Abstract
Separation of the life cycle of flowering plants into two distinct growth phases, vegetative and reproductive, is marked by the floral transition. The initial floral inductive signals are perceived in the leaves and transmitted to the shoot apex, where the vegetative shoot apical meristem is restructured into a reproductive meristem. In this study, we report cloning and characterization of the maize (Zea mays) flowering time gene delayed flowering1 (dlf1). Loss of dlf1 function results in late flowering, indicating dlf1 is required for timely promotion of the floral transition. dlf1 encodes a protein with a basic leucine zipper domain belonging to an evolutionarily conserved family. Three-dimensional protein modeling of a missense mutation within the basic domain suggests DLF1 protein functions through DNA binding. The spatial and temporal expression pattern of dlf1 indicates a threshold level of dlf1 is required in the shoot apex for proper timing of the floral transition. Double mutant analysis of dlf1 and indeterminate1 (id1), another late flowering mutation, places dlf1 downstream of id1 function and suggests dlf1 mediates floral inductive signals transmitted from leaves to the shoot apex. This study establishes an emergent framework for the genetic control of floral induction in maize and highlights the conserved topology of the floral transition network in flowering plants.
The population of undifferentiated stem cells comprising the shoot apical meristem (SAM) gives rise to all aboveground vegetative and reproductive structures of higher plants. Early during vegetative growth, the SAM produces leaves and axillary meristems, while later, during reproductive growth, the SAM produces inflorescences and flowers (McSteen et al., 2000). The switch from vegetative to reproductive development is called the floral transition. This critical point in plant development enables plants to time their reproductive phase of growth to coincide with optimal environmental conditions, thus ensuring reproductive success. The timing of the floral transition is affected by many inputs, including both endogenous signals and environmental cues (Mouradov et al., 2002; Boss et al., 2004; Bernier and Perilleux, 2005).
It is well accepted that the floral transition is triggered in the shoot apex by a leaf-derived mobile signal that is produced under inductive conditions (Knott, 1934; Zeevaart, 1976; Colasanti and Sundaresan, 2000; Corbesier and Coupland, 2005). Classic experiments with photoperiod-sensitive plants have shown that exposure of a single leaf to inductive photoperiods can produce a graft-transmissible signal capable of promoting flowering at the shoot apex. This mobile signal is termed “florigen” (Knott, 1934; Chailakhyan, 1936). Although the exact molecular nature of florigen is unknown, recent studies in Arabidopsis (Arabidopsis thaliana) identify FLOWERING LOCUS T (FT) mRNA as being a component of the mobile signal, as FT transcript is produced in leaves but moves to the shoot apex to activate floral identity genes (Abe et al., 2005; Huang et al., 2005; Wigge et al., 2005).
Molecular genetic analyses in Arabidopsis have established detailed models to explain the regulation of flowering time in this dicot species (Koornneef et al., 1998; Mouradov et al., 2002; Simpson and Dean, 2002). In Arabidopsis, the time to flower is regulated by the integration of four basic floral promotion pathways: photoperiod, autonomous, vernalization, and gibberellic acid pathways (Mouradov et al., 2002; Boss et al., 2004; Bernier and Perilleux, 2005). Less is known about what controls the floral transition in monocots, but a few genes have been identified from rice (Oryza sativa) and wheat (Triticum aestivum; Colasanti and Sundaresan, 2000; Hayama et al., 2003; Yan et al., 2003, 2004). In rice, three key regulatory genes are conserved between Arabidopsis and rice: GIGANTEA, CONSTANS (CO), and FT, although, under long-day photoperiods, rice FT expression is suppressed, leading to suppression of flowering, which is the opposite regulation of FT in Arabidopsis (Yano et al., 2000; Hayama et al., 2002; Kojima et al., 2002). In wheat, genes with similarity to APETALA1 (AP1) and CO have been cloned and shown to regulate the dependence of winter wheat cultivars on vernalization, a prolonged period of cold temperature required to promote flowering in spring (Yan et al., 2003, 2004).
Although maize (Zea mays) is one of the most widely grown monocot crops, very little is known about the molecular control of flowering time. In maize, genetic analyses have defined two separate loci that affect flowering time based on their loss of function: indeterminate1 (id1) and delayed flowering1 (dlf1; Neuffer et al., 1997). Mutations of these loci delay the floral transition to various degrees, prolonging vegetative growth to produce a late flowering phenotype. Mutations in id1 have the most severe effects on the floral transition, resulting in mutant plants flowering many weeks later than normal (Singleton, 1946; Colasanti et al., 1998). Mutations in dlf1 have a less dramatic effect on flowering, producing mutants that flower 1 to 2 weeks later than their wild-type sibs. These late flowering phenotypes define each locus as being required for normal floral transition. Of these two loci, only the id1 gene has been cloned and molecularly characterized. id1 encodes a zinc-finger DNA-binding protein expressed specifically in immature leaves (Colasanti et al., 1998; Kozaki et al., 2004). The floral induction pathway defined by id1 may be unique to monocots, as no clear id1 ortholog exists in the Arabidopsis genome (Colasanti et al., 2006). Consistent with its pattern of mRNA accumulation in leaves, id1 is thought to regulate the production or transmission of an inductive floral signal from the leaves to the shoot apex.
In this study, we report cloning and molecular characterization of the maize flowering locus dlf1. Loss of dlf1 function results in late flowering, indicating dlf1 is required for timely promotion of the floral transition. dlf1 encodes a putative basic Leu zipper (bZIP) transcription factor that is a member of a conserved protein family. Three-dimensional (3D) modeling of normal and missense mutant protein bound to a consensus bZIP DNA target suggests DLF1 functions through DNA binding. dlf1 is expressed in the shoot apex and transcript accumulation peaks near the time of the floral transition, indicating a threshold level of dlf1 is required for promotion of the floral transition. Analysis of dlf1 transcript accumulation in late flowering dlf1 and id1 mutant backgrounds connects regulation of dlf1 expression to a downstream factor. Analysis of dlf1 id1 double mutants places dlf1 downstream of id1 function. Therefore, we have cloned the dlf1 floral induction gene, which acts downstream of id1-derived leaf-produced signals in the shoot apex to elicit flowering.
RESULTS
The dlf1 Mutant Phenotype
Two dlf1 mutations (dlf1-N2389A and dlf1-N2461A) were isolated through ethyl methane sulfonate (EMS) mutagenesis by M.G. Neuffer (Neuffer et al., 1997) and obtained from the Maize Genetic Cooperation Stock Center (http://w3.ag.uiuc.edu/maize-coop/mgc-home.html). The dlf1 mutant phenotype is distinguished by an increase in leaf number (node no.) and a delay in flowering time (no. of days to shed pollen and exsert silks) by about 10 to 14 d compared to wild-type sib plants (Fig. 1A). The mutant phenotype suggests the normal function of dlf1 is to promote the floral transition and, when nonfunctional, results in late flowering. The dlf1 mutation segregates as a single Mendelian recessive mutation (Supplemental Table S1).
Figure 1.
Phenotype of dlf1 mutant plants. A, A wild-type sib plant (left, red arrow) compared to a dlf1 mutant plant (right, red arrowhead) in the field after flowering of the wild-type plant. Note the dlf1 mutant is taller, with extra leaves, and the tassel has not yet fully extended from the upper whorl of leaves. B, Close-up of a dlf1 tassel with increased branch number and lower branches bearing pistillate flowers enclosed in husk-like leaves (white arrow). Compare to the wild-type tassel in A. C, Close-up of dlf1 with extra ears born at the top ear-bearing node. Wild-type plants bear only single ears at one or two nodes.
