Abstract
When Manduca sexta attacks Nicotiana attenuata, fatty acid-amino acid conjugates (FACs) in the larvae's oral secretions (OS) are introduced into feeding wounds. These FACs trigger a transcriptional response that is similar to the response induced by insect damage. Using two-dimensional gel electrophoresis, matrix-assisted laser desorption ionization-time of flight, and liquid chromatography-tandem mass spectrometry, we characterized the proteins in phenolic extracts and in a nuclear fraction of leaves elicited by larval attack, and/or in leaves wounded and treated with OS, FAC-free OS, and synthetic FACs. Phenolic extracts yielded approximately 600 protein spots, many of which were altered by elicitation, whereas nuclear protein fractions yielded approximately 100 spots, most of which were unchanged by elicitation. Reproducible elicitor-induced changes in 90 spots were characterized. In general, proteins that increased were involved in primary metabolism, defense, and transcriptional and translational regulation; those that decreased were involved in photosynthesis. Like the transcriptional defense responses, proteomic changes were strongly elicited by the FACs in OS. A semiquantitative reverse transcription-PCR approach based on peptide sequences was used to compare transcript and protein accumulation patterns for 17 candidate proteins. In six cases the patterns of elicited transcript accumulation were consistent with those of elicited protein accumulation. Functional analysis of one of the identified proteins involved in photosynthesis, RuBPCase activase, was accomplished by virus-induced gene silencing. Plants with decreased levels of RuBPCase activase protein had reduced photosynthetic rates and RuBPCase activity, and less biomass, responses consistent with those of herbivore-attacked plants. We conclude that the response of the plant's proteome to herbivore elicitation is complex, and integrated transcriptome-proteome-metabolome analysis is required to fully understand this ubiquitous ecological interaction.
The majority of studies examining the induced defense responses of plants after insect attack have focused on the dynamics of the specific genes, proteins, and metabolites that are thought to be responsible for changes in resistance elicited by attack. More recently, large-scale transcriptional analyses with microarrays have broadened the scope of the analysis and revealed coordinated changes in hundreds of transcripts, suggesting that large-scale shifts in metabolism accompany the activation of defense responses (Reymond et al., 2000, 2004; Halitschke et al., 2001, 2003; Hermsmeier et al., 2001; Schittko et al., 2001; Kessler and Baldwin, 2002; Ralph et al., 2006a, 2006b). How the changes in the transcriptome translate into changes in the proteome and metabolome that eventually account for the phenotype of increased resistance is unknown, largely because the proteomic changes elicited by herbivore attack remain unstudied. It is becoming increasingly clear that proteomic changes cannot be directly predicted from changes in the transcriptome. Furthermore, in a given cellular microenvironment, both proteins and transcripts interact with other molecules in specific ways, and these interactions determine the regulation, expression, activity, and stability of specific mRNA and protein molecules. Parallel large-scale transcription and protein analyses promise to illuminate the function and regulation of genes and proteins (Bertone and Snyder, 2005; Lange and Ghassemian, 2005; Peck, 2005). Recent developments in mass spectrometric peptide sequencing now allow for protein identification on the proteomic scale (Aebersold and Mann, 2003; Glinski and Weckwerth, 2006). Information obtained with such tools will provide valuable insights into the mechanisms attacked plants use to shift their metabolic priorities from growth to defense (Walling, 2000; Hahlbrock et al., 2003).
Nicotiana attenuata Torr. Ex Wats. (Solanaceae), a post-fire annual inhabiting the Great Basin Desert, has a number of well-described herbivore-induced direct and indirect defenses (Baldwin, 2001). Direct defenses include nicotine and several other metabolites, and defense proteins such as trypsin proteinase inhibitors (TPIs) and Thr deaminase (TD; Hermsmeier et al., 2001; Schittko et al., 2001; Kang et al., 2006). Indirect defenses comprise the terpenoid and green leaf volatile alarm signals that provide information about the activity and location of feeding herbivores to the community of predators (Kessler and Baldwin, 2002). Several such combinations of direct and indirect defenses appear to be especially effective in reducing pest populations in an evolutionarily stable manner (Baldwin, 2001). The detailed information gained from such biological systems may lead to the development of valuable tools for managing insect infestation of crops more wisely (Anderson et al., 2005).
High-throughput transcriptional analysis of the interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host N. attenuata identified a large-scale transcriptional reconfiguration, which entailed decreases in photosynthetic-related processes and increases in defense-related processes (Walling, 2000; Halitschke et al., 2001, 2003; Hui et al., 2003). The standardized and synchronized treatment of elicited responses, namely, the application of M. sexta oral secretions (OS) to leaves punctured by a pattern wheel, a treatment that was found to mimic the transcriptional and metabolic responses of M. sexta larvae attack (Schittko et al., 2001; Halitschke et al., 2003; Hui et al., 2003; Roda et al., 2004), is crucial for understanding plant-herbivore interactions. The major constituents in OS from M. sexta and several other lepidopteran insects responsible for the differential activation of N. attenuata genes have been identified as fatty acid-amino acid conjugates (FACs). More than 70% of the OS-elicited transcriptional changes in N. attenuata's wound response as determined by microarray analysis can be mimicked by introducing chemically synthesized FACs into puncture wounds (Schittko et al., 2001; Halitschke et al., 2003; Hui et al., 2003; Roda et al., 2004).
As a follow-up to transcriptional analysis, we undertook a comparative proteome analysis of the M. sexta-N. attenuata interaction. Proteomic analysis was carried out by comparing the patterns of leaf proteins in the leaves of undamaged plants to those in elicited and attacked plants by two-dimensional gel electrophoresis (2-DE). We performed two types of proteomic analysis, addressing two main questions. First, how does a plant respond to the different elicitors found in M. sexta OS? To answer this question, we compared the patterns of protein accumulation observed when punctured wounds were treated with OS to those observed when the punctured wounds were treated with water, FACs, OS that had their FACs removed by ion-exchange chromatography (OS-FAC-free), and feeding M. sexta larvae. Second, how do these responses change over the time when leaves are known to increase their resistance to insect attack? To answer this question, we measured the accumulation of identified proteins at 6, 12, 30, 48, and 72 h after OS treatment of puncture wounds. We used a reverse transcription (RT)-PCR approach to determine the association between candidate proteins showing differential accumulation patterns and the abundance of their encoding mRNAs. In addition, the functional analysis of one of the proteins identified as being involved in photosynthesis, RuBPCase activase (RCA), was accomplished by gene silencing. This study identifies several well-characterized proteins whose direct and indirect roles in insect-elicited responses were not previously known, as well as several proteins of unknown function.
RESULTS
Comparison of 2-DE Protein Profiles of Control and Elicited Leaves of N. attenuata
Source leaves +1, +2, and +3 of rosette-stage plants of N. attenuata were punctured parallel to the midvein with a fabric pattern wheel six times at 30-min intervals (Fig. 1). To induce the plants with different elicitors, various solutions were applied to the punctured leaves (W + OS, W + FAC, and W + OS-FAC-free) or larvae were released on these leaves (Fig. 1). The leaf proteins were extracted using different methods and analyzed.
Figure 1.
Leaf numbering and elicitation procedures. The left section depicts the numbering system of leaf nodes of 30-d-old rosette-stage plants, and the upper-right section the leaf-wounding procedure with the pattern wheel, according to which three leaves (+1, +2, and +3) were wounded and elicitor solutions applied six times at 30-min intervals. The elicitor solutions were water, 0.005% Triton X-100, M. sexta OS, OS without FACs (OS-FAC-free), and chemically synthesized FACs, and, in the lower-right section, a feeding first-instar M. sexta larva.
Phenolic extracts of N. attenuata leaf proteins yielded approximately 600 protein spots on a 2-DE. Protein spots exhibiting variations in extracts from control and elicited leaves were identified and compared among three to five biological replicates. One such biological replicate represents two to four plants and three elicited or adjacent unelicited (systemic) leaves on each plant. Representative 2-DE gel images of leaf proteins from the 30 h harvest of control and W + OS-elicited leaves are presented in Figure 2. Each protein preparation was analyzed on at least three parallel 2-DE gels. In total, we generated and analyzed 72 gels for phenol extracts (three biological replicates, six treatments, and five time points) and 14 gels for nuclear extracts (three biological replicates and two time points). Analysis of the nuclear protein fractions revealed approximately 100 protein spots with no significant variation between control and OS-elicited leaf extracts (Supplemental Fig. S1). Although the total leaf protein profiles (phenolic extracts) responded to elicitation, the nuclear protein fraction did not, so we focused our efforts on characterizing the elicited changes in the total protein extracts. The similar protein patterns observed in control leaves from five harvests that were removed 6 to 72 h after elicitation indicated that the developmental or leaf maturation-related changes that occurred during this time did not significantly influence the part of the proteome that was detectable in our analysis (Fig. 2; Supplemental Figs. S2 and S3). In contrast, we identified 90 spots that exhibited differential accumulation patterns from 6 to 72 h after OS elicitation when wounded leaves were compared with the respective control leaves. These proteins were successfully identified using peptide mass fingerprints and/or peptide sequencing (Table I). Among these 90 proteins, 35 showed consistent accumulation trends in induced leaves at 30 h after OS elicitation (Fig. 2; Table II). Changes in these proteins were meticulously compared in four induction treatments (Table II). Fifty-one other proteins identified in this study, whose accumulation patterns consistently increased or decreased at specific harvest times in at least two replicates, were identified. The mass spectrometry (MS) data of four proteins did not yield information for the database search or de novo sequencing.
Figure 2.
Proteomic maps of two-dimensionally separated control and elicited leaf proteins. Locally treated leaves were collected 30 h after the first OS elicitation and proteins were extracted using the phenol extraction protocol. Approximately 550 μg of protein was resolved on 24-cm IPG strips of pH range 3 to 10 NL (isoelectric focusing), further separated on 12% acrylamide SDS-PAGE gels, and stained with Bio-Safe Coomassie G-250 stain. Gels were scanned and images analyzed using PDQuest software. Representative protein profiles (from nine gels) of control (A) and W + OS-elicited (B) leaves from different plants were used to compare protein accumulation patterns. Numbers and arrows indicate protein spots experiencing up-regulation (red arrows), down-regulation (black arrows), or no change (green arrows) in response to OS elicitation at this time point. For identities of the protein spots and their accumulation patterns in response to several elicitation treatments from 6 to 72 h time points, see Tables I and II.
Table I.
