Abstract
Alterations in the proteome of Arabidopsis (Arabidopsis thaliana) leaves during responses to challenge by Pseudomonas syringae pv tomato DC3000 were analyzed using two-dimensional gel electrophoresis. Protein changes characteristic of the establishment of disease, basal resistance, and resistance-gene-mediated resistance were examined by comparing responses to DC3000, a hrp mutant, and DC3000 expressing avrRpm1, respectively. The abundance of each protein identified was compared with that of selected transcripts obtained from comparable GeneChip experiments. We report changes in three subcellular fractions: total soluble protein, chloroplast enriched, and mitochondria enriched over four time points (1.5–6 h after inoculation). In total, 73 differential spots representing 52 unique proteins were successfully identified. Many of the changes in protein spot density occurred before significant transcriptional reprogramming was evident between treatments. The high proportion of proteins represented by more than one spot indicated that many of the changes to the proteome can be attributed to posttranscriptional modifications. Proteins found to show significant change after bacterial challenge are representative of two main functional groups: defense-related antioxidants and metabolic enzymes. Significant changes to photosystem II and to components of the mitochondrial permeability transition were also identified. Rapid communication between organelles and regulation of primary metabolism through redox-mediated signaling are supported by our data.
The response of plant cells to wounding or microbial challenge is rapid and specific. Unlike animal immune systems, each individual plant cell must orchestrate its own defense, as well as respond to cues from its neighbors. The paradigm for plant-microbe interactions is that conserved features of microbes, such as bacterially derived flagellin, harpin, or fungal chitin, are recognized by specific receptors and are designated pathogen-associated molecular patterns (PAMPs). Upon flagellin recognition, the flagellin-sensing receptor triggers a rapid signaling cascade that ultimately results in transcription of defense-related genes and gross morphological changes such as cytoskeletal rearrangement and thickening of the cell wall through papilla deposition. Recognition and signaling events occur extremely rapidly. For example, specific proteins are transiently phosphorylated minutes after elicitation of Arabidopsis (Arabidopsis thaliana) cell cultures with the 22-amino acid flagellin peptide, flg22 (Peck et al., 2001; Peck, 2003), and a transient increase in cytosolic calcium is observed within 15 min of leaf inoculation with bacteria (Grant et al., 2000). Generic responses such as these contribute to basal defense and are believed to underlie the observation that most plants are resistant to invasion by the majority of microbes.
Successful pathogens appear to have evolved mechanisms to interfere with or suppress basal defense. Gram-negative bacterial pathogens, such as Pseudomonas syringae pv tomato (Pst), introduce effector proteins to the host cell through a type III secretion system (T3SS). Assembly of the T3SS is induced by nutrient conditions typically associated with the apoplastic space (van Dijk et al. 1999; Collmer et al., 2000) and occurs rapidly (Thwaites et al., 2004). A functional T3SS is absolutely required for pathogenicity, as demonstrated by the asymptomatic response of the hrpA mutant of the normally highly virulent Pst strain DC3000 on Arabidopsis. The hrp mutant strongly elicits specific sets of genes indicative of basal defense (de Torres et al., 2003; Truman et al., 2006).
Although Pst DC3000 possesses up to 40 T3SS-delivered effector proteins that we describe as type 3 effectors (T3E), the majority are of unknown function and structure (Buell et al., 2003). Of the few that have been functionally characterized, most were discovered as avirulence determinants, where they were directly or indirectly recognized by their cognate resistance (R)-gene products (Dangl and Jones, 2001). It was only later that their essential roles in promoting virulence were uncovered, thus resolving the paradox of why a pathogen would maintain such a potentially detrimental cargo. Bacterial transcription of effectors in the plant is rapid; for example, the Pseudomonas effector, avrRpm1, is induced within an hour of leaf inoculation (Grant et al., 2000; Thwaites et al., 2004). Given the diverse variety of pathogens and their equally large repertoire of effectors, it has become apparent that plants do not possess enough R genes to intercept all potential avirulence determinants. Instead, it is hypothesized that effector proteins target key components of the cellular machinery and R proteins guard these effector targets, either directly or by sensing modification of the target (Mackey et al., 2002). R-protein activation triggers a specific defense reaction typified by a form of programmed cell death (PCD) known as the hypersensitive reaction (HR) and pathogen restriction (Grant and Mansfield, 1999).
The R gene, RPM1 (Grant et al., 1995), enables recognition of both avrRpm1 and avrB, triggering a rapid HR. The HR is usually associated with rapid increases in cytosolic calcium, acidification of the apoplast, prolonged oxidative burst, changes in phosphorylation, and increases in nitric oxide (NO). These responses appear to be conserved; only the timing differs for each R-avr gene combination (Bennett et al., 2005). The intracellular balance between reactive oxygen species (ROS) and reactive nitrogen species (RNS) is critical to trigger cell death in the HR (Delledonne et al., 2001) and differentiate from other ROS-mediated stresses, such as cold shock and auxin treatment (Laloi et al., 2004; Delledonne 2005). Consequently, cytosolic antioxidant enzymes are essential in controlling ROS generated by plant organelles (Davletova et al., 2005). Both chloroplasts and mitochondria are additional potential sources of ROS and possess well-developed mechanisms to rapidly detoxify free radicals. The chloroplast plays a central role in the generation of intracellular ROS through PSII and the plastoquinone pool (Ivanov and Khorobrykh, 2003). In contrast to animal cells, in plants mitochondria appear to contribute less to cellular ROS (Maxwell et al., 1999; Robson and Vanlerberghe, 2002), although there is widespread acceptance of the importance of mitochondria in plant PCD (Yao et al., 2004).
Changes to the proteome of chloroplasts are of particular interest because this organelle is implicated in the defense response in several ways. Components of both the salicylic acid (SA) and jasmonic acid biosynthetic pathways are located in this organelle (Wildermuth et al., 2001; Wasternack et al., 2006). SA-dependent induction of pathogenesis-related (PR) genes and the HR is often dependent on light, but not functional chloroplasts (Genoud et al., 2002; for review, see Karpinski et al., 2003). Photorespiration can modulate defense responses by dissipating excessive ROS, mitigating photooxidative damage (Zeier et al., 2004; Moreno et al., 2005).
To date, no global proteomic study has examined plant tissues actively responding to bacterial challenges or investigated associated posttranslational events occurring prior to major transcriptional changes (de Torres et al., 2003; Thilmony et al., 2006; Truman et al., 2006). This is in part due to significant technical difficulties associated with utilizing proteins and organelles extracted from green leaves for two-dimensional electrophoresis (2DE) and is further complicated by the necessity to infect synchronously sufficient quantities of tissue for replicate analyses. Nevertheless, it is essential to undertake in planta studies because the continued presence of the bacterium in the apoplast could modify defense responses and bacterial effector proteins are correctly delivered to the host cell. We have previously reported changes to two families of antioxidant enzymes, glutathione-S-transferases (GSTs) and peroxiredoxins (Prxs), during the defense response (Jones et al., 2004). Here, we present complete data on changes in proteomic expression in Arabidopsis leaves challenged with Pst DC3000 (disease inducing), hrpA (activating basal defenses), and DC3000 (avrRpm1), which triggers R-gene-mediated resistance, or mock inoculation (10 mm MgCl2). Total soluble protein, chloroplast-enriched, and mitochondria-enriched fractions were isolated and proteins were visualized by 2DE. The presence and identity of proteins corresponding to stained gel spots showing significant differences between treatments across three replicate gels were verified by liquid chromatography (LC)-tandem mass spectrometry (MS/MS).
RESULTS
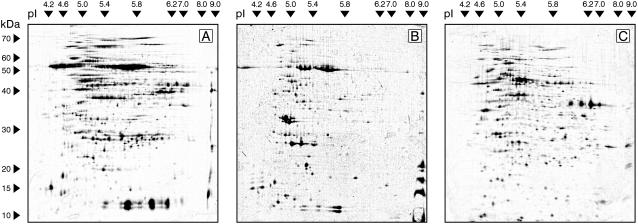
Proteins were extracted from leaves 1.5, 3, 4, and 6 h postinoculation (hpi) and resolved on 2DE gels as described in Jones et al. (2004). Representative gel images for each of the three subcellular fractions are shown in Figure 1. The numbers of spots resolved in each fraction, the stain employed, and the numbers of proteins identified are summarized in Table I. Spots showing significant change in density (P ≤ 0.05) between any of the four treatments were identified by LC-MS/MS.
Figure 1.
Representative 2DE gels visualizing proteins from three subcellular fractions extracted from Arabidopsis leaf tissue after challenge with DC3000 hrpA (3-hpi time point displayed). The first dimension was focused across a 3 to 9 pH unit range and the second electrophoretic dimension resolved proteins between 10 and 70 kD. A, Soluble fraction. B, Chloroplast-enriched fraction. C, Mitochondria-enriched fraction.
Table I.
Summary of amounts of protein used for gels, numbers of spots resolved, and proteins identified
| Feature | Soluble | Chloroplast | Mitochondria |
|---|---|---|---|
| Protein/gel | 75 μg (analytical); 600 mg (preparative) | 150 μg | 75 μg |
| Staining | Silver/Coomassie | Colloidal Coomassie | Silver |
| Total spots | 800 | 450 | 1,000 |
| Spots showing significant difference between treatments | 69 (8.6%) | 34 (7.5%) | 92 (9.2%) |
| Mixtures or not identified | 15 | 11 | 52 |
| Valid identified spots | 54 | 23 | 40 |
| Unique proteinsa | 39 | 18 | 33 (14 from Pseudomonas) |
| Proteins presented herea | 34 | 8 | 13 |
Some of the spots showed significant difference between the control (MgCl2) and DC3000 (avrRpm1) treatments, but these data are not classified in detail here.
Many proteins that differentiated between treatments were identified in more than one spot, indicating probable posttranslational modification (PTM). In total, 36 proteins (51 spots) were identified from the soluble fraction, eight proteins (nine spots) from the chloroplast, and 13 proteins (13 spots) from the mitochondrial fraction. Characteristics of the differential proteins are presented in Table II, including the number of unique peptide hits to the protein (indicating the robustness of the identification). The apparent Mr was used to differentiate between precursor and processed forms of proteins. Although some proteins migrate in an aberrant manner on SDS-PAGE gels, for the majority of proteins, especially those of unexceptional pI, the observed Mr is considered to be reasonably accurate. The value of enrichment of organelles was apparent from the recovery of proteins that were not detected in the total soluble fraction.
Table II.