One measure of flowering time is to count the number of leaves produced by the SAM before it transitions to reproductive growth. Thus, flowering time mutants that transition late remain in the vegetative stage longer and produce a greater number of leaves. To characterize the effect of the dlf1 mutation more fully, both dlf1-N2389A and dlf1-N2461A were backcrossed into several maize inbred backgrounds of different maturities and segregating families produced. The late flowering phenotype of both dlf1-N2389A and dlf1-N2461A is similar and independent of the maturity of the inbred background into which each was introgressed (Table I). In addition to the increase in leaf number, both dlf1 mutants produce plants with more biomass, having additional nodes and a much thicker stalk (Fig. 1A). Mutant plants also have an increased number of tassel branches and visible ear shoots (Fig. 1, B and C). Often, the tassel morphology is perturbed such that many of the lower tassel branches carry pistillate florets and are enclosed in husk-like leaves. Additionally, dlf1 mutant plants bear ears at three to four nodes with each node carrying up to two to three ears compared to wild-type plants, which bear single ears at one or at most two nodes. These effects are more commonly seen with the mutation introgressed into later maturity inbred backgrounds. Taken together, these phenotypes define dlf1 function as a promoter of the floral transition with pleiotropic effects on inflorescence development. Previous reports placed the dlf1 gene on the long arm of chromosome 7 (Neuffer et al., 1997). We mapped the dlf1 gene to the distal tip of the long arm of chromosome 7 in bin 7.06 between markers phi051 and phi116 (data not shown).
Table I.
Different dlf1 mutations delay flowering in different inbred backgrounds
| Mutant Allele | Population | Average Leaf No. Wild Typea | Average Leaf No. dlf1 |
|---|---|---|---|
| dlf1-N2389A | CM37 BC5F2bd | 14.1 ± 0.9 (34) | 21.5 ± 0.7 (16) |
| PHRE1 BC5F2bd | 17.1 ± 0.6 (50) | 22.3 ± 1.0 (8) | |
| PHN46 BC4F2be | 18.1 ± 0.6 (54) | 25.6 ± 0.6 (17) | |
| Mo17 BC4F2c | 17.6 ± 1.1 (73) | 24.0 ± 1.3 (25) | |
| dlf1-N2461A | PHN46 BC3F2be | 18.6 ± 0.8 (52) | 26.7 ± 1.0 (13) |
| B73 BC4F2b | 21.1 ± 0.7 (46) | 27.9 ± 1.3 (12) |
Average leaf number ± sd (no. of plants).
Grown in field 2002.
Grown in greenhouse fall 1997.
CM37 and PHRE1 are early maturity inbreds.
PHN46 is a midmaturity inbred.
Isolation of Transposon-Tagged dlf1 Alleles, Gene Cloning, and Gene Structure
To clone the dlf1 gene, new Mutator (Mu) transposon-induced alleles were isolated by crossing the original reference allele, dlf1-N2389A, in a standard transposon-tagging strategy (Chomet, 1994; Brutnell, 2002). From the Mu-tagging population, six new Mu-induced dlf1 mutations were recovered and confirmed as heritable (Supplemental Table S2).
A modified PCR-based cloning method (selected amplification of insertion flanking fragments [SAIFF]) was used to isolate genomic fragments that cosegregated with the dlf1 mutant phenotype from bulked DNA isolated from 10 homozygous wild-type and 10 homozygous dlf1-mu468 plants from BC2F2 families (Frey et al., 1998; da Costa e Silva et al., 2004). An amplification product of 800 bp was found in the dlf1-mu468 bulked DNA, which cosegregated perfectly with all 10 mutant individuals but was absent in all 10 homozygous wild-type sib plants (data not shown). An oligonucleotide primer specific to the 800-bp Mu-adjacent genomic DNA fragment was paired with a Mu-specific primer and used to amplify a single PCR product from dlf1-mu453, dlf1-mu710, and dlf1-mu461 families. Perfect cosegregation of this Mu-adjacent fragment with four independent dlf1 mutations proved this fragment was part of the dlf1 gene.
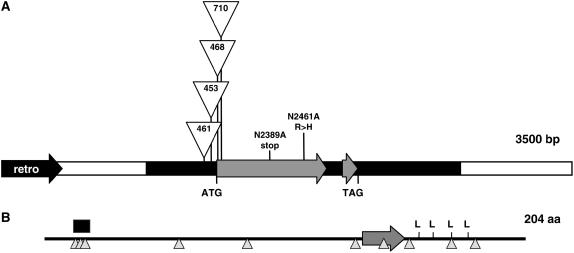
To obtain the genomic structure of the dlf1 gene, bacterial artificial chromosome (BAC) libraries were screened with over-go probes homologous to the Mu-adjacent fragment that cosegregated with the dlf1 phenotype. The dlf1 gene was localized within 3,486 bp of unique sequence surrounded by repetitive DNA with homology to retroelements (Fig. 2A). BLAST comparisons of the genomic sequence to maize expressed sequence tags (ESTs) in GenBank only identified two partial EST sequences (accessions CB885390 and DN209699). One partial EST sequence contained a poly(A) tail that set the boundary for the 3′ end of the transcription unit. A full-length cDNA was obtained by utilizing a reverse transcription (RT)-PCR primer-scanning technique (see “Materials and Methods”). The longest cDNA amplified was approximately 1.3 kb, with the start of transcription located between −192 and −183 from the A (=+1) of the ATG from the longest predicted open reading frame (ORF). The alignment between the genomic sequence and cDNA showed that the dlf1 gene contains a 612-bp-long ORF that encodes a putative protein of 204 amino acids and is interrupted by a single 80-bp intron. The poly(A) site is located about 540 bp downstream of the stop codon. Hybridization of genomic DNA from both B73 and Mo17 inbreds with a dlf1 3′-untranslated region (UTR) probe revealed a single HindIII band, indicating that dlf1 is a single-copy gene in the maize genome (data not shown).
Figure 2.
dlf1 genomic organization and protein structure. A, Diagram of the genomic organization of dlf1. The white line indicates the unique 5′ and 3′ sequences flanking the dlf1 coding sequence (gray arrows). The black arrow (retro) denotes the closest 5′ retroelement. The thick black line indicates, from left to right, the 5′-UTR, the single intron, and the 3′-UTR. The positions of the start (ATG) and stop (TAG) codons are noted. The insertion sites for four of the dlf1-mu alleles are indicated as are the position of the two EMS alleles. B, Organization of DLF1 protein motifs. The basic domain is indicated by the large gray arrow, the Leu zipper by the four Leu, a Ser-rich domain by the black box, and the position of putative Ser/Thr phosphorylation sites by upward-pointing triangles.
Cloning of the four independent Mu-insertion alleles revealed that two alleles had insertions in the ORF of exon 1, while the other two had insertions upstream of the start codon within the 5′-UTR (Fig. 2A). Of the two upstream insertions, the insertion further from the start codon (dlf1-mu461) has a weak delayed flowering phenotype, while the insertion closer to the start codon (dlf1-mu453) has a stronger mutant phenotype, indistinguishable from the two ORF insertions (dlf1-mu468, dlf1-mu710; Supplemental Table S2). The nature of the defect of the two EMS alleles was also investigated. The entire dlf1 coding region was amplified by PCR using DNA isolated from the EMS mutants and sequenced. The dlf1-N2389A allele has a C-to-T transition that converts a CAG codon to a TAG stop codon terminating the protein prematurely at amino acid position 88 (Fig. 2A). The mutation in dlf1-N2461A is a transition from G to A, changing an Arg (CGC) to His (CAC) at amino acid position 143 (Fig. 2A).
DLF1 Protein Features and Phylogenetic Analyses
According to InterProScan analysis (Zdobnov and Apweiler, 2001), the DLF1 protein is annotated as a bZIP transcription factor with a nuclear localization signal. The DLF1 protein contains a typical bZIP domain with the consensus N142-x7-R150-x9-L160-x6-L167-x6-L174-x6-L181 (Jakoby et al., 2002). A basic region between amino acids 133 and 156 includes an invariant DNA-binding motif N-x7-R/K, followed by a heptad repeat of four Leu. Nine Ser and one Ser/Thr predicted phosphorylation sites (Fig. 2B) were detected in the DLF1 protein using the program NetPhos 2.0 server (Blom et al., 1999). Therefore, activity of DLF1 might be modulated by phosphorylation and, like other bZIP transcription factors, it potentially participates in a signal transduction pathway (Siberil et al., 2001).