Protein spots identified with MALDI-TOF and/or LC-MS/MS
Mass spectrometric identification of protein spots with differential accumulation after OS elicitation using comparison of the measured sequences or peptide mass fingerprints with the NCBI green plant protein database. Type of response, as indicated by Roman numerals, is described in the text. No. Pep., Number of peptides.
| Spot No. | Accession No. | Name of Protein | Protein Identification Data
|
|||||
|---|---|---|---|---|---|---|---|---|
| MALDI (Score/No. Pep.) | LC-MS/MS(Score/No. Pep.) | Theoretical kD/pI | Observed kD/pI | Peptide Sequence (MS/MS) | Response | |||
| 1 | XP_472335 | OSJNBa0039C07.4 protein | 9.7/18 | 12.71/1 | 98.4/5.98 | 87.2/5.79 | (K)TAIAEGLAQR(I) | II |
| 2 | S58083 | TK | 9.79/12 | 12.07/6 | 75/6 | 72.3/7.05 | (R)FLAIDAVEK(A) | III |
| (K)NGNTGYDEIR(A) | ||||||||
| (K)VTTTIGFGSPNK(A) | ||||||||
| (K)ANSYSVHGSGLGAK(E) | ||||||||
| (K)ALPTYTPESPADATR(N) | ||||||||
| (R)KTPSILALSR(Q) | ||||||||
| 4 | BAA77604 | Plastidic ALD | 5.69/4 | 12.77/3 | 42.5/7.48 | 35.4/7.23 | (K)YTGEGESEEAK(K) | No change |
| (R)SAAYYQQGAR(F) | ||||||||
| (K)YTGEGESEEAKK(G) | ||||||||
| 5 | AAO23981 | Vacuolar H+-ATPase A2 subunit | 8.71/11 | 10.28/2 | 68.6/5.37 | 68.7/5.54 | (R)NLEDETR(-) | I |
| (R)SGDVYIPR(G) | ||||||||
| 6 | Q40565 | RCA | 10.13/12 | 11.62/5 | 48.3/8.17 | 40.3/5.02 | (R)TDNVPEEAVIK(I) | II |
| (R)VYDDEVRK(W) | ||||||||
| (K)WVSGTGIEAIGDK(L) | ||||||||
| (K)LLNSFDGPPTFEQPK(M) | ||||||||
| (R)VQLAETYLK(E) | ||||||||
| 7 | AAG59585 | TD | 12.43/10 | 12.72/6 | 65.6/6.21 | 64.5/5.94 | (R)VYDVAIDSPLQNAAK(L) | I |
| (R)EDMQSVFSFK(L) | ||||||||
| (K)DVYDEGR(N) | ||||||||
| (K)QALVLYSVGVNTK(S) | ||||||||
| (K)SSQLNTVNLTNNNLVK(E) | ||||||||
| (K)FLDAFSPR(W) | ||||||||
| 8 | AAM62737 | QR-like protein | 12.77/1 | 32.7/6.08 | 63/5.56 | (K)VVAAGLNPVDAKR(R) | III | |
| AAB39828 | Chaperonin | 11.51/1 | 18.10/10.2 | (K)IVNDGVTVAR(E) | ||||
| 9 | Q40565 | Rubisco activase | 11.62/11 | 48.3/8.17 | 39.1/5.25 | (K)GLVQDFSDDQQDIAR(G) | II | |
| (R)QYNMDNTLDGFYIAPSFMDK(L) | ||||||||
| (K)VPLILGMWGGK(G) | ||||||||
| (R)KMGINPIMMSAGELESGNAGEPAK(L) | ||||||||
| (K)MGINPIMMSAGELESGNAGEPAK(L) | ||||||||
| (R)VYDDEVR(K) | ||||||||
| (R)KWMSGTGIEAIGDK(L) | ||||||||
| (K)LLNSFDGPPTFEQPK(M) | ||||||||
| (K)LLEYGNMLVQEQENVKR(V) | ||||||||
| (K)RVQLAETYLK(E) | ||||||||
| (K)EAALGDANADAINTGNF(-) | ||||||||
| 12 | 2002393A | Oxygen-evolving complex protein | 5.17/4 | 11.46/3 | 26.48/49 | 29.8/4.83 | (R)RLTFDEIQSK(T) | II |
| (K)FCLEPTSFTVK(A) | ||||||||
| (R)GGSTGYDNAVALPAGGR(G) | ||||||||
| 13 | AAL71857 | DHAR | 4.07/4 | 23.6/8.2 | 22.8/8.3 | I | ||
| 14 | BAB64833 | PGM | 6.02/7 | 10.4/2 | 61/6.3 | 63.7/6.17 | (K)LVDLALASGK(I) | III |
| (R)LDQVQLLLK(G) | ||||||||
| 16 | AAP04394 | Glutathione S-transferase | 12.69/1 | 1/4.07 | 23.6/8.37 | (-)LLDVYETR(-) | I | |
| 17 | Q42961 | Phosphoglycerate kinase precursor | 5.2/12 | 50.1/8.75 | 41.6/6.99 | V | ||
| 18 | NP_199203 | Myosin heavy-chain MYA2 | 1.9/4 | 169.8/7.9 | 20.5/8.08 | I | ||
| 19 | BAA77604 | Plastidic ALD | 5.71/5 | 42.5/7.4 | 35.9/6.37 | V | ||
| Unspecified | 12.36/1 | 1.4/6.93 | (-)GAVQLWKYLEDR(-) | |||||
| 20 | Q40565 | Rubisco activase | 8.95/16 | 14.54/7 | 48.3/8.4 | 37.8/7.19 | (K)LVVHITK(N) | I |
| (K)SFQCELVFR(K) | ||||||||
| (R)EAAEIIR(K) | ||||||||
| (R)EAAEIIRK(G) | ||||||||
| (R)TDNVPEEAVIK(I) | ||||||||
| (R)VYDDEVRK(W) | ||||||||
| (R)VQLAETYLK(E) | ||||||||
| 21 | AAT38067 | Putative glycoprotease | 1.2/4 | 40.8/6.19 | 45.1/<4.55 | I | ||
| Unspecified | 12.72/1 | 0.9/11.7 | (-)VVKKKGFR(-) | |||||
| 22 | AAS90324 | 3-Dehydroquinate dehydratase | 5.31/4 | 56.1/6.08 | 32.4/7.93 | I | ||
| Unspecified | 12.72/1 | 1.1/9.71 | (-)NAGANAYAALTK(-) | |||||
| 23 | BAD28273 | Hypothetical protein | 5.38/4 | 22.2/9.7 | 35.3/>9.3 | IV | ||
| 24 | AAM97013 | Unknown protein Arabidopsis | 7.37/5 | 113.5/9.06 | 26.6/4.67 | I | ||
| BAD32968 | RNA-binding protein-like | 12.77/1 | 35.6/10.4 | (R)YADPVQR(E) | ||||
| 25 | Q9M5M6 | 60S ribosomal protein L30 | 3.62/4 | 12.2/9.6 | 87.2/8.48 | IV | ||
| Putative subtilase | 11.6/1 | 1.2/9.71 | (-)TVNTVGAGAATYK(-) | |||||
| 27 | Unspecified | 12.72/1 | 1.5/8.89 | 23.9/4.8 | (-)LKPDPNTCGGKDFK(-) | IV | ||
| 28 | S72477 | Probable cinnamyl-alcohol dehydrogenase | 12.72/1 | 38.3/5.86 | 38.9/8.51 | (R)DTSGVLSPFNFSR(R) | I | |
| 29 | AAB33843 | Aldehyde dehydrogenase | 4.95/4 | 52.6/5.64 | 55/7.14 | I | ||
| 30 | CAA79702 | Mitochondrial formate dehydrogenase precursor | 8.54/7 | 11.59/6 | 41.8/7.08 | 37/8.5 | (K)IVGVFYK(A) | I |
| (K)TVGTVGAGR(I) | ||||||||
| (K)MDSELENQIGAK(F) | ||||||||
| (K)FEEDLDK(M) | ||||||||
| (K)GVLIVNNAR(G) | ||||||||
| (K)DGELAPQYR(-) | ||||||||
| 32 | AAM28874 | ACS7 | 4.53/4 | 77.2/6.51 | 35.3/8.89 | IV | ||
| 34 | P43282 | S-adenosylmethionine synthetase | 8.4/4 | 11.38 /2 | 42.6/6.07 | 46.3/7.14 | (K)SVVASGLAR(R) | I |
| (K)TQVTVEYK(N) | ||||||||
| 35 | CAF22093 | Glyceraldehyde-3-P dehydrogenase | 12.21/2 | 19.59/8.82 | 35/8.84 | (R)AASFNIIPSSTGAAK(A) | II | |
| (K)VVISAPSK(D) | ||||||||
| 37 | BAD52936 | Putative homocysteine S methyltransferase 4 | 4.73/5 | 34.5/6.09 | 17.9/8.85 | IV | ||
| Unspecified | 12.72/1 | 1.2/6.75 | (-)QLMDAATTTNAK(-) | |||||
| 38 | AAM65239 | Unknown protein | 5.74/4 | 32.3/10.1 | 32.6/7.96 | V | ||
| 1SM4A | Ferredoxin Nadp reductase | 11.22/2 | 33.3/6 | (R)LYSIASSAIGDFGDSK(T) | ||||
| (K)DNTFVYMCGLK(G) | ||||||||
| 39 | NP_054979 | Hypothetical protein SpolCp075 | 2.32/4 | 7/10.8 | 35.3/>9.3 | I | ||
| 40 | AAF27063 | F4N2.23 | 4.38/4 | 95.3/6.22 | 37.6/5.68 | V | ||
| 42 | AAM46780 | Latex plastidic ALD | 6.51/4 | 42.6/8.6 | 35.1/5.91 | V | ||
| 43 | BAA01279 | O-acetylserine thiol lyase | 3.89/4 | 34.1/5.61 | 19.6/8.08 | I | ||
| 45 | CAA75381 | Translation elongation factor | 5.34/10 | 13.28/1 | 27.2/5.09 | 42/4.64 | (R)NTTVTGVEMFQK(I) | I |
| 46 | Q40565 | Rubisco activase | 7.82/13 | 12.72/13 | 34.36/6.76 | 35.1/6.14 | (K)IVVHITK(N) | I |
| (K)SFQCELVFR(K) | ||||||||
| (K)SFQCELVFRK(M) | ||||||||
| (R)YREAAEIIR(K) | ||||||||