Classification of differential proteins into functional groups and details of observed and predicted Mr and pI
Where a targeting peptide has been predicted, the Mr and pI of the processed protein is indicated in parentheses. The number of unique peptides sequenced by MS/MS is provided to indicate robustness of identification.
| Gene | Protein | Spot | Changea | Fractionb | Targetc | Observed Mr | Predicted Mrd | Observed pI | Predicted pI | Numbers of Peptides |
|---|---|---|---|---|---|---|---|---|---|---|
| Defense related | ||||||||||
| At1g20020 | FNR | 7,309 | T↑ and P↑ | Sol | Cpl | 34.5 | 41.48 (35.27) | 5.98 | 8.51 (6.20) | 5 |
| At2g30860 | GST AtGSTF9 | 7,215 | A↑ | Sol | No | 28.0 | 24.13 | 5.98 | 6.17 | 7 |
| At3g10920 | MSD1 | 6,110 | P↑A↓ | Sol | Mit | 26.1 | 25.44 (22.53) | 5.97 | 8.47 (6.06) | 8 |
| At3g27890 | NQR | 8,101 | A↓ | Sol | No | 25.6 | 21.51 | 5.99 | 7.7 | 5 |
| At4g23670 | MLP-related | 6,004 | P↑ and A↓ | Sol | No | 17.2 | 17.62 | 5.8 | 5.91 | 8 |
| At5g66190 | FNR | 4,303 | A↑ | Sol | No | 34.4 | 40.64 | 5.38 | 8.32 | 7 |
| At1g09340 | UDP-Glc 4-epimerase | 7,507 | P↑ | Cpl | No | 38.3 | 42.75 | 6.51 | 7.17 | 3 |
| At4g01050 | Hyp-rich glycoprotein | 2,802 | P↑T↓A↑ | Cpl | Cpl | 59.9 | 49.36 (47.8) | 4.70 | 5.22 (5.21) | 3 |
| At1g02920 | GST AtGSTF7 | 7,211 | P↑ P↑ | Sol | No | 27.7 28.0 | 23.58 | 6.00 5.99 | 6.14 | 5 |
| 8,201 | 3 | |||||||||
| At1g02930 | GST AtGSTF6 | 6,109 | P↑ P↑ | Sol | No | 27.4 27.5 | 23.4 | 6.00 5.80 | 6.17 | 5 |
| 6,101 | 4 | |||||||||
| At2g47730 | GST AtGSTF8 | 6,108 | P↑A↓ A↑ | Sol Sol | Cpl | 26.8 26.9 | 29.2 (24.1) | 6.0 5.7 | 8.90 (6.09) | 4 |
| 5,104 | 6 | |||||||||
| At4g02520 | GST AtGSTF2 | 7,213 | P↑ | Sol | No | 28.4 | 24.11 | 6.00 | 5.92 | 5 |
| At3g11630 | PrxA | 1,104 | P↓T↑ | Sol | Cpl | 24.8 | 29.1 (22.8) | 4.79 | 4.91 (5.01) | 8 |
| At3g52960 | PrxIIE | 1,007 | P↑ | Sol | Cpl | 19.3 | 24.67 (17.3) | 4.9 | 9.12 (5.02) | 14 |
| At5g06290 | 2-cys-PrxB | 111 | P↑ and A↓ | Sol | Cpl | 26.6 | 29.6 (24.6) | 4.55 | 4.71 (5.07) | 8 |
| Metabolism | ||||||||||
| Primary carbon metabolism | ||||||||||
| At1g13440 | G3PDH | 8,410 | P↓ | Sol | No | 39.0 | 37.08 | 6.00 | 6.34 | 7 |
| At1g32060 | PRK | 2,502 | P↓T↑ | Sol | Cpl | 42.1 | 44.72 (38.5) | 5.09 | 5.71 (5.22) | 17 |
| 2,613 | T↓ | Cpl | 42.9 | 5.00 | 2 | |||||
| At1g53240 | NAD-dependent MDH | 6,316 | P↓ | Sol | Mit | 35.1 | 35.75 (32.37) | 5.97 | 8.54 (5.73) | 6 |
| 8,403 | P↑T↓ | Sol | 35.3 | 5.99 | 10 | |||||
| At3g01500 | CA | 2,207 | P↓T↑ | Sol | Cpl | 28.8 | 36.12 (33.38) | 5.21 | 5.46 (5.43) | 7 |
| At3g04790 | Rib-5-P isomerase putative | 1,207 | T↓ | Sol | Cpl | 29.1 | 29.30 (25.44) | 5.00 | 5.72 (4.98) | 4 |
| At3g15020 | MDH | 5,202 | P↑ | Mit | Mit | 33.9 | 36.02 (32.6) | 5.67 | 8.30 (6.24) | 7 |
| At5g50850 | PDH E1β | 4,301 | P↑ | Mit | Mit | 36.7 | 39.45 (35.9) | 4.85 | 5.67 (5.11) | 2 |
| At5g14740 | CA2 | 4,217 | P↑ | Mit | No | 29.7 | 28.68 | 4.99 | 5.36 | 9 |
| 4,302 | A↓ | Sol | 31.9 | 5.36 | 7 | |||||
| At2g39730 | Rubisco activase | 1,609 | T↓ | Sol | Cpl | 47.4 | 51.98 (46.26) | 4.97 | 5.87 (5.09) | 10 |
| 2,421 | P↑ | Mit | 45.3 | 4.63 | 4 | |||||
| At3g55800 | Sedoheptulose-bisphosphatase | 1,512 | P↓ | Sol | Cpl | 39.2 | 42.79 (36.09) | 4.96 | 6.71 (5.21) | 8 |
| Other metabolic roles | ||||||||||
| At4g08900 | Putative arginase | 5,352 | P↑ | Mit | Mit | 35.6 | 37.55 (35.4) | 5.48 | 6.11 (5.5) | 1 |
| At5g08280 | Hydroxymethylbilane synthase | 8,409 | P↑T↓ | Sol | Cpl | 36.7 | 41.53 | 5.99 | 8.63 | 7 |
| At3g14210 | Lipase/acylhydrolase, | 8,510 | P↓ | Sol | Sec | 43.5 | 44.37 | 6.00 | 7.59 | 7 |
| PSII | ||||||||||
| At1g06680 | OEC23 | 1,404 | A↑ | Cpl | Cpl | 27.4 | 28.08 (24.90) | 4.57 | 6.90 (5.79) | 2 |
| At5g66570 | OEC1 | 2,417 | T↓A↑ | Cpl | Cpl | 33.2 | 35.11 (32.10) | 4.80 | 5.68 (5.08) | 6 |
| 1,417 | A↑ | Cpl | 33.3 | 4.73 | 6 | |||||
| Transcription | ||||||||||
| At1g20980 | SPB-like 2 | 4,307 | T↑ | Sol | TMR | 33.0 | 79.75 | 5.45 | 8.38 | 3 |
| At4g36690 | Splicing factor-like protein | 3,309 | P↑ | Mit | No | 34.4 | 28.86 | 4.72 | 5.17 | 2 |
| At5g19510 | Elongation factor 1B-α | 311 | T↑ | Sol | No | 32.4 | 24.24 | 4.39 | 4.42 | 8 |
| Protein folding and turnover | ||||||||||
| At1g79210 | 20S Proteasome PAB1 | 4,222 | A↓ | Mit | No | 28.1 | 25.71 | 5.07 | 5.53 | 2 |
| At3g01480 | Putative thylakoid lumen Rotamase (cyclophilin) | 503 | T↑ | Sol | Cpl | 39.3 | 47.95 | 4.50 | 5.06 | 14 |
| At3g27850 | 50S ribosomal protein L12-C. | 1,309 | A↑ | Cpl | Cpl | 20.8 | 19.67 (14.1) | 4.52 | 5.99 (4.68) | 4 |
| At3g62030 | Peptidylprolyl isomerase ROC4 cyclophilin | 1,105 | P↓T↑ | Sol | Cpl | 23.4 | 28.18 (20.0) | 4.9 | 8.83 (5.4) | 6 |
| At5g10450 | 14-3-3 protein GF14-λ | 206u | T↑A↓ | Sol | No | 30.3 | 28.72 | 4.59 | 4.77 | 11 |
| At3g63190 | Ribosome recycling factor | 9,201 | A↑ | Sol | Cpl | 27.7 | 32.22 (28.50) | 7.46 | 9.39 (9.33) | 5 |
| Cellular transport and ion homeostasis | ||||||||||
| At3g26520 | Aquaporin tip2.1 | 4,102 | P↓ | Mit | Mit Tmr | 25.2 | 25.89 (24.31) | 4.88 | 4.92 (4.92) | 1 |
| At4g09650 | H+-ATPase-related protein CF1δ | 2,109 | P↓ | Sol | Cpl | 23.1 | 25.65 (20.4) | 5.2 | 9.04 (5.31) | 10 |
| At4g11150 | V-type H+-ATPase subunit E | 7,212 | A↓ | Sol | No | 31.3 | 26.28 | 5.98 | 6.04 | 5 |
| At4g20260 | Endomembrane-associated protein F1C12.180 | 1,404 | A↓ | Sol | Pm | 35.4 | 24.57 | 5.02 | 4.99 | 2 |
| At4g32260 | H+-ATPase (ATPG) | 1,203 | T↑ | Cpl | Cpl | 18.2 | 23.90 (16.24) | 4.61 | 5.80 (4.78) | 7 |
| AtC00120 | ATPase α-chain | 4,508 | A↑ | Mit | Cpl | 52.5 | 55.35 | 4.97 | 5.19 | 3 |
| ArthCp029 | ATPase CF1β | 4,711 | A↑ | Cpl | No | 52.7 | 53.90 | 5.20 | 5.38 | 19 |
| At5g09650 | Inorganic pyrophosphatase-like protein | 1,304 | T↑A↓ | Sol | Cpl | 32.9 | 33.3 | 4.70 | 5.72 | 2 |
| Mitochondrial permeability transition | ||||||||||
| At5g48810 | Cytochrome b5 | 3,116 | A↓ | Mit | No | 18.7 | 15.15 | 4.82 | 4.87 | 2 |
| At5g15090 | Outer mitochondrial membrane protein porin 2 (VDAC 2) | 8,222 | P↓ | Mit | Mit | 29.1 | 29.00 | 7.91 | 7.97 | 8 |
| At4g03280 | Rieske Fe-S protein precursor | 7,127 | P↑ | Mit | Mit | 19.3 | 22.80 (19.0) | 5.72 | 8.58 (6.07) | 2 |
| Unclassified | ||||||||||
| At3g16640 | TCTP-like protein | 109 | T↑A↓ | Sol | Sec | 23.7 | 18.96 | 4.6 | 4.52 | 5 |
| 2,107 | A↓ | Mit | 24.1 | 3 | ||||||
| At2g37660 | 3-β-hydroxysteroid dehydrogenase/isomerase putative | 3,209 | T↓ | Sol | Cpl | 31.1 | 36.18 (27.62) | 5.28 | 6.23 (5.31) | 2 |
| 3,218 | A↓ | Sol | 31.1 | 5.29 | 3 | |||||
Significant differences in density (P ≤ 0.05) were recorded in response to PAMP (i.e. DC3000 hrpA mutant compared with mock inoculation; P); differentials between DC3000 hrpA and wild-type DC3000 inoculations (T); and difference between DC3000 wild type and DC3000 (avrRpm1) challenge (A).
Subcellular fraction from which the protein was recovered. Sol, Soluble, Cpl, chloroplast, Mit, mitochondria.
Predicted localization of protein based on N-terminal sequence. Sec, Secreted; Mit, mitochondria targeted; Cpl, chloroplast targeted; Pm, plasma membrane; TMR, predicted transmembrane region; No, no targeting motif recognized.
Mr and pI of processed protein in parentheses. Failure to discriminate between related genes is indicated by a “u” beside the spot number.
Proteins were assigned to seven functional categories (Table II) based on sequence homology, annotated function, or presence of characteristic motifs. These classifications are predominantly predictive and we emphasize in the text whether protein function has been experimentally validated.