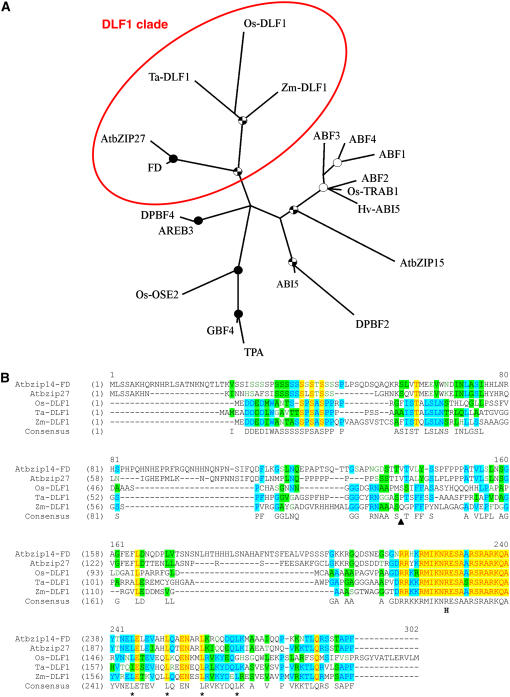
dlf-like genes were found in rice and wheat. The rice dlf-like gene is predicted from genomic sequence (GenBank accession BAC79182.1, chromosome 9). The wheat EST (CK206464) encodes a complete DLF-like protein with a 612-bp coding region and a 500-bp-long 3′-UTR. The relative sizes of the ORF and 3′-UTR are similar to the maize dlf1 gene, 612 bp and 550 bp, respectively. Maize, rice, and wheat DLF-like proteins share 48% to 54% identity over their entire amino acid sequence.
To understand the evolutionary relationship of the maize DLF1 protein with other bZIP transcription factors, we compared DLF1 with other plant proteins having a predicted bZIP signature. The bZIP family of proteins has been divided into 10 subgroups based on sequence similarity of their basic region and additional conserved domains (Jakoby et al., 2002). Phylogenetic analysis performed with the PHYLUP program places DLF1 and other monocot DLF-like proteins into group A. The maize DLF1 protein and monocot DLF-like proteins grouped with the Arabidopsis proteins AtbZIP14 and AtbZIP27, defining a DLF1 clade (Fig. 3A). The amino acid alignment of these five proteins within the DLF1 clade (Fig. 3B) identifies a Ser-rich domain at the N-terminal end of the proteins and the homologous basic domain followed by four Leu in the zipper domain. Loss-of-function mutations in Arabidopsis AtbZIP14, also known as FLOWERING LOCUS D (FD; Koornneef et al., 1991; Abe et al., 2005; Wigge et al., 2005; GenBank accession AB105818), result in late flowering, confirming that members of the DLF1 clade of group-A bZIP proteins promote the floral transition in both monocots and dicots.
Figure 3.
Phylogeny and sequence comparison of DLF1 and other bZIP proteins. A, Phylogenetic relationship of DLF1 and other bZIP proteins. The unrooted consensus tree built with the PHYLIP program indicates the relationship of maize DLF1 to Arabidopsis, rice, wheat, and barley (Hordeum vulgare) group-A bZIP proteins based on the classification of Jakoby et al. (2002). The DLF1 clade is circled. Details of the proteins used are listed in Supplemental Table S6. One thousand bootstrap replicates were used to assess the confidence of the branching, which is indicated by a black circle for bootstrap values of 90% to 100%, a checked circle for values of 70% to 90%, and a white circle for values of 50% to 70%. B, Alignment of members of the DLF1 clade. Members of the DLF1 clade were aligned with the VectorNTI alignment tool (Invitrogen). Yellow highlights indicate amino acid identity is conserved across all the proteins. Blue highlights indicate 60% amino acid identity and green highlights indicate 40% amino acid identity across the aligned proteins. The black triangle indicates the position of the premature STOP in the dlf1-N2389A nonsense mutation. The “H” indicates the position of the R>H substitution in the dlf1-N2461A missense mutation. The four Leu in the canonical zipper domain are denoted by an asterisk (*). AtbZIP14, the FD gene, Arabidopsis, BN000021; AtbZIP27, Arabidopsis, BN000022; Os-DLF1, rice, AB109206.2; and Ta-DLF1, wheat, EST CK206464.
3D Modeling of the Deduced Protein Encoded by the dlf1 Missense Mutation
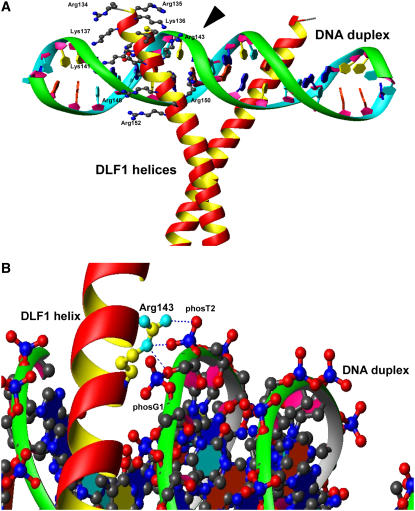
The wild-type DLF1 protein differs from the missense DLF1-N2461A protein by a single conservative amino acid substitution (Arg-143>His), yet the dlf1-N2461A mutation is probably nonfunctional. We constructed a 3D structure model of the DLF1 basic region and DNA complex by homologous modeling. Of the known protein structures, the basic region of DLF1 is best matched to a cAMP response element-binding (CREB; pdbcode: 1dh3) bZIP protein from mouse (Schumacher et al., 2000). The crystal structure of the CREB bZIP contains two α-helices in a scissors-grip shape, dimerized by the Leu zipper motif and bound to a 21-bp DNA segment. The CREB bZIP recognizes the bound DNA segment through interactions with the major groove defined by the sequence ATGACGTCAT. Plant bZIP proteins also consistently favorably bind to DNA sequences with an ACGT core (Jakoby et al., 2002). The CREB bZIP was used as the modeling template and sequence alignment was constructed by a profiling match. As the DLF1-N2461A mutation Arg-143>His occurred in the basic region, we only modeled the protein-DNA interaction portion and left the Leu zipper motif intact.
In the DLF1 structure model, the basic region helices fit into the major groove of the target DNA formed by the sequence ATGACGTCAT (Fig. 4A). Similar to other bZIP proteins, the DLF1 structure model demonstrated that DNA recognition was through two sets of interactions. First, side chains of invariant residues Asn-142 and Arg-150, together with conserved small residues Ser-145 and Ala-146, make direct contact to the edges of the nucleotide bases at the bottom of the major groove through either hydrogen bonds or van der Waals forces. Second, a number of basic Lys and Arg residues open their long arms gripping the ridges of the phosphodiester chain on both sides of the major groove (Fig. 4A). Arg-137, Arg-150, Arg-152, Lys-136, Lys-141, and the missense mutation site Arg-143 directly contact the phosphate groups of DNA, while Lys-135, Lys-153, and Arg-152 form water-mediated interactions to DNA. The backbone atoms of Arg-143 are buried in a valley formed by the DLF1 helix and DNA chain while the guanidinium group on its flexible long side chain protrudes out, forming direct interactions with DNA phosphate groups phosT2 and phosG1 (Fig. 4B). In modeling the DLF1-N2461A Arg-143>His mutant, we found that the space normally occupied by Arg-143's aliphatic side chain was unable to accommodate the bulky imidazole ring of His without severely distorting the conformation of the DNA backbone. The interaction energy between the phosphate group and His is much weaker compared to that for Arg-143. Therefore, the Arg-143>His mutation significantly reduces the binding affinity of DLF1-N2461A to DNA. Moreover, in CREB bZIP, two basic residues, Arg-294 and Arg-298, are aligned and directly contact the phosphate groups. Conversely, in this region of DLF1, only one of the two corresponding residues, Arg-143 and Ala-147, respectively, is basic. The disruption of this basic residue by the Arg-143>His mutation might not be tolerated. Multiple sequence alignments constructed by pfam's bZIP_1 profile revealed that Arg-143 is almost invariant in plant bZIP proteins. Of 202 plant sequences, AtbZIP31 (Jakoby et al., 2002) is the only exception in which Arg is replaced by a similar residue, Gln. The nonplant bZIP proteins show slightly more variation at this position with substitution of Lys, Gln, Asn, Thr, but not His. The bZIP structural analysis and structural modeling demonstrated all these allowable substitutions (Lys, Gln, Asn, Thr) are able to fit into the space occupied by Arg and maintain hydrogen bonding to phosphate group phosT2. Taken together, the contact between Arg-143 and DNA is critical for bZIP's function.
