Abstract
The U-box motif is a conserved domain found in the diverse isoforms of E3 ubiquitin ligase in eukaryotes. From water-stressed hot pepper (Capsicum annuum L. cv Pukang) plants, we isolated C. annuum putative U-box protein 1 (CaPUB1), which encodes a protein containing a single U-box motif in its N-terminal region. In vitro ubiquitination and site-directed mutagenesis assays revealed that CaPUB1 possessed E3 ubiquitin ligase activity and that the U-box motif was indeed essential for its enzyme activity. RNA gel-blot analysis showed that CaPUB1 mRNA was induced rapidly by a broad spectrum of abiotic stresses, including drought, high salinity, cold temperature, and mechanical wounding, but not in response to ethylene, abscisic acid, or a bacterial pathogen, suggesting its role in the early events in the abiotic-related defense response. Because transgenic work was extremely difficult in hot pepper, in this study we overexpressed CaPUB1 in Arabidopsis (Arabidopsis thaliana) to provide cellular information on the function of this gene in the development and plant responses to abiotic stresses. Transgenic Arabidopsis plants that constitutively expressed the CaPUB1 gene under the control of the cauliflower mosaic virus 35S promoter had markedly longer hypocotyls and roots and grew more rapidly than the wild type, leading to an early bolting phenotype. Microscopic analysis showed that 35S∷CaPUB1 roots had increased numbers of small-sized cells, resulting in disordered, highly populated cell layers in the cortex, endodermis, and stele. In addition, CaPUB1-overexpressing plants displayed increased sensitivity to water stress and mild salinity. These results indicate that CaPUB1 is functional in Arabidopsis cells, thereby effectively altering cell and tissue growth and also the response to abiotic stresses. Comparative proteomic analysis showed that the level of RPN6 protein, a non-ATPase subunit of the 26S proteasome complex, was significantly reduced in 35S∷CaPUB1 seedlings as compared to the wild type. Pull-down and ubiquitination assays demonstrated that RPN6 interacted physically with CaPUB1 and was ubiquitinated in a CaPUB1-dependent manner in vitro. Although the physiological function of CaPUB1 is not yet clear, there are several possibilities for its involvement in a subset of physiological responses to counteract dehydration and high-salinity stresses in transgenic Arabidopsis seedlings.
Higher plants are continuously faced with various environmental stresses during their entire life cycle. Drought, high salinity, heavy metals, and extreme temperatures are common abiotic stresses that seriously impair the growth and development of soil plants. Water deficiency resulting from dehydration and high salinity is one of the most severe environmental factors responsible for the reduction of crop yield on as much as one-half of the world's irrigated land (Boyer, 1982; Cushman and Bohnert, 2000). To cope with such unfavorable growth conditions, plants have developed unique defense mechanisms and processes for acclimation that enhance their tolerance to detrimental conditions. A number of genetic and cellular events that occur under such stress have been widely documented (Bray, 1997; Ishitani et al., 1997; Zhu, 2002). Diverse sets of genes induced by drought and/or salt stress have been identified recently with the aid of combined molecular and genetic approaches (Ingram and Bartels, 1996; Cushman and Bohnert, 2000; Seki et al., 2001, 2002; Ozturk et al., 2002; Oono et al., 2003). However, the biological functions of these genes in terms of stress tolerance or sensitivity are still largely unknown in higher plants. Thus, it is of immense importance to study the functions of stress-inducible genes to understand the molecular mechanisms of stress tolerance in crop plants.
The ubiquitin (Ub) 26S proteasome system is a crucial regulatory mechanism for protein degradation in all eukaryotic cells. This system degrades a wide range of proteins in the nucleus and cytoplasm and plays a key role in the control of cellular functions as diverse as cell cycle progression, endocytosis, protein sorting, embryogenesis, hormone responses, defense against pathogens, and senescence (Frugis and Chua, 2002; Vierstra, 2003; Moon et al., 2004; Smalle and Vierstra, 2004). Ub is a highly conserved 76-amino acid polypeptide. In the Ub 26S proteasome pathway, multiple Ub chains serve as degradation tags, leading to proteolysis of target proteins by the 26S proteasome complex. Ub is attached to substrate proteins in three consecutive steps, catalyzed by the set of enzymes E1, E2, and E3 (Kraft et al., 2005; Stone et al., 2005). In the initial step, E1 activates Ub in an ATP-dependent fashion. The activated Ub is transferred to the active site of the Ub-conjugating enzyme, E2. The Ub ligase E3 binds E2 and the substrate protein and catalyzes the formation of an isopeptide linkage between the carboxyl-terminal Gly of Ub and a Lys residue of the target protein (Vierstra, 2003; Moon et al., 2004). Additional Ub monomers are then sequentially added to the substrate-bound Ub moieties. In higher plants, E3 Ub ligases are encoded by a large gene family composed of diverse isoforms. On the basis of subunit composition, E3s can be divided into two groups. The homologous to the E6-AP carboxyl terminus and really interesting new gene (RING)/U-box E3 classes consist of a single subunit, whereas the SKP1, cullin/CDC53 F-box protein and anaphase-promoting complex E3 ligases consist of multiple polypeptides (Vierstra, 2003; Moon et al., 2004). Regardless of their subunit types, E3 Ub ligases are responsible for identifying proteins that should be ubiquitinated, thereby determining target specificity (Smalle and Vierstra, 2004).
Hot pepper (Capsicum annuum L. cv Pukang), a solanaceous species closely related to tobacco (Nicotiana tabacum), is one of the most economically important crops and is cultivated widely in East Asia for its hot-tasting fruits. We are interested in elucidating the adaptive response of hot pepper plants against abiotic stresses, such as water deficit. Recently, we have isolated and characterized a broad spectrum of cDNAs from hot pepper plants whose expression is enhanced rapidly in response to drought (Choi et al., 2002; Park et al., 2003; Hong and Kim, 2005). Among the identified cDNAs, pCaDRS3 (GenBank accession no. CK327619) encodes a partial protein that is homologous to a U-box-containing enzyme with putative Ub ligase activity (Hong and Kim, 2005). In this study, we have isolated a full-length CaDRS3 for C. annuum putative U-box protein 1 (CaPUB1) and analyzed the detailed expression pattern of its mRNA in response to various abiotic stresses, hormones, and wounding. We also present results indicating that overexpression of CaPUB1 alters plant response to abiotic stress as well as cell growth and differentiation in transgenic Arabidopsis (Arabidopsis thaliana) plants.
RESULTS
Isolation and Identification of Full-Length CaPUB1 cDNA
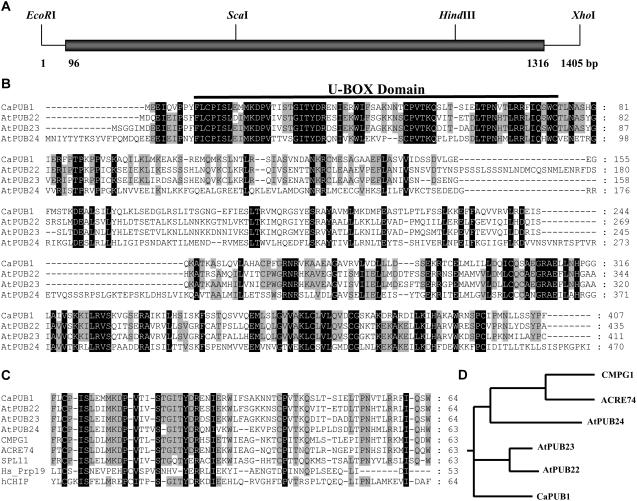
Using differential display PCR and subtractive hybridization analysis, we previously identified a broad spectrum of partial cDNA clones from hot pepper seedlings, which were rapidly induced by dehydration (Choi et al., 2002; Park et al., 2003; Hong and Kim, 2005). One of the isolated clones, pCaDRS3 (GenBank accession no. CK327619), renamed CaPUB, encoded a partial polypeptide homologous to a U-box-containing protein (Hong and Kim, 2005) and was used to isolate a full-length pCaPUB cDNA clone. The partial cDNA fragment, pCaDRS3, was radioactively labeled and used as a probe to screen the λ-uni-Zap II cDNA library constructed from water-stressed leaves of hot pepper plants. Numerous putative CaPUB cDNA clones were isolated. Restriction enzyme mapping and DNA-sequencing analysis indicated that these clones represented a single group of overlapping sequences. Among the isolated clones, pCaPUB1 contained the longest insert. The pCaPUB1 insert was 1,405 bp, consisting of a 95-bp 5′-untranslated region, a 1,221-bp coding region encoding 407 amino acids, and an 89-bp 3′-untranslated region (Fig. 1A). The sequence was deposited in GenBank (accession no. DQ211901). The predicted molecular mass of the protein encoded by pCaPUB1 was 45.1 kD, with a calculated pI of 9.02. The complete pCaPUB1 sequence was compared with other U-box proteins. A database search revealed that CaPUB1 was most closely related to the Arabidopsis AtPUB22 (At3g52450) and AtPUB23 (At2g35930) U-box proteins, whose cellular functions have not been identified, with 51% and 52% amino acid identities, respectively (Fig. 1B). On the other hand, CaPUB1 shared a relatively low degree of sequence identity with Arabidopsis AtPUB24 (At3g11840; 32% identity), tobacco Avr9/Cf-9 elicited protein 74 (GenBank accession no. AAP03884; 34% identity), and parsley (Petroselinum crispum) CMPG1 (31% identity; Kirsch et al., 2001), consistent with the notion that U-box proteins are encoded by a large multigene family (Azevedo et al., 2001; Mudgil et al., 2004). As was found in other U-box-containing proteins, CaPUB1 possessed a single U-box domain near the N-terminal region. The U-box motif of CaPUB1 was 56% to 73% conserved with regard to the Arabidopsis AtPUBs and rice (Oryza sativa) spotted leaf11 (SPL11) proteins (Zeng et al., 2004), and was 27% to 37% identical to the corresponding domain in human Hs_Prp19 and hCHIP (Ohi et al., 2003; Fig. 1C). This suggests that the domain is critical for Ub ligase activity in hot pepper plants.
