Abstract
Durum wheat (Triticum turgidum L. subsp. durum Desf.) Line 149 contains two novel major genes for excluding Na+ from leaf blades, named Nax1 and Nax2. The genes were separated into families containing a single gene and near-isogenic homozygous lines were selected. Lines containing either Nax1 or Nax2 had lower rates of Na+ transport from roots to shoots than their near-isogenic pairs due to lower rates of net loading of the xylem, not to lower rates of net uptake from the soil or higher rates of retranslocation in the phloem. Nax1 and Nax2 lines also had higher rates of K+ transport from root to shoot, resulting in an enhanced discrimination of K+ over Na+. Lines containing Nax1 differed from those containing Nax2 by unloading Na+ from the xylem as it entered the shoot so that Na+ was retained in the base of the leaf, leading to a high sheath to blade ratio of Na+ concentration. Gradients in tissue concentrations of Na+ along the leaf suggested that Na+ was continually removed from the xylem. The Nax2 line did not retain Na+ in the base of the leaf, suggesting that it functioned only in the root. The Nax2 gene therefore has a similar function to Kna1 in bread wheat (Triticum aestivum).
Salt tolerance in wheat (Triticum aestivum) and many other species is associated with the ability to exclude Na+ so that high Na+ concentrations do not occur in leaves, particularly in the leaf blade (Läuchli, 1984; Munns, 2005). High leaf Na+ concentrations can cause premature leaf senescence and loss of photosynthetic activity (James et al., 2002), which reduces the rate of carbon assimilation and ultimately grain yield (Husain et al., 2003).
Durum (pasta) wheat (Triticum turgidum L. subsp. durum Desf.) is more salt sensitive than bread wheat, probably because of its poorer ability to exclude Na+ from the leaf blade (Gorham et al., 1990). In monocotyledonous species, the leaf is composed of the blade (top part) and the sheath (bottom part). The bulk of the leaf's photosynthesis and transpiration occurs in the blade. An unusual source of Na+ exclusion was found in durum wheat Line 149 (Munns et al., 2000). A screen of a large number of durum-related genotypes for leaf blade Na+ accumulation showed that Line 149 had an unusually low concentration, as low as bread wheat (Munns et al., 2000). However, the concentration in shoots as a whole was not as low as bread wheat (Husain et al., 2004), suggesting that Na+ was retained in the leaf sheath. The Na+ transport characteristics of Line 149 were compared with the durum cv Tamaroi, which has the high Na+ concentrations typical of durum wheat, by measurement of 22Na+ transport and net Na+ accumulation (Davenport et al., 2005). The genotypes did not differ in unidirectional root uptake of Na+. The major differences in Na+ transport between the genotypes were in the rate of transfer to the shoot (net root xylem loading) and the preferential accumulation of Na+ in the leaf sheath versus the leaf blade (Davenport et al., 2005).
Genetic analysis of a cross between Line 149 and cv Tamaroi indicated two genes of major effect for Na+ exclusion (Munns et al., 2003). A quantitative trait locus for low Na+ concentration in leaf blades was mapped to the distal region on the long arm of chromosome 2A and named Nax1. This quantitative trait locus accounted for 38% of the phenotypic variation in the F2 generation, suggesting that it was associated with one of the two major genes. A microsatellite marker, gwm312, was closely linked to the trait and has been used to accelerate the transfer of this trait into commercial varieties of durum wheat (Lindsay et al., 2004). The presence of a second gene for Na+ exclusion was confirmed by the observation that some plants without the Line 149 allele of gwm312 still had moderately low Na+ concentrations in the leaf blade. A second gene, independent of Nax1, was suggested to contribute to the full expression of the Na+ exclusion trait (Lindsay et al., 2004). It was named Nax2.
To distinguish the physiological mechanisms of Nax1 and Nax2, families were developed containing only one of these genes. This article describes the separation of the two genes, their different functions, and their possible origin. We show that both Nax genes restrict the transport of Na+ from roots to shoots and result in enhanced K+-Na+ discrimination in the leaf blade. Nax2 has a similar mechanism to that described for Kna1 in bread wheat. Nax1 differs from Nax2 in removing Na+ from the xylem in the lower part of the leaf, as well as in the root, and represents a function not present in bread wheat. A wild wheat ancestor, Triticum monococcum, is the original source of both Nax genes.
RESULTS
Separation of Nax1 and Nax2
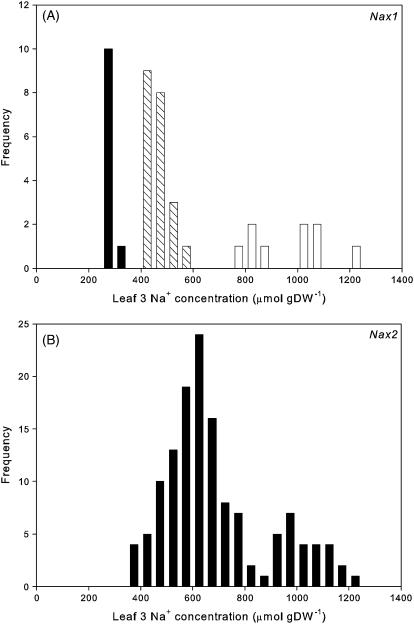
Near-isogenic lines with and without the Nax1 or Nax2 gene were developed by backcrossing Line 149 with cv Tamaroi to obtain single-gene BC5F2 families. These were selected using the phenotype of Na+ concentration in the leaf blade and a codominant microsatellite marker gwm312 that is closely linked to Nax1 (Lindsay et al., 2004). Putative single-gene Nax1 families showed a 1:2:1 distribution for leaf blade Na+ concentration (Fig. 1A), indicating the segregation of a single codominant gene. Progeny testing of the BC5F3 lines validated the single plant F2 phenotype (data not shown). Homozygous BC5F3 lines with or without Nax1 are designated [+]Nax1 and [−]Nax1, respectively.
Figure 1.
Frequency distributions for Na+ concentrations in the leaf blade of single-gene BC5F2 families grown at 150 mm NaCl for 10 d. A, Nax1 family (n = 41). Individuals in the low Na+ class (black bars) were homozygous for the Line 149 allele of gwm312; individuals in the high Na+ class (white bars) were homozygous for the cv Tamaroi allele of gwm312; and individuals in the intermediate class (hatched bars) were heterozygous. Parental means were 141 ± 14 μmol g−1 dry weight for Line 149 and 811 ± 31 for cv Tamaroi (n = 6). B, Nax2 family (n = 140). Parental means were 278 ± 37 μmol g−1 dry weight for Line 149 and 1,193 ± 48 for cv Tamaroi.