Figure 4.
3D structure model of the DLF1 basic domain complexed to DNA. A, 3D structure model of the DLF1 basic domain and Leu zipper bound to its target DNA duplex. The DLF1 protein dimerizes at the Leu zipper and binds the DNA duplex with its basic domain in a scissor-like grip. Arg-143 is indicated by the black arrowhead. B, Close-up of the 3D structure model indicating the potential linkage of Arg-143 to two phosphate groups of the DNA backbone.
Expression of dlf1 in the Shoot Apex
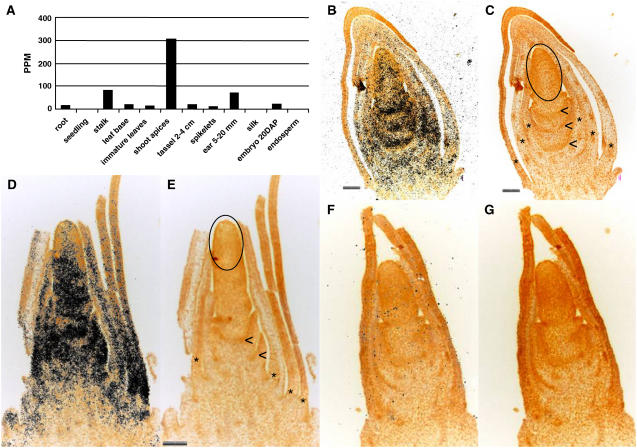
Temporal and spatial specific expression patterns of the dlf1 gene were identified through analysis of expressed dlf1 17-mer sequence tags using massively parallel signature sequencing (MPSS; Brenner et al., 2000). The most abundant expression of dlf1 was found in shoot apices collected near the time of the floral transition, accumulating to about 300 ppm (Fig. 5A). For comparison, maize tubulin (GenBank accession P18025) transcripts accumulate to about 1,000 ppm in the same shoot apex samples (data not shown). Expression of dlf1 was also detected in several other tissues, including immature ears, stalk, root tip meristems, embryos, and leaves, but at a much reduced level, accumulating only to 20 to 30 ppm. To confirm the in silico results, poly(A+) mRNA was isolated from shoot apices of wild type (B73), mutant dlf1-N2389A (in the CM37 inbred background), and mutant dlf1-N2461A (in the B73 inbred background) at the V3-V5 seedling stage (growth stages defined according to http://maize.agron.iastate.edu/corngrows.html). RNA gel-blot hybridization (Supplemental Fig. S1A) detected a transcript in all samples of approximately 1200 bp in length, which is consistent with the predicted transcript length (1.2–1.3-kb; Fig. 2A).
Figure 5.
Transcript localization of dlf1. A, Relative dlf1 expression abundance in ppm in different maize tissues determined by MPSS technology. DAP, Days after pollination. B and D, Localization of dlf1 transcript by in situ hybridization to longitudinal sections of a V5 stage early floral transition shoot apex (B) and V6 stage late floral transition shoot apex (D) with dlf1 antisense probe. Hybridization is indicated by black dots. Sense strand hybridization is shown in F. C, E, and G, Autofluorescence images of the same images in B, D, and F, respectively, to show details of the structures in the shoot apex. Asterisks mark the base of the immature leaves surrounding the SAM, arrowheads indicate developing stem, and the oval encircles the SAM proper. Scale bars = 100 microns and paired images are the same magnification.
To understand the spatial distribution of dlf1 expression in the shoot apex, in situ hybridizations were performed. Hybridization with antisense dlf1 shows transcript accumulation throughout longitudinal sections of the shoot apex during the floral transition (Fig. 5, B and D). The signal is less intense near the SAM and more intense in regions beneath the SAM, including the base of the nascent leaf primordia and the developing stem. Hybridization of similar stage shoot apices with dlf1 sense probe produced no signal (Fig. 5F).
To understand the dynamics of dlf1 expression through different stages of development, quantitative RT-PCR (qRT-PCR) was conducted on RNA isolated from shoot apices (including the SAM, leaf primordia, and subtending stem tissue) collected from wild-type (B73), dlf1-N2461A (missense mutant), and id1-m1 (late flowering Ds2-insertion mutant) plants at representative growth stages. Transcript abundance was measured and quantified by multiplex gene expression analysis (Johnson et al., 2002) conducted by Althea Technologies. Quantitative levels of amplification products for dlf1 were normalized to the internal control α-tubulin and expressed as a ratio (see “Materials and Methods”). For wild-type samples, dlf1 transcript is present in the shoot apex during vegetative growth, increases and peaks near the time of the floral transition, and then decreases to undetectable levels after the transition in early reproductive growth (Fig. 6). Such a pattern mirrors the two-phase expression pattern of FD in transitioning Arabidopsis shoot apices (Wigge et al., 2005; Searle et al., 2006). Furthermore, the pattern of expression is consistent with dlf1 functioning as a promoter of the floral transition after accumulating beyond a threshold level.
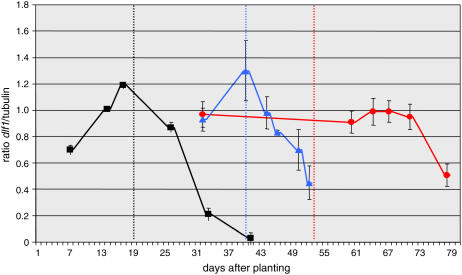
Figure 6.
Quantitative expression levels of dlf1 before, during, and after the floral transition in three genotypes. Quantitative expression of dlf1 is expressed as a ratio in arbitrary units (y axis) at different days after planting (x axis). Expression in wild type (inbred B73) is indicated by the black squares, in dlf1-N2461A by the blue triangles, and in id1-m1 by the red circles. Ratios are the average of two biological replicates and three technical replicates. Errors bars are sds. The time of the floral transition is indicated by the dotted line for each genotype; B73, black; dlf1-N2461A, blue; and id1-m1, red.
We also examined the pattern of dlf1 transcript accumulation in two late flowering mutant backgrounds. In the dlf1-N2461A missense mutant, as in wild type, the peak of expression occurred near the time of the floral transition, but the transition itself was delayed by approximately 20 d (Fig. 6). This pattern of dlf1 mRNA accumulation suggests the pattern and timing of expression is linked to an alternate signaling pathway that functions downstream of dlf1. In id1-m1 mutant apices, dlf1 transcript accumulates to levels comparable to late vegetative stage wild-type levels (compare to B73 14 d after planting) during the greatly expanded vegetative growth stage of this late flowering mutant but does not peak near the floral transition. After the floral transition occurs in id1-m1 mutants, dlf1 transcript levels remain relatively constant for many days. Only later when the id1-m1 mutants are further in the reproductive stage of growth does dlf1 transcript decline (Fig. 6).
Epistatic Analysis of id1 and dlf1
To clarify if dlf1 and id1 function within separate or the same floral inductive pathway, single and double mutants introgressed into the Mo17 inbred background were generated and characterized for their effects on flowering time. If dlf1 and id1 reside in separate pathways, then dlf1 id1 double mutant plants will flower later than either single mutant. On the other hand, if the dlf1 and id1 pathways converge, then double mutant plants will flower no later than the single, severe late flowering id1-m1 mutant. Two strong delayed flowering alleles, dlf1-mu468 and dlf1-mu453, were used to produce families segregating single dlf1, single id1-m1, and double dlf1 id1-m1 mutants. dlf1 and id1 mutants display distinct late flowering phenotypes with regard to leaf number, tassel morphology and plant vigor (Fig. 1A; Supplemental Fig. S2). Therefore, identification of each genotype is clear, based on its late flowering phenotype in the field. In segregating families, wild-type plants produced 16 to 17 leaves, dlf1 mutant plants had 25 to 27 leaves, and id1-m1 mutant plants had 34 to 42 leaves (Supplemental Fig. S2). All late flowering plants were genotyped at both dlf1 and id1 loci by PCR using primers designed specifically for each allele. dlf1 mutant plants defined by phenotype were all homozygous for the loss-of-function allele at dlf1, confirming their phenotypic classification. Furthermore, all the dlf1 mutant plants were either homozygous wild type or heterozygous at the id1 locus. None were homozygous id1-m1. Conversely, id1-m1 mutant plants were homozygous wild type, heterozygous, or homozygous mutant at the dlf1 locus (Supplemental Table S3). The same result was obtained for both mutant dlf1 alleles tested. Thus, dlf1 id1 double mutants have a phenotype indistinguishable from the single id1 homozygous mutants. These data indicate that id1 is epistatic to dlf1, and dlf1 must function within the same pathway but downstream of id1.