| (R)EAAEIIRK(G) | ||||||||
| (K)FYWAPTR(E) | ||||||||
| (R)VYDDEVR(K) | ||||||||
| (R)VYDDEVRK(W) | ||||||||
| (K)WVSGTGIEAIGDK(L) | ||||||||
| (K)LLNSFDGPPTFEQPK(M) | ||||||||
| (K)LLEYGNMLVQEQENVK(R) | ||||||||
| (K)LLEYGNMLVQEQENVKR(V) | ||||||||
| (R)VQLAETYLK(E) | ||||||||
| 47 | P49319 | Catalase isozyme 1 salicylic acid-binding protein | 5.15/18 | 56.7/7.02 | 54.9/8.64 | V | ||
| 48 | AAD39604 | F23M19.3 | 2.69/4 | 67/9.5 | 73.2/9.09 | V | ||
| 49 | CAD27635 | Maturase | 4.22/4 | 61.2/9.68 | 24.5/>9.3 | IV | ||
| 50 | BAA24535 | Spermidine synthase | 5.81/8 | −13.9/1 | 34.4/5.3 | 35/4.55 | (K)ASFCLPSFAK(R) | I |
| 51 | CAA78703 | Rubisco activase | 8.99/8 | 14.54/6 | 48.3/8.4 | 33/6.16 | (K)SFQCELVFR(K) | I |
| (R)TDNVPEEAVIK(I) | ||||||||
| (R)VYDDEVR(K) | ||||||||
| (K)LLNSFDGPPTFEQPK(M) | ||||||||
| (R)VQLAETYLK(E) | ||||||||
| (K)EAALGDANADAINTGNF(-) | ||||||||
| 53 | CAE05740 | OSJNBb0017I01.20 | 1.68/4 | 40.8/5.7 | 24.5/>9.3 | IV | ||
| 54 | AAG40343 | Hydroxymethyltransferase | 5.94/4 | 51.7/7.6 | 55/8.73 | IV | ||
| 55 | NP_918618 | OSJNBa0094H06.18 | 2.33/4 | 5.6/10.1 | 22/>9.3 | IV | ||
| 57 | AAF97728 | Potassium-sodium symporter | 2.52/4 | 61.9/9.13 | 38.9/7.53 | I | ||
| 58 | NP_915483 | P0446B05.25 | 5.04/4 | 38.6/9.6 | 42.1/8.5 | I | ||
| 60 | XP_470450 | Unknown protein | 3.86/4 | 48.2/10.5 | 37/7.37 | I | ||
| 61 | T07032 | S-2-hydroxy acid oxidase | 6.05/7 | 31.2/9.57 | 44.5/>9.3 | I | ||
| 62 | XP_468473 | Putative cytochrome P450 | 5.2/4 | 58.1/9.4 | 41.7/5.89 | V | ||
| 64 | BAD03386 | Hypothetical protein | 3.33/4 | 20.8/10 | 107.3/<4.5 | IV | ||
| 65 | CAA96433 | PME | 5.1/4 | 11.46/1 | 20.1/9.29 | 37/8.31 | (R)VGADMSVINR(C) | IV |
| 66 | CAC80376 | Glyceraldehyde-3-P dehydrogenase | 5.81/4 | 12.71/5 | 34/5.56 | 36.2/8.33 | (K)TLLFGEK(S) | II |
| (K)KVVISAPSK(D) | ||||||||
| (R)AASFNIIPSSTGAAK(A) | ||||||||
| (K)EASYDDIK(A) | ||||||||
| (K)AAIKEESEGK(L) | ||||||||
| 67 | AAN40027 | Hypothetical protein | 3.48/5 | 64.7/9.09 | 37.9/<4.5 | V | ||
| 68 | P29790 | ATP synthase γ-subunit | 9.47/7 | 41.4/8.4 | 35/8.64 | IV | ||
| CAB39974 | Glyceraldehyde-3-P dehydrogenase | 12.72/5 | 34.4/6.15 | (K)DAPMFVVGVNEK(E) | ||||
| (R)AASFNIIPSSTGAAK(A) | ||||||||
| (K)VLPSLNGK(L) | ||||||||
| (K)EATYDEIK(A) | ||||||||
| (K)AAIKEESEGK(L) | ||||||||
| 69 | AAF03099 | NADP-dependent glyceraldehyde phosphate dehydrogenase | 11.71/1 | 11.8/10.8 | 31/7.93 | (K)DSPLDVIAINDTGGVK(Q) | IV | |
| 70 | AAG23799 | Glyceraldehyde-3-P dehydrogenase | 12.66/3 | 18.2/6.1 | 35.1/8.73 | (R)AAALNIVPTSTGAAK(A) | V | |
| (K)AVALVLPSLK(G) | ||||||||
| (K)TFAEEVNAAFR(E) | ||||||||
| 71 | AAM62933 | Putative chloroplast 50S ribosomal protein L6 | 14.54/4 | 24.6/10.2 | 17.4/>9.3 | (R)TLTDNMVVGVSK(G) | V | |
| (R)ITVSGYDK(S) | ||||||||
| (K)SEIGQFAATVR(K) | ||||||||
| (K)YADEIVR(R) | ||||||||
| 72 | 2119042 | Gly-rich RNA-binding protein | 10.74/9 | 14.54/3 | 15.6/5.62 | 15/5.11 | (R)NITVNEAQSR(G) | II |
| (R)EGGYGGGGGYGGGR(R) | ||||||||
| (R)EGGYGGGGGGGYGGGR(R) | ||||||||
| 74 | P81665 | Oxygen-evolving enhancer protein 1 | 11.12/9 | 12.72/2 | 26.39/4.31 | 30.2/<4.55 | (R)GSSFLDPK(G) | II |
| (R)GGSTGYDNAVALPAGGR(G) | ||||||||
| 75 | P81665_1 | Oxygen-evolving enhancer protein | 12.49/1 | 1.09/4.07 | 30.2/<4.55 | (K)NAPPEFQK(T) | II | |
| 77 | T45856 | Hypothetical protein F3A4.110 | 4.26/4 | 57.2/9.46 | 30/<4.55 | V | ||
| AAP03871 | Oxygen-evolving complex 33-kD PSII protein | 12.71/6 | 35.17/5.4 | (K)RLTFDEIQSK(T) | ||||
| (R)LTFDEIQSK(T) | ||||||||
| (K)FCLEPTSFTVK(A) | ||||||||
| (K)DGIDYAAVTVQLPGGER(V) | ||||||||
| (R)GGSTGYDNAVALPAGGR(G) | ||||||||
| (R)GDEEELEKENVK(N) | ||||||||
| 79 | B24430 | Glyceraldehyde-3-P dehydrogenase | 6.51/8 | 10.85/4 | 41.78/8.56 | 41.6/8.45 | (K)YDSMLGTFK(A) | V |
| (K)IVDNETISVDGK(H) | ||||||||
| (K)GTMTTTHSYTGDQR(L) | ||||||||
| (R)AAALNIVPTSTGAAK(A) | ||||||||
| 80 | AAR96006 | 4-Nitrophenylphosphatase-like protein | 11.68/1 | 16.5/5.38 | 29.6/5.18 | (R)LVFVTNNSTK(S) | V | |
| 82 | 1909374A | Rubisco activase | 7.01/11 | 42.7/5.57 | 41/5.59 | II | ||
| 83 | AAO62992 | Chloroplast Gln synthetase | 12.73/2 | 20.9/5.82 | 41.7/5.89 | (K)AAQIFSDSK(V) | V | |
| (R)DISDAHYK(A) | ||||||||
| 84 | AAA34111 | Rubisco | 7.71/4 | 10.1/5.36 | 14.8/6.37 | V | ||
| 85 | AAG14029 | NADH dehydrogenase | 3.86/4 | 37.4/9.69 | 27/>9.3 | II | ||
| 87 | BAA31510 | Ribosomal protein L4 chloroplast | 2.85/4 | 31.2/6.59 | 27.6/8.71 | II | ||
| 89 | BAD35631 | Putative zinc-finger and C2 domain protein | 3.56/4 | 35.6/6.47 | 30.9/>9.3 | II | ||
| 90 | AAM64724 | Unknown protein | 3.22/4 | 61.7/5.23 | 87.9/<4.5 | V | ||
| 91 | BAD15634 | Mitochondrial transcription termination factor-like | 3.65/4 | 42.6/9.9 | 35.9/6.37 | V | ||
| 92 | CAB87677 | Putative protein | 4.78/5 | 34.7/6.56 | 42.2/<4.5 | II | ||
| 93 | AAR20278 | Maturase | 6.32/5 | 55.1/10.4 | 42.9/4.55 | V | ||
| 94 | AAO32311 | Putative chloroplast 50S ribosomal protein L6 | 5.29/6 | 24.9/10.3 | 18.9/>9.3 | III | ||
| 95 | CAA78703 | Rubisco activase | 9.14/12 | 11.62/6 | 48.3/8.4 | 41.3/4.71 | (K)GLVQDFSDDQQDIAR(G) | II |
| (R)EAAEIIR(K) | ||||||||
| (R)TDNVPEEAVIK(I) | ||||||||
| (R)VYDDEVR(K) | ||||||||
| (K)WVSGTGIEAIGDK(L) | ||||||||
| (R)VQLAETYLK(E) | ||||||||
| 97 | NP_174644 | Hypothetical protein | 2.74/4 | 21.6/7.7 | 19.7/<4.5 | V | ||
| 98 | BAD28931 | Hypothetical protein | 2.39/5 | 7.4/10.3 | 39.7/8.6 | V | ||
| 99 | AAF76363 | I box-binding factor | 2.77/4 | 23.9/8.31 | 42.7/>9.3 | V | ||
| 100 | BAD53039 | Hypothetical protein | 2.88/4 | 7.3/6.47 | 41/8.73 | V | ||
| 101 | XP_466540 | Putative speckle-type POZ | 4.05/8 | 38.8/10.5 | 76.2/5.25 | V | ||
| 103 | P29302 | PSI chain II D2 precursor | 5.71/5 | 22.4/10.1 | 41.5/9 | III | ||
| 105 | CAA71589 | Protein g5bf | 3.82/4 | 42.6/3.82 | 37/8.48 | III | ||
| 108 | AAU03361 | PS2 oxygen-evolving complex protein 3 | 14.51/1 | 0.95/10.9 | 15/>9.3 | (-)GKLKGSGKGK(-) | II | |
| 109 | O23787 | Thiazole biosynthetic enzyme precursor | 9.53/11 | 11.62/1 | 37.5/5.2 | 32.9/4.8 | (K)ALDMNSAEDAIVR(L) | IV |
Table II.