Putative single-gene Nax2 families showed a 3:1 distribution for leaf blade Na+ concentration (Fig. 1B), indicating the segregation of a single dominant gene. This was confirmed by progeny testing (C. Byrt, unpublished data). Homozygous Nax2 BC5F3 lines with or without Nax2 are called [+]Nax2 and [−]Nax2, respectively.
Effects of Nax1 and Nax2 on Na+ and K+ Concentrations in Leaf Blade and Sheath
The presence of either Nax1 or Nax2 reduced the Na+ concentration in the leaf blade and the leaf sheath (Table I). The leaf sheath in a cereal is the lower part of the leaf and is delineated from the blade by the ligule. Neither gene on its own reduced the Na+ concentration as much as when present together in parent Line 149 (Table I). The physiological mechanism by which the two genes achieved low Na+ concentrations in the blade differed. Nax1 was distinguished by a higher Na+ concentration in the leaf sheath than the leaf blade so that the sheath to blade ratio was similar to that of parent Line 149 (Table I). All lines lacking Nax1 had the same Na+ concentration in sheath and blade. Nax1 was also associated with a higher root Na+ concentration, as was Line 149 (Table I). The sheath is only a small proportion of the total leaf, making up only 23% of the total dry weight of the leaves. However, the amount of Na+ sequestered in this tissue was considerable and, in lines containing the Nax1 gene, 50% of the Na+ in the leaf as a whole was retained in this tissue (Table I).
Table I.
Na+ concentration in leaf blades, leaf sheaths, and total shoot and roots in Line 149, cv Tamaroi, and Nax1 and Nax2 near-isogenic lines grown in 50 mm NaCl for 10 d
Values are means ± se (n = 6). Fresh weight:dry weight ratios are 7.2 for leaf blades and 8.8 for leaf sheaths.
| Plant Material | Category | Na+ Concentration
|
Sheath Na+ % of Total Shoot | ||||
|---|---|---|---|---|---|---|---|
| Leaf Blades | Leaf Sheaths | Sheath to Blade Ratio | Total Shoot | Roots | |||
| μmol g−1 dry weight | μmol g−1 dry weight | μmol | |||||
| Parents | Line 149 | 104 ± 7 | 271 ± 12 | 2.6 | 160 ± 9 | 983 ± 23 | 50 |
| cv Tamaroi | 579 ± 17 | 559 ± 15 | 1.0 | 572 ± 16 | 811 ± 26 | 28 | |
| Nax1 lines | [+]Nax1 | 183 ± 6 | 427 ± 8 | 2.3 | 260 ± 6 | 886 ± 17 | 51 |
| [−]Nax1 | 599 ± 15 | 581 ± 21 | 1.0 | 593 ± 16 | 836 ± 17 | 26 | |
| Nax2 lines | [+]Nax2 | 210 ± 8 | 209 ± 20 | 1.0 | 209 ± 11 | 819 ± 20 | 28 |
| [−]Nax2 | 583 ± 18 | 529 ± 12 | 0.9 | 565 ± 16 | 805 ± 13 | 29 | |
| lsd(0.05) | 34 | 43 | 0.1 | 35 | 58 | 3 | |
Nax2 greatly reduced Na+ concentrations in both leaf blade and sheath (Table I). There was no preferential retention of Na+ in the leaf sheath, so the reduced Na+ uptake into the leaf blade was determined predominantly by the roots.
K+ concentration in both leaf blade and sheath was enhanced by the presence of either Nax1 or Nax2 (Table II). In the blade, the presence of both genes in Line 149 had a greater effect on K+ concentration than either gene alone, but in the sheath Nax2 alone produced as high a K+ concentration as Line 149. Thus, the Nax2 gene resulted in a higher concentration of K+ in the total shoot than did Nax1 (data not shown). The K+ to Na+ discrimination ratio was enhanced by both genes in the leaf blade, but, in the leaf sheath, the presence of Nax2 resulted in a much higher K+ to Na+ ratio than did Nax1 (Table II).
Table II.
K+ concentration in leaf blades, leaf sheaths, and roots in Line 149, cv Tamaroi, and Nax1 and Nax2 near-isogenic lines grown in 50 mm NaCl for 10 d
Values are means ± se (n = 6).
| Plant Material | Category | K+ Concentration
|
K to Na Ratio
|
|||
|---|---|---|---|---|---|---|
| Leaf Blades | Leaf Sheaths | Roots | Leaf Blades | Leaf Sheaths | ||
| μmol g−1 dry weight | ||||||
| Parents | Line 149 | 1,390 ± 14 | 1,386 ± 16 | 730 ± 23 | 13.8 ± 1.0 | 5.2 ± 0.3 |
| cv Tamaroi | 941 ± 15 | 1,099 ± 20 | 949 ± 48 | 1.6 ± 0.0 | 2.0 ± 0.1 | |
| Nax1 lines | [+]Nax1 | 1,180 ± 18 | 1,217 ± 17 | 842 ± 21 | 6.5 ± 0.2 | 2.9 ± 0.1 |
| [−]Nax1 | 901 ± 11 | 1,072 ± 12 | 828 ± 35 | 1.5 ± 0.0 | 1.9 ± 0.1 | |
| Nax2 lines | [+]Nax2 | 1,110 ± 19 | 1,324 ± 20 | 844 ± 36 | 5.4 ± 0.2 | 6.5 ± 0.6 |
| [−]Nax2 | 874 ± 19 | 1,075 ± 12 | 918 ± 33 | 1.5 ± 0.1 | 2.0 ± 0.1 | |
| lsd(0.05) | 40 | 49 | 75 | 1.4 | 0.7 | |
In the absence of NaCl, there was no difference in either Na+ or K+ concentration in the leaf blade between Line 149 and cv Tamaroi (Rivelli et al., 2002). Cl− concentrations were not measured because previous experiments had shown there was little difference between Line 149 and cv Tamaroi (Rivelli et al., 2002).
Nax1 and Nax2 Control of Na+ Transport from Root to Shoot
Time-course experiments using 22Na+ showed that the Nax1 and Nax2 genes reduced the rate of unidirectional transport from root to shoot and thereby accounted for the difference in total accumulation of Na+ over time in the shoot as a whole in the experiments described above. Reduced transport in the xylem, rather than higher retranslocation in the phloem, was therefore the likely function of the Nax genes.