DISCUSSION
dlf1 Promotes the Floral Transition in Maize
Previous EMS mutagenesis studies identified dlf1 as a recessive late flowering mutation (Neuffer et al., 1997). Late flowering mutants delay the timing of the floral transition and, as a result, prolong the vegetative stage of growth. As expected for a late flowering mutation, dlf1 mutants have an extended vegetative growth stage and initiate more leaves before transitioning to reproductive growth. Regardless of genetic background, on average dlf1 mutants produce five to nine more leaves and flower 1 to 2 weeks later than wild-type sibs. Such a phenotype indicates that the product of the dlf1 gene promotes the floral transition. Additional defects affecting inflorescence development also mark the dlf1 mutation. Both tassel and ear development are perturbed in dlf1 mutant plants. The detection of dlf1 transcript in developing ears (Fig. 5A) hints at a functional role for dlf1 in inflorescence development.
It is interesting to note that the sole other recessive late flowering mutation in maize, id1, also has inflorescence development defects in certain backgrounds (Singleton, 1946; Colasanti et al., 1998). Similar to dlf1, id1 mutant tassels have increased branching, but, distinct from dlf1, id1 tassels can revert to a vegetative state of development, displaying a “ball of shoots” phenotype. Whereas dlf1 mutants bear more ears than normal, id1 mutants never produce ears. This might be due to the more severe late flowering resulting from loss of id1 function. Although both dlf1 and id1 function as promoters of the floral transition, disruption of their function has distinctly different effects on inflorescence development. These differences may reflect the specific mechanistic function each gene plays in the hierarchy of floral inductive signaling or differences in the nature of their downstream target genes. Identification of downstream targets for both dlf1 and id1 will help to clarify this issue.
dlf1 Encodes a Putative bZIP Transcription Factor
Using a standard targeted transposon-tagging strategy and cosegregation analysis of four independent Mu alleles and two independent EMS alleles proved we cloned the dlf1 gene. dlf1 encodes a typical bZIP protein with a canonical basic DNA-binding domain and a Leu zipper motif important for protein dimerization (Jakoby et al., 2002). bZIP proteins function as transcription factors in the regulation of diverse biological processes such as pathogen defense, seed germination, flower development, and light, stress, and hormone signaling. In Arabidopsis, 75 putative bZIP proteins were predicted from the genomic sequence as containing a canonical bZIP domain. These 75 bZIP proteins were further subdivided into 10 groups based on sequence similarities of their basic region and other conserved domains (Jakoby et al., 2002). Phylogenetic analysis places DLF1 into group A (Fig. 3A). The group-A bZIPs have a C-terminal basic domain, three to four Leu repeats, and additional conserved phosphorylation motifs scattered throughout the protein. In general, group-A bZIPs appear to function in abscisic acid or stress signaling in seeds and vegetative tissues (Jakoby et al., 2002).
Homology comparisons with other plant databases identified two predicted proteins in monocots—one rice and one wheat—with significant sequence similarity to DLF1 (Fig. 3B). The two monocot DLF proteins group with the Arabidopsis bZIPs AtbZIP14 and AtbZIP27. AtbZIP14 has recently been identified as the floral activator FD (Abe et al., 2005; Wigge et al., 2005), suggesting DLF1 function is conserved in the regulation of the floral transition between monocots and dicots. Although outside the scope of this study, complementation of the Arabidopsis fd mutation with dlf1 would substantiate the presumed orthologous relationship of these two genes. Further, whether the rice and wheat DLF-like proteins function as flowering regulators will require additional functional studies, but it is tempting to speculate that all the members of the DLF1 clade define a floral transition family of bZIP proteins.
The activity of bZIP proteins is known to be regulated by phosphorylation (Siberil et al., 2001), and the DLF1 protein has 10 putative phosphorylation sites (Fig. 2B). The basic domain of bZIPs also contains a bipartite nuclear localization signal that is required for efficient translocation of the protein from the cytoplasm to the nucleus. The putative phosphorylation site within the basic domain of DLF1 is within the nuclear localization signal, and modification of this site might affect import of the protein into the nucleus. Alternatively, phosphorylation may reduce DNA-binding affinity of the DLF1 protein as phosphorylation of Ser residues within the basic domain introduces a negative charge near positions that interact directly with DNA. Therefore, the activity of DLF1 has the potential to be regulated by its phosphorylation state.
Our 3D structure model of DLF1 is compatible with previously modeled bZIPs and highlights critical contacts and interactions between the basic region residues and the DNA backbone. Our model provides a mechanistic explanation for the late flowering phenotype of the Arg-143>His missense mutation in DLF1-N2461A. This amino acid substitution produces a strong dlf1 mutant phenotype that is indistinguishable from dlf1-N2389A, which carries a premature stop codon truncating most of the protein including the bZIP domain (Fig. 4B). The structure model predicts that Arg-143 forms direct contact with two phosphate groups of the target DNA. Substitution of His at this position is predicted to not fit into the space normally occupied by Arg-143 without severe distortion of the DNA backbone. Thus, DLF1-N2461A might no longer bind its DNA target with comparable affinity. The structure model also predicts that the size and charge of amino acids at position 143 within the basic domain are critical for dlf1 function. Additionally, the model supports the idea that DLF1 must bind DNA to be functional.
dlf1 Promotes the Floral Transition at the Shoot Apex
Similar to FD in Arabidopsis, dlf1 is expressed in the shoot apex before, during, and after the floral transition (Supplemental Fig. S1B; Wigge et al., 2005). As vegetative growth proceeds, dlf1 mRNA abundance increases and peaks near the time of the floral transition (Fig. 6). After the floral transition, as reproductive development ensues, dlf1 mRNA abundance declines sharply, and later reaches undetectable levels. These results suggest that a critical level of dlf1 is required to promote the floral transition, and after the transition dlf1 expression is down-regulated as it is no longer needed during inflorescence development. Our in situ results show dlf1 transcripts localize preferentially beneath the SAM at the base of leaf primordia and in the developing stem (Fig. 5, B and D). The temporal and spatial expression of dlf1 is similar to the expression pattern of FD in Arabidopsis (Wigge et al., 2005). These data hint that dlf1 function may be associated with transducing the floral signal from leaves to the apical meristem. In Arabidopsis, FD mediates inductive floral signals transmitted from the leaves to the shoot apex through interaction with FT to activate expression of the meristem identity genes AP1, FUL, and CAL (Abe et al., 2005; Huang et al., 2005; Wigge et al., 2005). Since a maize FT gene has yet to be identified, we do not know if DLF1 interacts with an FT ortholog in the shoot apex. However, we hypothesize that since FD/DLF1 function is conserved between dicots and monocots, DLF should interact with an FT ortholog in maize to activate expression of meristem identity genes, thereby acting as an integrator of leaf-derived floral inductive signals at the shoot apex.
dlf1 Expression Is Linked to a Downstream Factor
The null dlf1-N2461 allele provides an opportunity to study expression of dlf1 in a dlf1 loss-of-function background. Our qRT-PCR results indicate that mutant dlf1-N2461A mRNA accumulation increases and peaks near the time of the floral transition, a pattern similar to wild type but shifted later in time to coincide with the delayed transition (Fig. 6). These results indicate that the timing and pattern of dlf1 transcript accumulation are not solely dependent on signals upstream of dlf1 since these signals are expected to be functional in the dlf1-N2461A mutant. Additionally, since the dlf1-N2461A missense mutation flowers as late as the dlf1-N2389A premature stop mutation, dlf1-N2461A is probably nonfunctional. Accordingly, we would expect no increase in transcript accumulation in dlf1-N2461A if dlf1 feedback regulates its own expression. Therefore, the timing and pattern of dlf1 transcript accumulation in dlf1-N2461A is not dependent on upstream signals or on dlf1 function. We hypothesize another gene (x) exists that functions downstream of dlf1 and is regulated by dlf1 and input from an alternate floral inductive pathway. We reason that the timing and pattern of dlf1 transcript accumulation must be dependent on gene x, which is linked to the timing of the floral transition through an alternate induction pathway and also feedback regulates dlf1 since dlf1 transcript accumulation reports the later floral transition of the dlf1-N2461A mutant.
