Abstract
Background
Pandemic influenza strains originate in nonhuman species. Pigs have an important role in interspecies transmission of the virus. We examined multiple swine-exposed human populations in the nation's number 1 swine-producing state for evidence of previous swine influenza virus infection.
Methods
We performed controlled, cross-sectional seroprevalence studies among 111 farmers, 97 meat processing workers, 65 veterinarians, and 79 control subjects using serum samples collected during the period of 2002–2004. Serum samples were tested using a hemagglutination inhibition assay against the following 6 influenza A virus isolates collected recently from pigs and humans: A/Swine/WI/238/97 (H1N1), A/Swine/WI/R33F/01 (H1N2), A/Swine/Minnesota/593/99 (H3N2), A/New Caledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), and A/Nanchang/933/95 (H3N2).
Results
Using multivariable proportional odds modeling, all 3 exposed study groups demonstrated markedly elevated titers against the H1N1 and H1N2 swine influenza virus isolates, compared with control subjects. Farmers had the strongest indication of exposure to swine H1N1 virus infection (odds ratio [OR], 35.3; 95% confidence interval [CI], 7.7–161.8), followed by veterinarians (OR, 17.8; 95% CI, 3.8–82.7), and meat processing workers (OR, 6.5; 95% CI, 1.4–29.5). Similarly, farmers had the highest odds for exposure to swine H1N2 virus (OR, 13.8; 95% CI, 5.4–35.4), followed by veterinarians (OR, 9.5; 95% CI, 3.6–24.6) and meat processing workers (OR, 2.7; 95% CI, 1.1–6.7).
Conclusions
Occupational exposure to pigs greatly increases workers' risk of swine influenza virus infection. Swine workers should be included in pandemic surveillance and in antiviral and immunization strategies.
Influenza A viruses infect a wide variety of species, including birds, pigs, humans, and horses. Pigs likely play important roles in interspecies transmission. Having receptors for both human and avian viruses, pigs may serve as a mixing vessel host for creation of novel reassortant progeny virus [1]. During the 1918 influenza pandemic, the US swine population experienced a widespread concomitant influenza epidemic, leading some to suggest a common viral cause and to question which species initiated the epidemics [2]. Although thought to be rare, cross-species infections with influenza A viruses have been documented from both pigs to humans [3, 4] and from humans to pigs [5, 6]. Recent reports indicate that the H5N1 avian influenza epidemic in Asia has involved pigs on farms in China [7, 8] and Indonesia [9]. Because pigs may play such important roles in human influenza epidemiology, we sought to serologically examine workers with occupational swine exposure, with a goal of identifying those at highest risk of a zoonotic influenza infection.
METHODS
Study subjects
Four adult populations were studied during the period of 2002–2004. Three populations consisted of persons who were exposed to swine (farmers, meat processing workers, and veterinarians or veterinary technicians, hereafter called “veterinarians”); the fourth population (control subjects) comprised volunteers associated with the University of Iowa (Iowa City) who had no occupational exposure to swine. All studies were conducted after institutional review board approval and with signed informed consent. Study participants completed occupational risk factor questionnaires.
The Iowa farmers belonged to a large rural cohort [10] and lived in a county that, in 1997, was ranked 91st nationally in swine production. The meat processing workers were employed by a pork-producing facility. The veterinarians were enrolled while attending a conference of the Iowa Veterinary Medical Association in the spring of 2004. Only veterinarians who reported a history of exposure to pigs were included in analyses. The meat processing workers, veterinarians, and control subjects were only permitted to participate if they had no immunocompromising conditions, were >18 years of age, and were not pregnant.