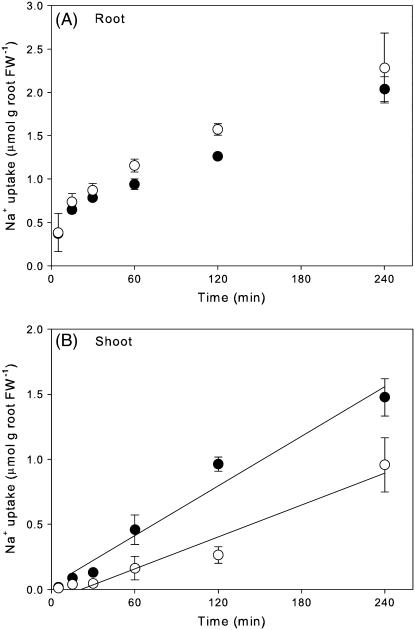
In the Nax1 lines, net 22Na+ uptake by roots was higher in [+]Nax1 than [−]Nax1 lines after 15 min (Fig. 2A), and this difference was maintained after 48 h of 22Na+ feeding (data not shown). This difference was consistent with the root total Na+ concentrations shown in Table I, with roots of the [+]Nax1 line containing more Na+ than those of the [−]Nax1 line. 22Na+ appeared in the shoots after 15 min and, after 30 min, the uptake by the [+]Nax1 and [−]Nax1 lines differed significantly (Fig. 2B). At 4 h, the calculated Na+ uptake rate of [+]Nax1 was two-thirds that of [−]Nax1 (Fig. 2B). The rapid appearance of 22Na+ in the shoot and the time course of root uptake established previously (Davenport et al., 2005) indicated rapid labeling of the root cytoplasmic pool with the external solution, leading to a steady rate of transport to the shoot by 30 min. If maintained over 10 d, this would completely account for the differences in total shoot Na+ concentration between lines with and without Nax1 (Table I).
Figure 2.
Rate of 22Na+ accumulation in Nax1 near-isogenic lines grown in 25 mm NaCl in root (A) and shoot (B); [+]Nax1 (○), [−]Nax1 (•). Values are means ± se (n = 4). Fitted linear regressions for 22Na+ uptake by the shoot are [+]Nax1: y = 4.08 × 10−3x − 0.09 (r2 = 0.96); [−]Nax1: y = 6.36 × 10−3x + 0.03 (r2 = 0.96).
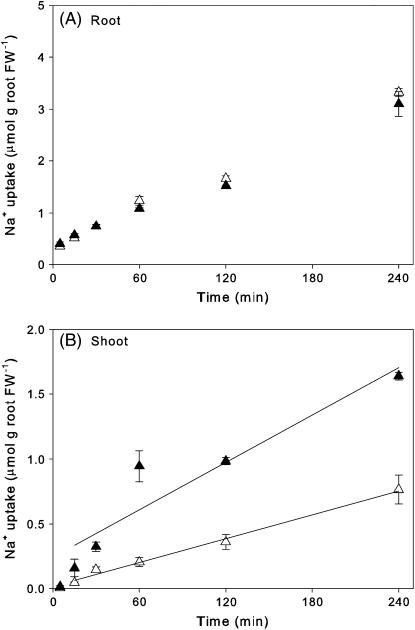
In the Nax2 lines, root 22Na+ uptake rates were the same for the [+]Nax2 and [−]Nax2 lines over the whole period studied (Fig. 3A). This finding was consistent with their having the same total Na+ concentrations in roots after 10 d in 50 mm NaCl (Table I). Shoot uptake of 22Na+ was apparent at 15 min, by which time [+]Nax2 plants had a lower rate of uptake than [−]Nax2 plants (Fig. 3B). The difference was statistically significant at 30 min. At 4 h, the shoot Na+ uptake rate for the [+]Nax2 line was one-half that for the [−]Nax2 line (Fig. 3B) and was sufficient to account for differences in shoot Na+ concentrations (Table I). Because the rates of root Na+ uptake were identical in the lines with and without Nax2, differences in shoot uptake are due to the net rate of xylem loading in the root.
Figure 3.
Rate of 22Na+ accumulation in Nax2 near-isogenic lines grown in 25 mm NaCl in root (A) and shoot (B); [+]Nax2 (▵); [−]Nax2 (▴). Values are means ± se (n = 4). Linear regressions for 22Na+ uptake by the shoot are [+]Nax2: y = 3.07 × 10−3x + 0.02 (r2 = 0.99); [−]Nax2: y = 6.07 × 10−3x + 0.25 (r2 = 0.88).
Nax1 and Nax2 Increase Withdrawal of Na+ from the Root Xylem
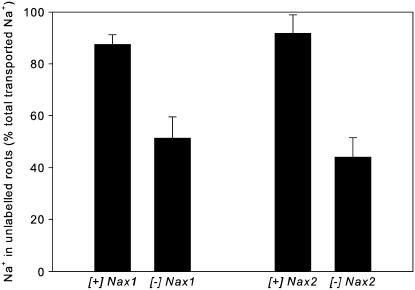
Control of shoot Na+ uptake could be due to either tight control of xylem loading or high rate of withdrawal of Na+ from the transpiration stream into the upper part of the roots. Evidence for xylem withdrawal of Na+ in the roots of both [+]Nax1 and [+]Nax2 lines was obtained in a separate compartmental loading experiment. When the lower part of the root was exposed to 22Na+, the [+]Nax1 line withdrew more of the total transported 22Na+ into the upper roots (88%) than the [−]Nax1 line (51%; Fig. 4). Similarly, the [+]Nax2 line withdrew more 22Na+ into the upper roots (91%) than the [−]Nax2 line (44%). These differences were associated with a 4-fold higher shoot 22Na+ content in both the [−]Nax1 and [−]Nax2 lines than their respective isogenic pairs (data not shown).
Figure 4.
Withdrawal of 22Na+ from the xylem by roots of Nax1 and Nax2 near-isogenic lines grown in 25 mm NaCl. Withdrawal was calculated as the amount of 22Na+ in the unlabeled upper roots as a percentage of total 22Na+ transported from the labeled lower roots after 2 h. Bars are means ± se (n = 5).
Net Na+ and K+ Transport Rates from Root to Shoot and Xylem Concentrations
The net Na+ and K+ transport rates from root to shoot were quantified from the increase in Na+ and K+ in roots and shoots between 6 and 10 d after exposure to 50 mm NaCl. Very low net Na+ transport rates were found in Line 149 and the [+]Nax1 line, with slightly higher rates in the [+]Nax2 line. Lines lacking Nax1 and Nax2 had high net Na+ transport rates similar to the recurrent parent cv Tamaroi (Table III). There was less variation in K+ transport rates between genotypes; however, the highest K+ transport rates were found in Line 149 and the [+]Nax2 line, suggesting that the Nax2 mechanism may involve the exchange of K+ and Na+ in net xylem loading in the root, possibly a replacement of K+ for Na+ in removal of Na+ from the xylem.
Table III.