Classification of Identified Proteins
Proteins were classified into three behavioral groups according to the observed changes in their spot density, PAMP responsive (differing in hrpA challenge compared to mock infiltration), T3E responsive (showing differential spot abundance between hrpA and DC3000 challenges), and AvrRpm1 responsive (differential in spot density between DC3000 [avrRpm1] and hrpA inoculations). The assignment of a spot to a particular behavioral group was based on a significant difference in density (P ≤ 0.05) between treatments and this cutoff forms the basis of our study. Overall patterns of spot changes are summarized in Figure 2 and individual proteins are presented in Tables III to V. Some proteins showed different patterns of behavior, depending upon the inducing challenge, and are therefore assigned to more than one category, as illustrated by the overlaps in Figure 2. For example, a protein may initially exhibit PAMP-induced behavior, but subsequently be suppressed by T3Es. Such behavior is entirely consistent with virulence factors targeting components of active plant defense as seen at the transcriptional level (Thilmony et al., 2006; Truman et al., 2006) and validates our approach to combine transcriptomic studies (Truman et al., 2006) with a comparative proteomics approach.
Figure 2.
Changes in spot density during basal (PAMP; P), susceptible (T3E; T), or resistant (RPM1/AvrRpm1; A) interactions. Each portion of the Venn diagram represents the number of spots showing either an increase (↑) or decrease (↓) in spot density. Overlaps contain spots that responded differently to two or more treatments.
Table III.
Protein spots responding to PAMPs
A change in spot density is classified as PAMP associated if the DC3000 hrpA inoculation is significantly different from the MgCl2-treated control regardless of the other two treatments. A protein spot is classified as PAMP associated and modulated by T3E if there were significant differences between DC3000 and DC3000 hrpA mutant inoculation and the latter was also significantly different from MgCl2 treatment.
| Gene | Proteina | Fraction Timeb | Spot Density Treatmentc
|
|||
|---|---|---|---|---|---|---|
| Mg | hrpA | DC3000 | avrRpm1 | |||
| Proteins decreasing in spot density in response to PAMPs | hpi | |||||
| At4g09650 | H+-ATPase-related protein | Sol 1.5 | 6,357a | 4,479b | 5,695ab | 4,442b |
| At3g55800 | Sedoheptulose-bisphosphatase | Sol 4 | 9,844a | 3,464b | 5,563b | 6,739b |
| At1g13440 | G3PDH | Sol 4 | 8,188a | 6,050b | 4,229b,c | 3,044c |
| At3g14210 | Lipase | Sol 4 | 15,398a | 9,977b | 10,067ab | 11,383ab |
| At3g14210 | Lipase | Sol 6 | 12,974a | 5,343 b | 8,765b | |
| At1g53240 | MDH (NAD) | Sol 6 | 1,587a | 1134b | 1,167ab | |
| At3g26520 | Aquaporin TIP1.2 | Mit 3 | 6,179a | 1,062b | 1,197b | 2,083b |
| At5g15090 | Outer mitochondrial membrane protein porin 2 | Mit 3 | 11,377a | 5,788b | 6,866b | 6,606b |
| Proteins decreasing in response to PAMPs modulated by T3E | ||||||
| At3g11630 | 2-cys-PrxA | Sol 1.5 | 1,760a | 280b | 1,071ac | 654c |
| At3g01500 | CA, chloroplast | Sol 1.5 | 1,504a | 750b | 1,310a | 1,676ab |
| At3g62030 | Peptidylprolyl isomerase (ROC4 cyclophilin ) | Sol 6 | 1,764a | 1,017b | 1,754a | |
| At1g32060 | PRK | Cpl 6 | 1,910a | 509b | 1,706a | |
| Proteins increasing in response to PAMPs | ||||||
| At1g20020 | FNR | Sol 1.5 | 515a | 910b | 879ab | 512a |
| At1g02930 | GST | Sol 1.5 | 1,363a | 2,373b | 2,071ab | 1,462ab |
| At5g06290 | 2-cys-PrxB | Sol 3 | 975a | 1,212ab | 1,330b | 1,318b |
| At4g23670 | MLP (related, MLP) | Sol 3 | 1,372a | 2,151b | 2,535b | 2,094b |
| At1g02930 | GST (GSTF6) | Sol 3 | 1,823a | 2,736b | 2,976c | 2,663b |
| At1g02920 | GST (GSTF7) | Sol 3 | 1,223a | 2,578b | 2,521ab | 2,258ab |
| At1g02920 | GST (GSTF7) | Sol 4 | 910a | 2,861b | 3,430b | 3,074b |
| At3g52960 | PrxIIE | Sol 4 | 1,815a | 2,449b | 2,595b | 2,677b |
| At1g02930 | GST (GSTF6) | Sol 4 | 2,210a | 3,640b | 3,822b | 3,491ab |
| At4g02520 | GST (GSTF2) | Sol 4 | 1,080a | 2,433b | 1,550ab | 2,081b |
| At1g02920 | GST (GSTF7) | Sol 6 | 2,244a | 4,444b | 4,161b | |
| At1g02930 | GST (GSTF6) | Sol 6 | 2,757 | 4,985b | 4,633b | |
| At4g02520 | GST (GSTF2) | Sol 6 | 1,704a | 2,659b | 2,100ab | |
| At1g09340 | UDP-Glc 4-epimerase-like | Cpl 3 | 584a | 1,478b | 1,335ab | 522ab |
| At3g15020 | MDH | Mit 3 | 1,472a | 2,492b | 2,530ab | 2,643ab |
| At4g08900 | Putative arginase | Mit 3 | 0a | 411b | 3,26b | 759b |
| At5g14740 | CA2 | Mit 3 | 4,748a | 7,784b | 6,431ab | 3,852ab |
| At4g36690 | Splicing factor-like protein | Mit 3 | 397a | 1,400b | 1,288b | 1,299ab |
| At5g50850 | PDH E1β-subunit | Mit 3 | 77a | 1,322b | 1,544b | 1,325ab |
| At4g03280 | Rieske Fe-S protein | Mit 3 | 0a | 1,287b | 807b | 1,164b |
| At2g39730 | Rubisco activase | Mit 3 | 0a | 1,795b | 1,867b | 1,566b |
| Proteins increasing in response to PAMPs modulated by T3E | ||||||
| At3g10920d | MSD1 | Sol 1.5 | 375a | 662b | 748b | 307a |
| At2g47730d | GST (GSTF8) | Sol 1.5 | 334a | 639b | 725b | 344a |
| At1g53240 | MDH [NAD] | Sol 3 | 599a | 1,330 b | 755a | 801ab |
| At5g08280 | Hydroxymethylbilane synthase | Sol 4 | 0a | 1,248b | 561a | 379a |
| At4g01050 | Hyp-rich glycoprotein | Cpl 1.5 | 853a | 1,499b | 1,045a | 1,501b |
Proteins are sorted by fraction, time after inoculation, and modulation by T3Es.
Subcellular fraction from which the protein was recovered. sol, Soluble; cpl, chloroplast; mit, mitochondria enriched. Time (hpi) at which the significant change was determined.
Averaged spot density over three replicates. Significant differences (labeled a, b, and c) were determined by t test (P ≤ 0.05). Mg, 10 mM MgCl2 treatment; hrpA, DC3000 hrpA treated; D3000, DC3000 treated; avrRpm1, DC3000 (avrRpm1) treated.
Proteins that appear to be modulated by AvrRpm1 but not DC3000 T3E. They are included in this table to facilitate discussion in the text.
Table V.
Proteins responding to RPM1/AvrRpm1
A change in spot density is classified as an AvrRpm1 response if the DC3000 (avrRpm1) inoculation causes a significant difference from the wild-type DC3000 inoculation.
| Gene | Proteina | Fraction Timeb | Spot Densityc
|
|||
|---|---|---|---|---|---|---|
| Mg | hrpA | DC3000 | avrRpm1 | |||
| Proteins increasing in spot density | hpi | |||||
| At5g66190 | FNR | Sol 1.5 | 479b | 420ab | 272a | 653b |
| At2g47730 | GST (GSTF8) | Sol 4 | 837a | 819a | 735a | 1,387b |
| At3g63190 | Ribosome recycling factor | Sol 4 | 673ab | 696a | 527a | 1,080b |
| At2g30860 | GST (GSTF9) | Sol 4 | 6,362ab | 6,704ab | 6,972a | 9,481b |
| At5g66570 | OEC33 K protein | Cpl 1.5 | 77,756ab | 91,780ab | 71,226b | 93,405a |
| At1g06680 | OEC23 K protein | Cpl 1.5 | 8,969ab | 5,867a | 6,279a | 8,640b |
| At3g27850 | 50S ribosomal protein L12-C | Cpl 1.5 | 8,969ab | 5,867a | 6,279a | 8,640b |
| ArthCp029 | ATPase CF1β-chain | Cpl 4 | 25,831a | 30849ab | 22,700a | 39,552b |
| ATCG00120 | ATPase-α | Mit 3 | 644a | 897a | 906a | 29,80b |
| Proteins decreasing in spot density | ||||||
| At5g06290 | 2-cys-PrxB | Sol 1.5 | 1,017ab | 926ab | 1,324a | 1,039b |
| At3g27890 | NQR | Sol 1.5 | 848ab | 872ab | 890a | 565b |
| At4g11150 | V-type proton-ATPase subunit E | Sol1.5 | 300ab | 335ab | 332a | 127b |
| At5g14740 | CA2 | Sol 1.5 | 2,412ab | 3,308a | 3,223a | 1,937b |
| At4g23670 | MLP | Sol 1.5 | 1,458a | 2,013ab | 2,352b | 1,615a |
| At4g20260 | Endomembrane-associated protein F1C12.180 | Sol 4 | 1,812ab | 2526ab | 1,302b | 1,993a |
| At2g37660 | 3-β-hydroxysteroid dehydrogenase/isomerase | Sol 4 | 1,406ab | 1,643a | 1,818a | 542b |
| At1g79210 | 20S proteasome α-subunit B | Mit 3 | 770ab | 892ab | 9,67a | 703b |
| At3g16640 | TCTP | Mit 3 | 394ab | 1,061ab | 1,000a | 455b |
| At5g48810 | Cytochrome b5 | Mit 3 | 1,476ab | 1,312ab | 1,701a | 843b |
Proteins are sorted by fraction, time after inoculation, and modulation by T3Es.
Subcellular fraction from which the protein was recovered. sol, Soluble; cpl, chloroplast; mit, mitochondria enriched. Time (hpi) at which the significant change was determined.
Averaged spot density over three replicates. Significant differences (labeled a, b) were determined by t test (P ≤ 0.05). Mg, 10 mM MgCl2 treatment; hrpA, DC3000 hrpA treated; D3000, DC3000 treated; avrRpm1, DC3000 (avrRpm1) treated.
Table IV.