Figure 1.
Sequence analysis of hot pepper CaPUB1. A, Restriction enzyme map analysis of the hot pepper CaPUB1 cDNA clone. Solid bar depicts the coding region. Solid lines represent 5′- and 3′-untranslated regions. The sequence of pCaPUB1 has been deposited in the GenBank database under accession number DQ211901. B, Comparison of the derived amino acid sequence of hot pepper CaPUB1 with those of the Arabidopsis AtPUB22 (At3g52450), AtPUB23 (At2g35930), and AtPUB24 (At3g11840) U-box proteins. Amino acid residues that are conserved in at least three of the four sequences are shaded, whereas amino acids that are identical in all four proteins are shown in black. The solid line represents the U-box motif, which is essential for E3 Ub ligase activity. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment. C, Sequence alignment of the U-box domain of CaPUB1 and other U-box proteins. The sequences of U-box motifs in hot pepper CaPUB1, Arabidopsis AtPUB22, AtPUB23, and AtPUB24, tobacco ACRE74 (GenBank accession no. AAP03884), parsley CMPG1 (Kirsch et al., 2001), rice SPL11 (Zeng et al., 2004), and human hCHIP and Prp19 (Ohi et al., 2003) are shown. Amino acid residues that are conserved in at least seven of the nine sequences are shaded. Amino acids that are identical in all nine proteins are shown in black. The numbers on the right indicate the amino acid residues. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment. D, Phylogenetic relationship of U-box Ub-ligase homologs from hot pepper (CaPUB1), Arabidopsis (AtPUB22, AtPUB23, and AtPUB24), parsley (CMPG1), and tobacco (ACRE74).
CaPUB1 Possesses E3 Ub Ligase Activity in Vitro and the U-Box Motif Is Essential for Enzyme Activity
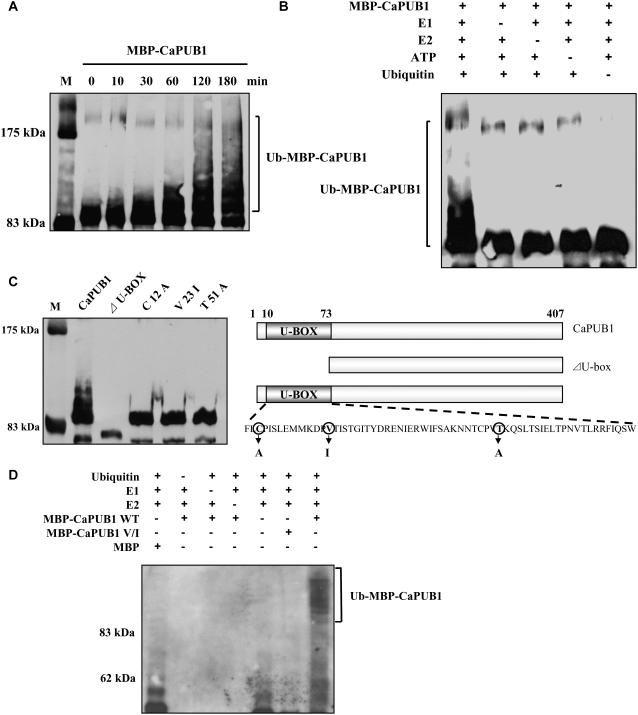
Many U-box-containing proteins function as E3 Ub ligases (Hatakeyama et al., 2001; Hatakeyama and Nakayama, 2003). To test whether CaPUB1 also contained E3 Ub ligase activity, full-length CaPUB1 was expressed in Escherichia coli as a fusion protein with the maltose-binding protein (MBP). The purified protein was used for in vitro ubiquitination analysis. The MBP-CaPUB1 fusion protein was incubated at 30°C in the presence or absence of Ub, ATP, human E1, and the human E2 protein UbcH5B for various time points and subjected to immunoblot analysis with the anti-MBP antibody. As shown in Figure 2A, MBP-CaPUB1 gave rise to high-molecular-mass ubiquitinated smear ladders in a time-dependent manner, whereas the MBP control showed no such modification. In addition, we detected no ubiquitinated signal in the absence of E1, E2, ATP, or Ub (Fig. 2B). We next constructed two versions of MBP-CaPUB1 mutants. The first version was a deletion mutant with a truncated N-terminal U-box domain (Fig. 2C). The second version contained point mutations in which Cys-12, Val-23, and Thr-51 residues, which are highly conserved among different U-box proteins, were replaced with Ala, Ile, and Ala, respectively. The functional groups of Cys-12 and Thr-51 have been shown to be involved in hydrogen bonding for the stable three-dimensional structure of the U-box motif, whereas Val-23 is a part of the well-pecked hydrophobic core (Ohi et al., 2003). Thus, these mutations could cause structural defects in the U-box domain of CaPUB1. In vitro ubiquitination assays revealed that all of the mutants were almost completely deficient in Ub ligase activity (Fig. 2C), indicating that destabilization of the U-box structure of CaPUB1 resulted in the loss of ligase activity. In addition, immunoblot assay with an anti-Ub antibody revealed that high-molecular-mass smear ladders were indeed ubiquitinated MBP-CaPUB1 (Fig. 2D). These ubiquitinated MBP-CaPUB1 smear bands were not detected with the V → I mutant protein. Taken together, it seems most likely that CaPUB1 possesses E3 Ub ligase enzyme activity.
Figure 2.
In vitro ubiquitination assay of CaPUB1. A, The MBP-CaPUB1 fusion protein expressed in E. coli was incubated at 30°C for the indicated time points in the presence of E1, E2, ATP, and Ub. Samples were resolved by 8% SDS-PAGE and subjected to immunoblot analysis with the anti-MBP antibody. B, MBP-CaPUB1 was incubated at 30°C for 120 min in the presence or absence of E1, E2, ATP, and/or Ub. Samples were identically analyzed as described above. Ub ligase enzyme activity was not detected in the absence of any of the E1, E2, ATP, or Ub. C, The U-box motif is essential for E3 Ub ligase activity. Wild-type MBP-CaPUB1 and its various mutants were used in the Ub ligase enzyme assays. C12A, V23I, and T51A are mutants of MBP-CaPUB1 in which the conserved Cys-12, Val-23, and Thr-51 residues are replaced with Ala, Ile, and Ala, respectively. ΔU-BOX is a deletion mutant in which the N-terminal U-box domain is truncated. D, In vitro ubiquitination assay of the wild-type and V → I mutant MBP-CaPUB1 was repeated using an anti-Ub antibody.
Organization and Expression of the CaPUB1 Gene
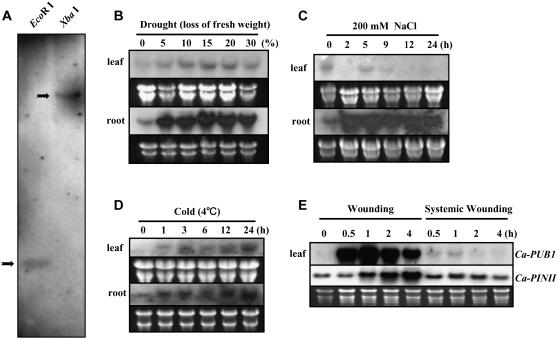
From the results described above, it appears that CaPUB1 is a U-box-containing E3 Ub ligase. We therefore wanted to characterize the CaPUB1 gene in more detail at the molecular level. To assess the exact copy number of CaPUB1 in the hot pepper genome, genomic Southern-blot analysis was carried out using pCaPUB1 as a probe. Genomic DNA isolated from mature leaves of hot pepper plants was digested with EcoRI or XbaI and hybridized with 32P-labeled pCaPUB1 under high-stringency conditions. This hybridization detected only one clear band (Fig. 3A), implying that the CaPUB1 gene is present as a single copy in the haploid hot pepper genome.
Figure 3.
Hybridization analysis of CaPUB1 genomic DNA and mRNA. A, Genomic Southern-blot analysis of the CaPUB1 gene. Hot pepper genomic DNA (10 μg/lane) was isolated from leaf tissue, digested with either EcoRI or XbaI and resolved on a 0.7% agarose gel. DNA on the gel was transferred to a nylon membrane filter. The filter was hybridized with 32P-labeled pCaPUB1 under high-stringency conditions. B to E, Light-grown 2-week-old hot pepper plants were subjected to drought (0%–30% loss of fresh weight; B), NaCl (200 mm; C), cold temperature (4°C; D), or mechanical wounding (E). Treated tissues were harvested at the indicated time points and total RNAs were isolated. Total RNAs (20 μg) were separated by electrophoresis on a 1% formaldehyde-agarose gel and blotted to a Hybond-N nylon membrane. To ensure equal loading of the RNA, the gel was stained with ethidium bromide after electrophoresis. To confirm complete transfer of RNA to the membrane filter, both gel and membrane were viewed under UV light after transfer. The filter was hybridized with 32P-labeled pCaPUB1or pCaPINII under high-stringency conditions.