Na+ and K+ net transport rates to the shoot, Na+ and K+ concentration in the xylem stream, and percentage exclusion of Na+ by the roots in Line 149, cv Tamaroi, and Nax1 and Nax2 near-isogenic lines grown in 50 mm NaCl
Values are calculated over a 6- to 10-d period from two experiments and are means ± se (n = 12).
| Plant Material | Category | Transport Rate from Root to Shoot
|
Ion Concentration in Xylem
|
Exclusion by RootsNa+ | ||
|---|---|---|---|---|---|---|
| Na+ | K+ | Na+ | K+ | |||
| μmol groot fresh weight−1 d−1 | mm | % | ||||
| Parents | Line 149 | 6.0 ± 0.6 | 45 ± 3 | 0.9 ± 0.1 | 6.7 ± 0.3 | 98.3 ± 0.2 |
| cv Tamaroi | 22.7 ± 0.8 | 41 ± 3 | 2.8 ± 0.1 | 5.0 ± 0.4 | 94.4 ± 0.3 | |
| Nax1 lines | [+]Nax1 | 6.5 ± 0.3 | 35 ± 3 | 1.0 ± 0.1 | 5.2 ± 0.3 | 98.0 ± 0.1 |
| [−]Nax1 | 22.5 ± 1.5 | 37 ± 3 | 2.8 ± 0.3 | 4.7 ± 0.5 | 94.4 ± 0.6 | |
| Nax2 lines | [+]Nax2 | 8.7 ± 0.9 | 42 ± 2 | 1.3 ± 0.1 | 6.3 ± 0.4 | 97.4 ± 0.2 |
| [−]Nax2 | 17.9 ± 1.3 | 33 ± 3 | 2.5 ± 0.1 | 4.5 ± 0.3 | 95.0 ± 0.2 | |
| lsd(0.05) | 2.9 | 7 | 0.5 | 1.1 | 0.9 | |
Na+ concentrations in the xylem were calculated from the net Na+ transport rate and the transpiration rate, assuming that there was little retranslocation of Na+ in the phloem. Transpiration measured over 24 h varied from 0.55 to 0.60 mL h−1 plant−1 with no significant difference between lines. Na+ concentrations in the xylem were calculated to be about 1 mm for lines containing either Nax1 and Nax2, and 2.5 mm or more if both genes were lacking (Table III). The extent of exclusion of Na+ from the soil solution (50 mm) in lines containing either Nax1 or Nax2 was therefore 98%. However, lines without these genes still excluded 94% of the NaCl in the solution (Table III). Thus, relatively small percentage differences in Na+ exclusion capability led to profound differences in the accumulation of Na+ into the shoot.
Ignoring the extent of retranslocation of K+ in the phloem, K+ concentrations in the xylem were calculated to be 6.5 mm if Nax2 was present, but 5 mm or less if it was absent (Table III). The real concentrations are probably twice this because the phloem can retranslocate 50% of the K+ carried in the xylem to the shoots (Wolf et al., 1990). Presuming that the recirculation of K+ is similar for all lines, the data indicate that the Nax2 gene is associated with enhanced K+ uptake.
Gradients in Na+ and K+ along the Leaf in the [+]Nax1 Line
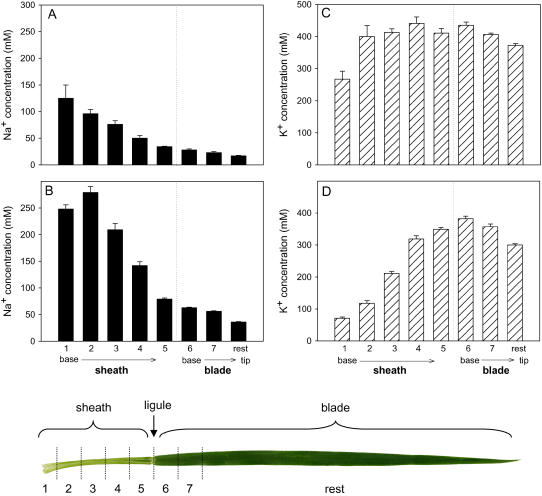
To examine in more detail the unusual phenotype of Nax1, namely, the high Na+ sheath to blade ratio, the distribution of Na+ along the entire length of a given leaf was measured. This would show whether the ability of cells lining the xylem to withdraw Na+ and sequester it was confined to the sheath (i.e. whether the property of xylem parenchyma cells was different in sheath and blade) or whether the ligule could act as a barrier to Na+ movement in the xylem from sheath to blade.
Leaves of the [+]Nax1 line, as well as parent Line 149, showed a gradient of Na+ concentrations, highest in the leaf base and lowest in the leaf tip, from the time the salt was added. There was no discontinuity at the junction between the sheath and the blade, indicating that there was no barrier to Na+ movement in the xylem at the ligule. Na+ concentrations in sheath and leaf blade segments of leaf 2 in the [+]Nax1 line after 2 and 5 d in 50 mm NaCl are shown in Figure 5. The increase in Na+ concentration with time in both sheath and blade indicated that the xylem parenchyma cells in the leaf sheath did not differ from those in the blade in the ability to withdraw Na+ from the xylem. After 5 d in 50 mm NaCl, Na+ concentrations in lower sheath segments reached a maximum of about 250 mm on a tissue-water content basis (Fig. 5) or 1,400 μmol g−1 dry weight, possibly indicating a threshold in the storage capacity of the cells. No gradient was found along the leaf in [−]Nax1 lines, which displayed an even profile of Na+ concentration across both the sheath and blade, with slightly higher concentrations in the upper (older) portions of both sheath and blade, probably reflecting the age of the tissue and a slightly longer exposure to salinity with the resultant increased deposition of Na+ (data not shown).
Figure 5.
Na+ (A and B) and K+ (C and D) concentrations in sheath and leaf segments of leaf 2 from [+]Nax1 seedlings grown in 50 mm NaCl for 2 d (A and C) and 5 d (B and D). Na+ and K+ concentrations were calculated on a tissue-water basis. The dotted line indicates the ligule. Bars are bulked (×3) means ± se (n = 4). The bottom diagram is an image of leaf 2 (total length 180 mm) with lines indicating where the sheath and blade were sectioned for the ion analysis shown above.
Measurements of K+ in leaves at 2 and 5 d indicated that K+ was displaced by Na+ over time. K+ was initially at high concentrations (400 mm) in all parts of the sheath and blade, but decreased over time as Na+ increased (Fig. 5). The decrease of K+ in the leaf sheath over time, while Na+ increased, indicated that K+ was either entering the xylem in exchange for Na+ and moving toward the leaf tip or entering the phloem and moving out of the leaf.