dlf1 Is Regulated by Signals Downstream of id1
To define epistatic interactions, double dlf1 id1 mutants were constructed and analyzed for their effects on flowering time. Our results showed dlf1 functions downstream of id1, as dlf1 id1 double mutants flower no later than single id1 mutants (Supplemental Table S3). It is not clear if dlf1 expression or another aspect of its activity is regulated downstream of id1. Our qRT-PCR analysis indicates that dlf1 transcript accumulates in id1-m1 mutants to levels similar to prefloral transition wild-type levels but does not appear to peak near the floral transition (Fig. 6). The lack of peak may be explained by the fact that the timing of the floral transition in id1-m1 mutants occurs over a broad period of time, often longer than 1 week (our unpublished data). Therefore, our expression results likely reflect an average accumulation of dlf1 transcript in pre-, post-, and floral transition stage id1-m1 shoot apices. This would also explain the minor but broad increase in dlf1 expression seen at 61 to 73 d after planting (Fig. 6). Alternatively, other aspects of dlf1 activity may be regulated downstream of id1, such as protein stability or protein modification (Gallie, 1993; Day and Tuite, 1998; Bailey-Serres, 1999; Nambara and McCourt, 1999; del Pozo and Estelle, 2000; Galichet and Gruissem, 2003; Laugesen et al., 2004; Novatchkova et al., 2004; Boisvert et al., 2005; Love and Hanover, 2005). In fact, bZIP proteins are known to participate in signal transduction pathways and are regulated by their phosphorylation state (Siberil et al., 2001). Therefore, the activity of the DLF1 protein may be dependent on signals downstream of id1 that affect its phosphorylation status, although such a possibility will require further investigation.
A Model for Floral Transition in Maize
In conclusion, we have cloned the dlf1 gene, a promoter of the floral transition in maize. The dlf1 gene encodes a putative bZIP transcription factor that likely functions through DNA binding. The spatial and temporal expression patterns of dlf1 in the shoot apex suggest a threshold level of dlf1 is required to mediate transmission of leaf-derived inductive signals for timely reprogramming of the SAM to reproductive growth. Expression analysis of dlf1 in late flowering mutant backgrounds and double mutant analyses suggest a model for control of the floral transition in maize.
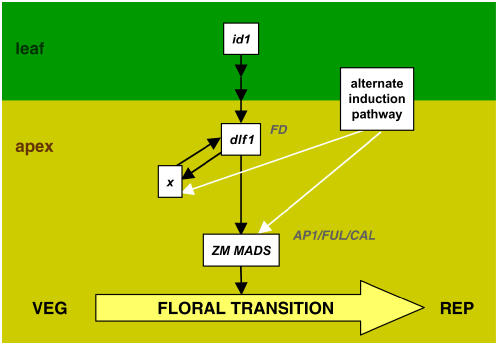
In our model (Fig. 7), id1 is a monocot-specific regulator of a leaf-derived floral stimulus that activates dlf1 in the shoot apex, possibly via transcriptional regulation or posttranscriptional modifications of the DLF1 protein. After a critical level of DLF1 is reached, the floral transition is induced, presumably through up-regulation of downstream meristem identity genes to restructure the meristem. In Arabidopsis, direct and indirect targets of the dlf1 putative ortholog FD include the MADS-box transcription factors SUPPRESSOR OF OVEREXPRESSION OF CONSTANS, SEPALLATA3, AP1, FUL, and CAL (Abe et al., 2005; Teper-Bamnolker and Samach, 2005; Wigge et al., 2005; Searle et al., 2006). Similar to Arabidopsis, maize MADS-box homologs (ZM MADS) may be targets of dlf1. Several candidates are currently being tested. Based on our qRT-PCR studies, we hypothesize the increase and peak of dlf1 expression are regulated through a positive feedback loop by the downstream gene x. Since dlf1 and id1 mutants eventually transition, an alternate induction pathway is proposed to function in parallel to the id1-dlf1 pathway. In the absence of dlf1 or id1 function, signaling through this alternate pathway would offer another route to floral induction and would also allow expression of the downstream targets genes x and ZM MADS. This model proposes a molecular genetic framework for the floral transition in maize. Future experiments designed to test the relationships proposed in this model and to identify additional flowering time regulatory genes will confirm and extend this framework, furthering our understanding of the molecular basis of flowering in diverse plant species.
Figure 7.
Proposed model of the genetic network for floral transition in maize. The switch from vegetative (VEG) growth to reproductive (REP) growth is marked by the floral transition (yellow arrow). The floral inductive signal initiates in leaves (leaf, top) and its production or transmission is regulated by id1. The activity of dlf1 in the shoot apex (apex, bottom) is indirectly (multiple arrows) regulated downstream of id1 function. Expression of dlf1 activates an early downstream target (gene x) that feeds back to up-regulate dlf1. This positive feedback loop results in the increase and peak of dlf1 expression to sufficient levels to promote the floral transition. Similar to known targets of FD in Arabidopsis (AP1/FUL/CAL), a maize MADS-box gene (ZM MADS) is also proposed as a direct target of dlf1. An alternate induction pathway is hypothesized (white arrows) to activate expression of genes downstream of dlf1 (x and ZM MADS). Induction through the alternate pathway would explain the ability of mutant dlf1 or mutant id1 plants to eventually flower. Additionally, signaling through the alternate pathway explains the increase and peak of dlf1 mRNA coincident with the delayed floral transition in a dlf1 mutant background. The relevant Arabidopsis homologs are indicated at the appropriate nodes but not boxed.
MATERIALS AND METHODS
Mutant Stocks and Mutant Characterization
Both dlf1 EMS alleles were obtained from the Maize Genetic Cooperation Stock Center (http://w3.ag.uiuc.edu/maize-coop/mgc-home.html), stock numbers 716C (dlf1-N2389A) and 716D (dlf1-N2461A). All plants were grown in the Pioneer nursery under standard conditions with supplemental irrigation. Mutant characterization was conducted during the summer of 2002. Measurement of total leaf number was done by marking the fifth leaf produced at the V4-V5 growth stage either with an indelible marker or by cutting the leaf with serrated scissors. At the V9-V10 stage, the 10th leaf was similarly marked, as leaf number 5 usually had senesced before the total number of leaves had been produced. For the late flowering mutants, leaf number 15 was typically marked as well at V14-V15. After tassel emergence of wild-type and mutant plants, total leaf number was calculated based on counting leaves from the uppermost marked leaf (either leaf no. 10 or no. 15). The floral transition was determined by visual inspection after dissecting immature leaves from shoot apices to expose the SAM. Vegetative stage SAMs are proportionally domed shaped and initiate leaf primordia. Meristems that have elongated to a size approximately twice as tall as wide or have initiated visible branch meristems are postfloral transition stage.
Mapping of the dlf1 Locus
The dlf1 locus was already placed on chromosome 7 (Neuffer et al., 1997). To confirm this location and identify close markers, a modified bulk segregant mapping protocol was employed. The original Coop dlf1-N2389A stock was crossed to CM37 and the F1 self-pollinated to create an F2 mapping population. One hundred F2 plants were phenotyped as either wild type or dlf1, and DNA from 20 wild-type siblings (1 μg/plant) was bulked and used as template along with DNA from 15 individual dlf1 mutants for SSR mapping. Fifty-one SSR markers distributed across the 10 maize (Zea mays) chromosomes were assessed. Linkage was assigned based on the presence of two alleles in the wild-type bulk sample but segregation of one allele preferentially with the dlf1 mutant samples. The tightest linkage was detected with phi116 (two recombinants/30 chromosomes tested), which was used to distinguish plants heterozygous for the dlf1 reference allele from plants heterozygous for newly tagged dlf1-mu alleles isolated in the directed Mu-tagging screen.