To examine the spatial expression pattern of the CaPUB1 gene, we investigated the level of corresponding mRNA in different hot pepper vegetative tissues by RNA gel-blot analysis. Total RNA samples were isolated from leaves, stems, and roots of 2-week-old light-grown plants and hybridized with the 32P-labeled gene probe under high-stringency conditions. The results revealed that the amount of mRNA was very low in all tissues examined, with its level being barely detectable after prolonged exposure of the blot to x-ray film (data not shown). Because CaPUB1 was identified initially in water-stressed leaves, we considered the possibility that the expression of CaPUB1 is modulated by abiotic stresses. To test this possibility, its mRNA accumulation profile was monitored under various abiotic stress conditions. As a first step, 2-week-old light-grown hot pepper plants harvested from agar plates were dehydrated on Whatman 3MM filter paper at room temperature and approximately 60% humidity under dim light. The degree of water stress was determined by the decrease in the fresh weight of the plants. Figure 3B shows that the low, basal level of the transcript (approximately 1.5 kb) begins to elevate pronouncedly in response to 5% water loss. Although this increase was seen in every tissue examined, including leaves and roots, the highest induction was in the root tissue. Expression of CaPUB1 mRNA was further stimulated as the plants were exposed to more severe water loss (15%–20%). Next, the expression profile of CaPUB1 was examined under salt stress. Induction of the CaPUB1 transcript was clearly detected after a 2-h treatment of the root tissue with 200 mm NaCl (Fig. 3C). This marked increase in mRNA level was continuously maintained for at least 24 h. In contrast, expression of the gene was unaffected by high salinity in the leaves. This indicates that root tissue is a major part of the plant to induce the CaPUB1 gene in response to water and salt stresses. Likewise, cold temperature (24 h at 4°C) also enhanced CaPUB1 gene expression in root and leaf tissues, although the induction was less evident than with water or salt stresses (Fig. 3D). Thus, these results demonstrate that the CaPUB1 gene is activated in response to a broad spectrum of abiotic stresses.
To further understand the regulation of CaPUB1, we analyzed its expression in response to mechanical wounding, one of the most common stresses that plants encounter during their life cycle. The CaPUB1 transcript in leaves was induced rapidly (within 30 min) upon wounding, attaining a maximal level at 1 h, and declining thereafter (Fig. 3E). A positive control for wound induction, the proteinase inhibitor II homolog, CaPINII, showed a distinct pattern of activation compared with CaPUB1. The transcript began accumulating at 1 h and increased continuously for at least 4 h after wounding. This indicated that the induction of CaPUB1 by wounding was not an artifact. Intriguingly, as was the case for CaPINII, slight activation of CaPUB1 was also observed in unwounded systemic leaves (Fig. 3E). On the other hand, expression of the CaPUB1 gene was unaffected by abscisic acid (ABA), ethylene, or the bacterial pathogen Xanthomonas axonopodis (data not shown). Overall, these results are consistent with the view that the CaPUB1 gene is subject to control by diverse environmental factors, but not by biotic stresses.
Overexpression of CaPUB1 Alters Cell Growth and Differentiation in Transgenic Arabidopsis Plants
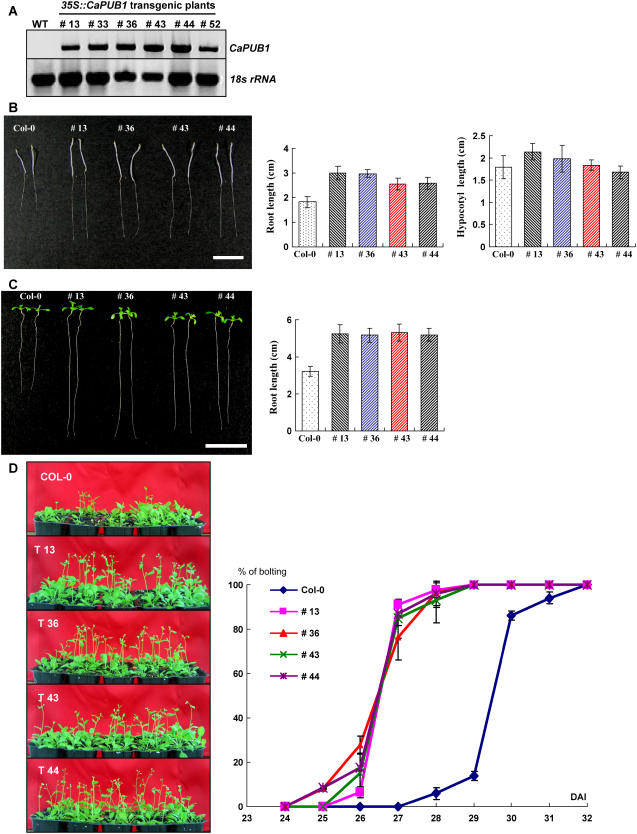
To address the cellular function of CaPUB1, attempts were made to establish transgenic hot pepper plants that constitutively expressed the CaPUB1 gene. Unfortunately, this approach turned out to be unsuccessful due to technical difficulties. Transformation and regeneration yield was extremely low so that we could not obtain enough independent transgenic lines. Instead, we established transgenic Arabidopsis plants that overexpressed CaPUB1 under the control of the cauliflower mosaic virus 35S constitutive promoter. Numerous independent T4 transgenic lines that exhibited markedly enhanced levels of the CaPUB1 transcript under normal growth conditions were chosen for further analysis (Fig. 4A).
Figure 4.
Molecular characterization and phenotype of CaPUB1-overexpressing transgenic Arabidopsis plants. A, RT-PCR analysis of wild-type and 35S∷CaPUB1 transgenic lines. B and C, Morphological comparisons of the wild-type and CaPUB1-overexpressing lines under dark (B) or light (C) growth conditions. Bar = 15 mm. D, Early bolting phenotype of 35S∷CaPUB1 relative to the wild-type plant. [See online article for color version of this figure.]
Figure 4, B and C, show the morphological comparison of 35S∷CaPUB1 and wild-type seedlings at an early stage of development. During our search for phenotypic differences, we observed that most of the independent 35S∷CaPUB1 seedlings displayed significantly longer hypocotyls and roots than the control seedlings under dark growth conditions, with the root length being predominantly different (Fig. 4B). Morphology of cotyledons, however, was somewhat similar in the dark. Under greenhouse conditions, root growth could be more readily distinguishable. We found that the roots of 35S∷CaPUB1 seedlings were 1.6- to 1.7-fold longer than in control plants (Fig. 4C). A more unique phenotype specific for the 35S∷CaPUB1 construct was that the transgenic lines grew more rapidly than the controls, thereby resulting in early bolting (Fig. 4D). Under our experimental conditions, the majority of wild-type Arabidopsis plants bolted at 29 to 30 d after imbibition. On the other hand, most independent CaPUB1-overexpressing plants exhibited earlier bolting at 26 to 27 d after imbibition (Fig. 4D).
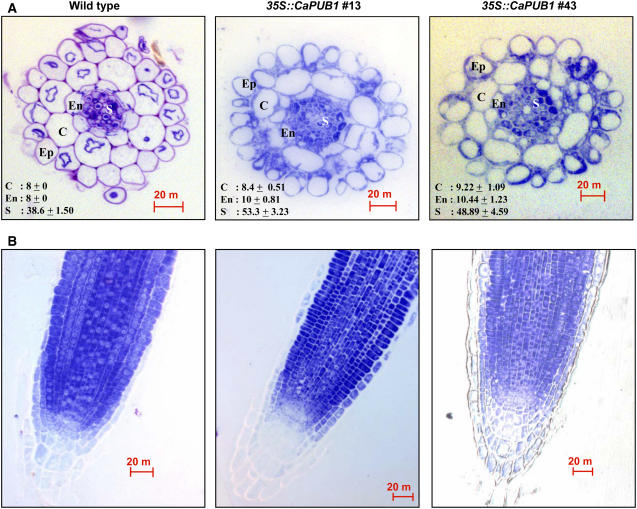
The detailed cellular phenotype was further investigated by comparing roots of 5-d-old light-grown 35S∷CaPUB1 and wild-type Arabidopsis seedlings. Analysis of transverse sections showed that the organization of the cortex, endodermis, and stele cell layers of CaPUB1-overexpressing roots appeared to be abnormal. In most sections, transgenic roots had increased numbers of small-sized cells, resulting in disordered, highly populated cell layers, as compared with the control roots (Fig. 5A). Furthermore, discontinuity in the endodermis cell layer was found frequently, which we have not been able to observe in wild-type roots (Fig. 5A). The cortex cell layer also was discontinuous in some transgenic roots. Altered cell layers were also observed in the longitudinal sections of roots; additional cell layers were seen clearly around the endodermis and cortex in the transgenic lines (Fig. 5B).
Figure 5.
Cellular phenotypes of wild-type and 35S∷CaPUB1 roots. A, Transverse sections of roots from 5-d-old light-grown wild-type and 35S∷CaPUB1 seedlings (lines 13 and 43) stained with toluidine. Average number of cells in the cortex, endodermis, and stele is indicated. Ep, Epidermis; C, cortex; En, endodermis; S, stele. Bar = 20 μm. B, Longitudinal sections of roots from 5-d-old light-grown wild-type and 35S∷CaPUB1 seedlings (lines 13 and 43) stained with toluidine. Bar = 20 μm. [See online article for color version of this figure.]