Laboratory methods
Serum samples were tested using a hemagglutination inhibition (HI) assay [11] against 6 isolates of recently circulating swine and human influenza A viruses, consisting of 3 each of the H1 and H3 subtypes and including A/Swine/WI/238/97 (H1N1), A/Swine/WI/R33F/01 (H1N2), A/Swine/Minnesota/593/99 (H3N2), A/New Caledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), and A/Nanchang/933/95 (H3N2). A/Swine/WI/238/97 (H1N1) is a wholly classic swine H1N1 virus, representative of the overall phylogenetic lineage of viruses that have circulated among pigs in the United States for at least 70 years [12]. A/Swine/WI/R33F/01 (H1N2) is a reassortant H1N2 swine virus representative (unpublished data) of the viruses that appeared among swine in the United States in 1999. These viruses have classic swine H1 HA, NP, M, and NS genes; human NA and PB1 genes; and avian PA and PB2 genes [13-16]. A/Swine/Minnesota/593/99 (H3N2) is representative of the triple-reassortant H3N2 viruses that appeared in the swine population in the United States beginning in 1998 [17-19]. These viruses also have a mixture of human (HA, NA, and PB1), classic swine (M, NP, and NS), and avian (PA and PB2) influenza virus genes.
The Centers for Disease Control and Prevention HI serologic protocol was followed. The 3 human antigen strains, swine H3N2, and swine H1N2 were grown in embryonated chicken eggs. Swine H1N1 was grown in Madin-Darby canine kidney cells. Serum samples were pretreated with receptor-destroying enzyme (1 part serum to 3 parts enzyme) from Vibrio cholerae overnight, and they were then hemadsorbed with guinea pig blood (for human strains) or turkey erythrocytes (for swine strains). HI titer results are reported as the reciprocal of the highest dilution of serum that inhibited virus-induced hemagglutination of a 0.5% (from guinea pigs) or 0.65% (from turkeys) solution of erythrocytes. To examine potential confounding through cross-reactivity, HI titers from control antisera were determined against the reference virus strains (e.g., antisera to swine H1 influenza virus was examined against human H1 virus).
Statistical methods
HI test results were first evaluated as dichotomous outcomes (HI titers of ≥ 1:40 were considered to be evidence of previous infection [20, 21]) and later as ordinal outcomes, with the goal of examining the entire distribution of antibody titer levels.
In dichotomous outcome examinations, the χ2 statistic or 2-sided Fisher's exact test, with 95% CIs calculated for ORs, were used. Geometric mean HI titers were also calculated for each virus strain and compared by risk factor using the Wilcoxon rank sum test, with normal approximation. Unconditional logistic regression was used to examine multiple independent variables for their association with the outcomes. Covariates with bivariate P values <.1 were considered for inclusion in all logistic regression models.
Proportional odds modeling [22] was used to examine the entire spectrum of serologic test results for associations with potential risk factors. Final multivariable models were designed using a saturated model and manual backwards elimination.
RESULTS
Demographic characteristics and seroprevalence
Farmers, meat processing workers, and veterinarians were older than control subjects and were more likely to be male and white (table 1). In dichotomous comparisons, farmers had much greater odds than did control subjects of being seropositive (titer, ≥ 1:40) against both the swine H1N1 virus (17.4% vs. 0%; OR, 22.9; 95% CI, 3.9–∞) and the swine H1N2 virus (20.7% vs. 1.3%; OR, 20.7; 95% CI, 2.5–172.1). Veterinarians also had increased odds of being seropositive for the swine H1N1 virus (10.9% vs. 0%; OR, 12.8; 95% CI, 1.9–∞) and the swine H1N2 virus (19.1% vs. 1.3%; OR, 18.1; 95% CI, 2.3–138.8). Meat processing workers had no increased odds of seropositivity against any swine virus (data not shown). All 3 exposure groups had a high prevalence of antibodies against the swine H3N2 isolate, but none of these prevalence values were significantly different than the controls' (data not shown). None of the exposure groups demonstrated increased odds of seropositivity against any of the human strains (data not shown). Geometric mean antibody titers were elevated for the swine H1N1 and swine H1N2 isolates for farmers and veterinarians and were found to differ from the titers for control subjects by the Wilcoxon rank sum test with normal approximation (table 2).
Table 1.