Proteins responding to T3Es
A change in spot density is classified as T3E if the DC3000 inoculation was significantly different from the DC3000 hrpA inoculation regardless of the other two treatments. A protein spot is classified as T3E and modulated by AvrRpm1 if the DC3000 inoculation was significantly different from the DC3000 (avrRpm1) treatment and DC3000 was significantly different from DC3000 hrpA.
| Gene | Proteina | Fraction Timeb | Spot Densityc
|
|||
|---|---|---|---|---|---|---|
| Mg | hrpA | DC3000 | avrRpm1 | |||
| Proteins increasing in spot density | hpi | |||||
| At3g01480 | Putative thylakoid lumen rotamase (cyclophilin) | Sol 1.5 | 1,360ab | 872a | 2,167b | 1,576ab |
| At5g19510 | Elongation factor-1B α-subunit | Sol 1.5 | 1,454ab | 1,238a | 1,613b | 1,110ab |
| At1g20980 | SPB-like 2 | Sol 3 | 448a | 449a | 630b | 528ab |
| At1g32060 | PRK | Sol 3 | 3,517ab | 2,058a | 3,528b | 2,808ab |
| At1g20020 | FNR | Sol 6 | 728ab | 397a | 657b | |
| At4g32260 | H+-ATPase-like protein | Cpl 6 | 1,723ab | 425a | 1,566b | |
| Proteins decreasing in spot density | ||||||
| At2g39730 | Rubisco activase | Sol 4 | 3,308ab | 3,728a | 2,118b | 2,547b |
| At2g37660 | 3-β-Hydroxysteroid dehydrogenase/isomerase | Sol 6 | 1,096ab | 1,784a | 1,162b | |
| At3g04790 | Rib-5-P isomerase | Sol 6 | 4,194ab | 5,846a | 3,347b | |
| Protein increasing in spot density modified by RPM1/AvrRpm1 | ||||||
| At3g16640 | TCTP homolog | Sol 1.5 | 1,176ab | 704a | 1,178b | 637a |
| At5g10450 | 14-3-3-like protein AFT1 | Sol 1.5 | 459a | 413a | 900b | 541a |
| At5g09650 | Inorganic pyrophosphatase-like protein | Sol 3 | 697ab | 498a | 893b | 642a |
| Protein decreasing in spot density modified by RPM1/AvrRpm1 | ||||||
| At5g66570 | OEC33 K protein | Cpl 1.5 | 12,512ab | 10,150a | 6,614b | 11,780a |
Proteins are sorted by fraction, time after inoculation, and modulation by T3Es.
Subcellular fraction from which the protein was recovered. sol, Soluble; cpl, chloroplast; mit, mitochondria enriched. Time (hpi) at which the significant change was determined.
Averaged spot density over three replicates. Significant differences (labeled a, b) were determined by t test (P ≤ 0.05). Mg, 10 mM MgCl2 treatment; hrpA, DC3000 hrpA treated; D3000, DC3000 treated; avrRpm1, DC3000 (avrRpm1) treated.
Changes Associated with Basal Defense and Recognition of PAMPs
We classified 41 spots as PAMP responsive (30 unique proteins), of which 29 increased in density in all three fractions (Fig. 2). Notably, the majority of the increases in density (19 spots) were observed in the soluble fraction. Table III presents quantitative data for each protein spot classified in this group and a graphic representation of these data is presented in Supplemental Figure S1.
PAMP-Responsive Proteins—Soluble Fraction
Twenty-eight spots isolated from the soluble fraction showed significant differences in density between MgCl2 and hrpA treatments. Preliminary studies of the soluble fraction (Jones et al., 2004) identified 13 spots representing GST, GSTF2 (At4g02520), GSTF6 (At1g02930), GSTF7 (At4g02520), and GSTF8 (At2g47730) to be PAMP responsive with significant increases after bacterial challenge. A similar PAMP response was observed for the Prxs (PrxIIE [At3g52960]) and 2-cys-Prx (PrxB [At5g06290]). In contrast, 2-cys-Prx (PrxA [At3g11630]) decreased significantly in response to PAMPs. All of these proteins are implicated in defense-related redox signaling.
Three additional defense-related proteins increased in density in response to PAMPs: manganese superoxide dismutase (MSD1 [At3g10920]), a ferredoxin (Fd) NADP(+) reductase (FNR [At1g20020]), and a major latex protein (MLP [At4g23670]). MLP has similarity to the PR BetvI protein family, which includes well-characterized pollen allergens and intracellular PR proteins. Tsunezuka et al. (2005) found an intracellular PR protein (PBZ1) induced in the rice (Oryza sativa) lesion mimic mutant cdr2, which exhibits enhanced resistance to rice blast (Midoh and Iwata, 1996), probably through inappropriate activation of basal defense mechanisms.
Four metabolic enzymes involved in glycolysis and the pentose phosphate shunt decreased in spot density in a PAMP-associated manner: sedoheptulose bisphosphatase (At3g55800), chloroplast carbonic anhydrase (CA; At3g01500), glyceraldehyde-3-P dehydrogenase (G3PDH; At1g13440), and malate dehydrogenase (MDH; At1g53240). Consistent with the decreases of primary metabolic enzymes, an ATP synthase CF1δ-subunit (At4g09650) rapidly decreases in abundance by 1.5 hpi. The predicted Mr suggests that it corresponds to the precursor of this nuclear-encoded chloroplast protein. CF1δ, a component of the peripheral CF1 complex of two-sector ATPases, plays a central role in oxidative or photosynthetic phosphorylation.
Other proteins showing PAMP-associated changes include a cyclophilin (ROC4 [At3g62030], decrease 6 hpi), which plays a role in protein synthesis and folding, and two proteins of unclear function, a lipase (At3g14210, decrease) and a putative hydroxymethylbilane synthase (At5g08280, increase).
Potential targets of T3Es may be identified by comparison between DC3000 hrpA treatment (PAMP responsive) and DC3000- or DC3000 (avrRpm1)-responsive spots. Eight of the 25 PAMP-responsive proteins were further modulated by T3Es, as indicated by a significant difference between hrpA treatment and DC3000 or, in one case, hrpA and DC3000 versus DC3000 (avrRpm1). The effect of T3Es was antagonistic to PAMP-induced change, reducing DC3000 spot intensity back to that of the MgCl2 control. The proteins targeted by T3Es were PrxA (At3g11630), CA (At3g01500), ROC4 cyclophilin (At3g62030), MDH (At1g53240), hydryoxymethylbilane synthase (At5g08280), GSTF2 (At4g02520), and, by AvrRpm1 alone, MSD1 (At3g10920) and GSTF8 (At2g47730).
PAMP-Responsive Proteins—Chloroplast
Three proteins from the chloroplast-enriched fraction had modified behavior in response to PAMPs. Two proteins increased in spot density, a UPD-Glc 4-epimerase (At1g09340) and a Hyp-rich protein (At4g01050), and both may be associated with papilla formation. The putative UDP-Glc 4-epimerase is predicted to be involved in synthesis of cell wall components. Notably, the observed Mr of the epimerase was smaller than predicted, suggesting some posttranslational processing, and this may account for its unexpected localization to the chloroplasts.
Similarly, Hyp-rich proteins are defense-related glycoproteins proposed to reinforce cell wall appositions during defense responses (Bestwick et al., 1995). Interestingly, At4g01050 does not accumulate following DC3000 challenge, supporting the hypothesis that T3Es function to suppress basal defense responses, and one such associated macroscopic change is suppression of cell wall alterations. In accordance with our data, the PR protein ELI9 is also a Hyp-rich glycoprotein and is induced following inoculation with Pseudomonas (Almeras et al., 2003).
In common with the pattern emerging from the soluble fraction, another component of glycolysis, phosphoribulokinase (PRK; At1g32060), decreased in a PAMP-associated manner at 6 hpi. This protein was also targeted by T3Es as spot density returned to control levels after inoculation with DC3000.
PAMP-Responsive Proteins—Mitochondria
The mitochondrial fraction was only examined at 3 hpi, yet a relatively large number of PAMP-related changes were identified. Seven proteins increased in spot density: a different isoform of MDH (At3g15020) from that described in the soluble fraction, a putative arginase (At4g08900), pyruvate dehydrogenase (PDH; At5g50850), CA2 (At5g14740), mRNA splicing-like factor (At4g36690), Rubisco activase (At2g39730), and the Rieske Fe-S (At4g03280) protein. Two porins decreased in spot density: aquaporin (At3g26520) and the outer mitochondrial membrane porin (voltage-dependent anion-selective channel protein [VDAC 2]; At5g15090).
As in the other two subcellular fractions, glycolytic enzymes showed PAMP-related changes. MDH has a role in the tricarboxylic-acid pathway, glycolysis, gluconeogenesis, and lipid, fatty acid, and isoprenoid metabolism. This protein is targeted to mitochondria and appears to be the mature form of the protein. A spot of similar Mr to the mature form of the β-subunit of PDH is the first (E1) component of the PDH complex that converts pyruvate to acetyl-CoA in the mitochondrial matrix. PDH has a role in the metabolism of the pyruvate family (Ala, Ile, Leu, Val) and d-Ala. In the TCA cycle, MDH and PDH activity can be coupled to form citrate from malate.
Arginase (At4g08900), which accumulated following bacterial challenge, is targeted to mitochondria and hydrolyzes the first step of Arg degradation to produce l-Orn, leading to metabolism of Pro, His, and polyamines. Tomato (Lycopersicon esculentum) arginase is regulated in plants by wounding, jasmonate, and coronatine (Chen et al., 2004), but does not appear until 2 d after inoculation. Arginase may be involved in remobilization of nitrogen during defense responses. Alternatively, arginase may play an important role in regulating the metabolism of l-Arg to NO, thought to play a signaling role in both compatible and incompatible interactions.
A spot containing the mature form of the mitochondrial Rieske Fe-S protein, a component of the mitochondrial cytochrome bc1 complex (Emmermann et al., 1994), increased after all bacterial challenges. Remarkably, this spot was not present in the mock MgCl2 treatment, suggesting that this represents a rapid and highly specific mitochondrial PAMP response.
Aquaporin (also known as γ-tonoplast intrinsic protein [TIP] 1.2), a putative transmembrane protein targeted to mitochondria, decreased in spot density with all bacterial treatments. Aquaporin facilitates the transport of water across cell membranes, playing a critical role in short-term regulation of plant water status in response to various environmental perturbations (Maurel et al., 2002). Water status is considered to be important to successful pathogenesis (Wright and Beattie, 2004) and we interpret these dynamics as a host basal response to limit pathogen ingress. The outer mitochondrial membrane porin 2 (VDAC 2 or high sugar-responsive 2) also decreased in spot density with bacterial challenge. This protein can be phosphorylated and behaves as a general diffusion pore for small hydrophilic molecules, such as ATP and ADP, between the cytosol and mitochondria. It is a component of the permeability transition pore complex (Bernardi et al., 1994).
T3E-Responsive Proteins
Twenty-two proteins were associated with the establishment of pathogenicity (i.e. displaying significant difference between the hrpA mutant and DC3000). Eight of these proteins have already been discussed, highlighting how T3Es can impact upon basal defense responses (Fig. 2; Table III); the remainder are listed in Table IV (graphic representation in Supplemental Fig. S2). No proteins associated with T3E responses were identified from the mitochondria-enriched fraction.