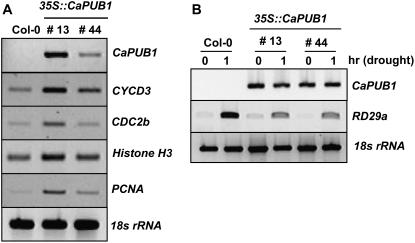
Extra cells in the cortex, endodermis, and steles in the 35S∷CaPUB1 transgenic lines probably arose from additional cell divisions. This hyperproliferative phenotype prompted us to examine the expression of cell cycle-controlled genes by reverse transcription (RT)-PCR. As shown in Figure 6A, the transcript level of the D-type cyclin CycD3 (Riou-Khamlichi et al., 1999), the cell cycle-dependent kinase-related gene CDC2b (Yoshizumi et al., 1999), the S-phase-specific proliferating cell nuclear antigen (PCNA), and the chromosome assembly histone H3 (Kosugi and Ohashi, 2003) was significantly up-regulated in CaPUB1-overexpressing lines 13 and 44. Thus, increased numbers of cells in 35S∷CaPUB1 plants correlate with increased expression of cell cycle progression genes. Taken together, these results, along with those in Figures 4 and 5, led to the suggestion that, under these experimental conditions, CaPUB1 was functionally relevant in heterologous Arabidopsis cells and, hence, might participate in a subset of the positive control of cell and tissue growth in transgenic lines.
Figure 6.
Expression of cell cycle-related and drought-induced genes in the 35S∷CaPUB1 plants. A, Transcription of cell-cycle-regulated genes was determined by RT-PCR. Wild-type and two independent 35S∷CaPUB1 transgenic lines (13 and 44) were analyzed. The cell cycle-related genes are the D-type cyclin CycD3, cell cycle-dependent kinase-related gene CDC2b, S-phase-specific PCNA, and chromosome assembly histone H3. As a negative control, the 18S rRNA transcript level was shown. B, Induction level of the RD29a gene in the wild type and two independent 35S∷CaPUB1 transgenic lines (13 and 44) in response to drought stress. Light-grown 10-d-old wild-type and 35S∷CaPUB1 transgenic seedlings were dehydrated for 1 h on Whatman 3MM filter paper at room temperature. Induction level of RD29a, a typical drought stress-induced gene (Liu et al., 1998), was examined by RT-PCR. The 18S rRNA transcript level was used as a loading control.
Overexpression of CaPUB1 Changes Plant Response to Water and Salt Stresses
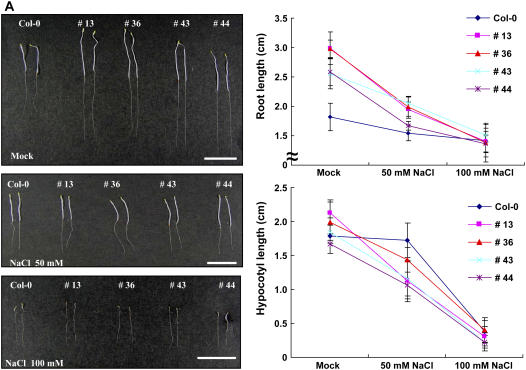
The aforementioned results concerning the RNA expression profile led us to hypothesize that the hot pepper CaPUB1 might function in the defense mechanism against abiotic stresses. Therefore, the effects of CaPUB1 overexpression on the response of Arabidopsis to water and salt stresses were examined. First, root and hypocotyl growth assays were performed with 5- to 7-d-old transgenic and wild-type seedlings that had been incubated with 50 to 100 mm NaCl. As illustrated in Figure 7, significant differences were observed between the 35S∷CaPUB1 and wild-type plants after being exposed to NaCl treatment. Whereas elongation of etiolated control seedlings was generally unaffected by the presence of 50 mm NaCl, growth of the dark-grown transgenic roots and hypocotyls was severely impaired by this mild salinity, indicating increased sensitivity to salt stress (Fig. 7A). Consequently, the clearly distinct phenotype of wild-type and 35S∷CaPUB1 seedlings became quite similar in the presence of 50 to 100 mm NaCl (Fig. 7A). Under light conditions, the growth of control roots was enhanced slightly in the presence of 50 mm NaCl. In contrast, the light-grown 35S∷CaPUB1 roots displayed a significant reduction of growth in response to mild salinity (Fig. 7B). This also reveals enhanced sensitivity to salinity. In the presence of 100 mm NaCl, elongation of wild-type roots was approximately 75% of normal, whereas elongation of transgenic roots was greatly reduced to 36% to 44% of the control, depending on the independent transgenic line (Fig. 7B).
Figure 7.
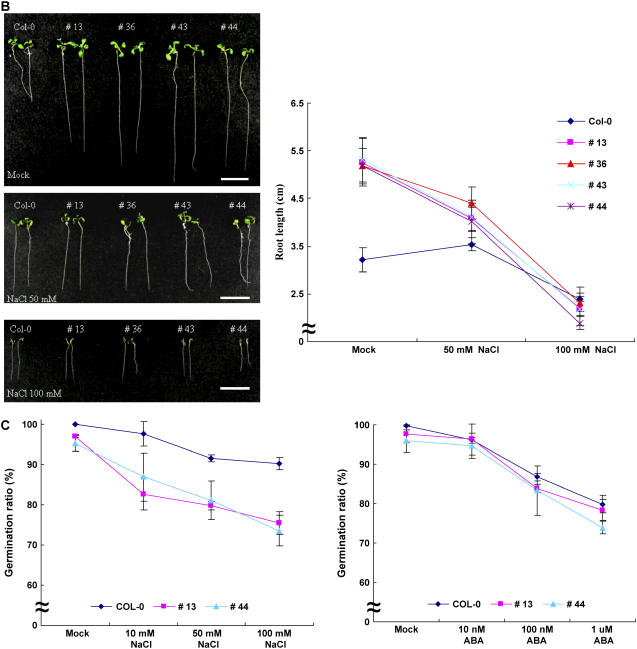
Increased sensitivity of 35S∷CaPUB1 transgenic lines to salt stress. A, Five-day-old etiolated wild-type and transgenic seedlings were incubated with 50 to 100 mm NaCl and root and hypocotyl growth assays were carried out. The values are means ± sd (n = 4). Bar = 15 mm. B, Seven-day-old light-grown wild-type and transgenic seedlings were subjected to 50 to 100 mm NaCl and the growth patterns of roots and hypocotyls were monitored. The values are means ± sd (n = 4). Bar = 15 mm. C, Germination ratio of wild-type and 35S∷CaPUB1 transgenic lines in the absence or presence of NaCl (10–100 mm; left); germination ratio of wild-type and 35S∷CaPUB1 transgenic lines in the absence or presence of ABA (0.01–1 μm; right). [See online article for color version of this figure.]
High concentrations of salts inhibit germination of Arabidopsis (Quesada et al., 2000; Zhu, 2000). Under mild salt stress (10–100 mm NaCl), however, the germination efficiency of wild-type plants was only slightly affected (Fig. 7C). In contrast, germination of 35S∷CaPUB1 plants was reduced 9% to 15% by 10 mm NaCl, 15% to 17% by 50 mm NaCl, and 22% to 24% by 100 mm NaCl, indicating that CaPUB1-overexpressing plants are hypersensitive to salt at the germination stage. Thus, both the germination (Fig. 7C) and postgermination (Fig. 7, A and B) growth of 35S∷CaPUB1 transgenic plants are more sensitive to mild salinity than wild-type plants. On the other hand, wild-type and transgenic plants exhibited similar sensitivity to exogenously applied ABA (0.011 μm) in germination (Fig. 7C). This is in line with the result that expression of the CaPUB1 gene is not responsive to ABA, suggesting that the function of CaPUB1 might be independent of ABA.
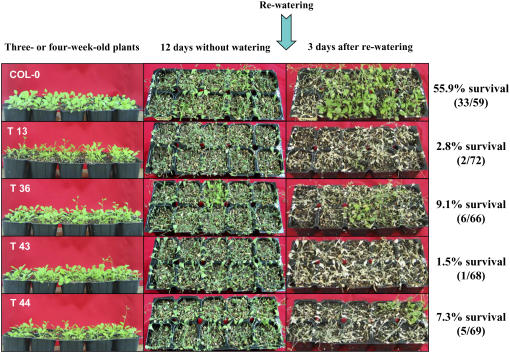
In the next experiment, we addressed the capacity of wild-type and 35S∷CaPUB1 plants to respond to dehydration. Dehydration sensitivity was scored as the capacity of plants to resume growth after water stress when returned to normal conditions. Three- or 4-week-old Arabidopsis plants were grown in pots. When the soil was allowed to dry by withholding water for 12 d, all plants displayed wilting. After rewatering for 3 d, 33 of 59 wild-type plants survived and continued to grow (55.9% survival; Fig. 8). Under these experimental conditions, however, most of the CaPUB1-overexpressing lines examined were unable to recover after rewatering, as opposed to wild type, and eventually died (1.5%–9.1% survival; Fig. 8). In addition, the expression level of RD29a, a typical drought stress-induced gene (Liu et al., 1998), was markedly lower in 35S∷CaPUB1 lines compared to the wild-type plant in the water stress condition (Fig. 6B). These results, in conjunction with the data in Figure 7, A and B, indicate that 35S∷CaPUB1 transgenic plants were more sensitive to water deficit and NaCl (50–100 mm) as compared with wild-type plants. Overall, we interpreted these results to suggest that CaPUB1 might be involved in the control of plant responses to counteract unfavorable growth conditions.
Figure 8.