Nucleic Acid Isolations
DNA was isolated from frozen mature leaves for Mu cosegregation analysis from 10 wild-type and 10 dlf1 plants each from families mu468, mu461, mu710, and mu453 using a standard urea extraction (Liu et al., 1995).
Total RNA was extracted from 1 g of material using a hot-phenol extraction procedure and selective precipitation with 4 m LiCl to remove traces of DNA and small RNA species (Verwoerd et al., 1989; Brugiere et al., 1999). RNA was quantified using a spectrophotometer (Beckman Instruments) at 260 nm. Poly(A) was prepared from total RNA (400 μg) using the Oligotex poly(A) purification kit using the manufacturer's recommendations (Qiagen).
The SAIFF Method to Clone dlf1
Genomic DNA (approximately 0.3 μg) from Mu+ (mutant) and Mu− (homozygous wild type) mu468 plants (10 each) were digested with MseI in 1× RL buffer (10 mm Tris-HCl, pH 7.5, 10 mm MgOAc, 50 mm KOAc, 5 mm DTT) at 37°C for 3 h in a final volume of 25 μL. Following denaturation at 65°C for 20 min, 5 μL of ligation mix (0.3 μL 100 mm rATP, 0.5 μL 10× RL buffer, 1 μL 40 μm adaptor, 1 μL T4 ligase [3 U/μL; Promega], and 2.2 μL of water) were added to each digestion reaction. The MseI adaptor is a mixture of 5′-TACTCAGGACTCATCGACCGT and 5′-GTGAACGGTCGATGAGTCCTGAG. After overnight incubation at 4°C, the ligation reactions were purified with the Qiagen PCR purification kit to remove the excess adaptor.
The Mu-flanking fragments were amplified with Mu TIR primer MuExt22D (5′-CCAACGCCAWSGCCTCYATTTC) and MseI adaptor primer MseExt18 (5′-GTGAACGGTCGATGAGTC) with Qiagen's HotStartTaq DNA polymerase. A 2-μL aliquot of the purified ligation reaction was used in a 10-μL PCR reaction, with a final concentration of 5% DMSO. The cycling conditions were 95°C 15 min; 94°C 30 s, 55°C 30 s, 72°C 2 min 30 s × 20 cycles; and 72°C 7 min. The PCR reaction was diluted 1:10 by adding 90 μL of water. Equal volumes from each reaction were bulked to make the Mu+ and Mu− pools (10 plants/pool).
Mu-flanking fragments from both Mu+ and Mu− pools were amplified with nested Mu TIR primer MuInt19 (5′-GCCTCYATTTCGTCGAATC) and +2 selective adaptor primers with Takara's Ex Taq DNA polymerase. A total of 16 +2 selective primers were used (Supplemental Table S4). A 1-μL aliquot of the pooled PCR reaction was used in a 10-μL PCR reaction, with a final concentration of 5% DMSO. The touchdown cycling conditions were 95°C 2 min; 94°C 30 s, 65°C to 0.8°C/cycle 30 s, 72°C 2 min 30 s × 11 cycles; 94°C 30 s, 56°C 30 s, 72°C 2 min 30 s × 24 cycles; and 72°C 7 min.
PCR products of MuInt19 paired with 16 +2 selective adaptor primers were separated on 1.5% agarose gel. A fragment segregating with the phenotype was identified with +2 primer MseIntATC. The nested PCR was repeated with MseIntATC on all the individual plants (10 Mu+ and 10 Mu−). A perfect cosegregation was observed for a fragment of 800 bp. The cosegregating fragment was sequenced, and a fragment-specific primer, 468R (5′-AGCTGCACCTTCGTCTCC), was designed to pair with the MuTIR primers. Mu insertion in the candidate gene was confirmed in family mu468 as well as three more independent mutant families, mu461, mu710, and mu453. To identify mutations in the EMS-induced reference alleles N2389A and N2461A, a set of primers was designed to cover the coding region, including the 5′- and 3′-UTRs of the dlf1 gene. 5′-CGCCGACAGACATGTCGTCCTCGAGCAC, 5′-CATCTCCACGCAGCTGAGCCTCAACTCC, 5′-TACTCGCTTTAGGAGAGCCTTTGACACG, 5′-GTTCTGAGGACATTGACCGGAGATGAG, 5′-ACCTGCTTCGACTCATCTCCGGTCAA, and 5′-GCGGTCTCTGGTGTCATTTGACCAGT. PCR products were directly sequenced.
Full-Length cDNA Isolation
To obtain full-length cDNA, a RT-PCR primer-scanning technique was used. Forward oligonucleotide primers were designed at positions −580, −512, −322, −219, −163, and −102 upstream of the putative start site of translation (A = +1 of the ATG; Supplemental Table S5). Each forward primer was paired with a reverse primer designed from sequences in the 3′-UTR just downstream of the STOP codon. Amplification was tested against two templates, genomic B73 DNA, as a positive primer control, and cDNA reverse transcribed from prefloral transition B73 shoot apex mRNA. Actin primers were used as a positive control to assess amplification competence of the cDNA template and B73 genomic templates. Additional forward primers were designed between the most 5′ forward primer that amplified a product from the cDNA template and the next 5′ primer (see Supplemental Table S5). Reiterating this process, the start of the longest dlf1 mRNA was established.
RNA Gel-Blot Analysis
For RNA gel-blot analysis, electrophoretic separation of poly(A+) RNA was performed on 1.5% agarose gels containing 5% (v/v) of a solution of 37% formaldehyde in 3-(N-morpholino)-propanesulfonic acid buffer [0.02 m 3-(N-morpholino)-propanesulfonic acid, pH 7.0, 5 mm sodium acetate, and 1 mm EDTA]. Gels were blotted to nylon membranes (Roche Molecular Biochemicals) using the TurboBlotter (Schleicher & Schuell), with 20× SSC (1× SSC is 150 mm NaCl and 15 mm sodium citrate) as the transfer buffer. Blots were probed with 32P-labeled PCR-generated probes of the full-length dlf1 gene.
MPSS Analysis
The DuPont MPSS (Solexa) database consists of libraries of 17-bp sequence tags from 2 × 105 to 2 × 106 cDNAs isolated from over 200 diverse maize tissues and developmental stages. BLAST analysis allows for quantification of a specific 17-bp signature sequence corresponding to a unique cDNA. cDNA abundance is expressed in ppm and is the number of times a particular 17-bp sequence is found in a million sequences from a library.
qRT-PCR by Althea
For quantitative expression analysis, shoot apices were collected from field-grown (summer 2004) B73, homozygous dlf1-N2461A, and homozygous id1-m1 plants from vegetative, floral transition, and reproductive growth stages in two biological replicates. Representative plants of each genotype were dissected every few days to monitor progression through development of the SAM. As the plants reached the appropriate developmental stage, five shoot apices (including one to two leaf primordia) were collected, bulked, and frozen in liquid nitrogen.
Multiplexed, quantitative RT-PCR was done in triplicate using the eXpress Profiling method by Althea Technologies. Twenty-five nanograms of total RNA from each sample was reverse transcribed, followed by PCR, using the protocol established for eXpress Profiling multiplex RT-PCR (Johnson et al., 2002). PCR products were diluted in deionized water, mixed with GENESCAN 400HD (ROX), and run on an ABI 3100 capillary electrophoresis system according to the manufacturer's protocol (Applied Biosystems). Data was analyzed by Genescan for determination of product size and relative expression levels as defined by peak area. The resultant raw data was normalized against α-tubulin as the internal control within the same reaction.
In Situ Hybridization
In situ hybridizations were performed by the Phylogeny company (http://www.phylogenyinc.com) according the protocol of Jackson (Jackson, 1991) modified according to Bradley (Bradley et al., 1993).
Cloning and Sequencing of dlf1 Genomic Fragments
The Mo17 BAC genomic library was screened with over-go probes. Five BAC clones were identified and confirmed by DNA gel-blot hybridization with gene-specific probes. HindIII and EcoRI BAC fragments were subcloned into pBluescript II KS+ (Stratagene), hybridized with over-go probes, and positive clones were sequenced.