T3E-Responsive Proteins—Soluble Fraction
The vast majority of T3E-responsive proteins were isolated from the soluble fraction (18 proteins), six of which were also identified as PAMP responsive, as described previously. Once again, components associated with glycolysis showed significant change: MDH (discussed in the PAMP section), PRK, and Rubisco activase. Five spots showed increases specific to T3Es: a squamosa promoter binding protein (SPB; At1g20980), a second cyclophilin (At3g01480), an elongation factor (At5g19510), PRK (At1g32060), and FNR (At1g20020). Three proteins decreased in relative spot density: Rib-5-P isomerase (At3g04790), a putative 3-β-hydroxysteroid dehydrogenase/isomerase (At2g37660), and a different spot of Rubisco activase (At2g19730).
The majority of proteins identified in this study were metabolic or defense related; it was therefore notable that the only signaling components identified were regulated by T3Es. A 14-3-3-like protein showed a greater than 2-fold increase in DC3000-treated leaves at 1.5 hpi. 14-3-3 proteins are well known in mammalian systems as phosphopeptide binding proteins and are thus implicated in signaling and enzyme regulation through phosphorylation. Unfortunately, Arabidopsis contains many isoforms of 14-3-3 proteins and the peptide hits obtained could not unambiguously identify a specific candidate protein.
The other signaling protein was a putative plant transcription factor (SPB-box family [At1g20980]), originally identified as being important in floral development (Klein et al., 1996). Because the spot Mr was much smaller than that expected, we hypothesize that it represents an SPB-box breakdown product. Early modification of host transcriptional complexes represents one possible mechanism for T3E-mediated suppression of basal plant defenses. Alternatively, removal of a positive regulator of basal defense would contribute toward compatibility.
The significant increase in abundance of two cyclophilins (At3g01480 and At3g62030) in DC3000 relative to hrpA inoculations at 1.5 and at 6 hpi, respectively, is of particular interest. Cyclophilins, members of the peptidyl propyl cis-trans isomerases family, have recently been associated with in planta processing of T3Es (Coaker et al., 2005).
Three proteins that showed T3E-specific increases were also targeted by RPM1/AvrRpm1: an inorganic pyrophosphatase (At5g09650), a translationally controlled tumor protein (TCTP) homolog (At3g16640), and the 14-3-3 protein (discussed above). These three proteins are clearly implicated in the establishment of disease and suggest that an AvrRpm1-modulated signaling pathway may restrict T3E-mediated accumulation of these proteins.
T3E-Responsive Proteins—Chloroplast
Four chloroplast proteins were classified as T3E responsive. A PSII oxygen-evolving complex (OEC) protein (At5g66570) was identified in three spots, one of which decreased significantly at 1.5 hpi in DC3000 when compared to hrpA and avrRpm1. This behavior parallels transcriptional modulation of the chloroplast photosynthetic machinery reported in both basal and gene-for-gene responses (Zou et al., 2005; Thilmony et al., 2006; Truman et al., 2006).
The PAMP-induced Hyp-rich glycoprotein (At4g01050) discussed above decreased in abundance, whereas an H+-ATPase synthase protein (At4g32260) increased. PRK (At1g32060) increased in both the soluble and later in the chloroplast fractions.
AvrRpm1-Responsive Proteins
AvrRpm1 challenge leading to the HR induced a dynamic response in the proteome, with 27 spots (26 unique proteins) identified as significantly changing across all fractions (Fig. 2; Table V; Supplemental Fig. S3). Of these, 15 decreased relative to one or more treatments. Changes in this category could be due to AvrRpm1 directly or indirectly modifying host proteins or interfering with functions of other effector proteins, or could be caused by processes downstream of activated RPM1.
AvrRpm1-Responsive Proteins—Soluble Fraction
Seventeen soluble proteins were classified as AvrRpm1/RPM1 responsive. Five have been discussed in previous sections because they modify either PAMP-responsive proteins or other T3E-induced changes. Seven defense-related enzymes were targeted by AvrRpm1/RPM1: MSD1 (PAMP section; Table III), FNR (At5g66190), and two GSTs (GSTF8 [At2g47730] and GSTF9 [At2g30860]) increased in spot density, whereas PrxB (At5g06290), NADPH quinone reductase (NQR; At3g27890), and MLP (At4g23670) decreased. A putative 3-β-hydroxy-δ5-steroid dehydrogenase also decreased in spot density.
Notably, several enzymes that modulate redox systems in the cell showed specific responses to AvrRpm1/RPM1. As reported previously (Jones et al., 2004), GSTF8 and PrxB responded to AvrRpm1. In this more detailed analysis, we also found changes in a single spot of GSTF9. GSTF9 increases in spot density at the same time (4 hpi) as GSTF8. MSD1 was represented by two different spots of similar Mr, but with a different pI. One showed significantly decreased abundance at 1.5 hpi and the other at 3 hpi. MDS1 is targeted to mitochondria (Kliebenstein et al., 1998), but has also been reported in peroxisomes (Alscher et al., 2002) and the soluble fraction (Swidzinski et al., 2004). Similarly, the oxidoreductase NQR showed a specific decrease in spot density early in the resistance reaction. A FNR (At5g66190) increased in spot density relative to DC3000 inoculation. Unlike the other ferredoxin reductase (At1g20020), which accumulated following DC3000 challenge, At5g66190 does not contain any known targeting motifs and the Mr of this spot was smaller than predicted; therefore, it may represent a breakdown product. Normally FNR is located in the thylakoid membrane; however, oxidative stress causes increased membrane solubilization (Palatnik et al., 1997), which would account for the increased FNR abundance in the soluble fraction observed following DC3000 (avrRpm1) challenge.
The behavior of two proteins implicates cellular ion homeostasis as an early target of AvrRpm1 function. The E-subunit of vacuolar H+-ATPase (V-ATPase) showed an early decrease in spot density (1.5 hpi). V-ATPases are responsible for generating energy for transport of ions and metabolites and also play an important role as stress response enzymes. Another stress-responsive protein, a second form of extrachloroplastic CA2, as described by Fett and Coleman (1994), also decreased in DC3000 (avrRpm1)-challenged leaves at 1.5 hpi compared to DC3000 and the hrpA mutant.
AvrRpm1-Responsive Proteins—Chloroplast
Our previous gene-profiling studies suggested that AvrRpm1/RPM1 recognition rapidly targets cellular organelles (de Torres et al., 2003; Truman et al., 2006). In total, six proteins were classified as AvrRpm1 responsive in the chloroplast fraction. Two OECs (At1g06680 and At5g66570), ATP synthase CF1 (ArthCp0029), and a 50S ribosomal protein (At3g27850) all increased, whereas PRK (At1g32060) decreased.
AvrRpm1-Responsive Proteins—Mitochondria
In contrast to the lack of response to DC3000 in the mitochondria-enriched fraction, AvrRpm1 delivery modified the mitochondrial proteome with four proteins changing significantly. An ATPase (AtCg00120) showed the only increase in spot density in this class, whereas cytochrome b5 (At5g48810), a 20S proteasome subunit (At1g79210) and a derivative of the TCTP homolog (At3g16640), decreased in spot density. TCTP was previously seen in the soluble T3E responses at 1.5 hpi.
The low frequency of T3E responses found in the mitochondrial fraction suggests that, compared with other effectors, either AvrRpm1 specifically targets mitochondria or the organelle is a major downstream target of activated RPM1 signaling. Of particular interest is the decrease in spot density of the 20S proteasome α-subunit B peptidase (PAB1; Fu et al., 1998). 26S proteasome activity has been linked with both resistance and susceptible interactions, where negative regulators of cell death or basal defense responses are targeted for degradation (Austin et al., 2002; Suty et al., 2003). The mass of PAB1 was larger than expected and no recognizable motifs target this protein to mitochondria.
DISCUSSION
The strength of this proteomic analysis was the ability to separate components of basal defense (by inclusion of the hrpA mutant) from disease and resistance responses, DC3000, and DC3000 (avrRpm1) inoculations. Moreover, the experimental design not only enables the subcellular targets of the defense responses to be determined, but, in some cases, where precursor proteins were identified, also provides insight into the dynamics of the response. Finally, we were able to correlate proteome changes with transcriptomic analysis of exactly the same system (Truman et al., 2006). Two significant conclusions regarding plant-pathogen responses have emerged: (1) the general infection response targets multiple organelles; and (2) all fractions show specific differences between hrpA mutant and D3000 challenge at 4 hpi before statistically significant transcriptional differences have been recorded (Truman et al., 2006). Indeed, even by 12 hpi, there was only limited correlation between gene activation and spot abundance for the corresponding protein (Supplemental Table S1). For example, in contrast to increases in spot density, transcripts for MDH (At1g53240), CA2 (At5g14740), and Rieske Fe-S (At4g03280) all showed significant decreases at 12 hpi following bacterial challenge. Several studies have also shown poor overall correlation between transcript and protein levels (Gygi et al., 1999; Scherl et al., 2005), although, for the more abundant proteins, mRNA levels may account for approximately 60% to 70% of the amount of the corresponding protein (Gygi et al., 1999; Jones et al., 2004; Swidzinski et al., 2004; Peck, 2005). This work demonstrates that widespread change to the proteome occurs before significant transcriptional reprogramming during the defense response.
We attribute proteomic changes observed here either to selective amplification of small (not statistically significant) alterations in mRNA levels or to rapid and specific PTMs induced by PAMP, T3E activities, or R-gene-mediated signaling. The finding that several proteins were identified in two or more spots supports the case for some PTM. However, we do not exclude the possibility that the observed spot shifts arose from alternative splicing or processing at the mRNA level.
Our results show that changes to primary metabolism and antioxidant enzymes involved in basal defense are altered by the introduction of T3Es. We identified proteins specific to the establishment of disease and showed that components of PSII, mitochondrial permeability transition (MPT), and cytoplasmic antioxidant enzymes were modified during R-gene-mediated HR. Below we discuss potential mechanisms that may integrate the complex changes observed in the three subcellular fractions.
Primary Carbon Metabolism
Ten of the identified proteins are associated with primary carbon metabolism (Table II). Nine of these proteins showed PAMP-related responses and, in the soluble fraction, most spots decreased in density. These changes generally occurred at later time points (4 and 6 hpi) and correlate with the significant and strong down-regulation of many transcripts of primary carbon metabolism occurring during basal defense at 12 hpi (Truman et al., 2006).
Strikingly, one-half of the PAMP-responsive spots were modified in response to T3Es. In all cases, the trend was to reverse PAMP-induced response, returning protein abundance to control levels. From these data, it is not possible to determine whether the metabolic proteins are directly modified by T3Es or, as may be more likely, the observed changes are a response to T3E-driven modifications of the redox status of the cytoplasm. Significant PAMP-associated changes to antioxidant enzymes (GSTs, Prx) in this system have been reported previously (Jones et al., 2004) and additional redox-related enzymes were shown to change in the experiment described here (MSD, FNR, FQR, as discussed below). The Calvin cycle is strongly regulated by light, mediated through redox status of the Fd-thioredoxin system, which targets many of the enzymes reported here (for review, see Schürmann, 2003) and these modifications may account for observed spot shifts. In particular, two spots of PRK changed in density in response to T3Es, and the redox status of PRK directly regulates the activity of G3PDH, with which it forms a complex (Lebreton et al., 2003; Graciet et al., 2004). Furthermore, G3PDH is implicated in lipid-mediated defense signaling. Levels of jasmonic acid and SA are regulated by the balance between glycerol-3-P and oleic acid (Kachroo et al., 2005). Rubisco activase also showed changes in response to T3Es and, through control of Rubsico, may also regulate the Calvin cycle (Zhang and Portis, 1999).