Changes of proteins in N. attenuata leaves upon several induction treatments
The accumulations of N. attenuata leaf proteins exhibiting consistent up- or down-regulation (indicated by arrows) at a minimum of three time points (6–72 h) after OS treatment are listed with their P values of water control (W + W), OS (W + OS), FACs (W + FAC) induction, OS without FACs (W + OS-FAC-free), and caterpillar-feeding induction (Larvae) at 48 h. The protein names represent the database identification of the mass spectrometric analyses with the highest confidence. Numbers indicate P values; n/a, P value is not available; –, no change; and x, spot not detectable. qltv, Qualitative change in protein spots.
| Spot No. | Accession No. | Name of Protein | Induction Pattern and P Values for the Treatments
|
||||
|---|---|---|---|---|---|---|---|
| W + W | W + OS | W + FAC | W + OS-FAC-Free | Larvae | |||
| Photosynthesis and photorespiration-related metabolism | |||||||
| 6 | CAA78703 | Rubisco activase | – | ↓ 0.19 | ↓ 0.93 | ↓ 0.11 | ↓ 0.07 |
| 9 | XP_473501, Q40565 | OSJNBb0078D11.3, Rubisco activase | – | ↓ 0.04 | ↓ 0.21 | ↓ 0.003 | ↓ 0.05 |
| 12 | Q40459 | Oxygen-evolving enhancer protein 1 | – | ↓ 0.31 | – | ↓ 0.45 | – |
| 20 | CAA78703 | Rubisco activase | – | ↑ qltv | – | ↑ 0.18 | – |
| 46 | CAA78703 | Rubisco activase | ↑ 0.01 | ↑ qltv | ↑ 0.05 | ↑ 0.15 | ↑ 0.07 |
| 51 | CAA78703 | Rubisco activase | ↑ 0.02 | ↑ qltv | – | ↑ 0.18 | ↑ 0.19 |
| 74 | P81665 | Oxygen-evolving enhancer protein 1 | – | ↓ 0.05 | ↓ 0.11 | – | ↑ 0.06 |
| 75 | P81665_1 | Oxygen-evolving enhancer protein 1 | – | ↓ 0.11 | ↓ 0.15 | ↓ 0.62 | – |
| 77 | AAP03871 | Oxygen-evolving complex 33-kD PSII protein | – | ↓ 0.05 | ↓ 0.12 | – | – |
| 82 | 1909374A | Rubisco activase | – | ↓ 0.27 | ↓ 0.58 | ↓ 0.02 | ↓ 0.12 |
| 95 | CAA78703 | Rubisco activase | ↑ 0.06 | ↓ 0.27 | ↓ 0.41 | ↓ 0.02 | – |
| 108 | AAU03361 | Oxygen-evolving complex protein 3 | – | ↓ 0.01 | – | – | – |
| Primary metabolism | |||||||
| 1 | XP_472335 | OSJNBa0039C07.4 | – | – | – | ↑ 0.09 | ↑ 0.15 |
| 2 | S58083 | TK | ↑ 0.04 | ↑ 0.77 | – | ↑ 0.18 | – |
| 5 | AAO23981 | Vacuolar H+-ATPase A2 subunit | – | ↑ 0.13 | ↑ 0.08 | – | ↑ 0.02 |
| 7 | AAG59585 | TD | ↑ 0.17 | ↑ 0.24 | ↑ 0.34 | – | ↑ 0.02 |
| 21 | AAT38067 | Putative glycoprotease | – | ↑ qltv | – | – | – |
| 29 | AAB33843 | Aldehyde dehydrogenase | – | ↑ 0.12 | ↑ 0.28 | x n/a | – |
| 30 | CAA79702 | Mitochondrial formate dehydrogenase precursor | – | ↑ 0.35 | ↑ 0.18 | – | – |
| 32 | AAM28874 | ACS7 | – | ↑ 0.22 | ↑ 0.27 | ↑ n/a | ↑ n/a |
| 34 | AAO85809 | S-adenosylmethionine synthetase | – | ↑ 0.25 | ↑ 0.06 | ↑ 0.11 | ↑ 0.03 |
| 35 | CAF22093 | Glyceraldehyde-3-P dehydrogenase | – | ↓ 0.8 | – | - n/a | ↑ 0.004 |
| 43 | BAA01279 | O-acetylserine thiol lyase | – | ↑ qltv | ↑ 0.11 | ↑ 0.23 | ↑ 0.18 |
| 50 | BAA24535 | Spermidine synthase | ↑ 0.09 | ↑ 0.08 | ↑ 0.01 | ↑ n/a | – |
| 66 | BAC87864 | Glyceraldehyde-3-P dehydrogenase | – | ↓ 0.26 | – | – | ↓ 0.08 |
| 83 | AAO62992 | Chloroplast Gln synthetase | – | – | – | – | ↑ 0.03 |
| Transcription and translation | |||||||
| 45 | BAA02028 | Translation elongation factor | – | ↑ qltv | – | – | – |
| 72 | BAA03741 | Gly-rich RNA-binding protein | – | ↓ 0.02 | – | ↓ 0.03 | - 0.51 |
| 87 | BAA31510 | Ribosomal protein L4 chloroplast | – | ↓ qltv | – | - n/a | - n/a |
| Secretory pathway | |||||||
| 57 | AAF97728 | Potassium-sodium symporter | ↑ 0.08 | ↑ 0.11 | ↑ 0.04 | ↑ 0.008 | - n/a |
| Secondary metabolism | |||||||
| 8 | AAM62737 | QR-like protein | – | – | ↓ 0.37 | ↓ 0.005 | ↓ 0.21 |
| 22 | AAS90324 | 3-dehydroquinate dehydratase | ↑ 0.21 | ↑ 0.002 | – | – | ↑ 0.77 |
| 24 | BAD32968 | RNA-binding-like protein | ↑ 0.18 | ↑ 0.05 | ↑ 0.02 | ↑ 0.04 | ↑ 0.003 |
| Unspecified function and unknown proteins | |||||||
| 58 | NP_915483 | P0446B05.25 | – | ↑ 0.08 | ↑ 0.25 | – | – |
| 92 | CAB87677 | Putative protein | – | ↓ 0.04 | – | – | – |
Identification of Proteins by MALDI-TOF and LC-MS/MS
The tryptic peptide mass fingerprints of selected proteins were determined from matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectra using the green plant database of the National Center for Biotechnology Information (NCBI; http://www.ncim.nlm.nih.gov); protein spots showing identity with RuBPCase were omitted from further analysis. Peptide sequence data obtained from liquid chromatography (LC)-MS/MS were compared with green plant or nonredundant NCBI databases. Amino acid sequences from de novo data with unspecified protein hits were searched using MS-BLAST. Most of the 45 proteins identified by peptide sequences agree with the peptide mass fingerprints. An additional 42 proteins have been identified by their peptide mass fingerprints, which match at least four peptides. Table I presents detailed information about these 84 protein spots, such as identification scores, number of matching peptides, and peptide sequences, and identifies their putative function with accession numbers of database proteins, molecular masses, and isoelectric points. More than 20% of proteins remain unidentified due to the lack of a database match or a match only to proteins with unknown functions.
Patterns of Protein Accumulation Elicited by the Different Treatments
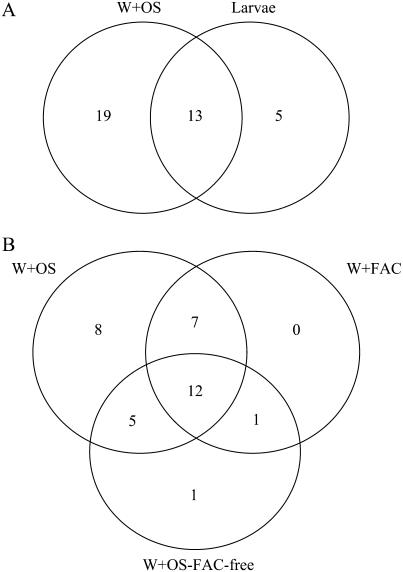
Accumulation patterns for 35 proteins were monitored in the four elicitation treatments and compared to patterns observed in control leaves. Differences among these treatments were seen in only a few spots. The larval feeding treatment was terminated after 48 h and the responses in this treatment were compared with those of the W + OS treatments. A similar but lower rate of protein accumulation was found in the larval treatment compared to the W + OS treatment, perhaps because the amount of damage and the amount of OS introduced into wounds after 48 h of feeding by five neonate larvae are likely less than those inflicted by the W + OS treatment. Of the 35 differentially regulated proteins in these two treatments, 37% (13 proteins) were commonly regulated (nine up- and four down-regulated; Fig. 3A; Table II). Interestingly, spot 83, a chloroplast Gln synthetase, was up-regulated in caterpillar-attacked leaves but not regulated in the other elicitation treatments (Table II). However, a vacuolar H+-ATPase A2 subunit, TD, and one spot with glyceraldehyde-3-P dehydrogenase of caterpillar-attacked leaves share accumulation patterns similar to those of leaves treated with OS.
Figure 3.
Comparison of protein accumulation patterns in N. attenuata leaves among different inducers. The Venn diagram presents the number of protein spots that exhibit differential accumulation patterns among different elicitor treatments compared to control leaves. Shown are a comparison between larvae- and W + OS-induced leaves (A) and a comparison among the W + OS, W + FACs, and W + OS-FAC-free leaves (B). The numbers in circles indicate the protein spots having differential accumulation and the numbers in the common area represent protein spots with similar patterns of accumulation.
Because the amount of mechanical damage and elicitor solutions introduced represented parameters that did not differ among the W + OS, W + FAC, and W + OS-FAC-free treatments, the cause of the differences is more easily inferred. Of the 35 proteins that were differentially regulated by these three treatments, 12 (seven up- and five down-regulated) were commonly regulated among all three treatments (Fig. 3B; Table II). The seven commonly up-regulated protein spots were RNA-binding protein (spot 24), acyl-CoA synthetase 7 (ACS7; spot 32), S-adenosylmethionine synthetase (spot 34), O-acetylserine thiol lyase (spot 43), RCA (spot 46), spermidine synthase (spot 50), and potassium sodium symporter (spot 57). Interestingly, four of the five down-regulated spots identified as RCA consistently accumulated in lower amounts (spots 6, 9, 82, and 95); the fifth protein was the oxygen-evolving enhancer protein 1 (spot 75). To determine which proteins were regulated by the FACs in OS, we compared the regulation elicited by the W + OS and W + FAC treatments. Seven proteins were commonly regulated and no proteins were uniquely regulated by the FAC treatments that were not changed by the W + OS treatment. Only one spot (OSJNBa0039c07.4, spot 1) was uniquely regulated by W + OS-FAC-free treatment, and one spot (quinone oxidoreductase [QR]-like protein, spot 8) was commonly regulated by this treatment and by the W + FAC treatment, demonstrating that FACs account for the majority of the elicitor activity of OS at the protein level. However, OS clearly contain factors other than FACs that elicit differential accumulations of proteins, as can be seen by comparing the responses elicited by the W + OS and W + OS-FAC-free treatments (Fig. 3B). These treatments commonly regulated the following five proteins, none of which were regulated by the W + FAC treatment: RCA (spots 20 and 51), transketolase (TK; spot 2), the oxygen-evolving enhancer protein 1 (spot 12), and the Gly-rich RNA-binding protein (spot 72). These results demonstrate that although FACs are the main elicitors of specific changes in protein accumulation in OS, other factors are clearly also active.