Demographic characteristics of the population in a study of zoonotic influenza virus infection.
| Characteristic | Farmersa (n = 111) | Meat processing workers (n = 97) | Veterinarians (n = 65) | Control subjects (n = 79) |
|---|---|---|---|---|
| Sex | ||||
| Male | 61 (55)b | 52 (54)b | 50 (77)b | 26 (33) |
| Female | 50 (45) | 45 (46) | 15 (23) | 53 (67) |
| Age, years | ||||
| <25 | 1 (1) | 15 (15) | 2 (3) | 12 (15) |
| 25–39 | 11 (10) | 32 (33) | 11 (17) | 41 (52) |
| >39 | 99 (89) | 50 (52) | 52 (80) | 26 (33) |
| Age, mean years | 47.0b | 39.5b | 48.4b | 35.3 |
| Race/ethnicity | ||||
| White | 111 (100) | 96 (99) | 62 (97) | 57 (72) |
| Black | 0 (0) | 1 (1) | 2 (3) | 18 (23) |
| Hispanic | 0 (0) | 5 (5) | 1 (1) | 2 (3) |
| Asian | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| American Indian | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| Pacific Islander | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
NOTE. Data are no. (%) of subjects, unless otherwise indicated.
Participants indicated they lived on a farm or worked on a farm for ≥10 each week in a leading swine–producing county. Individual swine exposure was not quantified.
Significantly different than controls at α = .05.
Table 2.
Geometric mean titers of antibodies against swine viruses.
| Geometric mean titer, by virus |
|||
|---|---|---|---|
| Study population | Swine H1N1 | Swine H1N2 | Swine H3N2 |
| Farmers | 10.3a | 12.5a | 22 |
| Meat processing workers | 6.2 | 6.8 | 54 |
| Veterinarians | 8.9a | 11.3a | 28.3 |
| Control subjects | 5.1 | 5.6 | 37.9 |
The geometric mean titers for farmers and veterinarians was found to differ from that for control subjects by the Wilcoxon rank sum analysis with normal approximation.
Bivariate analysis
In bivariate analysis (data not shown), among meat processing workers, none of the 38 possible risk factors were found to be strongly associated with elevated HI titers against any of the swine viral strains. Among veterinarians, we examined 28 potential occupational risk factors (data not shown) and found a positive association between self-reported receipt of the 1976 swine influenza vaccine and elevated HI titers against both swine H1N2 and swine H3N2 viruses. In addition, elevated HI titers against the swine H3N2 isolate were associated with having received a 2003–2004 influenza vaccination, as well as with the presence of others in the household. Among all of the groups (farmers, meat processing workers, veterinarians, and control subjects), elevated titers against swine H3N2 were associated with having elevated titers against human H3N2 strains, suggesting cross-reactivity. The association was stronger for the Nanchang than for the Panama strain. Multicollinearity was found between human H3N2 Nanchang seropositivity and history of receiving the 2003 influenza vaccine.
Logistic regression analysis
Because of multicollinearity between the Panama and Nanchang strains of human H3N2, only 1 strain was included in logistic regression modeling for swine H3N2 serology. The human H3N2 Nanchang isolate was chosen because it had a stronger positive association with the dependent variable. After adjusting for sex, age, and homologous human influenza strains, unconditional logistic regression revealed that farmers had elevated titers against both the swine H1N1 and H1N2 viruses (OR, 30.6 [95% CI, 4.3–∞] and 16.0 [95% CI, 1.9–776.4], respectively), veterinarians had elevated titers against the swine H1N2 virus (OR, 13.4; 95% CI, 1.5–670.5), and meat processing workers had elevated titers against the swine H3N2 virus (OR, 5.8; 95% CI, 1.7–23.0).
Proportional odds model
In the unadjusted proportional odds model, ORs for antibodies against swine H1N1 and swine H1N2 were elevated for all 3 exposure groups, compared with control subjects (table 3). Multivariable proportional odds modeling was then performed using data that were common to the 3 exposure groups and control subjects. Age (as a continuous variable) and sex were forced into the proportional odds models as potential confounders. Age was not found to be statistically significant in any of the models. However, male sex was associated with both swine H1N1 and swine H1N2 serologic outcomes. After controlling for confounders, ORs for elevated antibody titers against the swine H1N1 and swine H1N2 strains continued to be elevated in all 3 exposure groups, compared with control subjects. Farmers and veterinarians had particularly high odds of elevated titers against swine H1N1 virus (OR, 35.3 and 17.8, respectively). Farmers and veterinarians also had markedly increased odds of elevated titers against the recently circulating swine H1N2 virus (OR, 13.8 and 9.5, respectively). Meat processing workers had elevated ORs for both swine H1N1 and H1N2 viruses (6.5 and 2.7, respectively).