Increased sensitivity of 35S∷CaPUB1 transgenic lines to water stress. Wild-type and transgenic Arabidopsis plants were grown in pots for 4 weeks. Water was withheld for 12 d, followed by rewatering for 3 d. Dehydration sensitivity was assayed as the capability of plants to resume growth when returned to normal conditions following water stress. [See online article for color version of this figure.]
Two-Dimensional Gel Electrophoresis and Protein Identification by Matrix-Assisted Laser-Desorption Ionization-Time-of-Flight Mass Spectrometry in Transgenic Arabidopsis Plants
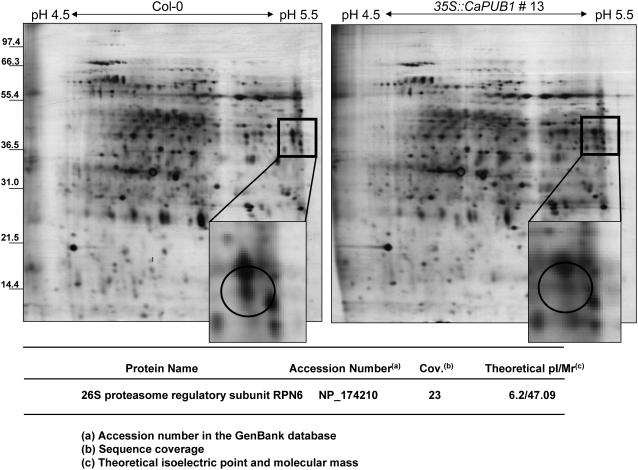
In vivo and in vitro results showed that hot pepper CaPUB1 was an E3 ligase and that perturbation of CaPUB1 expression resulted in changes in cell growth and differentiation and alteration of plant response to abiotic stress. This raised the question as to whether CaPUB1 exerted its cellular functions by participating in the mechanism by which certain proteins were degraded by the Ub 26S proteasome pathway. To unravel this question, we decided to employ two-dimensional gel electrophoresis (2-DE) analysis. Total proteins were extracted from 7-d-old whole seedlings of wild-type and 35S∷CaPUB1 line 13 and subjected to a linear pH 4 to 7 immobilized pH gradient strip in the first dimension. Subsequently, 9% to 16% gradient SDS-PAGE was carried out in the second dimension, as described in “Materials and Methods.” After electrophoresis, the resolved proteins were visualized by staining with silver nitrate and the number of individual protein spots on the gel was analyzed using Melanie III software (Fig. 9). Individual proteins with Mr values from 20,000 to 70,000 and pIs from 4.5 to 5.5 and from 5.5 to 7.0, respectively, were analyzed because maximal separation and reproducibility were obtained within these parameters. Among the numerous silver-stained proteins, we detected a spot whose abundance was significantly reduced in 35S∷CaPUB1 line 13 as compared to wild type (Fig. 9). This spot was excised from the gel, digested with trypsin, and then analyzed by matrix-assisted laser-desorption ionization (MALDI)-time-of-flight (TOF) mass spectrometry (MS). The protein identified by this method turned out to be the 26S proteasome regulatory subunit RPN6 (Kwok et al., 1999; Peng et al., 2001; Fig. 9). The same results were obtained using transgenic line 43 (data not shown). Taken together, these results supported the notion that the complex protein mixtures present in CaPUB1-overexpressing plants were well resolved by this 2-DE system with highly reproducible patterns.
Figure 9.
Silver-stained 2-DE map of wild-type and 35S∷CaPUB1 transgenic Arabidopsis seedlings. Protein samples were prepared from 7-d-old whole seedlings as described in “Materials and Methods.” Proteins (350 μg) were applied to an immobilized pH gradient strip by the in-gel rehydration method. Subsequently, 9% to 16% gradient SDS-PAGE was performed to separate proteins in the second dimension. The proteins were visualized with silver nitrate. The arrow in the magnified region of the 2-DE image shows a protein spot whose abundance is clearly reduced in the 35S∷CaPUB1 transgenic line as compared to the wild type. This spot was excised from the gel and analyzed by MALDI-TOF MS.
CaPUB1 Interacts with RPN6 Protein
RPN6 protein was identified initially as the Arabidopsis non-ATPase S9 (subunit 9) of the 19S regulatory particle (RP) from the 26S proteasome complex (Kwok et al., 1999). In yeast (Saccharomyces cerevisiae), RPN6, along with other non-ATPase subunits, forms the lid subcomplex of 19S RP, which directs substrate recognition and processing before substrates are funneled into the 20S core particle (CP) for degradation (Baumeister et al., 1998). The results in Figure 9 show that the level of RPN6 was reduced in CaPUB1-overexpressing Arabidopsis lines. This raised the tantalizing possibility that CaPUB1 Ub ligase might interact with RPN6 and participated in controlling the stability of the RPN6 protein.
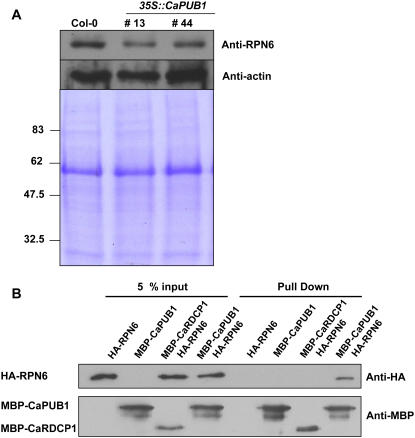
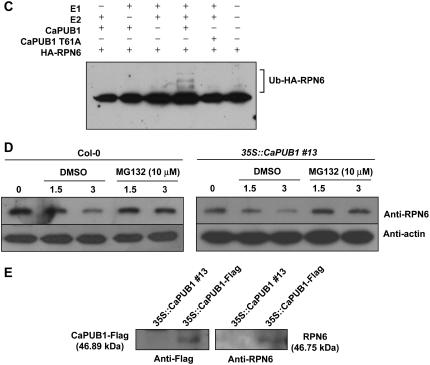
As a first step to investigate this possibility, immunoblot analysis was performed using an anti-RPN6 antibody in wild-type and transgenic plants. The results showed that the relative abundance of RPN6 protein decreased significantly in both transgenic lines 13 and 44 compared to that in wild-type plants (Fig. 10A). To obtain evidence indicating a specific interaction between CaPUB1 and RPN6, we performed an in vitro pull-down assay. Both CaPUB1 and RPN6 were expressed in E. coli cells as MBP and haemagglutinin (HA) fusion proteins, respectively. The purified fusion proteins were coincubated with an amylose affinity matrix, followed by extensive washing. The bound protein was eluted from the amylose resin by 10 mm maltose and immunoblotted with the anti-HA antibody. Figure 10B shows that HA-RPN6 was pulled down from the amylose affinity resin by the MBP-CaPUB1 protein, but not by HA-RPN6 alone. As a specificity control, we tested the interaction between RPN6 and CaRDCP1, a hot pepper RING-finger domain Ub ligase. As shown in Figure 10B, CaRDCP1 failed to bind RPN6. These results suggest that the CaPUB1 and RPN6 proteins physically interact in vitro. In the next experiment, the in vitro ubiquitination assay was carried out. In this experiment, CaPUB1 was used as a source of E3 Ub ligase. The bacterially expressed HA-RPN6 protein was incubated at 30°C in the presence or absence of Ub, ATP, E1, E2, and MBP-CaPUB1 for the appropriate time period and subjected to immunoblotting using the anti-HA antibody. Figure 10C shows the production of higher molecular weight ubiquitinated ladders when HA-RPN6 was incubated with MBP-CaPUB1 in the presence of Ub, ATP, E1, and E2. The exclusion of any E1, E2, or MBP-CaPUB1 abolished the ubiquitination signals (Fig. 10C). Furthermore, the CaPUB1 T51A mutant failed to ubiquitinate HA-RPN6, suggesting that the ubiquitination of RPN6 by CaPUB1 was not an experimental artifact, but was due to the specific interaction between these two proteins (Fig. 10C).
Figure 10.
CaPUB1 interacts with the RPN6 protein. A, Immunoblot analysis of RPN6 protein. Protein samples (20 μg of total proteins) were prepared from 7-d-old wild-type and two independent transgenic seedlings (lines 13 and 44), and subjected to immunoblot analysis using an anti-RPN6 antibody or anti-actin antibody as a negative control. The blot was visualized by the chemiluminescence detection method. The equivalence of protein loading among lanes of the SDS-PAGE gel was demonstrated by Coomassie Brilliant Blue R-250 staining of the proteins on the gel. B, In vitro pull-down assay. MBP-CaPUB1 and HA-RPN6 fusion proteins were coincubated with amylose affinity matrix. The bound protein was eluted from the amylose resin by 10 mm maltose, resolved by 12.5% SDS-PAGE, and transferred to a nitrocellulose membrane. The blot was probed with an anti-HA antibody or anti-MBP antibody. C, In vitro ubiquitination assay of RPN6 using CaPUB1 as the E3 Ub ligase. Recombinant HA-RPN6 protein was incubated in the presence or absence of Ub, ATP, E1, E2, and wild-type MBP-CaPUB1 or its mutant form (T51A), for the appropriate time periods, and subjected to immunoblotting using an anti-HA antibody. D, In vivo interaction of CaPUB1 and RPN6. The wild-type (left) and 35S∷CaPUB1 transgenic seedlings (right) were treated with 100 μm cycloheximide for different time periods (0–3 h) in the absence or presence of MG132, an inhibitor of the 26S proteasome. During incubation, changes in the level of RPN6 protein were monitored using an anti-RPN6 antibody. E, In vivo immunoprecipitation assay. Putative interacting proteins with CaPUB1 were immunoprecipitated from the 35S∷CaPUB1-Flag transgenic seedlings using an anti-Flag antibody. Immunoprecipitated proteins were subsequently analyzed by immunoblotting with an anti-RPN antibody. As a specificity control, 35S∷CaPUB1 seedlings were used in an identical experiment. [See online article for color version of this figure.]