Changes in the Patterns of W + OS-Elicited Protein Accumulation
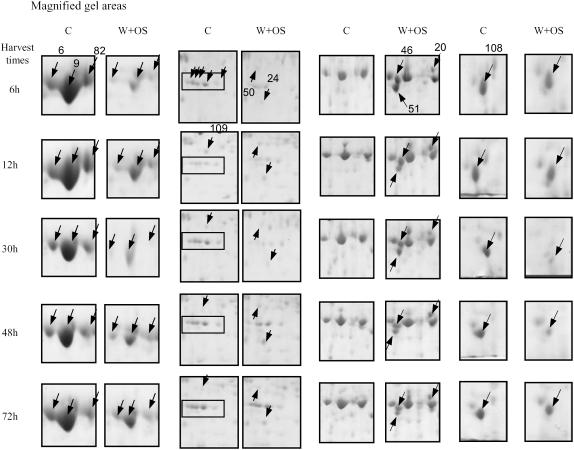
The early (6, 12, and 30 h) and later (48–72 h) changes in the W + OS-elicited proteome were characterized by the relative densitometric quantitation of 90 regulated protein spots (Table I; Fig. 4; Supplemental Figs. S2 and S3). Of these 90 regulated protein spots, 17 are involved in photosynthesis and photorespiration—28 in primary metabolism, 13 in transcription and translation processes—and nine function in secondary metabolism pathways; one protein in each case was identified in signal transduction, secretory pathway, and cytoskeleton formation (Table I; Fig. 2). Twenty proteins were not identified or were identified but had unknown functions. To simplify the analysis, we categorized the protein accumulation patterns as: (I) consistently high accumulation, (II) consistently low accumulation, (III) early high accumulation (up to 30 h) and no change or low later (30 h onward), (IV) no change or low early on and high accumulation or no change later, and (V) no consistent pattern (Table I). Twenty-seven proteins exhibited a type I response (Table I). Of these, four proteins belong to the photosynthesis and photorespiration category and eight to the primary metabolism category; four proteins were categorized as involved in secondary metabolism, seven as unknown proteins; one is a translation elongation factor; and one protein contributes to the cytoskeleton and one to the secretory pathway. Fifteen proteins exhibited a type II response and are present in the following categories: eight are related to photosynthesis and photorespiration, four to primary metabolism, one to transcription and translation, one to signal transduction, and one is a putative protein. For type I and II responses, we also included proteins that showed consistent accumulation in four harvests. We categorized six proteins with type III responses. One is involved in photosynthesis, three in primary metabolism, one in transcription and translation, and one in secondary metabolism. Sixteen proteins exhibited type IV responses, seven in primary metabolism, four in transcription and translation, and five with no known function. We identified 26 proteins with type V responses, indicating they either have very specific kinetics or were not detected at other harvests of several replicates. In summary, we observed most proteins with similar accumulation patterns during the five time harvests after applying OS to N. attenuata leaves. However, few proteins specific to one or two harvest times and exhibiting increased or decreased accumulation patterns were also identified.
Figure 4.
Magnified areas of two-dimensional gels. The four sections show magnified areas of control and W + OS-elicited leaves at different times after elicitation (6–72 h). The boxes in the gel area (column 2) encompass spots 74, 75, 77, 78, and 80 (left to right). Numbers of protein spots with arrows are indicated in the first gel of their appearance, whereas only the positions of protein spots are shown with arrows in subsequent gels. Proteins eluted from these spots were identified by MALDI-TOF and/or LC-MS/MS (see Table I).
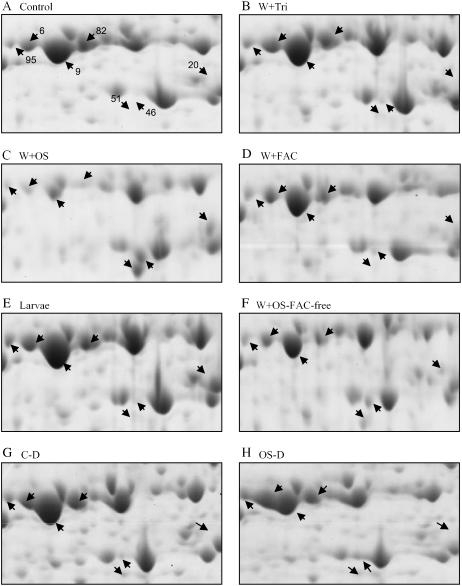
The Response of RCA Protein(s) to OS Elicitation
The changes in RCA protein spots were one of the most apparent OS-elicited responses, so much so that these spots reliably differentiated gels of control and elicited samples. We identified seven protein spots that have one or several peptides exhibiting homology with RCA (Tables I and II). In four spots, molecular weight and pI are very similar in terms of reported RCA proteins from several plant species. After W + OS in elicited leaves compared to controls, the levels of these proteins (spots 6, 9, 82, and 95) decreased (Fig. 5), whereas levels of RCA (spots 20, 46, and 51) strongly increased. Since these changes were not significant in W + W or W + FAC treatments, we infer that factors other than FACs in OS are responsible (Fig. 5D). This inference is supported by the observation that the responses observed in the W + OS-FAC-free treatment were similar to those observed in the W + OS treatment (Fig. 5F). Peptide data obtained from spots 6 and 95 match data from the C-terminal region of RCA protein, whereas spots 9, 20, and 46 yielded peptide sequences spanning almost the entire length of RCA. Protein spot 51 generated peptides from the N-terminal region of RCA. These results suggest that massive changes are occurring in RCA proteins due to proteolytic cleavage, differential subunit formation, or gene expression. RCA is a member of the chaperonin protein family, which is involved in RuBPCase activation, and RuBPCase is known to be transcriptionally down-regulated after elicitation (Hermsmeier et al., 2001).
Figure 5.
Magnified areas [see areas (a) and (b) in Supplemental Figure S4] of two-dimensional gels of protein extracts from control and elicited plants from treated and distal leaves for RCA spots. Leaf protein profiles of control (A) and punctured leaves treated with 0.0025% Triton X-100 (W + Tri; B), solutions of M. sexta OS (W + OS; C), chemically synthesized FACs (W + FAC; D), feeding M. sexta larva (E), OS without FACs (W+OS-FAC-free; F), a control of distal leaves (C-D; G) and W + OS of distal leaves (OS-D; H) are shown. Leaves were collected 30 h after the application of elicitor solution and 48 h after the release of larvae on local leaves. The accumulation patterns of RCA spots were compared among these treatments.
To determine whether the changes observed in RCA proteins are due to the proteolytic activity of the OS fraction, we analyzed the 2-DE protein patterns of undamaged systemic leaves on elicited plants. Interestingly, proteins in spots 46 and 51 were detectable in OS-elicited systemic leaves, whereas decreasing amounts of protein in spots 6, 9, 82, and 95 were noticed in the systemic leaves compared to leaves growing at the same nodes on control plants (Fig. 5, G and H). Furthermore, to confirm that these changes in RCA spots are not due to a plant protein fraction present in OS, we performed a 2-DE analysis of OS. Of the nearly 100 proteins we identified by peptide mass fingerprints, no protein spots were detected with similarity to RCA (data not shown). These results demonstrate that even if proteolysis of RCA is responsible for these changes, it occurs via plant metabolism and is not due to the proteolytic activity of OS.
Transcript Accumulation Patterns in N. attenuata-Elicited Leaves
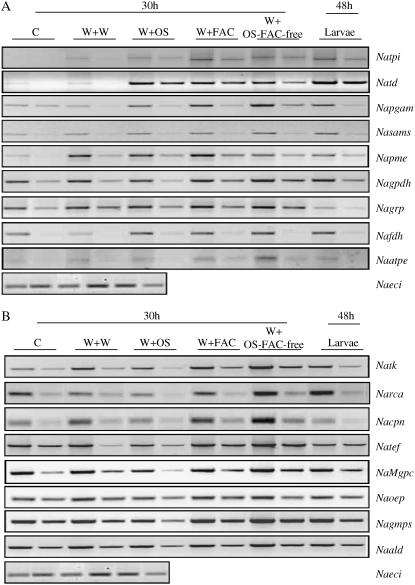
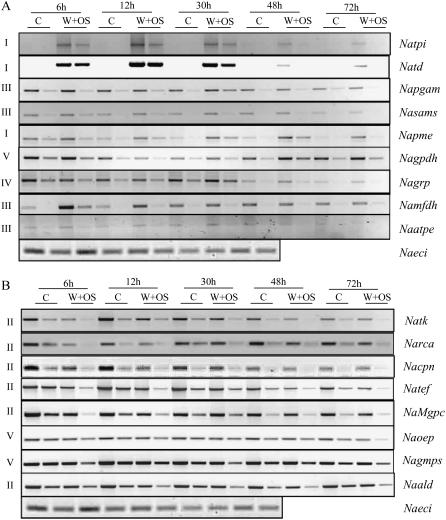
RT-PCR was used to quantify the transcripts of 17 genes coding for 17 differentially accumulated proteins to assess the correlations between protein and mRNA accumulation patterns (Figs. 6 and 7). Different peptide sequences obtained from protein spots were used to design primers to compare mRNA accumulation in W + OS-elicited leaves with that in control leaves 30 h after elicitation. The well-characterized insect-induced protein TPI was not identified in the protein gels; however, it was used as a positive control. TPI may not be detected in induced leaves in proteomic analysis due to its extractability and/or low levels. Patterns of mRNA accumulation were compared among 17 candidate genes 30 h after the different elicitation treatments (Fig. 6). Levels of mRNA increased after W + OS, W + FAC, W + OS-FAC-free, and larval treatments in seven transcripts, namely, tpi, td, pgam (phosphoglycerate mutase [PGM]), sams (S-adenosylmethionine synthetase), pme (pectin methyl esterase [PME]), gpdh (glyceraldehyde-3-P dehydrogenase), fdh (mitochondrial formate dehydrogenase), and atpe (vacuolar ATPase); however, treatment-specific increases were also observed (Fig. 6A). tpi, td, and sams transcripts responded maximally to larval feeding, whereas pgam and atpe accumulated maximally in response to the W + OS-FAC-free treatment. The W + FAC treatment elicited the largest response in pme mRNA. ald (aldolase [ALD]) transcripts decreased in response to W + OS and larval treatments, but increased after W + FAC and W + OS-FAC-free treatments. Levels of tef (translation elongation factor) increased after W + OS-FAC-free treatments compared to other elicitor treatments. Gln synthetase transcripts increased only after W + FAC and W + OS-FAC-free and larval treatments but not after W + OS treatment. Levels of grp (Gly-rich RNA-binding protein) and cpn (chaperonin) increased after W + FAC and W + OS-FAC-free treatments, but decreased in response to the presence of larvae. grp also showed increased accumulation after W + OS treatments. Levels of tk increased maximally after W + OS-FAC-free treatment. Transcripts of rca decreased after W + OS treatments and increased after larval and W + OS-FAC-free treatments, but remained unchanged after the W + FAC treatment (Fig. 6B). Transcript accumulation patterns of seven candidates correlated with their respective protein abundances. The specific mRNA accumulation patterns observed in OS-FAC-free-elicited N. attenuata leaves indicate that signaling molecules other than FACs exist in M. sexta OS and that these also elicit specific protein accumulation patterns.