Table 3.
ORs for increased serologic response against swine influenza virus by hemagglutination inhibition assay, determined by proportional odds modeling.
| Virus |
||||||
|---|---|---|---|---|---|---|
| Swine H1N1 |
Swine H1N2 |
Swine H3N2 |
||||
| Population | Unadjusted OR (95% CI) | Adjusted OR (95% CI)a | Unadjusted OR (95% CI) | Adjusted OR (95% CI)b | Unadjusted OR (95% CI) | Adjusted OR (95% CI)c |
| Occupation | ||||||
| Farmer | 32.7 (7.6–139.8) | 35.3 (7.7–161.8) | 15.9 (6.8–37.1) | 13.8 (5.4–35.4) | 0.4 (0.3–0.7) | 0.4 (0.2–0.8) |
| Meat processing worker | 7.5 (1.7–33.6) | 6.5 (1.4–29.5) | 3 (1.2–7.3) | 2.7 (1.1–6.7) | 1.5 (0.9–2.6) | 1.5 (0.9–2.6) |
| Veterinarian | 23.1 (5.2–102.4) | 17.8 (3.8–82.7) | 13 (5.3–31.8) | 9.5 (3.6–24.6) | 0.6 (0.3–1.1) | 0.8 (0.4–1.5) |
| Age: years | 1 (1–1.1) | 1 (1–1) | 1 (1–1.1) | 1 (1–1) | 1 (1–1) | 1 (1–1) |
| Sex: male vs. female | 3.3 (1.9–5.5) | 2.9 (1.6–5.2) | 2.7 (1.8–4.2) | 2.3 (1.4–3.7) | 1.1 (0.7–1.5) | 1.2 (0.8–1.8) |
| Human influenza strain | ||||||
| H1N1 | 1.8 (1.1–3) | 2.8 (1.6–5) | 1.8 (1.1–2.8) | 2.7 (1.6–4.5) | … | … |
| H3N2 Panama | … | … | … | … | 5.6 (3.8–8.5) | … |
| H3N2 Nanchang | … | … | … | … | 44.6 (24.6–81) | 45.5 (25–83.1) |
| Vaccine historyd | ||||||
| Received 2003 flu vaccine | …e | …d | 2.1 (1–4.4) | …d | 3.6 (2–6.7) | …d |
| Received 1976 swine flu vaccine | …e | …d | 12.2 (3.6–41.7) | …d | 2.9 (0.9–9.4) | …d |
NOTE. To satisfy the proportional odds assumption, swine H1N1 and H1N2 titers of >1:80 were combined into 1 category, as were swine H3N2 titers of >1:320. Values shown in boldface are statistically significant.
P = .23, by score test.
P = .53, by score test.
P = .27, by score test.
Variable available for veterinarians and control subjects only.
Does not meet proportional odds assumption.
Cross-reactivity
Serologic cross-reactivity between swine and human viral strains was assessed through cross-testing of reference antisera (e.g., swine antisera against H3 swine viruses was tested by HI assay against human H3 viruses). Human H1N1 antisera showed some cross-reactivity against swine H1N1 and H1N2 viruses, with HI titers of 1:20 and 1:80 respectively (table 4). Human H3N2 antisera demonstrated high cross-reactivity to swine H3N2 virus, with HI titers of>1:640. These data supported our need to control for antibodies against human influenza virus that cross-react in swine influenza virus assays.
Table 4.