Retranslocation of Na+ from Shoots to Roots
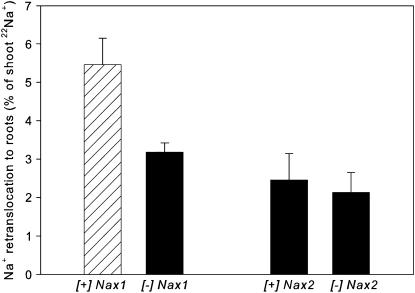
The rate of retranslocation of Na+ from the shoot to the root was estimated using a split-root system, where one-half of the roots were fed 22Na+ for 48 h and the appearance of radioactivity in the unlabeled roots and solution was used to estimate retranslocation. Retranslocation of Na+ was a relatively small component of shoot Na+ uptake (2%–6%; Fig. 6). These values are likely to be an underestimate of the total retranslocation because shoot 22Na+ had not yet come to an equilibrium with the feeding solution. Comparisons between lines showed that the [+]Nax1 line had twice the rate of retranslocation than [−]Nax1 and both [+]Nax2 and [−]Nax2 lines (Fig. 6). This difference may not be an intrinsic property of the Nax1 gene but a result of the higher Na+ in the sheath tissue of these lines (Table I; Fig. 5).
Figure 6.
Estimate of retranslocation of 22Na+ from shoot to roots in Nax1 and Nax2 near-isogenic lines after 48 h of labeling with 22Na+ in seedlings grown in a split-root system in 25 mm NaCl. Bars are means ± se (n = 4).
Origin of Nax1 and Nax2
Durum Line 149 was derived from a cross between durum cv Marrocos and an accession of the wheat progenitor T. monococcum, C68-101 (The, 1973). C68-101 also has the Line 149 allele of gwm312, indicating that it was the source of the Nax1 gene.
Na+ transport rates from root to shoot in C68-101 were similar to Line 149, but cv Marrocos had a much higher transport rate (Table IV). C68-101 had the same low Na+ concentrations in leaves as Line 149, the same high K+-Na+ discrimination, as well as the characteristic of Nax1, which is the high sheath to blade ratio (Table IV). Cultivar Marrocos had a sheath to blade ratio of 1:1. These data indicate that C68-101 is the source of both Nax1 and Nax2 genes.
Table IV.
Na+ and K+ concentrations and transport rates in Line 149 and its parents, the wheat progenitor T. monococcum C68-101, and durum cv Marrocos grown in 50 mm NaCl
Transport rates are calculated over 6 to 10 d and are means ± se (n = 6).
| Genotype | Leaf Blade Na+ | Leaf Blade K+ | K+ to Na+ Ratio in Blades | Leaf Sheath Na+ | Leaf Sheath K+ | K+ to Na+ Ratio in Sheaths | Na+ Ratio Sheath:Blade | Na+ Shoot Transport Rate |
|---|---|---|---|---|---|---|---|---|
| μmol g−1 dry weight | μmol g−1 dry weight | μmol groot fresh weight−1 d−1 | ||||||
| C68-101 | 116 ± 37 | 1,126 ± 29 | 9.7 | 342 ± 63 | 1,132 ± 38 | 3.3 | 2.9 | 8.6 ± 2.3 |
| cv Marrocos | 678 ± 34 | 1,158 ± 20 | 1.7 | 619 ± 27 | 1,235 ± 15 | 2.0 | 0.9 | 21.9 ± 0.5 |
| Line 149 | 104 ± 7 | 1,426 ± 9 | 13.7 | 404 ± 18 | 1,294 ± 8 | 3.2 | 3.9 | 6.2 ± 0.4 |
| lsd(0.05) | 75 | 62 | 6.2 | 101 | 61 | 1.2 | 0.6 | 4.4 |
DISCUSSION
The two genes for Na+ exclusion, Nax1 and Nax2, reduce Na+ transport from root to shoot, as evidenced by the time course of 22Na+ transport. Net Na+ transport in the xylem accounted for the differences in shoot concentrations, not retranslocation in the phloem. However, there were differences in the mechanisms of action of the two genes. First, Nax1 had a higher rate of deposition of Na+ in the leaf sheath than Nax2 and, consequently, a higher ratio of Na+ concentration in the sheath to blade ratio. Second, Nax1 had a lower rate of K+ transport from root to shoot than Nax2, but the displacement of K+ in the sheath led to an equal deposition in the leaf blade. Thus, both Nax1 and Nax2 lead to Na+ exclusion from the leaf blade and a high K to Na+ ratio, but by different mechanisms.
Nax1 and Nax2 Withdraw Na+ from the Xylem in the Roots
That Na+ can be withdrawn from the xylem in the roots was shown by feeding the lower parts of roots with 22Na+ and the appearance of a large proportion of the labeled Na+ in the upper roots. The proportion was 2-fold greater in lines containing Nax1 or Nax2 than their respective isogenic pairs. The Nax2 mechanism was confined to the roots and had the effect of reducing the transport of Na+ from root to shoot while increasing the transport of K+, and so resulted in a net exchange of Na+ for K+. Removal of Na+ from the xylem by the upper part of the root could induce a consequent influx of K+ into the xylem to restore the electrical potential. Alternatively, or additionally, higher K+ translocation in [+]Nax2 plants could indicate greater K+-Na+ selectivity in loading of xylem in the roots. Our data do not allow us to discriminate between these possibilities. The phenotype of Nax2 is very similar to that of Kna1 in bread wheat, which is considered to discriminate between Na+ and K+ at the point of xylem loading (Gorham et al., 1990).
Reabsorption of Na+ from the xylem in the upper part of the root system has been described for maize (Zea mays; Shone et al., 1969; Johanson and Cheeseman, 1983), soybean (Glycine max; Lacan and Durand, 1996), common bean (Phaseolus vulgaris; Jacoby, 1964), and scarlet runner bean (Phaseolus coccineus; Kramer et al., 1977). The studies with soybean indicated an exchange of K+ for Na+, energized by H+-ATPases, and the authors suggested that Na+-H+ and K+-H+ antiporters at the plasma membrane of the xylem parenchyma might be involved (Lacan and Durand, 1996).
Na+ reabsorption from the xylem in the upper part of scarlet runner bean roots was associated with cells having the appearance of transfer cells. These are xylem parenchyma cells with a wall labyrinth that increases the surface area of the plasma membrane, suggesting a function in transport processes (Kramer et al., 1977). Transfer cells have been described in the roots of other species (Kramer, 1983) but have not been found in wheat (A. Läuchli, personal communication).
Nax1 Also Withdraws Na+ from the Xylem in the Leaf
The function of Nax1 in removing Na+ was not restricted to the root because the transport of Na+ to the shoot as a whole was lower in the [+]Nax1 than the [−]Nax1 family.
The mechanism conferred by Nax1, which was characterized by the deposition of Na+ in the leaf sheath, is not confined to wheat germplasm containing the Nax1 gene. Preferential deposition of Na+ in the leaf base has been described for rice (Oryza sativa), common reed (Phragmites communis; Matsushita and Matoh, 1991), and sorghum (Lacerda et al., 2003). An equivalent of the Nax1 gene may be present in these other species. However, it may not be widespread. For instance, barley (Hordeum vulgare) does not have preferential retention of Na+ in the leaf sheath (Munns et al., 1988).