Figure 6.
Gene expression analysis using the RT-PCR approach on candidate proteins differentially elicited in N. attenuata leaves after various elicitor treatments. Quantitative RT-PCR analysis of 17 candidate genes in nonwounded and in wounded leaves treated with different elicitors, which show (A) pattern of nine genes: TPI (Natpi), TD (Natd), PGM (Napgam), S-adenosylmethionine synthetase (Nasams), PME (Napme), glyceraldehyde-3-P dehydrogenase (Nagpdh), Gly-rich RNA-binding protein (Nagrp), mitochondrial formate dehydrogenase (Namfd), and ATPase (Naatpe); and (B) pattern of eight genes: TK (Natk), RCA (Narca), chaperonin (Nacpn), translation elongation factor (Natef), Mg-protoporphyrin IX chelatase (NaMgpc), oxygen-evolving protein (Naoep), Gln synthetase (Nagmps), and ALD (Naald). Transcript abundance was checked after 48 h of larval feeding and 30 h for control (C), and the application of wounding with water (W + W), larval OS (W + OS), FACs (W + FAC), and OS devoid of FACs (W + OS-FAC-free). PCR reactions were carried out with two concentrations of cDNA for all primers, in at least three replicates. A single concentration of cDNA was used for ECI (AB010717, sulfite reductase) primers. ECI was used as internal standard to determine the equal amplification of cDNA. The gene names, accession numbers, primer sequences, sizes of amplified cDNA fragments, and respective peptide sequence maps of proteins are provided in Supplemental Table S1 and Supplemental Figure S5.
Figure 7.
Gene expression analysis of candidate proteins exhibiting differential accumulation in control and OS-induced N. attenuata leaves at different times after elicitation. Quantitative RT-PCR analysis of candidate genes in control leaves (C) and OS-induced leaves (W + OS) that show up-regulation (A; nine genes) and down-regulation (B; eight genes) between 6 and 72 h is shown. PCR reactions were carried out with two concentrations of cDNA for all primers, in at least three replicates. A single concentration of cDNA was used for ECI primers to determine the equal amount of the cDNA. The gene names, accession numbers, primer sequences, sizes of amplified cDNA fragments, and respective peptide sequence maps on proteins are provided in Supplemental Table S1 and Supplemental Figure S5.
The differential accumulation of mRNA was observed in all 17 genes. To simplify the data, we categorized accumulation patterns as we had done for the proteins (Fig. 7). In control plants, levels of tpi transcripts were barely detectable, but they were strongly elicited—a type I pattern—by W + OS treatment. This pattern was also found in td transcripts where both (tpi and td) had reached their peaks at 12 h and decreased slightly thereafter. pme transcripts were up-regulated at all time points (type I). mRNAs of pgam, sams, fdh, and atpe were consistently up-regulated up to 30 h, slightly down-regulated at 48 h, and unchanged at 72 h (type III; Fig. 7A). Transcripts of gpdh were slightly up-regulated at 6 h, down-regulated at 12 and 30 h, and up-regulated again at 48 and 72 h (type V). grp accumulated fewer transcripts in induced leaves at early compared to later time points (30–72 h; type IV). Levels of tk, rca, tef, mgpc (magnesium [Mg]-protoporphyrin IX chelatase), ald, and cpn (6-48 h) showed decreased mRNA accumulation at all time points (type II; Fig. 7B). Transcripts of gmps (Gln synthetase) and oep (oxygen-evolving protein) were down-regulated consistently up to 30 h, but at 48 h the levels of accumulation were the same as in control leaves; at 72 h after OS application (type V) the levels were down-regulated. In summary, mRNA accumulation patterns match protein accumulation patterns in several candidate proteins; however, cases in which protein and mRNA accumulation did not match need to be studied further. One possible explanation for such discrepancy might be that several isoforms of these proteins exist, which makes it difficult to identify specific ones.
Silencing rca in N. attenuata Results in Reduced Photosynthesis Rate and Biomass, Responses Consistent with Herbivore-Attacked Plants
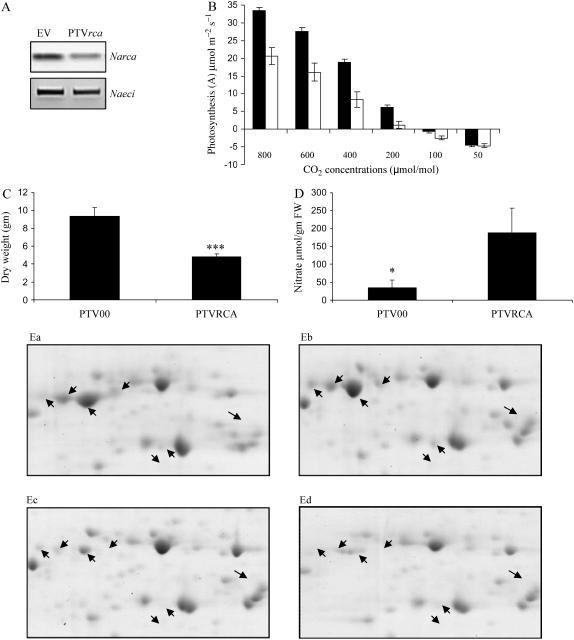
Massive changes in RCA protein levels after elicitation were found in the proteomic analysis. Transcriptional analysis of induced tissues indicated that the Narca gene is down-regulated after herbivory, supporting our earlier large-scale transcriptional analysis (Hermsmeier et al., 2001; Schittko et al., 2001). To understand the functional consequences of this herbivore-elicited down-regulation of RCA proteins in N. attenuata plants, we silenced its expression by virus-induced gene silencing (VIGS) in N. attenuata plants. Transcript levels of Narca were >50% reduced in rca-VIGS plants compared to empty vector (EV)-transformed plants (Fig. 8A). Proteomic analysis of these plants revealed significant reductions among all seven RCA protein spots in control and OS-induced leaves as compared to EV plants (Fig. 8, Ea–Ed). This analysis strongly validated the proteomic analytical procedures used in this study. The lower accumulation of RCA proteins correlated with decreased photosynthetic rates and biomass, and increased nitrate levels. The net photosynthesis rates of rca-VIGS plants were 40% to 50% lower compared to EV plants (Fig. 8B). The slope of a regression of photosynthetic rate against Ci provides an in vivo measure of RuBPCase activity (Manter and Kerrigan, 2004), and the slope of the A/Ci curve of EV plants is approximately 1.5 times greater than that of the rca-VIGS plants, which clearly demonstrates that silencing RCA reduced RuBPCase activity, which in turn decreased net photosynthesis. The biomass of rca-silenced plants was reduced by 48% to 50% (Fig. 8C). In contrast, nitrate concentrations were significantly higher in rca-VIGS plants than in EV plants (Fig. 8D).
Figure 8.
Effect of RCA reduction on photosynthesis, biomass, and nitrate content in N. attenuata. Three-week-old N. attenuata plants were inoculated with Agrobacterium tumefaciens harboring a TRV-based construct containing a 268-bp fragment of Narca or EV constructs for the VIGS experiments. A, Gene silencing was confirmed at transcript and (Ea–Ed) proteomic levels. RCA spots are from rca-silenced plants compared with EV-transformed plants; shown are EV control leaves (Ea), EV leaves treated with OS (Eb), rca-VIGS control leaves (Ec), and rca-VIGS leaves treated with OS (Ed). B, Net photosynthetic rates and intercellular CO2 concentrations of rca-VIGS plants (white bars) were measured and compared to EV control plants (black bars) under saturating light (approximately 1,400 μm m−2 s−1) intensities using a LI-COR 6400 portable photosynthesis system. C, Seven-week-old plants were harvested and dried in the oven at 60oC for 48 h for dry mass measurements. D, The nitrate content of these plants was measured spectrophotometrically.
DISCUSSION
We conducted a proteomic analysis of a natural plant-herbivore interaction for which previous research had characterized the host plant's transcriptional responses and the larval elicitors that activate them. From this analysis of 90 herbivore-elicited protein spots in over 84 2-DE gels, we not only developed the techniques for extracting, separating, and analyzing complex protein mixtures reproducibly, but we also characterized proteomic responses to elicitation and learned that the larval elicitors responsible for the transcriptional response account for a majority of the specific changes in the proteome. Previous transcriptional analyses of this particular interaction, as well as of other plant-herbivore interactions, can be broadly seen as representing a shift in metabolism from growth- to defense-related processes (Walling, 2000; Reymond et al., 2004; Bostock, 2005; Leitner et al., 2005; Schmidt et al., 2005; Thompson and Goggin, 2006). We organize the discussion into five sections: the change in proteins associated with (1) photosynthesis; (2) primary and (3) secondary metabolism as these proteins relate to supporting known herbivore-elicited defense responses; (4) the kinetics and elicitors of the transcripts for a subset of 17 elicited proteins; and (5) the signals from the feeding larvae responsible for changes in the proteome.
Changes in the Photosynthesis-Related Proteome
We identified seven different functional photosynthesis-related proteins (from the analysis of 20 protein spots), which accumulate differentially in herbivore-induced leaf tissues. We excluded RuBPCase from the analysis because its transcripts are known to degrade in response to herbivore and pathogen attack (Hermsmeier et al., 2001; Hahlbrock et al., 2003; Hui et al., 2003). Western-blot analysis of 2-DE-separated leaf proteins with two different RuBPCase antibodies revealed extensive fragmentation of this protein in response to chilling rice (Oryza sativa) plants (Yan et al., 2006). Given these stress-associated dynamics, it is not surprising that we found seven RCA protein spots that responded to herbivore elicitation. Yan et al. (2006) identified five RCA spots in rice leaf proteins that responded to chilling. Since the RCA protein is involved in activating RuBPCase and maintaining its active state, the dynamics of these two very abundant proteins are clearly interdependent.