To provide additional evidence for the interaction between CaPUB1 and RPN6, two in vivo experiments were performed. First, wild-type and 35S∷CaPUB1 transgenic seedlings were treated with 100 μm cycloheximide for different time periods (0–3 h) in the absence or presence of MG132 (10 μm), an inhibitor of the 26S proteasome. During incubation, changes in the level of RPN6 protein were monitored using an anti-RPN6 antibody. As shown in Figure 10D, the amount of RPN6 protein was gradually decreased in the absence of MG132 in a time-dependent fashion, with the protein level being lower and slightly more rapidly degraded in the 35S∷CaPUB1 seedlings compared to the wild type. In contrast, the protein level was unchanged when wild-type and transgenic seedlings were incubated with MG132. These results indicate that RPN6 was degraded in a proteasome-dependent manner in CaPUB1-overexpressing seedlings. Second, transgenic Arabidopsis plants expressing the CaPUB1-Flag fusion protein were constructed. The putative interacting proteins with CaPUB1 were immunoprecipitated using an anti-Flag antibody from the 35S-CaPUB1-Flag seedlings. The immunoprecipitated proteins were subsequently analyzed by immunoblotting with an anti-RPN antibody. The result in Figure 10E shows that RPN6 was immunoprecipitated by an anti-Flag antibody, indicating the in vivo interaction between CaPUB1-Flag and RPN6 proteins. This suggests that RPN6 indeed interacted with CaPUB1 and degraded in a CaPUB1-dependent manner in CaPUB1-overexpressing transgenic plants. Overall, the results in Figures 9 and 10 are consistent with the hypothesis that hot pepper CaPUB1 Ub ligase interacts with RPN6, thereby affecting the turnover of the RPN6 protein in 35S∷CaPUB1 transgenic Arabidopsis plants.
DISCUSSION
The molecular and cellular processes underlying the acclimation of hot pepper to abiotic stresses have attracted much interest because it is an economically important crop and its response to adverse environmental factors is not well understood. From water-stressed hot pepper plants, we isolated the CaPUB1 gene that encodes a protein containing a single U-box motif, which is a highly conserved domain found in the diverse isoforms of eukaryotic E3 Ub ligase (Fig. 1). With the aid of in vitro ubiquitination and site-directed mutagenesis assays, we were able to confirm that bacterially expressed CaPUB1 possessed E3 Ub ligase activity and that the N-terminal U-box motif was essential for its enzyme activity (Fig. 2). RNA gel-blot studies showed that, in hot pepper seedlings, CaPUB1 mRNA was highly inducible in response to diverse environmental stresses, including dehydration, high salinity, and cold temperature (Fig. 3). It was worth noting that CaPUB1 was also rapidly activated (within 30 min) in both local and systemic leaves by mechanical wounding, although induction in systemic leaves was less pronounced than in local leaves. In contrast, CaPUB1 was not induced by ethylene, ABA, or infection by a bacterial pathogen. Thus, it would be reasonable to consider that, in hot pepper, CaPUB1 may not play a role in the biotic stress response, but rather functions specifically in early events in the abiotic-related defense response to deal with the effective plant adaptation process.
Although a number of U-box-containing proteins have been known to function as E3 Ub ligases (Hatakeyama et al., 2001; Hatakeyama and Nakayama, 2003), our understanding of their biological roles in higher plants is still rudimentary (Azevedo et al., 2001; Mudgil et al., 2004). A previous study showed that the Arabidopsis U-box AtCHIP transcript was induced significantly by extreme temperatures (Yan et al., 2003). Overexpression of AtCHIP rendered Arabidopsis plants more sensitive to both low and high temperatures (Yan et al., 2003). AtCHIP was subsequently reported to interact with an A-subunit of protein phosphatase 2A and suggested to be involved in the ABA-related stress-signaling pathway (Luo et al., 2006). Zeng et al. (2004) isolated a rice spl11 mutant that exhibited a spontaneous cell death phenotype and increased resistance to bacterial and fungal pathogens. The SPL11 protein was identified as a U-box-containing E3 Ub ligase. This suggests a role for the ubiquitination system as a negative regulator in the control of cell death and pathogen defense in rice plants (Zeng et al., 2004). On the other hand, the two most recent studies implicate tobacco U-box E3 Ub ligase NtCMPG1 and tobacco ACRE276 and its functional homolog Arabidopsis AtPUB17, respectively, as essential positive mediators for plant defense and disease resistance (Gonzalez-Lamothe et al., 2006; Yang et al., 2006). These results prompted us to create transgenic Arabidopsis plants that constitutively expressed the hot pepper CaPUB1 gene (Fig. 4A). Our search for phenotypic differences indicates that CaPUB1 is functionally relevant in heterologous Arabidopsis cells, as evidenced by the following observations. Ectopic expression of CaPUB1 caused a clearly distinct phenotype in comparison with control plants, including markedly enhanced growth of hypocotyls and roots (Figs. 4 and 5), early bolting (Fig. 4), and increased sensitivity to water and salt stresses (Figs. 7 and 8). The more sensitive phenotype of 35S∷CaPUB1 lines to drought and salinity is reminiscent of the Arabidopsis U-box AtCHIP-overexpressing plants, which are hypersensitive to extreme temperatures, indicating that increased AtCHIP or CaPUB1 alone might not be enough for enhancing stress tolerance (Yan et al., 2003). Closer inspection revealed that the individual transgenic roots had increased numbers of small-sized cells around the cortex, endodermis, and stele, resulting in disordered, highly populated cell layers relative to control roots (Fig. 5). Expression of several cell cycle-controlled genes, including CycD3, CDC2b, PCNA, and histone H3, are up-regulated in transgenic seedlings (Fig. 6). Thus, the increase in transgenic root growth was associated with the changes in cell numbers. Because CaPUB1 was identified as a stress-inducible gene (Fig. 3), such phenotypic properties were intriguing. Taken together, it is attractive to speculate that the E3 Ub ligase CaPUB1 is actively involved in the Ub-mediated degradation of as-yet unidentified proteins, thereby effectively altering a subset of cell and tissue growth control factors, as well as the response to abiotic stresses in transgenic lines.
Nevertheless, we could not rule out the possibility that overexpression of a heterologous CaPUB1 gene with a strong cauliflower mosaic virus 35S promoter in Arabidopsis may create a pleiotropic phenotype that is difficult to interpret. In this regard, it is critical to consider the link between the cellular function of CaPUB1 and the disordered cell layers with small-sized cells and hypersensitivity to drought and salt in CaPUB1-overexpressing plants. We speculate on the possibility that the early bolting phenotype of the 35S∷CaPUB1 plants, which results in early production of seeds, could be one of the possible mechanisms by which plants escape from drought or salt stress. Indeed, it was reported that there was a large positive genetic correlation between dehydration avoidance and flowering time (drought escape) among 39 different naturally occurring genotypes of Arabidopsis (Mckay et al., 2003). In Impatiens capensis plants, correlative selection was detected for early flowering combined with high stomatal conductance in water-limiting conditions, suggesting that early flowering and stress avoidance may be key mechanisms behind adaptation to early season drought (Heschel and Riginos, 2005). Thus, there is a tantalizing possibility that CaPUB1, which causes an early flowering phenotype in 35S∷CaPUB1, is associated with a drought escape mechanism.
However, it should be noted that overexpression of CaPUB1 results in hypersensitivity to drought and salt. It has been known that the normal response of plants to abiotic stresses is growth arrest rather than promotion. Also, faster growth of vegetative organs does not necessarily result in early flowering. Thus, hypersensitivity to drought and salt and a rapid-growing phenotype of 35S∷CaPUB1 plants could indicate that CaPUB1 probably functions as a negative regulator that inhibits the adaptive stress response. In this regard, the early-flowering phenotype of stressed plants might not be because of enhanced growth, but because of other flowering-time control mechanisms. Negative function of U-box E3 Ub ligase SPL11 has been known in the control of cell death and pathogen defense in rice plants (Zeng et al., 2004).
High-resolution 2-DE has recently been shown to be a powerful tool for studying complex patterns of protein expression in various tissues of higher plants (Roberts, 2002; Rose et al., 2004). Thus, to identify possible target proteins of CaPUB1, 2-DE analysis was carried out to compare the protein profiles of the 35S∷CaPUB1 transgenic and wild-type Arabidopsis plants. This comparative proteomic approach demonstrates reduced levels of the RPN6 protein in 35S∷CaPUB1 as compared with the wild type (Fig. 9). In vitro pull-down and ubiquitination assays show that RPN6 directly interacts with CaPUB1 and is ubiquitinated in a CaPUB1-dependent manner (Fig. 10, B and C). The level of RPN6 protein became higher when both wild-type and transgenic seedlings were incubated with MG132, a 26S proteasome complex inhibitor, compared to without MG132 (Fig. 10D). Furthermore, an in vivo pull-down assay shows that the RPN6 and CaPUB1 proteins interact in CaPUB1-overexpressing plants (Fig. 10E).