The gradients in Na+ and K+ concentration along the leaf and their change over time (Fig. 5) indicate that the cells lining the xylem were removing Na+ from the xylem stream, storing it in parenchyma cells in the sheath, and causing a displacement of the K+ there. It is possible to explain the gradient of Na+ along the leaf with a model incorporating the passive movement of Na+ from the xylem and possibly a subsequent active scavenging of Na+ as the concentration falls. Na+ can move passively from the xylem into the xylem parenchyma cells, against a concentration gradient, due to the negative electrical potential of the cells, which might be −100 to −200 mV. Na+ in the xylem could initially move passively via a Na+-permeable channel or a Na+ uniporter into the xylem parenchyma in the basal sheath tissue, leading to high rates of retrieval in these cells compared to the cells in the upper sheath and leaf blade, which would experience progressively lower Na+ concentrations in the xylem stream and subsequently lower rates of Na+ retrieval. Active uptake might be necessary to scavenge Na+ at very low concentrations, depending on the cytosolic Na+ concentration of cells near the leaf tip. Alternatively, passive uptake could act to maintain apoplastic leaf Na+ at low levels.
Candidate Genes
The major candidates for transporters that could withdraw Na+ from the xylem are nonselective cation channels and high-affinity K+ transporters (HKTs) that function as Na+ uniporters (Tester and Davenport, 2003). A nonselective cation channel can be ruled out for Nax1 because it appears to be Na+-selective in withdrawing cations from the xylem in the sheath. The concentration of Na+ in the sheath increased over time, whereas that of K+ diminished (Fig. 5). This selectivity for Na+ over K+ was also shown by Davenport et al. (2005), where unidirectional uptake of 22Na+, but not 86Rb+, into leaf sheaths was elevated in Line 149 compared to cv Tamaroi. A nonselective cation channel cannot be ruled out for Nax2.
HKT transporters characterized so far are Na+-selective or function in Na+-coupled K+ symport (although the latter may be an artifact of heterologous expression in at least some cases; Haro et al., 2005). The rice OsHKT8 transporter is Na+-selective and is proposed to withdraw Na+ from the xylem (Ren et al., 2005). The Arabidopsis (Arabidopsis thaliana) Na+-selective ortholog AtHKT1 appears to withdraw Na+ from the xylem along the length of the transpiration stream (Sunarpi et al., 2005). It is possible that Nax1 is a HKT transporter involved in Na+ withdrawal from the xylem and expressed in root and leaf vasculature. Na+-selective HKTs have been implicated in Na+ withdrawal from the xylem with concomitant enhancement of K+ uptake to the shoot, but it is not clear whether the HKTs affect K+ transport directly, or indirectly, via an influence on cation homeostasis (Rus et al., 2004; Ren et al., 2005).
Other Mechanisms Control Root Na+ Concentrations
Although lines containing either Nax1 or Nax2 excluded 98% of the Na+ from entering the shoot, in the absence of both genes, 94% was excluded. This means that other genes control the net uptake of Na+ from the soil solution and possibly the net loading of the xylem. Control of Na+ concentrations in wheat roots is quite remarkable. In experiments when the external NaCl concentration ranged up to 150 mm, the maximal Na+ concentration in roots was no more than 50 mm, even in durum wheat lines lacking Nax1 and Nax2 (Husain et al., 2004). There was little genotypic difference in root concentration but a large difference in shoot concentration. This was also observed by Gorham et al. (1990) for a wider range of wheat species.
The physiological mechanism for this control of root Na+ concentrations is not just restriction of unidirectional uptake, which is quite high in relation to the net rates of Na+ uptake (Davenport et al., 1997), but to Na+ efflux, as shown by a significant amount of 22Na+ efflux found in roots of both Line 149 and cv Tamaroi (Davenport et al., 2005). Lines without Nax1 and Nax2 also withdrew one-half of Na+ from xylem (Fig. 4), which presumably was then effluxed.
Retranslocation of Na+ from Shoot to Root Is Small
The experiment involving labeling of a split-root system with 22Na+ indicates that shoot export of Na+ was only a small proportion of the import, likely to be no more than 10%. This is similar to the value of 10% for barley grown in 100 mm NaCl, obtained from direct measurements of phloem sap collected by aphid stylets (Wolf et al., 1990).
Nax2 clearly does not control loading of Na+ into the phloem because the rate of retranslocation was the same for [+]Nax1 and [−]Nax2 lines. However, the [+]Nax1 line had twice the rate of retranslocation as the [−]Nax1 line (Fig. 6). This result was surprising because we expected higher rates of retranslocation in the phloem to be associated with higher shoot Na+ concentrations and, consequently, to be greater in the [−]Nax1 and [−]Nax2 lines, which had higher Na+ concentrations in both blade and sheath than their isogenic pairs (Table I). It is possible that higher rates of retranslocation of Na+ to the roots in the [+]Nax1 line are a function of high tissue concentrations at the base of the leaf sheath (Fig. 5) and possibly specific localization of Na+ in the cells in the vascular bundles that might be involved in loading of the phloem. Recirculation of 22Na+ that was deposited in the shoot base was shown in common bean (Jacoby, 1979) and common reed (Matsushita and Matoh, 1991). We conclude that enhanced recirculation in the [+]Nax1 family is an indirect effect of the Nax1 gene, not the primary effect.
Relationship of Nax Genes to Other Na+-Excluding Genes in Wheat
It is apparent that both Nax1 and Nax2 genes come from T. monococcum C68-101, a diploid A genome species, and not from the durum parent of Line 149, cv Marrocos.
The function of the Nax genes is generally similar to the Kna1 gene in bread wheat, which is on chromosome 4D (Dubcovsky et al., 1996). However, Nax1 varies from Kna1 in phenotype as well as in homoeologous chromosomal location. Nax1 is located on chromosome 2A (Lindsay et al., 2004) and carries the phenotype of retention of Na+ in the leaf sheath and a high sheath to blade Na+ concentration ratio. In contrast, the phenotype of Nax2 is the same as Kna1, as described by Gorham et al. (1990). Like Kna1, Nax2 results in low Na+ and high K+ concentrations in the leaf blades of plants growing in 50 mm NaCl and does not cause preferential deposition of Na+ in the leaf sheath. We have examined several bread wheat cultivars and found the same concentration in sheath and blade (R. Munns and R. James, unpublished data). It is possible that Nax2 and Kna1 may be homoeologous genes.
In summary, both Nax genes restrict the transport of Na+ from roots to shoots with a high selectivity for K+ over Na+. Both result in enhanced K+-Na+ discrimination in the leaf blade, although by different mechanisms. The Nax1 gene promotes withdrawal of Na+ from the xylem in the base of the leaf as well as the root. This gene could serve a unique function in reducing the movement of Na+ into the leaf blade at high salinity or in conditions when root function is impaired, such as in waterlogged soil. The Nax2 gene is likely to perform a similar function to Kna1.