RCA belongs to the AAA+ protein super family, whose members generally function as chaperonins, and uses the Sensor-2 domain to recognize its substrate (Vargas-Suarez et al., 2004; Kirch et al., 2005). Our analysis revealed substantial changes in RCA protein spots by herbivore elicitation, which suggests that the regulation of the RCA subunit content and its composition may contribute to optimizing plant performance during attack. How this occurs requires further study. Chaperonins assist in folding a wide range of structurally and functionally unrelated proteins, and it is possible that RCA interacts with proteins other than RuBPCase when plants are attacked. As is consistent with the existence of alternative functions, RCA is expressed in the nonphotosynthetic seeds of monocots and dicots that lack RuBPCase (Vargas-Suarez et al., 2004).
RCA modulates the activity of RuBPCase, a key regulatory enzyme of photosynthetic carbon assimilation, by facilitating the removal of sugar phosphates (ribulose bisphosphate) that prevent substrate binding and carbamylation of the protein's active site. The VIGS experiment demonstrated that reducing RCA protein and transcript levels, as occurs in OS-elicited plants, reduced net photosynthetic rates of N. attenuata plants, which in turn was reflected in the reduced biomass of these plants. The role of RCA in regulating the activity of RuBPCase (Portis, 1995) was clearly seen in the in vivo measures of RuBPCase activity, which were reduced in rca-silenced lines. RCA-mediated reductions in carbon gain also inhibited the plant's ability to reduce and assimilate nitrate, the concentrations of which increased in rca-silenced plants. Numerous studies have shown that genetic or environmental manipulations that decrease photosynthesis also strongly inhibit nitrate assimilation (Matt et al., 2002). It has been shown that herbivore-induced reductions in photosynthetic rates are greater than the amount of canopy area removed by the herbivore (Zangerl et al., 2002), a result that points to herbivore-induced reductions in RCA protein as a potential explanation for the unexpectedly large decrease in photosynthetic rate in attacked leaves.
In addition to RCA, five other photosynthetic proteins were found to be down-regulated during all harvests after elicitation: three oxygen-evolving enhancer proteins and two oxygen-evolving complex proteins. In contrast, PSI chain II D2 precursors and S2 hydroxy acid oxidase proteins increased after OS elicitation. These results confirm our earlier transcription analysis of N. attenuata-M. sexta interaction, which revealed that most photosynthesis-related genes are down-regulated after herbivore attack (Halitschke et al., 2001; Hui et al., 2003).
The Potential Defensive Role of the Elicited Proteins Involved in Primary Metabolism
Of the 28 primary metabolism proteins that were differentially regulated, eight are involved in amino acid metabolism and were up-regulated in response to OS elicitation: TD, S-adenosylmethionine synthetase, O-acetylserine thiol lyase, spermidine synthase, Gln synthetase, 3-dehydroquinate dehydratase, homo-Cys-S-methyltransferase, and hydroxymethyltransferase. Chen et al. (2005) recently demonstrated a defensive role for tomato (Solanum esculentum) TD, the enzyme that functions as an antinutritional protein in the first committed step in isoleucine biosynthesis by degrading Thr and presumably limiting the availability of this essential amino acid to the feeding insect. It is possible that the other up-regulated amino-acid-related proteins in N. attenuata leaves play a similar role, disturbing the amino acid metabolism of insects and thereby functioning as antinutritional defenses.
A protein spot identified as an ACS had a type III response. Several acs genes reportedly played diverse roles in plant metabolism (Hayashi et al., 2002; Schnurr et al., 2004). A member of the acs gene family (opcl1) in Arabidopsis (Arabidopsis thaliana) is reported to be responsive to wounding and involved in jasmonic acid biosynthesis (Koo et al., 2006). Jasmonic acid is a well-characterized lipid-derived signal molecule that regulates defense-related processes in plants. The acs gene is also expressed in the epidermal layers of young and rapidly growing leaves of Arabidopsis, suggesting that the ACS enzyme may act to synthesize cutin or cuticular waxes (Hayashi et al., 2002; Schnurr et al., 2004). Cutin-reinforced leaves could decrease the palatability of leaves for insects. Another protein similar to glyceraldehyde-3-P dehydrogenase protein exhibited differential accumulation in elicited leaves. Pathogen elicitation of maize (Zea mays) cell suspension cultures elicits the accumulation levels of GAPDH (Chivasa et al., 2005) in Arabidopsis and is known to be regulated by redox (Hancock et al., 2005) and reversibly inhibited by nitric oxide (Lindermayr et al., 2005). These characteristics suggest that in addition to its catalytic role in glycolysis, GAPDH may be involved in reactive oxygen species signaling, which is important in plant-herbivore interactions.
We found proteins similar to TK and plastidic ALD to be differentially regulated by OS elicitation. High levels of accumulation of the TK protein after OS elicitation might provide substrates for the synthesis of phenolic-based defenses. Proteomic analysis of germinating maize embryos infected by Fusarium verticilliodes showed the increased accumulation of ALD proteins (Campo et al., 2004). Gene expression of ALD is regulated by light and environmental factors, and thus might be influenced by photosynthetic activity and/or regulated by other photosynthesis-related genes (Yamada et al., 2000) as we also observed in N. attenuata. Another protein similar to PGM is up-regulated in OS-induced leaves. Expression of the Arabidopsis PGM is known to be regulated by hormones and Glc and induced by sedentary plant-parasitic nematodes (Mazarei et al., 2003; Bourgis et al., 2005). After elicitation, proteins with similarity to L6, L9, and L30 type ribosomal proteins increased, while proteins similar to the L4 and L6 types decreased. Ribosomal proteins contribute to the structures required for protein biosynthesis, as do translation elongation factors, which catalyze the translocation of the two tRNAs and the mRNA after peptidyl transfer to the 80 S ribosome (Jorgensen et al., 2006). In rice, translation elongation factor genes are reported to be inducible in response to several environmental stresses and to abscisic acid treatment (Li et al., 1999). The changes observed in the N. attenuata translation elongation factor protein and several ribosomal proteins suggest a massive reorganization occurs in protein biosynthesis after herbivore damage to leaf tissues.
The Potential Defensive Role of the Elicited Proteins Involved in Secondary Metabolism
Seven out of eight proteins involved in the production of secondary metabolites showed increased accumulation levels at at least one time point after OS elicitation. Levels of a QR-like protein increased after OS elicitation. QRs are multisubunit enzymes that are involved in the generation of free radical semiquinones. Recently, QRs genes have been shown to be up-regulated in the epidermis during powdery mildew infection to wheat (Triticum aestivum; Greenshields et al., 2005). A protein spot similar to dehydroascorbate reductase (DHAR) showed a type I response after OS induction. DHAR functions in the regeneration of ascorbate, a potent antioxidant protecting plants against oxidative damage imposed by environmental stresses. It has been demonstrated in Arabidopsis that ascorbate deficiency can lead to the modification of defense pathways (Kiddle et al., 2003). Another protein similar to 3-dehydroquinate dehydratase, which was up-regulated, is known to be involved in the synthesis of lignin, chlorogenic acid, alkaloids, indole, other aromatic compounds, and tannins (Bischoff et al., 2001). The increased levels of this protein confirm results from our earlier studies, which show that several genes involved in the shikimate pathway are up-regulated in response to herbivory in N. attenuata (Baldwin, 2001). Importantly, a protein with similarity to cinnamyl-alcohol dehydrogenase, which is further downstream of the shikimate pathway, was also up-regulated. This enzyme catalyzes the reduction of phenylpropenyl aldehydes to monolignols, the last monomers before lignin synthesis (Kim et al., 2004). Lignification is an important process that facilitates tissue repair, increasing the toughness of leaves, which in turn inhibits bacterial growth or can make feeding difficult for chewing insects (Chittoor et al., 1997).
A protein spot showing similarity to spermidine synthase is up-regulated from 6 to 72 h after OS elicitation. Spermidine synthase is involved in polyamine metabolism and thereby the production of secondary metabolite defense compounds (Franceschetti et al., 2004). This enzyme also catalyzes the first specific step in the biosynthesis of the tropane and nicotine alkaloids (Franceschetti et al., 2004; Stenzel et al., 2006). High levels of nicotine in N. attenuata leaves in response to herbivory have been demonstrated in several studies (Baldwin, 2001; Steppuhn et al., 2004). Transgenic N. attenuata plants with reduced levels of constitutive or induced nicotine were preferred by insects in choice assays, and, when these plants are grown in their native habitat, they are highly susceptible to several other insect pests (Steppuhn et al., 2004). Another protein similar to S-adenosylmethionine synthetase accumulates in elicited leaves and might be responsible for the supply of adenosylmethionine in ethylene biosynthesis (Urao et al., 2000). The increase in S-adenosylmethionine synthetase corresponds to the dramatic burst of ethylene and nicotine production that occurs when M. sexta larvae feed on N. attenuata leaves (Baldwin, 2001; Steppuhn et al., 2004). Here, we were able to confirm these findings by demonstrating that S-adenosylmethionine synthetase and spermidine synthase proteins accumulate after herbivore attack.
Comparing Changes in mRNA and Protein Accumulation
It is evident from several studies that the proteomic analysis of a plant's response does not necessarily match the transcriptional analysis of the same response. One of the major constraints to linking protein accumulation with mRNA in this study is the presence of several isoforms and/or subunits of certain proteins, especially when candidate proteins belong to a multigene family. The difficulty of differentiating between isoforms when using the RT-PCR approach remains a limitation for comparing protein and transcript accumulation. In the well-studied eukaryotic system of yeast, similar mRNA expression levels were accompanied by a wide range of protein abundance levels and vice versa (Gygi et al., 1999). Chilling rice plants results in changes in the abundance of several mRNA transcripts, which are not reflected in changes in their corresponding proteins (Yan et al., 2006). We compared accumulation levels in leaves of 17 gene transcripts with those of their corresponding proteins at five time points after elicitation with different insect-derived signals. Six genes showed transcript patterns that correlated strongly with the patterns of protein abundance in the time series analysis. We used the Natpi gene for expression analysis, as it is known to respond to OS elicitation and insect damage at both transcript and protein levels (Zavala et al., 2004). For TPI, TD, Gly-rich RNA-binding protein, PGM, RCA, and Gln synthetase, the abundance of mRNA was a rough predictor of protein abundance, a relationship that has been shown for other proteins (Futcher et al., 1999). ald (spot 19) showed an inconsistent pattern of protein accumulation, whereas in its transcript level we found a type II pattern of expression (always down). The mRNA and protein levels of the remaining 10 proteins were differently regulated when compared in the time series analysis after OS elicitation. Comparison of protein and mRNA accumulation after different elicitation treatments identified seven members, namely, tpi, td, pgm, sams, fdh, gpdh, and atpe, which shared similar patterns. For the cpn, protein and mRNA levels were similarly regulated only after larval feeding but not in the other elicitation treatments. pme and ald were not regulated by the different elicitors at the protein level, but all treatments elicited strong increases at the transcript level.
