ, Lane WS, Stillman DJ, Kornberg RD. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fondell JD, Ge H, Roeder RG. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik S, Roeder RG. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Genes Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taatjes DJ, Tjian R. Mol Cell. 2004;14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Taatjes DJ, Schneider-Poetsch T, Tjian R. Nat Struct Mol Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 8.Xiao H, Tao Y, Roeder RG. J Biol Chem. 1999;274:3937–3940. doi: 10.1074/jbc.274.7.3937. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 10.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 11.Yang F, DeBeaumont R, Zhou S, Naar AM. Proc Natl Acad Sci USA. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 13.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 17.Malik S, Baek HJ, Wu W, Roeder RG. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik S, Gu W, Wu W, Qin J, Roeder RG. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 19.Ryu S, Zhou S, Ladurner AG, Tjian R. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 20.Ryu S, Tjian R. Proc Natl Acad Sci USA. 1999;96:7137–7142. doi: 10.1073/pnas.96.13.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Baek HJ, Malik S, Qin J, Roeder RG. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 24.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 25.Fondell JD, Guermah M, Malik S, Roeder RG. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taatjes DJ, Naar AM, Andel F, III, Nogales E, Tjian R. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 27.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Washburn MP, Wolters D, Yates JR., III Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 29.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Sadygov RG, Yates JR., III Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 31.Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard M, Allaire PD, McPherson PS, Blondeau F. Mol Cell Proteomics. 2005;4:1145–1154. doi: 10.1074/mcp.M500043-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Blondeau F, Ritter B, Allaire PD, Wasiak S, Girard M, Hussain NK, Angers A, Legendre-Guillemin V, Roy L, Boismenu D, et al. Proc Natl Acad Sci USA. 2004;101:3833–3838. doi: 10.1073/pnas.0308186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Zybailov B, Coleman MK, Florens L, Washburn MP. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 36.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aso T, Conaway JW, Conaway RC. J Biol Chem. 1994;269:26575–26583. [PubMed] [Google Scholar]
- 38.Chadick JZ, Asturias FJ. Trends Biochem Sci. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Koleske AJ, Young RA. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 40.Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Florens L, Washburn MP. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 43.Xie H, Griffin TJ. J Proteome Res. 2006;5:1003–1009. doi: 10.1021/pr050472i. [DOI] [PubMed] [Google Scholar]
- 44.Tabb DL, McDonald WH, Yates JR., III J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elias JE, Haas W, Faherty BK, Gygi SP. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 46.Brower CS, Sato S, Tomomori-Sato C, Kamura T, Pause A, Stearman R, Klausner RD, Malik S, Lane WS, Sorokina I, et al. Proc Natl Acad Sci USA. 2002;99:10353–10358. doi: 10.1073/pnas.162424199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.