MATERIALS AND METHODS
Plant Materials
Parental material used in crossing and in Na+ uptake and flux experiments were durum wheat (Triticum turgidum) Line 149 and cv Tamaroi, and the parents of Line 149, Triticum monococcum C68-101 and durum cv Marrocos. Seeds were provided by Dr. Ray Hare of the Tamworth Agricultural Institute, New South Wales Department of Primary Industries.
Development of Near-Isogenic Nax1 and Nax2 Lines
An F2 family derived from a cross between Line 149 and cv Tamaroi previously identified a microsatellite marker (gwm312) closely linked to the Nax1 gene (Lindsay et al., 2004). From this F2 family, individuals with leaf Na+ concentrations as low as parent Line 149 were selected and backcrossed to cv Tamaroi four times. Each backcross was selfed, and individual F2 plants with the lowest leaf Na+ concentrations were used for the next backcross. The BC4F2 family of 100 individuals was used to isolate the Nax1 and Nax2 genes into separate BC5F2 single-gene families. Selections were based on allelic variation of the gwm312 marker in combination with a Na+ phenotype screen. The presence of Nax2 was evident in lines that carried the cv Tamaroi allele for gwm312 but were intermediate for Na+. Whereas plants that were homozygous for the Line 149 gwm312 allele usually had a low Na+ phenotype, some plants were intermediate for Na+, indicating the possible absence of Nax2.
To develop Lines containing only Nax1, BC4F2 individuals were selected that were homozygous for the Line 149 allele of gwm312 but had an intermediate Na+ concentration. To develop lines containing only Nax2, BC4F2 individuals were selected that were homozygous for the cv Tamaroi allele of gwm312 but had an intermediate Na+ concentration. These selections were backcrossed to the recurrent parent cv Tamaroi and selfed in the BC5F1. The resulting BC5F2 families were scored for leaf Na+ concentration. Plants were grown in supported hydroponics as described previously (Munns and James, 2003). At approximately 6 d after emergence, 25 mm NaCl was added twice a day to a final concentration of 150 mm, and CaCl2 was added to give a final concentration of 10 mm. Plants were grown in a controlled environment chamber with a 10-h photoperiod and photosynthetic photon flux density of 800 μmol m−2 s−1 at 25°C during the day and 18°C during the night. After 10 d, the blade of leaf 3 was harvested and Na+ concentration was measured (Lindsay et al., 2004). Progeny testing of the resulting BC5F3 lines using leaf Na+ concentration score confirmed zygosity.
Homozygous BC5F3 low and high Na+ near-isogenic Nax1 and Nax2 lines used in Na+ uptake studies were given the annotations [+]Nax1, [−]Nax1, [+]Nax2, and [−]Nax2, respectively.
Na+ and K+ Transport Rates and Gradients in Na+ Concentrations along the Leaf
Plants were grown as described above, except that the final NaCl concentration was 50 mm NaCl, and CaCl2 was added to give a final concentration of 4 mm. To measure transport rates, plants were harvested after 6 and 10 d in 50 mm NaCl, six replicates per harvest. Previous studies had shown that Na+ net uptake rates reached steady state in parental lines cv Tamaroi and Line 149 at 5 d in 50 mm NaCl (Davenport et al., 2005). Shoots were separated into leaf blades and leaf sheaths. Roots were washed in a cold solution of 10 mm Ca(NO3)2 for 10 to 15 s, blotted, and weighed. All plant material was then dried at 70°C for 3 d, weighed, and extracted in 500 mm HNO3 at 80°C for 1.5 h and analyzed for Na+ and K+ by an inductively coupled plasma-atomic emission spectrometer (Varian Vista Pro). Net Na+ and K+ transport rates (roots to shoots) were calculated on a root fresh-weight basis according to Pitman (1988) and Storey (1995). The rate of net ion uptake, J (mol groot fresh weight−1 d−1) was calculated as:
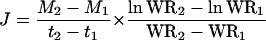 |
(1) |
and ion transport rates from roots to shoots, Js (mol groot fresh weight−1 d−1), as:
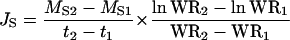 |
(2) |
where M2 and M1 are the ion contents of the whole plant (mol), Ms2 and Ms1 are the shoot ion contents (mol) at times t1 and t2 (d), and WR1 and WR2 are the corresponding root fresh weights (g). Rates and standard errors were calculated on paired plants after ranking the six plants at each harvest in order of increasing root dry weight.
Transpiration rates, E (gwater groot fresh weight−1 d−1), were estimated from the measured leaf area and whole-plant water loss of a corresponding set of seedlings grown in pots containing coarse sand, which were watered and flushed daily with 50 mm NaCl in one-half-strength modified Hoagland solution. The xylem concentration (mol L−1) was estimated from the uptake rate and the transpiration rate as:
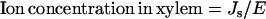 |
(3) |
This estimation presumes there is no retranslocation from shoots to roots.
For the analysis of ion gradients along leaves, plants were harvested at 2 and 5 d in 50 mm NaCl. Leaf sheaths were divided into five equal segments (between 1–3 mg dry weight) from the basal tissue (connecting to the root-shoot junction) to the upper tissue (connecting to the ligule). Leaf blades were divided into three segments: two basal segments, similar in size to sheath segments, and the remainder of the leaf blade.
22Na+ Uptake
Na+ uptake to the root and shoot was measured using 22Na+ as described previously (Davenport et al., 2005). Seeds were germinated and then transferred to Eppendorf tubes with the base removed and suspended over hydroponic solution. Seedlings were exposed to one-half-strength modified Hoagland solution (P concentration reduced from 1 mm to 100 μm) for 5 d and then transferred to one-half-strength modified Hoagland plus 25 mm NaCl and 2 mm CaCl2 for 5 d before experiments.
22Na+ Retranslocation
22Na+ retranslocation into roots was measured with a split-root system. Three-day-old seedlings were transferred to a pretreatment solution of 25 mm NaCl and 2 mm CaCl2 in one-half-strength modified Hoagland solution. After 7 d, the roots were divided evenly and placed in two beakers that were covered with foil and connected by tape, each containing 120 mL of pretreatment solution. The shoot was supported between the two beakers in an upright position and placed on a rotating shaker under a light bank with a photosynthetic photon flux density of 150 μmol m−2 s−1 and a 16-h photoperiod for 20 h before solutions were refreshed and 22Na+ added to one beaker. Labeled and unlabeled roots and shoots were harvested after 48 h and 22Na+ was measured in the unlabeled root and surrounding solution. Na+ retranslocation was calculated as a percentage of total shoot 22Na+, taking into account the size of the labeled and unlabeled roots.