CA has a well-defined role in carbon fixation in C4 photosynthetic plants, but its role in C3 plants is less clear. It participates in mitochondrial complex 1, but occurs in two additional forms: cytosolic, where it is involved in CO2 transport, and in the chloroplast stroma, where it has been proposed to expedite CO2 diffusion, partner Rubisco in a Calvin cycle enzyme complex (Jebanathirajah and Coleman, 1998), and/or buffer pH (Savchenko et al., 2000). Here, we identified a rapid PAMP-related change (1.5 hpi) in CA, which was modulated by T3Es, whereas CA2 was modified in response to AvrRpm1. The chloroplast CA functions as a SA-binding protein 3 exhibiting antioxidant activity and may play a role in the HR because silencing of the tobacco (Nicotiana tabacum) CA gene in tobacco leaves suppressed the HR (Slaymaker et al., 2002). We propose that the suppression of CA by T3Es may represent a mechanism of attenuating antioxidant responses associated with basal defense.
Components Implicated in the Establishment of Disease
Nine proteins were directly or indirectly modified by the introduction of T3Es, but did not show PAMP-associated changes. The T3E-specific changes include several proteins of unclear function, cyclophilins, and the only signaling proteins identified in this study, a 14-3-3 protein and an SBP protein. 14-3-3 proteins are implicated in regulating primary carbon metabolism (Moorhead et al., 1999) and are central to the integration of signaling, including metabolism and cell fate (Pozuelo Rubio et al., 2004). Therefore, targeting of this protein by T3E may be consistent with the modulation of PAMP-associated changes to primary carbon metabolism. 14-3-3 proteins are known to bind and regulate nitrate reductase (MacKintosh and Meek, 2001) and thus may also interfere with the NO-ROS balance critical for defense (Delledonne et al., 2001). Notably, induction of the 14-3-3 spot in the susceptible interaction was abrogated in the presence of AvrRpm1, suggesting that this protein is clearly involved in disease establishment.
Two cyclophilins were also targeted by T3Es. Cyclophilins are generally associated with trafficking pathways, protein folding, and chaperone activity. The T3E-induced increase in AtCYP38 is significant in light of recent findings that cyclophilins appear to play an important role in modification of T3Es. AvrRpt2 is activated by a host cyclophilin facilitating its cleavage of RIN4 (Coaker et al., 2005). Cyclophilins are also virulence targets; the Agrobacterium VirD2 protein interacts with Arabidopsis cyclophilin ROC1 and CyPA (Deng et al., 1998). A mammalian cyclophilin has been implicated in opening of the MPT pore during Bax-mediated apoptosis (Halestrap et al., 2002; Li et al., 2004). One interpretation of these data is that T3Es may induce accumulation of host proteins that facilitate their processing and contribution to pathogenesis.
Proteins Responsive to AvrRpm1 Delivery and Recognition
The characteristic oxidative burst of the HR consists of the rapid generation of superoxide and the accumulation of peroxide (Lamb and Dixon, 1997). A concomitant peak of NO has been detected in cell cultures (Clarke et al., 2000) and the exact balance and nature of ROS and NO is critical to trigger the HR (Delledonne et al., 2001; Romero-Puertas et al., 2004). The RPM1-regulated HR is light dependent, making it likely that the chloroplast and photosynthetic machinery participate in ROS generation. Mitochondria are also a potential source of ROS and this organelle is known to play a determinative role in triggering PCD in mammalian systems (Yao et al., 2004). Finally, cytoplasmic antioxidant enzymes and the glutathione-ascorbate cycle regulate the redox status of the cell (Noctor and Foyer, 1998). Whereas these responses are by no means exclusive to, or dependent upon, the HR triggered by AvrRpm1/RPM1, in our model system we observed specific changes to all three systems: antioxidant enzymes in the soluble fraction, components of the mitochondrial permeability pore, and PSII.
Cytoplasmic Antioxidant Enzymes
The majority of the defense-related proteins identified in this screen and in Jones et al. (2004) showed modifications associated with the resistance reaction. We identified 13 soluble proteins, which can be classified as defense related; two FNRs, MSD1, NQR, MLP, three Prxs, and five GSTs. Nine increased in spot density during basal defense, whereas six were modified by AvrRpm1 delivery and two changed during the susceptible interaction. Most of the differences occurred by the earliest time point, 1.5 hpi, and thus represent rapid changes to these proteins without significant transcriptional differences. We suggest that, whereas bacterial challenge may generally induce antioxidant enzymes, modification of specific forms allows fine tuning of redox-associated signaling in response to the strain of DC3000 and orchestrates the outcome of the interaction, leading to disease or PCD.
Identification of several GSTs and Prx proteins in this study may be due to their relative abundance and amenability to analysis by 2DE. Our data suggest that bacterial challenge generally induces both families of enzymes in accordance with their widely accepted roles as stress-responsive proteins. However, specific GSTs and Prxs may act in concert with other antioxidant enzymes, such as MSD1 and FNR. The mitochondrial protein MSD1 showed a rapid increase in basal defense and equally rapid suppression in the resistance reaction. One spot of GSTF8 showed exactly the same response. Changes to both of these proteins were observed in the soluble fraction despite the predicted subcellular localization of MSD1 to mitochondria and GSTF8 to the chloroplast. Superoxide dismutase has been implicated in signaling cascades that utilize ROS (Delledonne et al., 2001; Desikan et al., 2002) with MSD controlling the amount of peroxide formed. Superoxide dismutase activity is essential for the oxidative burst in suspension cultures of Arabidopsis cells treated with the bacterial T3E, harpin (Desikan et al., 1996). Similarly, NQR catalyzes the reduction of quinone substrates to hydroquinones and, in mammalian systems, participates in the phase II detoxification pathway involved in cellular defense against oxidative stress (Merk et al., 1991). An NQR (P1-Zcr) induced by oxidative stress in Arabidopsis scavenges lipid peroxide-derived α-β-unsaturated aldehydes (Mano et al., 2002). Like MSD, NQR likely functions to protect plant cells from oxidative damage (Sparla et al., 1999) and therefore a reduction in these enzymes suggests that one function of AvrRpm1 is to attenuate the host's ability to neutralize ROS- or RNS-generated signals, including lipid peroxides, during the AvrRpm1-mediated HR.
Prxs have an important role in regulating the ROS-RNS balance (Dietz et al., 2006) and a floodgate theory for peroxide signaling has been proposed based upon the inactivation of these enzymes by overoxidation (Wood et al., 2003). The specific modulation of other redox-related enzymes as described above may further contribute to a floodgate mechanism. Many of these enzymes also have the ability to modify lipids and it is likely that there is a link between redox and sphingolipid signaling during plant PCD (Gechev and Hille, 2005). In common with GSTF2 and GSTF8 (Wagner et al., 2002), heterologously expressed 2-cys-Prx reduces linolenic acid hydroperoxide and also phosphatidylcholine dilinoleoyl hydroperoxide (Dietz, 2003).
Two GSTs (a second spot of GSTF8 and GSTF9) showed specific increases in the resistance reaction, as did FNR. FNR catalyzes the final step of electron transport in photosynthesis, reducing NADP(+) to NADPH, and is involved in cellular defense against oxidative stress in Escherichia coli (Krapp et al., 1997) and humans (Liu and Chen, 2002). FNR-suppressed tobacco plants are susceptible to photooxidative damage (Palatnik et al., 2003). We predict that alteration of this protein during the resistance response may be associated with modification of components of PSII and chloroplast systems as discussed below.
PSII
Two components of PSII were modified: two spots corresponding to OEC1 (At5g66570 PsbO1 or OEC33) and one spot of OEC23 (PsbP1, At1g06680). All three spots showed changes at the earliest time point and increased in density in tissue undergoing the HR rather than disease (DC3000). This result suggests that resistance mechanisms utilize or modify PSII. Suppression of transcripts encoding proteins localized to the chloroplast was reported from global expression profiling of the AvrRpm1-RPM1 interaction (Truman et al., 2006), but these changes occurred much later in the infection process. Similarly, Zou et al. (2005) reported R-gene-specific down-regulation of PSII transcription during HR in soybean (Glycine max) and suggested physical damage to the reaction centers resulted in decreased photosynthetic activity.
The photosystem centers are a potential source of ROS for the second, sustained oxidative burst during an incompatible interaction and their involvement could explain the light dependence of the HR. Other studies have shown that PSII core proteins are depleted by tobacco mosaic virus infections (Lehto et al., 2003) and protein levels of the 23 subunit decrease with viral infection (Rahoutei et al., 2000; Perez-Bueno et al., 2004). Importantly, OEC33 is essential for stability of the OEC complex (Yamamoto et al., 1998) and PSII cannot function without it. OEC33 was predicted to interact with the RNA helicase of Tobacco mosaic virus, suggesting that it could be targeted to suppress host plant basal defense (Abbink et al., 2002).
MPT
In mammals, a critical step in triggering PCD is MPT. Two pathways to mammalian MPT appear to be regulated through the electron carrier cytochrome bc1: the first involving ROS and movement of the Rieske factor; and the second mediated through Ca2+ ions (Armstrong et al., 2004). The three subunits of cytochrome bc1 directly involved with electron transport are cytochromes b and c and the Rieske Fe-S protein (Armstrong et al., 2004). We identified two components of the cytochrome bc1 complex showing significant changes; cytochrome b5 and the Rieske protein, and VDAC 2, a component of MPT (Bernardi et al., 1994). Two mitochondrial porins, aquaporin and VDAC 2, showed strong decreases in spot density in the mitochondrial fraction associated with basal defense. VDAC 2 increased in spot density in soluble extracts of cells undergoing PCD triggered by senescence or heat shock (Swidzinski et al., 2004). Combined with our data, this suggests that VDAC 2 is released from mitochondria into the cytoplasm. Aquaporin shows a similar change. One of the first responses to harpin in Arabidopsis suspension cells is a decrease in the mitochondrial membrane potential and rapid release of cytochrome c into the cytosol (Krause and Durner, 2004). The involvement of Rieske protein may indicate that defense-related MPT occurs through the unregulated (cyclosporin A-independent) pathway (Armstrong et al., 2004) activated by depletion of cytosolic glutathione, and triggering permeability transition through cytochrome bc1 mediated increased ROS production.
Our results suggest that release of outer membrane proteins is a basal response rather than a proapoptotic signal, possibly representing an initial priming event that requires further modifications to tip the cell into PCD. We have previously shown complex modification of GSTs in early basal defense (Jones et al., 2004). Such changes may lead to depletion of glutathione and subsequent initiation of MPT through the cytochrome bc1 complex, releasing aquaporin and VDAC from mitochondria as priming factors. Further modification, such as AvrRpm1/RPM1-specific changes to cytochrome b5, could then trigger PCD from this potentiated state.