Hemagglutination inhibition titers of control serum samples to reference virus strains.
| Titer, by reference influenza A virus |
|||
|---|---|---|---|
| Antisera control | A/Swine/WI/238/97 (Swine H1N1 virus) | A/Swine/WI/R33F/01 (Swine H1N2 virus) | A/Swine/Minnesota/593/99 (Swine H3N2 virus) |
| A/New Caledonia/20/99 (human H1N1 virus)a | 1:20 | 1:80 | 1:40 |
| A/Swine/Indiana/1726188 (swine H1N1 virus)b | 1:80 | 1:80 | 1:10 |
| A/Panama/2007/99 (human H3N2 virus)a | 1:5 | 1:5 | >1:640 |
| A/Swine/Minnesota/593/99 (swine H3N2 virus)b | 1:5 | 1:5 | >1:640 |
| B/Sichuana | 1:5 | 1:5 | 1:10 |
| B/Hong Konga | 1:5 | 1:5 | 1:5 |
| Normal sheep seruma | 1:5 | 1:5 | 1:5 |
Sheep antisera.
Swine antisera.
Validation
Although we conducted considerable internal reliability testing of our HI assays, we also sought to validate our work. We sent a sample of 30 blinded serum samples representing all 4 study groups and various elevations in titers to an independent influenza virus research laboratory. We found 80% agreement (within 1 titer) for swine H1N2 and 94% agreement for swine H3N2. Because agreement was relatively low (43%) when swine H1N1 virus grown in embryonated eggs was used, we switched to a lower-passage H1N1 virus grown in Madin-Darby canine kidney cells and repeated our assays of the 30 serum samples. Because this interlaboratory agreement was much improved at 70%, we repeated all study HI assays using this lower-passage H1N1 virus grown in Madin-Darby canine kidney cells. Study HI swine H1N1 data reflect these repeated assays.
DISCUSSION
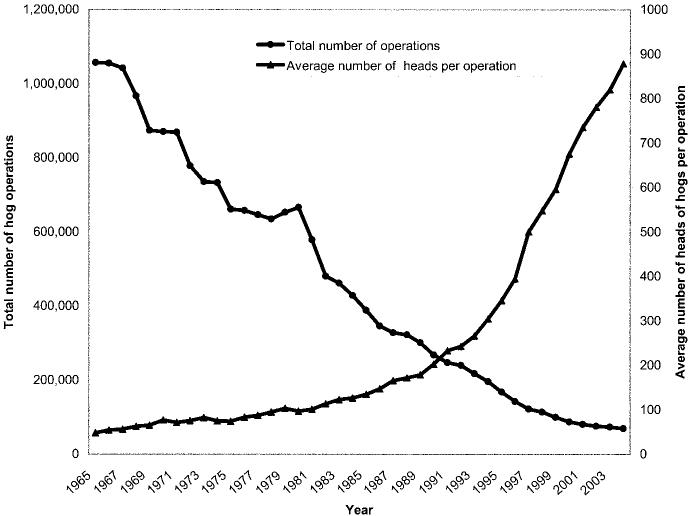
During the past 60 years, the US swine industry has changed in composition from primarily small herds on family farms to include immense herds in large, corporate facilities (figure 1). The US pork industry now generates $11 billion annually and employs an estimated 575,000 persons (2002 figures) [24]. Although pork production facilities today are larger, fewer, and more efficient and require fewer workers, it is estimated that, nationwide, at least 100,000 workers work in swine barns with live pigs (Dr. Liz Wagstrom, personal communication).
Figure 1.
Trends in hog operations in the United States. Adapted from [23].
Iowa is the leading swine-producing state in the United States, with 9300 farms [25] (2004 figure), raising 25 million hogs per year [25] (a rate of 8.6 swine per human resident [26]). Today's large herds are maintained through the frequent introduction of young swine into swine-producing facilities. This constant influx of potentially pathogen-susceptible animals makes swine pathogen eradication difficult to achieve. Therefore, swine influenza infections, which were formerly seasonal (like human influenza infections), now have become enzootic, and swine influenza transmission occurs year-round in much of the US swine industry. Although these influenza virus infections among pigs are generally thought to be mild, they provide a constant opportunity for zoonotic influenza virus infections among humans who are occupationally exposed. Continual swine influenza transmission in US swine herds also provides the opportunity for human influenza viruses to mix with swine or avian influenza viruses and generate novel progeny viruses.