Phylogenetic Analysis
A phylogenetic tree was produced by neighbor joining using protein distances derived from the alignment of the bZIP domain of the proteins (Supplemental Tables S6 and S7). One thousand bootstrap replicates were used to assess the confidence of the branching.
3D Homologous Modeling
The bZIP structural template was identified with HMMER against an HMM profile library, SUPERFAMILY, that represents all proteins of known structures (Madera et al., 2004). The survey of the variation of Arg-143 position among all the bZIP proteins was performed with HMMER 2.2g (Eddy, 1998) using Pfam profiles bzip_1 and bizp_2 (Bateman et al., 2002). Overall bZIP proteins were extracted from Uniprot, while the plant bZIP sequences were from the National Center for Biotechnology Information's nr database selected according to taxonomy IDs. The bZIP sequence identification cutoff was based on E value of <0.001 among HMMER 2.2g matches.
The DLF1 model was obtained by manually changing each mismatched residue from the template and subsequently searching various rotamers to remove steric conflict and position the side chain into a local minimum. The raw model was then under a series of energy minimizations, first Steepest Descent, followed by Conjugated Gradients, with constraints to fix the DNA heavy atoms and restrict the helical backbone movement. Structure analysis, energy minimization, manual adjustment, and sequence alignment were performed with InsightII (2001 release; Accelrys) on the Octane workstation of Silicon graphics.
Double Mutant Analyses
Families segregating single and double dlf1 id1 mutants were constructed by crossing heterozygous dlf1/+ plants to heterozygous id1-m1/+ plants in the Mo17 inbred background. Heterozygotes were confirmed by PCR genotyping. Primers used to genotype id1-m1 were as follows: F-5′-TGCTCCTGCATATATGCGAGGGAATGCT, R-5′-GATCCGTCCGGTGAGAGATTTAGGCT, and Ds2 TIR-5′-GCTTTTCTTGCATGGGATGGGCCTCAAA. Primers used to genotype the dlf1 gene were as follows: F-5′-GCGGTCTCTGGTGTCATTTGACCAGT, R-5′-CTCAGCTGCGTGGAGATGAAG, dlf1-mu453 MuTIR-5′-GTCTATAAATGACAATTATCTCGCATAGAGG, and dlf1-mu468 MuTIR-5′-CTTCCCTCTTCGTCCATAATGGCAA.
PCR amplification was performed using Expand Long Template DNA polymerase (Roche). The PCR conditions were 95°C for 2 min, followed by 35 cycles at 94°C for 30 s, 60°C for 45 s, 72°C for 45 s, and a final extension of 72°C for 10 min. Single and double mutant characterization was done during the summer 2005 season on field-grown plants.
Sequence data from this article can be found in the EMBL/GenBank data libraries for the genomic dlf1 sequence under accession number EF093788 and for the longest mRNA under accession number EF093789.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of dlf1 in the shoot apex.
Supplemental Figure S2. Phenotypes of wild type, dlf1 mutant, and id1 mutant.
Supplemental Table S1. Segregation ratio of dlf1-N2389A in different F2 backgrounds.
Supplemental Table S2. List of new Mu-induced dlf1 mutations and phenotypes.
Supplemental Table S3. Epistatic analysis of id1 and dlf1.
Supplemental Table S4. List of SAIFF 16 +2 selective primers.
Supplemental Table S5. List of dlf1 forward primers tested in primer-scanning method.
Supplemental Table S6. List of bZIP proteins used in the phylogenetic analysis.
Supplemental Table S7. Multiple sequence alignment of the bZIP domain used in constructing the phylogenetic tree.
Acknowledgments
We thank Pedro Hermon for tissue sampling, Zhanshan Dong for collecting field data for double mutants, Xin Meng and Sunita Chilakamari for genotyping the double mutants, and David Selinger for the phylogenetic analysis. We are grateful to Phylogeny for in situ hybridizations, Althea Technologies for quantitative RT-PCR, and Solexa for production of the MPSS expression data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Olga N. Danilevskaya (olga.danilevskaya@pioneer.com).
The online version of this article contains Web-only data.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J (1999) Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci 4: 142–148 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G, Perilleux C (2005) A physiological overview of the genetics of flowering time control. Plant Biotechnol J 3: 3–16 [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Chenard CA, Richard S (2005) Protein interfaces in signaling regulated by arginine methylation. Sci STKE 2005: re2. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell (Suppl) 16: S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Brugiere N, Dubois F, Limami AM, Lelandais M, Roux Y, Sangwan RS, Hirel B (1999) Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11: 1995–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP (2002) Transposon tagging in maize. Funct Integr Genomics 2: 4–12 [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK (1936) New facts in support of the hormonal theory of plant development. CR (Dokl) Acad Sci URSS 13: 79–83 [Google Scholar]
- Chomet PS (1994) Transposon Tagging with Mutator. Springer-Verlag, New York
- Colasanti J, Sundaresan V (2000) ‘Florigen’ enters the molecular age: long-distance signals that cause plants to flower. Trends Biochem Sci 25: 236–240 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AY, Coneva V, Kozaki A, Mable BK (2006) The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Coupland G (2005) Photoperiodic flowering of Arabidopsis: integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ 28: 54–66 [Google Scholar]
- da Costa e Silva O, Lorbiecke R, Garg P, Muller L, Wassmann M, Lauert P, Scanlon M, Hsia A-P, Schnable PS, Krupinska K, et al (2004) The Etched1 gene of Zea mays (L.) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. Plant J 38: 923–939 [DOI] [PubMed] [Google Scholar]
- Day DA, Tuite MF (1998) Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol 157: 361–371 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44: 123–128 [DOI] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Gierl A (1998) A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J 13: 717–721 [Google Scholar]
- Galichet A, Gruissem W (2003) Protein farnesylation in plants—conserved mechanisms but different targets. Curr Opin Plant Biol 6: 530–535 [DOI] [PubMed] [Google Scholar]
- Gallie DR (1993) Posttranscriptional regulation of gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 77–105 [Google Scholar]
- Hayama R, Izawa T, Shimamoto K (2002) Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol 43: 494–504 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Huang T, Bohlenius H, Eriksson S, Parcy F, Nilsson O (2005) The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309: 1694–1696 [DOI] [PubMed] [Google Scholar]
- Jackson D (1991) In situ hybridization in plants. In DJ Bowles, SJ Gurr, M McPherson, eds, Molecular Plant Pathology: A Practical Approach. Oxford University Press, Oxford, pp 163–174
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Johnson PH, Walker RP, Jones SW, Stephens K, Meurer J, Zajchowski DA, Luke MM, Eeckman F, Tan Y, Wong L, et al (2002) Multiplex gene expression analysis for high-throughput drug discovery. Screening and analysis of compounds affecting genes overexpressed in cancer cells. Mol Cancer Ther 1: 1293–1304 [PubMed] [Google Scholar]
- Knott JE (1934) Effect of a localized photoperiod on spinach. Proceedings of the Society of Horticultural Science 31: 152–154 [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ (1998) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen S, Bergoin A, Rossignol M (2004) Deciphering the plant phosphoproteome: tools and strategies for a challenging task. Plant Physiol Biochem 42: 929–936 [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA (2005) The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE 2005: re13. [DOI] [PubMed] [Google Scholar]
- Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J (2004) The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res 32: D235–D239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5: 61–66 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, McCourt P (1999) Protein farnesylation in plants: a greasy tale. Curr Opin Plant Biol 2: 388–392 [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Coe EH, Wessler SR (1997) Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220: 1–8 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Goodman RH, Brennan RG (2000) The structure of a CREB bZIP·somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J Biol Chem 275: 35242–35247 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siberil Y, Doireau P, Gantet P (2001) Plant bZIP G-box binding factors: modular structure and activation mechanisms. Eur J Biochem 268: 5655–5666 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Singleton WR (1946) Inheritance of indeterminate growth in maize. J Hered 37: 61–64 [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD (1976) Physiology of flower formation. Annu Rev Plant Physiol 27: 321–348 [Google Scholar]