Communication between Organelles
Recently, communication between chloroplasts and mitochondria has been shown to be involved in plant PCD and, unlike animal systems, the release of cytochrome c is not necessarily a trigger (Yao et al., 2004). Photorespiration can provide a route to regulate ROS production rapidly during the defense response. Photorespiration is initiated in the chloroplast through regulation of Rubisco by Rubisco activase producing glycolate-2-P, which is metabolized in mitochondria and peroxisomes. Production of glycolate-2-P links back to glycolysis and G3PDH (Kachroo et al., 2004). Several observations support a role for photorespiration in defense. For example, Taler et al. (2004) found that photorespiratory enzymes confer resistance to disease and excessive production of ROS in plants deficient in a photorespiratory enzyme (Ser hydroxymethyltransferase) can impair restriction of pathogen growth during the HR (Moreno et al., 2005). Photorespiration also has a significant impact upon glutathione levels. The ascorbate-glutathione cycle is central to cytoplasmic detoxification of ROS and involves both organelles (Dutilleul et al., 2003). Interestingly, several of the nuclear-encoded photorespiratory enzymes are dual targeted to both organelles (Chew et al., 2003). Both organelles are also involved in SA metabolism (chloroplasts [Wildermuth et al., 2001]; mitochondria [Chivasa and Carr, 1998]) and glutathione is also involved in triggering MPT.
Correlation with Transcription Profiling
Our recent global gene-profiling study revealed no significant differences in transcription between DC3000- and hrpA mutant-challenged leaves at 4 hpi. On the assumption that translational activity directly relates to transcript level, we interpret all significant changes in spots between PAMP and T3E treatments to be the result of PTM. Moreover, we also identified proteins where the presence of AvrRpm1 abrogated the T3E response, a result that was not observed in our expression-profiling data (Truman et al., 2006).
CONCLUSION
To the best of our knowledge, this is the first report to provide broad coverage of early changes to the defense proteome in response to inoculation with three strains of Pst DC3000. The immense dynamic range of proteins means that, without exhaustive fractionation, we could not dig very deeply into the proteome to observe changes in typically lower abundance proteins, such as protein kinases or transcription factors. Figure 3 provides a stylized overview of the major modifications reported here. Our data show that several chloroplast systems are modified during all aspects of the defense response. Components of the Calvin-Benson cycle are rapidly altered during basal defense and some of these changes are reversed by T3Es. PSII has emerged as a target of resistance signaling. Mitochondrial porins appear to be modified early in basal defense, with specific alterations to other components in response to AvrRpm1. Finally, the interplay between redox status and glycolysis with probable links to lipid signaling (through G3PDH, some GSTs, lipase, and NQR) may coordinate communication between organelles.
Figure 3.
A stylized schematic representing the major cellular processes discussed in the text. Systems that show alterations during basal defense are marked with yellow stars; for example, responses expected to be triggered by the flagellin-sensing receptor. Systems modified by the introduction of T3Es (blue hexagons) are marked with blue stars. Alterations associated with RPM1/AvrRpm1 recognition are represented by red stars. Possible routes for communication between organelles are indicated by broad arrows.
MATERIALS AND METHODS
General Procedures
All chemicals were purchased from Sigma, except where specified. Maintenance of bacteria, growth condition of plants, and treatments are detailed in de Torres et al. (2003). To minimize biological variation, plants were inoculated with each DC3000 derivative or the mock control (10 mm MgCl2) in an alternating block pattern so that every treatment contained the same number of plants from identical positions in a growth room tray. The use of an alternating inoculation pattern was designed to remove positional effects on the plants due to small variations in light intensity or humidity.
Soluble Fraction
Protein extraction, isoelectric focusing (IEF), SDS-PAGE, image analysis, and protein identification are detailed in Jones et al. (2004). Protein identifications described here were by LC-MS/MS as specified below.
Chloroplast-Enriched Fraction
Chloroplasts were isolated from fresh leaves immediately after harvesting. Leaves were homogenized in 10 mL of ice-cold grinding buffer (300 mm sorbitol, 25 mm Tricine/KOH, pH 7.8, 5 mm NaCl) in a chilled blender. The debris was filtered through two layers of miracloth and centrifuged at 4°C for 2 min at 2,500g. The supernatant (containing mitochondria) was discarded and the pellet gently resuspended in 3 mL of wash buffer (330 mm sorbitol, 40 mm HEPES/KOH, pH 7.6, 10 mm NaCl, 5 mm MgCl2, 1 mm KH2PO4). The suspension was centrifuged for 5 min at 2,500g. The resultant pellet was resuspended in 200 μL 10 mm Tris-Cl with protease inhibitors and snap frozen in liquid nitrogen. Chloroplasts were lysed by three freeze-thaw cycles in 10 mm Tris-Cl with protease inhibitors. Proteins were extracted in 100-μL aliquots using a 2D clean-up kit (Bio-Rad, 163–2,130) and solublized in 100 μL of SB310 buffer (5 m urea, 2 m thiourea, 2% CHAPS, 2% SB3-10, 0.5% Trition X 100) at room temperature for 1 h with intermittent vortexing. Extracts were centrifuged at 20,000g for 10 min at room temperature. Protein concentration was determined relative to bovine serum albumin using the Bradford assay (Bio-Rad). For IEF, 150 μg of protein was diluted to 350 μL with SB310 buffer and 2 mm tightly bound protein and 2% 3-10NL ampholytes (Amersham) were added. IEF strips (18 cm, no-linear, pI 3–10; Amersham) were rehydrated prior to IEF (program: 500 V for 1 h, 1,000 V for 1 h, 3,500 V for 40,000 V h, maximum 50 μA/strip). After focusing, the strips were either immediately equilibrated for SDS-PAGE or stored at −70°C. SDS-PAGE was performed as detailed in Jones et al. (2004). Protein spots were visualized with colloidal Coomassie. Gel images from each time point were analyzed together using PDQuest software (Bio-Rad). Spot detection parameters were the same as the soluble fraction. Spots were selected for identification based on significant difference between treatment pairs (t test, P < 0.05). The same gels were used for analysis and spot identification.
Mitochondria-Enriched Fraction
Mitochondria were extracted directly after harvesting inoculated material. Leaves were homogenized in a chilled blender with 10 mL of extraction buffer (450 mm Suc; 1.5 mm EGTA, pH 8.0; 0.2% [w/v] fatty acid-free bovine serum albumin; 0.6% [w/v] PVP40; 10 mm dithiothreitol; 15 mm MOPS KOH, pH 7.4; 1% [v/v] protease inhibitor cocktail). The debris was filtered through two layers of miracloth and centrifuged at 3,000g for 5 min at 4°C (to remove chloroplasts in the pellet). The supernatant was collected and centrifuged at 12,000g for 15 min at 4°C. The supernatant was discarded and the pellet resuspended in 6 mL of washing buffer (300 mm Suc; 1.0 mm EGTA, pH 8.0; 10 mm MOPS KOH, pH 7.4; 1% [v/v] protease inhibitor cocktail.). The differential centrifugation was repeated two more times to reduce contamination from other organelles and the pellet was finally resuspended in 200 μL 10 mm Tris-Cl, with protease inhibitors snap frozen in liquid nitrogen and stored at −70°C. Mitochondria were lysed in 10 mm Tris-Cl with three freeze-thaw cycles and proteins were precipitated with 3 volumes of 10% TCA in acetone at −20°C for 2 h. Proteins were pelleted by centrifugation at 20,000g for 10 min at 4°C and the pellet was washed with acetone. Proteins were solubilized in SB310 buffer and concentration was determined as detailed in the chloroplast section. Seventy-five micrograms of protein were used for each gel. Gels were silver stained using the method of Santoni et al. (1994), but omitting glutaraldehyde entirely and formaldehyde until the development step. This modification had minimal effects of the quality of staining, but rendered the protein spots more compatible for MS/MS. Image analysis was as detailed for the chloroplast fraction. The same gels were used for analysis and identification; faint spots were pooled from several gels for identification.
Protein Identification
Most of the proteins in the soluble fraction were identified as described previously (Jones et al., 2004). Protein spots from the chloroplast and mitochondrial fractions were excised manually, destained, reduced, and carboxyamidomethylated. Gel pieces were washed with ammonium bicarbonate and treated with trypsin (Promega) overnight at 37°C. Peptides were collected and gel pieces washed with 50% acetonitrile. Washes were pooled and the volume reduced. Acidified (formic acid) peptides were analyzed by LC-MS/MS. Proteins from the chloroplast fraction were identified using capillary LC; a Zorbax C18 0.5 × 35 mm (Agilent) trap column, a Zorbax 300SB C18 0.3 × 150 mm (Agilent) analytical column. Proteins from the mitochondrial fraction, and low abundance spots from the other fractions, were identified using nanoflow LC; Zorbax 300SB C18 0.3 × 5 mm and 5-μm trap columns (Agilent) and 75-μm i.d. 15-cm packed with 3 μm C18 100a analytical column (Hichrome Ltd). The mass spectrometer in both cases was a QTrap (Applied Biosystems/Sciex). Fragmentation data were used to search the Matrix Science database using MASCOT (http://www.matrixscience.com/home.html). Probability-based scores were used to evaluate identifications and at least two unique peptide hits to the same protein were required.
Bioinfomatics
Chloroplast localization predictions used the ChloroP 1.1 server (Wilkins et al., 1998). Predicted transit peptide lengths were removed and used to calculate the mature protein characteristics. Domain searches from Citing CD-Search (Marchler-Bauer et al., 2003) and MitoProt II 1.0a4 (Claros and Vincens, 1996) were used to predict mitochondrially imported proteins, associated transit peptides, and cleavage sites. The Mr and pI were then recalculated from Expasy.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphic representations of data presented in Table III. Protein spots responding to PAMPs.
Supplemental Figure S2. Graphic representations of data presented in Table IV. Proteins responding to T3Es.
Supplemental Figure S3. Graphic representations of data presented in Table IV. Proteins responding to RPM1/AvrRpm1.
Supplemental Table S1. Direct comparison of the fold changes observed in transcriptiomic and proteomic datasets.
Acknowledgments
We would like to thank Wendy Byrne for dedicated assistance with plant growth and William Truman for assistance with transcriptomic data. We gratefully acknowledge Dr. Paul Dupree and Dr. Kathryn Lilley from the Cambridge Centre for Proteomics for access to Biotechnology and Biological Science Research Center GARNet supported mass spectrometry facilities for some of the soluble fraction spot identifications.