The potential for animal-to-animal transmission (reflected in the basic reproductive number, R0) among pigs in a swine confinement operation will be much greater than on a traditional farm because of the pigs' crowding (resulting in prolonged and more frequent contact). In addition, virus-laden secretions from pigs may be more concentrated, and reductions in ventilation and sunshine exposure may prolong viral viability. Thus, a confinement operation worker's probability of acquiring influenza virus infection may be increased, compared with that of a traditional swine worker, and certainly increased when compared with the risk among non–swine workers exposed only to human-to-human influenza activity. This risk is even greater if the virus does not kill pig hosts and if new susceptible animals are frequently introduced to the farm, sustaining transmission. Swine workers may initiate epidemics by enhancing the mixing of viral strains that may lead to reassortment and novel progeny influenza viruses of pandemic potential. They may serve as a conduit for a novel virus to move from swine to man or from man to swine. One might envision that, once a novel virus is introduced into a densely populated swine barn, the viral loads swine workers would experience could overwhelm any partial immunity they might possess. After work, they may readily communicate that virus to their family members and neighbors. Similarly, they might introduce a novel human virus to crowded swine populations, which could have tremendous economic impact on the US agricultural industry. Recall that during the 1918–1919 human influenza pandemic, there were concomitant US swine herd die-offs (likely from the virus) that caused much economic hardship [27].
Human infections with swine influenza virus have been previously described. In 1988, swine influenza virus exposures at a Wisconsin county fair led to infections in at least 50 swine exhibitors and 3 of their family members, and infection resulted in the death of a previously healthy woman who attended the fair [28]. Although many swine influenza virus infections in humans are likely mild or asymptomatic, the potential for a virulent, highly communicable novel virus to move from swine to man seems to be great.
There are 3 developments that heighten our awareness of this threat. First is our new understanding of how common these infections must be—thus, the importance of this report. Second is the recent change in animal husbandry in the US pork and poultry industries. Today, tens of thousands of susceptible pigs or poultry are housed in confinement facilities, serving as a tremendous potential reservoir of susceptible animals, whose dense populations may hasten viral mutation and reassortment. The third development of concern is the sudden emergence and rapid spread of very virulent H5N1 virus that is now endemic in many parts of Asia and that has spread via migrating birds to Russia. Fortunately, although this virus infects swine, as yet, it is not readily transmissible between pigs [29]. However, the H5N1 virus is readily transmitted among domestic birds and waterfowl. Therefore, if migrating birds were to introduce the virus to North American pork production facilities, and if the virus were to prove to be efficient in transmission, the resultant swine-to-human swine virus infections could result in considerable human morbidity. Even with modern industry biosecurity measures, transmission would be difficult to control. Additionally, as much of the pork industry is rural, detection of such a novel virus invasion could be delayed, especially if the virus does not readily kill swine.
The elevated serologic responses to swine H1N1 virus among farmers and veterinarians observed in this study are consistent with a previous study of these occupational groups [20]. Our study found seropositivity to swine H1N2 among farm workers that had not been previously detected. All 3 swine exposure groups had elevated titers against the swine H3N2 virus. Strong association of this virus with human H3N2 virus (A/Nanchang/933/95) suggests cross-reactivity between swine and human strains, which was not unexpected, because the swine H3N2 virus HA gene is of human origin [17-19]. Although meat processing workers were at increased risk for antibodies against swine H1N1 and swine H1N2 virus, their ORs were lower than those for farmers and veterinarians. We speculate that this may be the result of limited exposure to live pigs.
Our study is unique, because to our knowledge, this is one of the largest such serologic assessment to date of occupational transmission of swine virus to human infections. Serologically, we studied the antibody response to 2 recently emerged circulating swine viruses in a state with year-round, active virus transmission within swine herds. In addition, we controlled for the confounding of serologic response to human influenza virus as an explanation for increased serologic response to the swine viruses and employed proportional odds modeling to better discriminate the effect of potential risk factors to the swine virus serologic outcomes.
This study has a number of limitations. There was a lack of detailed exposure information for the farmer group, and the study design did not allow us to determine whether individuals developed clinical symptoms with seroconversion. It is possible that the elevated titers compared by proportional odds modeling do not correlate with infection. However, our exposed populations had statistically significant evidence of swine virus infection by both the proportional odds modeling and the more traditional use of a titer cutpoint. Our choice of a ≥1:40 cut-point is consistent with the literature [20, 21] but could also be viewed as conservative. In prospective cohort studies, Fox et al. [30] demonstrated that, among children, HI titers of ≥1:20 against human H1 virus were protective and that almost any titer detected among adults was evidence of protection. We are presently conducting a large, prospective study of swine-exposed workers to better understand occupational zoonotic influenza infection.