The 26S proteasome is a 2-MD ATP-dependent protease complex that removes many Ub-conjugated cellular proteins. The complex is composed of the 20S catalytic CP and two 19S RPs that form the base and lid at the ends of the CP (Voges et al., 1999). The 20S CP is a hollow cylinder containing proteolytic activity, whereas the function of the 19S RP is to direct substrate selection, unfolding, and entry into the 20S CP for destruction. The base multisubunit RP is in direct contact with the 20S CP and consists of six ATPase subunits (RPT1–RPT6) and three non-ATPase subunits (RPN1, RPN2, and RPN10; Baumeister et al., 1998; Glickman et al., 1998). The lid particle is assembled from eight non-ATPase subunits, RPN3, RPN5-9, RPN11, and RPN12. In Arabidopsis, the biological role of the 19S RPs is becoming increasingly apparent in the mediation of diverse cellular functions and metabolism. For example, the non-ATPase RPN12 subunit has been demonstrated to be involved in cytokinin responses. A T-DNA insertion mutant of RPN12 exhibits reduced leaf formation and root elongation, delayed skotomorphogenesis, and decreased growth responses to exogenous cytokinin (Smalle et al., 2002). On the other hand, a mutation in the RPN10 gene results in a pleiotropic phenotype in Arabidopsis. The rpn10-1 mutant is more sensitive to ABA, salinity, Suc stress, and DNA-damaging agents, whereas the mutant is less responsive to cytokinin and auxin (Smalle et al., 2003). Peng et al. (2001) found that the RPN6 protein, a non-ATPase regulatory subunit, exists in two distinct protein complexes of about 800 and 500 kD, respectively, in Arabidopsis and cauliflower (Brassica oleracea) plants. The large complex likely represents the proteasome 19S RP, whereas the small complex, designated PR500, is loosely associated with an hsp70 protein. Interestingly, the PR500 complex disappeared when plants were subjected to heat shock and canavanine treatment, indicating that the level of PR500 fluctuated in response to stresses in Arabidopsis (Peng et al., 2001). In this regard, it should be noted that the level of RPN6 is significantly lower in CaPUB1-overexpressing lines under normal growth conditions (Figs. 9 and 10A). Our results demonstrate that RPN6 physically interacts with CaPUB1 and is ubiquitinated in vitro in a CaPUB1-dependent manner (Fig. 10, B and C). Thus, it is tempting to propose that CaPUB1 is involved in the degradation of RPN6 in transgenic Arabidopsis plants. The decrease in the level of RPN6 protein, in turn, might result in the rapid-growing phenotype and increased sensitivity to abiotic stresses. This regulatory interaction between CaPUB1 and RPN6 would permit the plant to fine-tune its responses to diverse developmental and environmental cues. However, it is still possible that CaPUB1 might interact with other cellular proteins so that the increased level of CaPUB1 gave rise to the disturbed stoichiometry between CaPUB1 and interacting proteins in transgenic Arabidopsis plants. This hypothesis is in line with our results that there are several proteins whose levels are down-regulated in 35S∷CaPUB1 seedlings detected by 2-DE analysis (data not shown).
A database search identified that CaPUB1 shares significant sequence identity with two Arabidopsis U-box proteins, AtPUB22 (At3g52450; 51% amino acid identity) and AtPUB23 (At2g35930; 52% amino acid identity). Intriguingly, expression of both Arabidopsis U-box genes is highly inducible by various abiotic stresses (S.K. Cho, M.Y. Ryu, and W.T. Kim, unpublished data). Therefore, it is of immense importance to study cellular functions of these Arabidopsis CaPUB1 potential orthologs. In conclusion, the results presented in this study suggest that CaPUB1 acts in transgenic Arabidopsis plants to turn on a subset of physiological responses for coping with dehydration and high-salinity stresses. Further functional studies of the Arabidopsis PUB1 orthologs and characterization of the protein it interacts with will be required to further our understanding of the relationship between E3 Ub ligases and responses to abiotic stresses in higher plants.
MATERIALS AND METHODS
Plant Materials and Stress Treatments
Dry hot pepper seeds (Capsicum annuum L. cv Pukang) were soaked once with 70% ethanol and then rinsed extensively with sterilized water. Seedlings were grown in a mixture of soil and vermiculite or on Murashige and Skoog medium containing 1% Suc, vitamin B5 (12 mg L−1), and 0.8% agar (pH 5.8) in a 25°C growth chamber with a 16-h light/8-h dark photoperiod. Hot pepper plants were subjected to various biotic and abiotic stresses as described by Park et al. (2003).
Construction of MBP-CaPUB1 Recombinant Protein
The full-length pCaPUB1 cDNA clone was introduced to the plasmid pMAL-c2X (New England BioLabs) and used for expression of the MBP-CaPUB1 fusion protein. The Escherichia coli BL21 (DE3) strain containing the constructed plasmid was grown at 37°C in 100 mL of Luria-Bertani medium (10 g tryptone, 5 g yeast extract, and 10 g L−1 NaCl) supplemented with 50 μg mL−1 ampicillin. Cells were grown for an additional 4 h at 30°C after induction with 1 mm isopropylthio-β-galactoside at OD600 0.6 to 1.0. The pellet was resuspended in phosphate-buffered saline (PBS) containing 1 mm phenylmethylsulfonyl fluoride. The suspension was sonicated on ice with a Vibracell sonicator (Sonics and Materials). Triton X-100 was added to a final concentration of 1% (v/v). The fusion protein was purified by affinity chromatography using MBP Sepharose 4B from MBP purification modules (Pharmacia), as described previously (Lee et al., 2004).
In Vitro Ubiquitination Assay
The bacterially expressed MBP-CaPUB1 fusion protein (500 ng) was brought to 150 μL with ubiquitination reaction buffer (50 mm Tris-HCl, pH 7.5, 2.5 mm MgCl2, 0.5 mm dithiothreitol [DTT]) containing 4 mm ATP and 20 ng each of human E1 and E2 UbcH5B. The reaction mixture was incubated for the appropriate time periods (0–180 min) at 30°C. The reaction was stopped by the addition of SDS sample buffer. After boiling for 10 min, the sample was separated by 8% SDS-PAGE and subjected to immunoblotting using the anti-MBP antibody (New England BioLabs).
DNA and RNA Gel-Blot Analyses
Hot pepper leaf genomic DNA was isolated as described previously (Park et al., 2003). DNA (10 μg/lane) was digested with appropriate enzymes, separated by electrophoresis on 0.7% agarose gel, and blotted onto a nylon membrane filter (Amersham). The filter was hybridized with 32P-labeled pCaPUB1 under high-stringency conditions, as described by Lee and Kim (2003). Total RNAs of hot pepper and transgenic Arabidopsis (Arabidopsis thaliana) plants were obtained as described by Mang et al. (2004b). The RNA gel blots were hybridized with various 32P-labeled cDNA probes for CaPUB1 and CaPINII under high-stringency hybridization and washing conditions. The blot was visualized by autoradiography at −80°C using Kodak XAR-5 film and an intensifying screen. Hybridization signals were quantified with a phosphor imager (Fuji).
Generation of the 35S∷CaPUB1 Construct and Transformation of Arabidopsis
Full-length pCaPUB1 cDNA was inserted into the corresponding sites of the binary vector pMBP2. The fusion gene construct was transferred to Agrobacterium tumefaciens strain AGL1 by electroporation, as described by Joo et al. (2004). Arabidopsis transformation was accomplished by the floral-dip method (Clough and Bent, 1998). The seeds collected from the plants were selected on a 0.5× Murashige and Skoog plate containing 25 mg L−1 kanamycin to obtain independent transgenic lines. The presence of the transgene was confirmed by PCR and genomic Southern-blot analysis. Homozygous T4 lines were obtained by further self crossing according to Joo et al. (2004) and used in this study.
Measurement of Hypocotyl and Root Lengths of Wild-Type and Transgenic Arabidopsis Seedlings
Assay of hypocotyl and root growth under salt stress was performed as described by Cho et al. (2006). Briefly, wild-type and transgenic Arabidopsis seeds were surface sterilized and plated on 0.8% agar plates (select agar; Life Technologies) containing 0.5× Murashige and Skoog salts, 0.5 mm MES, pH 5.7, 1% Suc, and 1× vitamin B5. To test hypocotyl and root growth, vertically oriented agar plates were incubated at 22°C in the dark or in a 16-h light/8-h dark cycle for 5 to 7 d. During incubation, the advancing hypocotyl and root tips were recorded using a marker pen on the outside of the plates and the image of the marked plate was scanned using ScionImage software (Scion Corp).
Germination Assay
Wild-type and transgenic seeds collected at the same time were used. The germination ratio was monitored in the absence or presence of various concentrations of NaCl (10–100 mm) or ABA (0.01–1 μm; Cho et al., 2006).
Microscopy
Five-day-old light-grown wild-type and transgenic Arabidopsis seedlings were fixed in PBS buffer containing 4% paraformaldehyde and embedded in plastic resin (Technovit 8100; Heraeus Kulzer) following the manufacturer's manual. Samples were cut into 2-μm thicknesses using a microtome (Leica Instruments). The sections were stained with a 0.1% aqueous solution of Toluidine Blue O (Sigma) for 1 min and visualized by light-field microscopy.
RT-PCR
RT-PCR was performed in a total volume of 25 μL containing 1 μL of the first-strand cDNA reaction products, 1 μm primers, 10 mm Tris (pH 8.0), 50 mm KCl, 1.5 mm MgCl2, 0.01% gelatin, 200 μm deoxynucleotides, and 2.5 units of Taq polymerase (Promega). Twenty-five thermal cycles were carried out, each consisting of 45 s at 95°C, 1 min at 60°C, and 90 s at 72°C in an automatic thermal cycler (Perkin-Elmer/Cetus). PCR products were separated on a 1% agarose gel and then visualized under UV light.