This work was supported by the Biotechnology and Biological Science Research Council (grant no. P14635).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Murray Grant (m.grant@imperial.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abbink TE, Peart JR, Mos TN, Baulcombe DC, Bol JF, Linthorst HJ (2002) Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 295: 307–319 [DOI] [PubMed] [Google Scholar]
- Almeras E, Stolz S, Vollenweider S, Reymond P, Mene-Saffrane L, Farmer EE (2003) Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J 34: 205–216 [DOI] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 [PubMed] [Google Scholar]
- Armstrong JS, Yang H, Duan W, Whiteman M (2004) Cytochrome bc1 regulates the mitochondrial permeability transition by two distinct pathways. J Biol Chem 279: 50420–50428 [DOI] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Bennett M, Mehta M, Grant M (2005) Biophoton imaging: a nondestructive method for assaying R gene responses. Mol Plant Microbe Interact 18: 95–102 [DOI] [PubMed] [Google Scholar]
- Bernardi P, Broekemeier KM, Pfeiffer DR (1994) Recent progress on regulation of the mitochondrial permeability transition pore; a cyclosporin-sensitive pore in the inner mitochondrial membrane. J Bioenerg Biomembr 26: 509–517 [DOI] [PubMed] [Google Scholar]
- Bestwick CS, Bennett MH, Mansfield JW (1995) Hrp mutant of Pseudomonas syringae pv phaseolicola induces cell wall alterations but not membrane damage leading to the hypersensitive reaction in lettuce. Plant Physiol 108: 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279: 45998–46007 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Carr JP (1998) Cyanide restores N gene-mediated resistance to tobacco mosaic virus in transgenic tobacco expressing salicylic acid hydroxylase. Plant Cell 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B (2005) Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 22: 548–550 [DOI] [PubMed] [Google Scholar]
- Collmer A, Badel JL, Charkowski AO, Deng WL, Fouts DE, Ramos AR, Rehm AH, Anderson DM, Schneewind O, van Dijk K, et al (2000) Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc Natl Acad Sci USA 97: 8770–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection: the transition from basal to induced defense responses in RPM1-mediated resistance. Plant J 33: 665–676 [DOI] [PubMed] [Google Scholar]
- Delledonne M (2005) NO news is good news for plants. Curr Opin Plant Biol 8: 390–396 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Chen L, Wood DW, Metcalfe T, Liang X, Gordon MP, Comai L, Nester EW (1998) Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc Natl Acad Sci USA 95: 7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Coffey MJ, Neill SJ (1996) Generation of active oxygen in elicited cells of Arabidopsis thaliana is mediated by a NADPH oxidase-like enzyme. FEBS Lett 382: 213–217 [DOI] [PubMed] [Google Scholar]
- Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54: 93–107 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, de Paepe R (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmermann M, Clericus M, Braun HP, Mozo T, Heins L, Kruft V, Schmitz UK (1994) Molecular features, processing and import of the Rieske iron-sulfur protein from potato mitochondria. Plant Mol Biol 25: 271–281 [DOI] [PubMed] [Google Scholar]
- Fett JP, Coleman JR (1994) Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol 105: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD (1998) Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149: 677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T, Buchala AJ, Chua NH, Metraux JP (2002) Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J 31: 87–95 [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Gontero B (2004) Emergence of new regulatory mechanisms in the Benson-Calvin pathway via protein-protein interactions: a glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. J Exp Bot 55: 1245–1254 [DOI] [PubMed] [Google Scholar]
- Grant JJ, Yun BW, Loake GJ (2000) Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J 24: 569–582 [DOI] [PubMed] [Google Scholar]
- Grant M, Mansfield J (1999) Early events in host-pathogen interactions. Curr Opin Plant Biol 2: 312–319 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis Rpm1 gene enabling dual-specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP, McStay GP, Clarke SJ (2002) The permeability transition pore complex: another view. Biochimie 84: 153–166 [DOI] [PubMed] [Google Scholar]
- Ivanov B, Khorobrykh S (2003) Participation of photosynthetic electron transport in production and scavenging of reactive oxygen species. Antioxid Redox Signal 5: 43–53 [DOI] [PubMed] [Google Scholar]
- Jebanathirajah JA, Coleman JR (1998) Association of carbonic anhydrase with a Calvin cycle enzyme complex in Nicotiana tabacum. Planta 204: 177–182 [DOI] [PubMed] [Google Scholar]
- Jones AM, Thomas V, Truman B, Lilley K, Mansfield J, Grant M (2004) Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochemistry 65: 1805–1816 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Venugopal SC, Lapchyk L, Falcone D, Hildebrand D, Kachroo P (2004) Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc Natl Acad Sci USA 101: 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Venugopal SC, Navarre DA, Lapchyk L, Kachroo A (2005) Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol 139: 1717–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Gabrys H, Mateo A, Karpinska B, Mullineaux PM (2003) Light perception in plant disease defense signalling. Curr Opin Plant Biol 6: 390–396 [DOI] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp AR, Tognetti VB, Carrillo N, Acevedo A (1997) The role of ferredoxin-NADP+ reductase in the concerted cell defense against oxidative damage—studies using Escherichia coli mutants and cloned plant genes. Eur J Biochem 249: 556–563 [DOI] [PubMed] [Google Scholar]
- Krause M, Durner J (2004) Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol Plant Microbe Interact 17: 131–139 [DOI] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lebreton S, Graciet E, Gontero B (2003) Modulation, via protein-protein interactions, of glyceraldehyde-3-phosphate dehydrogenase activity through redox phosphoribulokinase regulation. J Biol Chem 278: 12078–12084 [DOI] [PubMed] [Google Scholar]
- Lehto K, Tikkanen M, Hiriart JB, Paakkarinen V, Aro EM (2003) Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Mol Plant Microbe Interact 16: 1135–1144 [DOI] [PubMed] [Google Scholar]
- Li Y, Johnson N, Capano M, Edwards M, Crompton M (2004) Cyclophilin-D promotes the mitochondrial permeability transition but has opposite effects on apoptosis and necrosis. Biochem J 383: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X (2002) The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 21: 7195–7204 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Meek SE (2001) Regulation of plant NR activity by reversible phosphorylation, 14-3-3 proteins and proteolysis. Cell Mol Life Sci 58: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano J, Torii Y, Hayashi S, Takimoto K, Matsui K, Nakamura K, Inze D, Babiychuk E, Kushnir S, Asada K (2002) The NADPH:quinone oxidoreductase P1-zeta-crystallin in Arabidopsis catalyzes the alpha,beta-hydrogenation of 2-alkenals: detoxification of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol 43: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31: 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Javot H, Lauvergeat V, Gerbeau P, Tournaire C, Santoni V, Heyes J (2002) Molecular physiology of aquaporins in plants. Int Rev Cytol 215: 105–148 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk H, Jugert F, Bonnekoh B, Mahrle G (1991) Induction and inhibition of NAD(P)H: quinone reductase in murine and human skin. Skin Pharmacol 4: 183–190 [DOI] [PubMed] [Google Scholar]
- Midoh N, Iwata M (1996) Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol 37: 9–18 [DOI] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, Deiting U, Stitt M, Scarabel M, Aitken A, et al (1999) Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Moreno JI, Martin R, Castresana C (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J 41: 451–463 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Tognetti VB, Poli HO, Rodriguez RE, Blanco N, Gattuso M, Hajirezaei MR, Sonnewald U, Valle EM, Carrillo N (2003) Transgenic tobacco plants expressing antisense ferredoxin-NADP(H) reductase transcripts display increased susceptibility to photo-oxidative damage. Plant J 35: 332–341 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Valle EM, Carrillo N (1997) Oxidative stress causes ferredoxin-NADP+ reductase solubilization from the thylakoid membranes in methyl viologen-treated plants. Plant Physiol 115: 1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC (2003) Early phosphorylation events in biotic stress. Curr Opin Plant Biol 6: 334–338 [DOI] [PubMed] [Google Scholar]
- Peck SC (2005) Update on proteomics in Arabidopsis: where do we go from here? Plant Physiol 138: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC, Nuhse TS, Hess D, Iglesias A, Meins F, Boller T (2001) Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13: 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Bueno ML, Rahoutei J, Sajnani C, Garcia-Luque I, Baron M (2004) Proteomic analysis of the oxygen-evolving complex of photosystem II under biotic stress: studies on Nicotiana benthamiana infected with tobamoviruses. Proteomics 4: 418–425 [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, MacKintosh C (2004) 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation, and trafficking. Biochem J 379: 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahoutei J, Garcia-Luque I, Baron M (2000) Inhibition of photosynthesis by viral infection: effect on PSII structure and function. Physiol Plant 110: 286–292 [Google Scholar]
- Robson CA, Vanlerberghe GC (2002) Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol 129: 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M (2004) Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol 6: 795–803 [DOI] [PubMed] [Google Scholar]
- Santoni V, Caboche M (1994) Use of 2-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192: 557–566 [Google Scholar]
- Savchenko G, Wiese C, Neimanis S, Hedrich R, Heber U (2000) pH regulation in apoplastic and cytoplasmic cell compartments of leaves. Planta 211: 246–255 [DOI] [PubMed] [Google Scholar]
- Scherl A, Francois P, Bento M, Deshusses JM, Charbonnier Y, Converset V, Huyghe A, Walter N, Hoogland C, Appel RD, et al (2005) Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J Microbiol Methods 60: 247–257 [DOI] [PubMed] [Google Scholar]
- Schürmann P (2003) Redox signaling in the chloroplast: the ferredoxin/thioredoxin system. Antioxid Redox Signal 5: 69–78 [DOI] [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99: 11640–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Tedeschi G, Pupillo P, Trost P (1999) Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett 463: 382–386 [DOI] [PubMed] [Google Scholar]
- Suty L, Lequeu J, Lancon A, Etienne P, Petitot AS, Blein JP (2003) Preferential induction of 20S proteasome subunits during elicitation of plant defense reactions: towards the characterization of plant defense proteasomes. Int J Biochem Cell Biol 35: 637–650 [DOI] [PubMed] [Google Scholar]
- Swidzinski JA, Leaver CJ, Sweetlove LJ (2004) A proteomic analysis of plant programmed cell death. Phytochemistry 65: 1829–1838 [DOI] [PubMed] [Google Scholar]
- Taler D, Galperin M, Benjamin I, Cohen Y, Kenigsbuch D (2004) Plant ER genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16: 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Thwaites R, Spanu PD, Panopoulos NJ, Stevens C, Mansfield JW (2004) Transcriptional regulation of components of the type III secretion system and effectors in Pseudomonas syringae pv phaseolicola. Mol Plant Microbe Interact 17: 1250–1258 [DOI] [PubMed] [Google Scholar]
- Truman W, de Torres Zabala M, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defense responses during pathogenesis and resistance. Plant J 46: 14–33 [DOI] [PubMed] [Google Scholar]
- Tsunezuka H, Fujiwara M, Kawasaki T, Shimamoto K (2005) Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol Plant Microbe Interact 18: 52–59 [DOI] [PubMed] [Google Scholar]
- van Dijk K, Fouts DE, Rehm AH, Hill AR, Collmer A, Alfano JR (1999) The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol 181: 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O (2006) The wound response in tomato—role of jasmonic acid. J Plant Physiol 163: 297–306 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez J-C, Williams KL, Appel RD, Hochstrasser DF (1998) Protein identification and analysis tools in the ExPASy server. In AJ Link, ed, 2-D Proteome Analysis Protocols. Humana Press, Totowa, NJ [DOI] [PubMed]
- Wood ZA, Poole LB, Karplus PA (2003) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653 [DOI] [PubMed] [Google Scholar]
- Wright CA, Beattie GA (2004) Pseudomonas syringae pv tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Ishikawa Y, Nakatani E, Yamada M, Zhang H, Wydrzynski T (1998) Role of an extrinsic 33 kilodalton protein of photosystem II in the turnover of the reaction center-binding protein D1 during photoinhibition. Biochemistry 37: 1565–1574 [DOI] [PubMed] [Google Scholar]
- Yao N, Eisfelder BJ, Marvin J, Greenberg JT (2004) The mitochondrion—an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 40: 596–610 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defense responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zhang N, Portis AR Jr (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA 96: 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18: 1161–1174 [DOI] [PubMed] [Google Scholar]