We have documented evidence for swine influenza virus infection among 3 different occupational groups with exposure to pigs. Each of the 3 groups had antibody evidence of infection with 2 different swine viruses, and their odds of elevated serologic titers were much greater than those for control subjects. Serologic risk factor data controlled for potential confounders, such as serologic response to human influenza virus and vaccine. Study laboratory findings were validated by a blinded external laboratory, and serologic assay results were corroborated by studies of virus-specific antisera. We argue that these data are compelling evidence that swine influenza virus infections frequently occur among swine workers. It seems prudent to consider swine workers for sentinel influenza surveillance and routine human inactivated influenza vaccine immunizations. Additionally, in the event of heightened pandemic threat, protection of swine-exposed workers with antiviral medications and pandemic strain vaccines seems to be an important strategy to limit the spread of influenza virus among human and susceptible domestic animals.
Acknowledgments
We thank Dr. Kevin M. Kelly (University of Iowa's Keokuk County Rural Health Study and the Great Plains Center for Agricultural Health & Safety, Iowa City), for assisting us with data preparation; Debbie Wellman and Kelly Lesher (University of Iowa's Center for Emerging Infectious Diseases, Iowa City), for providing serologic assay study support; Dr. Alexander Karasin (University of Wisconsin, Madison); the Union of Food and Commercial Workers (Washington, DC) and the Iowa Veterinary Medical Association (Ankeny), for helping us to engage their members; and Drs. Carolyn Bridges, Jackie Katz, and Alexander Klimov (Centers for Disease Control and Prevention, Atlanta, GA), for their advice and assistance with laboratory reagents and the adaptation of serologic procedures.
Footnotes
Financial support. University of Iowa Center for Health Effects of Environmental Contamination and University of Iowa Center for Emerging Infectious Diseases funds. Serologic assay work was made possible in part by a grant from the National Institutes of Allergy and Infectious Diseases (NIAID-R21 AI059214-01).
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Webster RG, Sharp GB, Claas EC. Interspecies transmission of influenza viruses. Am J Respir Crit Care Med. 1995;152:S25–30. doi: 10.1164/ajrccm/152.4_Pt_2.S25. [DOI] [PubMed] [Google Scholar]
- 2.Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: a continuing enigma. J Gen Virol. 2003;84:2285–92. doi: 10.1099/vir.0.19302-0. [DOI] [PubMed] [Google Scholar]
- 3.Kimura K, Adlakha A, Simon PM. Fatal case of swine influenza virus in an immunocompetent host. Mayo Clin Proc. 1998;73:243–5. doi: 10.4065/73.3.243. [DOI] [PubMed] [Google Scholar]
- 4.Dacso CC, Couch RB, Six HR, Young JF, Quarles JM, Kasel JA. Sporadic occurrence of zoonotic swine influenza virus infections. J Clin Microbiol. 1984;20:833–5. doi: 10.1128/jcm.20.4.833-835.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong JC, Paccaud MF, de Ronde-Verloop FM, et al. Isolation of swine-like influenza A(H1N1) viruses from man in Switzerland and The Netherlands. Ann Inst Pasteur Virol. 1988;139:429–37. doi: 10.1016/s0769-2617(88)80078-9. [DOI] [PubMed] [Google Scholar]
- 6.Hinshaw VS, Bean WJ, Jr, Webster RG, Easterday BC. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology. 1978;84:51–62. doi: 10.1016/0042-6822(78)90217-9. [DOI] [PubMed] [Google Scholar]
- 7.Haiyan LI, Chin J, Chin J. In Chinese. Prev Vet Med. 2004;24:304–9. [Google Scholar]
- 8.Haiyan LI, Chin J, Chin J. In Chinese. Prev Vet Med. 2004;26:1–6. [Google Scholar]
- 9.ProMED-mail. Avian influenza—eastern Asia. 2005 April 14; ProMED-mail archive number 20050414.1079. Available at: http://www.promedmail.org/pls/promed/f?pp2400:1001:::NO::F2400_P1001_BACK_ PAGE,F2400_p1001_PUB_MAIL_ID:1000%2C28683.