2-DE Analysis and Protein Identification by MALDI-TOF MS
The 35S∷CaPUB1 and wild-type Arabidopsis seedlings were harvested into lysis buffer containing 9 m urea, 4% CHAPS, and protease inhibitor cocktail (Roche Diagnostics). After homogenizing extensively for 5 min on ice, the cell lysates were centrifuged at 100,000 g for 15 min at 4°C in a Beckman TL-100 table ultracentrifuge. The supernatants were taken as the total protein extract and stored at −70°C until use. Protein extracts were normalized with a Bradford protein assay kit from Bio-Rad. Protein (350 μg) in 350 μL 2-DE sample buffer (9 m urea, 4% CHAPS, 100 mm DTT, 0.5% Pharmalyte [pH 4–7], and trace bromphenol blue) was loaded into 18-cm, pH 4 to 7 isoelectric focusing gel strips (Amersham-Pharmacia Biotech) by the in-gel rehydration method (Rabilloud et al., 1994). The proteins were focused for 70,000 V h using a Multiphor II system (Amersham-Pharmacia Biotech) and equilibrated for 15 min in buffer (6 m urea, 30% glycerol, 15% SDS, 50 mm Tris-Cl, pH 8.8) containing 1% DTT and, for another 15 min, in the same buffer containing 4.8% iodoacetamide. The isoelectric focusing gel strips were loaded on top of 9% to 16% gradient polyacrylamide gels (200 × 250 × 1.0 mm) for a second round of SDS-PAGE using the Hoefer IsoDALT apparatus (Amersham-Pharmacia Biotech) until the tracking dye reached the anode ends of the gels. The gels were stained with silver nitrate according to the modified procedure by Yan et al. (2000). Silver-stained gels were scanned using a high-resolution scanner (Bio-Rad GS-800 calibrated imaging densitometer). Scanned images were analyzed with Melanie III software according to the manufacturer's protocol.
Protein spots on silver-stained gels were excised and digested with sequencing-grade trypsin, as described by Mang et al. (2004a). For MALDI-TOF MS analysis, 0.5 μL tryptic peptide mixture was dispensed on the MALDI sample plate, along with 0.5 μL matrix solution consisting of 10 mg mL−1 α-cyano-4-hydroxycinnamic acid, 0.1% trifluoroacetic acid, and 50% acetonitrile, and analyzed with a PerSeptive Biosystem Voyager-DE STR MALDI-TOF mass spectrometer in reflector mode over a mass range of 1,000 to 3,000 D. Proteins were identified by peptide mass fingerprinting with the search programs ProFound (http://129.85.19.192/profound_bin/WebProFound.exe, Rockefeller University, version 4.10.5) and MASCOT (http://www.matrixscience.com). Searches for sequence homologies were performed at http://www.arabidopsis.org/Blast.
Pull-Down Assay and Immunoblot Analysis
For in vitro pull-down assay, the bacterially expressed MBP-CaPUB1 and HA-RPN6 fusion proteins were coincubated at 30°C for 30 min in a 750-μL reaction buffer (20 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 1 mm EDTA), containing 30 μL amylose resin (New England BioLabs) with gentle agitation. The resin was washed three times with 500 μL reaction buffer. MBP-CaPUB1 was eluted from the amylose affinity resin with 150 μL buffer containing 10 mm maltose and then subjected to immunoblotting with anti-HA antibody (New England BioLabs). For in vivo pull-down assay, putative interacting proteins with CaPUB1 were pulled down by an anti-Flag antibody from 35S∷CaPUB1-Flag transgenic seedlings. Pulled-down proteins were then analyzed by immunoblotting using an anti-RPN6 antibody.
Protein samples were separated by 12.5% SDS-PAGE and transferred to a nitrocellulose membrane as described (Lee et al., 2006). The membrane was blocked against nonspecific binding by incubation in PBS blocking buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, 0.01% [w/v] Tween 20, and 5% [w/w] dried nonfat milk) for 1 h at room temperature and then incubated with anti-HA antibody (1:10,000 dilution), anti-MBP antibody (1:10,000 dilution), anti-RPN6 antibody (1:5,000 dilution), or anti-actin antibody (1:5,000 dilution). The membrane was washed three times with PBS blocking buffer, incubated with horseradish peroxidase-conjugated anti-rabbit IgG (Amersham Life Sciences) for 1 h, and washed extensively with PBS buffer (Lee et al., 2006). Proteins were visualized with the ECL1 Plus western-blotting detection system (Amersham Life Sciences) and exposed to Kodak BioMax ML film.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number DQ211901.
This work was supported in part by grants from the Basic Research Program of the Korea Science and Engineering Foundation (project no. R01–2004–000–10487–0 to W.T.K. and H.S.P.), the Plant Diversity Research Center (Twenty-First Century Frontier Research Program funded by the Ministry of Science and Technology of the Korean government to W.T.K., H.S.P., and J.K.), and the Plant Metabolism Research Center at Kyung Hee University (Science Research Center project no. R11–2000–081 from the Korea Science and Engineering Foundation to W.T.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Woo Taek Kim (wtkim@yonsei.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends Plant Sci 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 [DOI] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2: 48–54 [Google Scholar]
- Cho SK, Kim JE, Park J-A, Eom TJ, Kim WT (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580: 3136–3144 [DOI] [PubMed] [Google Scholar]
- Choi KH, Hong CB, Kim WT (2002) Isolation and characterization of drought-induced cDNA clones from hot pepper (Capsicum annuum). J Plant Biol 45: 212–218 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117–124 [DOI] [PubMed] [Google Scholar]
- Frugis G, Chua N-H (2002) Ubiquitin-mediated proteolysis in plant hormone signal transduction. Trends Cell Biol 12: 308–311 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI (2003) U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun 302: 635–645 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Heschel MS, Riginos C (2005) Mechanisms of selection for drought stress tolerance and avoidance in Impatiens capensis (Balsaminaceae). Am J Bot 92: 37–44 [DOI] [PubMed] [Google Scholar]
- Hong J-P, Kim WT (2005) Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv Pukang). Planta 220: 875–888 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu J (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Park KY, Kim WT (2004) Light differentially regulates the expression of two members of the auxin-induced 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.) seedlings. Planta 218: 976–988 [DOI] [PubMed] [Google Scholar]
- Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K (2001) A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J 26: 217–227 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132: 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau O-S, Deng X-W, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng X-W (1999) Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with COP9 complex. J Mol Biol 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Deng XW, Kim WT (2006) Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem Biophys Res Commun 350: 484–491 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Hong J-P, Oh S-K, Lee S, Choi D, Kim WT (2004) The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol Biol 55: 61–81 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Kim WT (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46: 649–657 [DOI] [PubMed] [Google Scholar]
- Mang HG, Kang EO, Shim JH, Kim S-y, Park KY, Kim YS, Bahk YY, Kim WT (2004. a) A proteomic analysis identifies glutathione S-transferase isoforms whose abundance is differentially regulated by ethylene during the formation of early root epidermis in Arabidopsis seedlings. Biochim Biophys Acta 1676: 213–219 [DOI] [PubMed] [Google Scholar]
- Mang HG, Lee J-H, Park J-A, Pyee J, Pai H-S, Lee JH, Kim WT (2004. b) The CaPRP1 gene encoding a putative proline-rich glycoprotein in highly expressed in rapidly elongating early roots and leaves in hot pepper (Capsicum annuum L. cv. Pukang). Biochim Biophys Acta 1674: 103–108 [DOI] [PubMed] [Google Scholar]
- Mckay JK, Richards JH, Mitchell-olds T (2003) Genetics of drought adaptation in Arabidopsis thaliana. I. Pleiotropy contributes to genetic correlation among ecological traits. Mol Ecol 12: 1137–1151 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu S-H, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insight into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, Satou M, Sakurai T, Ishida J, Akiyama K, et al (2003) Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J 34: 868–887 [DOI] [PubMed] [Google Scholar]
- Ozturk NZ, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48: 551–573 [DOI] [PubMed] [Google Scholar]
- Park JA, Cho SK, Kim JE, Chung HS, Hong J-P, Hwang B, Hong CB, Kim WT (2003) Isolation of cDNAs differentially expressed in response to drought stress and characterization of the Ca-LEAL1 gene encoding a new family of atypical LEA-like protein homologue in hot pepper (Capsicum annuum L. cv. Pukang). Plant Sci 165: 471–481 [Google Scholar]
- Peng Z, Staub JM, Serino G, Kwok SF, Kurepa J, Bruce BD, Vierstra RD, Wei N, Deng X-W (2001) The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol Biol Cell 12: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T, Valette C, Lawrence JJ (1994) Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis 15: 1552–1558 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntly R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Roberts JK (2002) Proteomics and a future generation of plant biologists. Plant Mol Biol 48: 143–154 [PubMed] [Google Scholar]
- Rose JKC, Bashir S, Giovannoni JJ, Jahn MM, Saravanan RS (2004) Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J 39: 715–733 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carnici P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN 12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H (2003) AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol 132: 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ (2000) A modified silver staining protocol for visualization of proteins compatible with matrix assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21: 3666–3672 [DOI] [PubMed] [Google Scholar]
- Yang C-W, Gonzalez-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O'Donnell E, Jones JDG, Sadanandom A (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi T, Nagata N, Shimaga H, Matsui M (1999) An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11: 1883–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L-R, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang G-L (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]