Withdrawal of 22Na+ from Xylem by Upper Parts of Roots
Seedlings were grown as described for 22Na+ uptake and transferred to a flat Perspex chamber (15 × 2.5 × 2.5 cm) with two unequal-sized compartments separated by a movable Perspex barrier pierced with a hole for the root. The seedling was secured in the larger compartment so that the shoot was upright and the lower portion (4–5 cm) of a single root was sealed into the smaller compartment with silicon grease. The small compartment was filled with 5 mL of 25 mm NaCl in one-half-strength Hoagland solution and the rest of the root system was covered in filter paper wetted with the same solution (identical to the saline growth solution). Both compartments were sealed to maintain high humidity. Plants were placed under a light bank on a slowly rotating shaker for 1 h before the solution was replaced with 22Na+-labeled solution of the same composition. After 2 h, the plant was harvested and divided into three parts: the labeled root, the upper part of the root and contacting paper, and the shoot. Withdrawal of Na+ from xylem in the upper part of the root was calculated as the amount of 22Na+ there as a percentage of the total amount of 22Na+ transported from the labeled root (i.e. 22Na+ in the upper root and shoot).
Statistical Analysis
Data summarized in Tables I to IV were analyzed using ANOVA and lsds (P = 0.05) were used to compare genotype means.
Acknowledgments
We thank our colleague Ray Hare, durum breeder from New South Wales Department of Primary Industries, for provision of novel genetic material and for identifying the pedigree of Line 149. We are also grateful to Wolfgang Spielmeyer, Evans Lagudah, Mark Tester, and John Passioura for advice and support; Karen Glover for marker analysis; and Lorraine Mason for cation analysis.
This work was supported by the Commonwealth Scientific and Industrial Research Organization (study award to R.A.J.), the Royal Society (Dorothy Hodgkin fellowship to R.J.D.), and the Grains Research and Development Corporation (to R.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard A. James (richard.james@csiro.au).
References
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Reid RJ, Smith FAS (1997) Sodium-calcium interactions in two wheat species differing in salinity tolerance. Physiol Plant 99: 323–327 [Google Scholar]
- Dubcovsky J, Santa Maria G, Epstein E, Luo MC, Dvořák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92: 448–454 [DOI] [PubMed] [Google Scholar]
- Gorham J, Wyn Jones RG, Bristol A (1990) Partial characterization of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta 180: 590–597 [DOI] [PubMed] [Google Scholar]
- Haro R, Bañuelos MA, Senn ME, Barrero-Gil J, Rodríguez-Navarro A (2005) HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol 139: 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain S, Munns R, Condon AG (2003) Effect of sodium exclusion trait on chlorophyll retention and growth of durum wheat in saline soil. Aust J Agric Res 54: 589–597 [Google Scholar]
- Husain S, von Caemmerer S, Munns R (2004) Control of salt transport from roots to shoots of wheat in saline soil. Funct Plant Biol 31: 1115–1126 [DOI] [PubMed] [Google Scholar]
- Jacoby B (1964) Function of bean roots and stems in sodium retention. Plant Physiol 39: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby B (1979) Sodium recirculation and loss from Phaseolus vulgaris. Ann Bot (Lond) 43: 741–744 [Google Scholar]
- James RA, Rivelli AR, Munns R, von Caemmerer S (2002) Factors affecting CO2 assimilation, leaf injury and growth in salt-stressed durum wheat. Funct Plant Biol 29: 1393–1403 [DOI] [PubMed] [Google Scholar]
- Johanson JG, Cheeseman JM (1983) Uptake and distribution of sodium and potassium by corn seedlings. Plant Physiol 73: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D (1983) The possible role of transfer cells in the adaptation of plants to salinity. Physiol Plant 58: 549–555 [Google Scholar]
- Kramer D, Läuchli A, Yeo AR, Gullasch J (1977) Transfer cells in roots of Phaseolus coccineus: ultrastructure and possible function in exclusion of sodium form the shoot. Ann Bot (Lond) 41: 1031–1040 [Google Scholar]
- Lacan D, Durand M (1996) Na+-K+ exchange at the xylem/symplast boundary. Plant Physiol 110: 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda CF, Cambraia J, Oliva MA, Ruiz HA, Prisco JT (2003) Solute accumulation and distribution during shoot and leaf development in two sorghum genotypes under salt stress. Environ Exp Bot 49: 107–120 [Google Scholar]
- Läuchli A (1984) Salt exclusion: an adaptation of legumes for crops and pastures under saline conditions. In RC Staples, ed, Salinity Tolerance in Plants: Strategies for Crop Improvement. Wiley, New York, pp 171–187
- Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Matsushita N, Matoh T (1991) Characterization of Na+ exclusion mechanisms of salt-tolerant reed plants in comparison with salt-sensitive rice plants. Physiol Plant 83: 170–176 [Google Scholar]
- Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Gardner PA, Tonnet ML, Rawson HM (1988) Growth and development in NaCl-treated plants. 2. Do Na+ or Cl− concentrations in dividing or expanding tissues determine growth in barley? Aust J Plant Physiol 15: 529–540 [Google Scholar]
- Munns R, Hare RA, James RA, Rebetzke GJ (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51: 69–74 [Google Scholar]
- Munns R, James RA (2003) Screening methods for salt tolerance: a case study with tetraploid wheat. Plant Soil 253: 201–218 [Google Scholar]
- Munns R, Rebetzke GJ, Husain S, James RA, Hare RA (2003) Genetic control of sodium exclusion in durum wheat. Aust J Agric Res 54: 627–635 [Google Scholar]
- Pitman MG (1988) Whole plants. In DA Baker, JL Hall, eds, Solute Transport in Plant Cells and Tissues. Longman Scientific & Technical, Harlow, UK, pp 346–391
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rivelli AR, James RA, Munns R, Condon AG (2002) Effect of salinity on water relations and growth of wheat genotypes with contrasting sodium uptake. Funct Plant Biol 29: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Rus A, Lee BH, Muñnoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136: 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shone MGT, Clarkson DT, Sanderson J (1969) The absorption and translocation of sodium by maize seedlings. Planta 86: 301–314 [DOI] [PubMed] [Google Scholar]
- Storey R (1995) Salt tolerance, ion relations and the effect of root medium on the response of citrus to salinity. Aust J Plant Physiol 22: 101–114 [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan W-Y, Leung H-Y, Hattori K, et al (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The TT (1973) Transference of resistance to stem rust from Triticum monococcum L. to hexaploid wheat. PhD thesis. University of Sydney, Sydney
- Wolf O, Munns R, Tonnet ML, Jeschke WD (1990) Concentrations and transport of solutes in xylem and phloem along the leaf axis of NaCl-treated Hordeum vulgare. J Exp Bot 41: 1133–1141 [Google Scholar]