- 10.Stromquist AM, Merchant JA, Burmeister LF, Zwerling C, Reynolds SJ. The Keokuk County Rural Health Study: methodology and demographics. Journal of Agromedicine. 1997:243–8. [Google Scholar]
- 11.Kendal AP, Pereria MS, Shekel J. Concepts and procedures for laboratory-based influenza surveillance. World Health Organization; Geneva: 1982. [Google Scholar]
- 12.Olsen CW, Carey S, Hinshaw L, Karasin AI. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch Virol. 2000;145:1399–419. doi: 10.1007/s007050070098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karasin AI, Landgraf J, Swenson S, et al. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol. 2002;40:1073–9. doi: 10.1128/JCM.40.3.1073-1079.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol. 2000;38:2453–6. doi: 10.1128/jcm.38.6.2453-2456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi YK, Goyal SM, Farnham MW, Joo HS. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 2002;87:173–9. doi: 10.1016/s0168-1702(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 16.Choi YK, Goyal SM, Joo HS. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch Virol. 2002;147:1209–20. doi: 10.1007/s00705-002-0788-4. [DOI] [PubMed] [Google Scholar]
- 17.Karasin AI, Schutten MM, Cooper LA, et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. doi: 10.1016/s0168-1702(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhou NN, Senne DA, Landgraf JS, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–6. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. Evolution of swine H3N2 influenza viruses in the United States. J Virol. 2000;74:8243–51. doi: 10.1128/jvi.74.18.8243-8251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen CW, Brammer L, Easterday BC, et al. Serologic evidence of H1 swine influenza virus infection in swine farm residents and employees. Emerg Infect Dis. 2002;8:814–9. doi: 10.3201/eid0808.010474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayora-Talavera G, Cadavieco-Burgos JM, Canul-Armas AB. Serologic evidence of human and swine influenza in Mayan persons. Emerg Infect Dis. 2005;11:158–61. doi: 10.3201/eid1101.040554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullagh P. Regression models for ordinal data. J R Statis Soc Series B. 1980;42:109–42. [Google Scholar]
- 23.US Department of Agriculture, National Agricultural Statitstics Service Historical data. Available at: http://www.usda.gov/nass/pubs/histdata.htm. Accessed 18 August 2005.
- 24.National Pork Board . Pork facts 2002/2003. National Pork Board; Des Moines, IA: 2002. Available at: http://www.porkboard.org/docs/2002-3%20PORK%20FACTS%20BK.pdf. [Google Scholar]
- 25.Iowa Pork Producing Association . The Iowa pork industry. Iowa Pork Industry; 2005. Available at: http://www.iowapork.org/about_us/porkdata_nr.html. [Google Scholar]
- 26.US Census Bureau State and country quickfacts. Available at: http://quickfacts.census.gov/qfd/states/19000.html. Accessed 18 August 2005.
- 27.Crosby A. America's forgotten pandemic: the influenza of 1918. 2nd ed. University of Texas, Austin; Austin, TX: 2003. [Google Scholar]
- 28.Wells DL, Hopfensperger DJ, Arden NH, et al. Swine influenza virus infections: transmission from ill pigs to humans at a Wisconsin agricultural fair and subsequent probable person-to-person transmission. JAMA. 1991;265:478–81. doi: 10.1001/jama.265.4.478. [DOI] [PubMed] [Google Scholar]
- 29.Choi YK, Nguyen TD, Ozaki H, et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol. 2005;79:10821–5. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox JP, Cooney MK, Hall CE, Foy HM. Influenzavirus infections in Seattle families, 1975–1979. II. Pattern of infection in invaded households and relation of age and prior antibody to occurrence of infection and related illness. Am J Epidemiol. 1982;116:228–42. doi: 10.1093/oxfordjournals.aje.a113408. [DOI] [PubMed] [Google Scholar]