Insect-Derived Signals and Protein Accumulation Patterns in N. attenuata Leaves
The patterns of differential protein accumulation as well as the identity of the elicited proteins provide insights into the nature of the larval elicitors that are involved in regulating plant proteomes. Previous research using a detailed microarray and secondary metabolite analysis demonstrated that the two most abundant FACs in M. sexta OS can account for all measured direct (TPI and nicotine) and indirect (cis-α-bergamotene) defenses, the endogenous jasmonic acid burst that elicits them, and 65% to 86% of the induced transcriptional changes elicited by attack on N. attenuata leaves (Halitschke et al., 2001, 2003; Roda et al., 2004). Here we extend the analysis to the level of the proteome and find that the treatment of wounds with four synthetic FACs accounts for 19 of the 32 proteins elicited by W + OS treatment. Moreover, excluding FACs from OS by ion-exchange chromatography removes the regulation of eight proteins, and treating FAC-free OS elicits only one additional protein, which was not elicited by treatment of the synthetic FACs. It is interesting to note that the response of certain proteins can not be attributed to the presence of FACs (Table II; Fig. 5). For example, RCA spots 20, 46, and 51 exhibit distinctly higher accumulation levels in leaves after treatment with OS and FAC-free OS compared to levels in leaves after treatment with synthetic FACs, suggesting that unknown factors in OS modulate the elicitation activity of FACs.
FACs are plant defense elicitors that are synthesized in the larval midgut and elicit defense responses in the plant that appear to benefit the plant. However, other elicitors are produced by the plant during caterpillar attack and some of these may benefit the caterpillar. Recent work from our group has shown that treatment of wounds with OS and larval attack dramatically increases methanol (MeOH) emissions from attacked plants. This MeOH release is elicited not by the FACs in OS but, rather, by the high pH of OS (pH 9.3), and is associated with increases in transcripts and activity of leaf PME and decreases in the degree of pectin methylation (von Dahl et al., 2006). The OS elicitation of PME transcripts and activity is consistent with the hypothesis that methylated pectin accounts for the MeOH emission (von Dahl et al., 2006). The results of this study confirm these conclusions and extend the analysis to the levels of PME protein, which we found to increase after wounding, and to the application of OS at 30 and 72 h after elicitation (Table I). This MeOH emission results in decreased TPI levels, but how this works has not been discovered; the decreased TPIs enhance larval performance (von Dahl et al., 2006) and it appears that the feeding larvae generate a beneficial signal by exploiting an indispensable feature of cell wall pectin chemistry essential for plant growth.
In conclusion, FACs play a major role in organizing not only transcriptional responses but also proteomic responses. This dual role is not simply a result of an overlap of the transcriptional and proteomic responses to FACs because the responses of the transcriptome and proteome are clearly very different.
MATERIALS AND METHODS
Plant Material and Preparation of Elicitors
A 17th-generation inbred line of Nicotiana attenuata (ecotype Utah), originally collected from a natural population in Washington County, Utah, was used for all experiments. Seed germination and plant growth were conducted as described earlier (Halitschke et al., 2003). All elicitation experiments were conducted with plants in the rosette stage of growth. Manduca sexta eggs were collected from Lytle Preserve in Utah and bred in our laboratory. To elicit plants, freshly hatched first-instar caterpillars reared on N. attenuata wild-type plants were released on leaves growing at particular positions. OS were collected from fourth- and fifth-instar larvae. Collected OS were stored at −20°C under argon and diluted 1:1 (v/v) with sterile water prior to use (Halitschke et al., 2003).
The FAC solution was prepared as described earlier (Halitschke et al., 2001). To obtain OS without FACs (OS-FAC-free), OS were separated on ion-exchange Ultrafree-MC columns (Millipore) that were filled with 300 mg of basic anion-exchange Amberlite IRA-400 (Sigma). The removal of FACs from OS was verified by atmospheric pressure chemical ionization-HPLC-MS analysis using a Phenomenex C18(2) Luna, particle size 5 μm, 250- × 4.6-mm HPLC-separation column (Phenomenex) as described earlier (Halitschke et al., 2001).
Elicitation Experiments
Source leaves +1, +2, and +3 of rosette-stage plants of N. attenuata (Fig. 1) were punctured parallel to the central leaf vein with a fabric pattern wheel six times at 30-min intervals. Induction treatments were started between 9 and 10 am and performed every 30 min for 3 h. Around 20 μL of 1:1 water-diluted OS or OS-FAC-free or FACs (in 0.0025% Triton X-100) or sterile water or 0.0025% Triton X-100 were applied to the punctured leaves (Halitschke et al., 2001, 2003). Local leaves were harvested for protein and RNA extraction after 6, 12, 30, 48, and 72 h. Systemic leaf samples (leaves −1, −2, and −3) were collected 30 h after the initial induction treatment (Fig. 1). For the caterpillar-feeding treatments, 15 M. sexta neonate larvae (five larvae per leaf) were released on +1, +2, and +3 leaves and their feeding patterns were monitored frequently to ensure that only the designated leaves were attacked. Damaged leaves were harvested after 30 and 48 h. All harvested tissues were frozen in liquid nitrogen and stored at −80°C prior to protein extraction. The complete experiment was repeated at least three times with a minimum of three biological replicates.
Protein Extraction, Separation by 2-DE, and Analysis of Protein Spots
Total protein was extracted from elicited and control leaves with a modified phenolic extraction procedure (Schuster and Davies, 1983; Saravanan and Rose, 2004). For nuclear protein extraction, the ReadyPrep protein extraction kit was used according to the manufacturer's instructions (Bio-Rad). Protein quantification was carried out with a 2-DE Quant kit according to the manufacturer's instructions (GE Healthcare Bio-Sciences AB). For first-dimension separation, 24-cm IPG strips pH3-10 NL (Bio-Rad) were rehydrated with 550 μg (410 μL of rehydration buffer) of protein for 11 to 12 h at room temperature. The proteins were focused on a Protean IEF Cell (Bio-Rad) at 20°C, rapid voltage ramping, 63,000 volt h, and 50 μA current per IPG strip. Prior to second-dimension separation, the IPG strips were equilibrated with a DTT buffer (3.6 g urea, 5 mg dithiothreitol, 1 mL glycerol, 4.4 mL 10% SDS, and 1 mL 0.6 m Tris-HCl buffer, pH 6.8, in 10 mL) followed by an iodoacetamide buffer (similar to DTT buffer, with 0.25 g iodoacetamide instead of DTT), 20 min each at room temperature on a shaker. SDS-PAGE for second-dimension separation was performed with 12% gels and the Ettan Daltsix electrophoresis unit (GE Healthcare Bio-Sciences AB) at 25°C and 17 W/gel. The gels were stained with Bio-Safe Coomassie G-250 (Bio-Rad) according to the manufacturer's instructions. Gel images were created using a GS-800 calibrated densitometer (Bio-Rad). Spot detection, indexing, matching, normalization, and quantitation were done using the PDQuest Version 7.3.1 software (Bio-Rad). Protein spots of interest were excised and used for tryptic protein digestion. The P values were calculated with at least three 2-DE images of the corresponding biological replicates. The ratios of the relative spot quantities of the induced 2-DE images between changing spots and an unchanged reference spot from the same 2-DE image were calculated and compared with a two-tailed and paired t test against the relative spot quantity ratios from the control images for the respective treatments (Table II).
Protein Digestion for MS Analysis
Proteins of interest were processed on 96-well microtiter plates with an Ettan TA Digester running the Digester Version 1.10 software (both GE Healthcare Bio-Sciences AB) using the following protocol. The excised gel plugs were washed four times with 70 μL of acetonitrile/50 mm ammonium bicarbonate for 20 min each. The second wash was performed twice with 70 μL of 70% acetonitrile for 20 min and the gel plugs were air-dried for 1 h. Trypsin digestion was carried out overnight with 50 ng of trypsin (Porcine trypsin; Promega) in 15 μL of 50 mm ammonium bicarbonate at 37°C. The 15-μL solution from the preceding step was mixed with 25 μL of extraction buffer (50% acetonitrile and 0.1% trifluoroacetic acid), incubated for 20 min, and transferred to a 96-well plate. The gel plug was then incubated with 40 μL of extraction buffer for 20 min and transferred to the plate. The 60-μL solution containing the peptide mixture was then vacuum-dried for 1 h.
MALDI-TOF and LC-MS/MS Analysis of Protein Spots
Dry peptides were dissolved in 15 μL of aqueous 0.1% trifluoroacetic acid. One μL aliquot was mixed with 1 μL of α-cyano-4-hydroxycinnamic acid (alpha matrix, 10 mg/mL in ethanol:acetonitrile, 1:1 [v/v]), and 1 μL of this mixture was transferred onto a metal 96-spot MALDI target plate for cocrystallization. A MALDImicro MX mass spectrometer (Waters) was used in reflectron mode to analyze the tryptic peptides. A strong electrical field was created to accelerate the sample ions into the flight tube toward the detector (5 kV on the sample table, −12 kV on the extraction grid, pulse voltage of 1.95 kV, and 2.35-kV detector voltages). A nitrogen laser (337 nm, 5 Hz) was used for ionization, with energies of approximately 50 μJ per pulse. MassLynx Version 4.0 software served for data acquisition (Waters). Each spectrum was combined from 10 laser pulses. Human Glu-Fibrinopeptide B (1,570.6774 D) served as an external lock-mass reference. Bovine serum albumin tryptic digest was used to calibrate the mass spectrometer (MPrep; Waters). The MALDI-TOF spectra searches were performed in the PLGS Version 2.1.5 software (Waters) using the Green Plant Version 1.0 database of the NCBI. The search parameters were as follows: peptide tolerance of 80 ppm, one missed cleavage, carbamidomethyl modification of cysteins, and possible Met oxidation. An estimated calibration error of 0.05 D and a minimum of four peptide matches were the criteria for obtaining positive database hits. The MALDI-TOF peptide signal intensities were used to estimate the injection amount in the subsequent peptide sequence analysis.
The peptides, redissolved in aqueous 0.1% trifluoroacetic acid, were subsequently used for nanoLC-MS/MS analysis. The peptides were separated on a CapLC XE nanoLC system (Waters). A mobile phase flow of 0.1% aqueous formic acid (20 μL/min for 5 min) was used to concentrate and desalt the samples on a 5- × 0.35-mm Symmetry-300 C18 precolumn with 5-μm particle size. The samples were eluted and separated on a 150-mm × 75-μm NanoEase Atlantis C18 column, particle size 3 μm, using an increasing acetonitrile gradient (in 0.1% aqueous formic acid). Phases A (5% MeCN in 0.1% formic acid) and B (95% MeCN in 0.1% formic acid) were linearly mixed using a gradient program set to 5% phase B in A for 5 min, increased to 40% B in 25 min, and to 60% A in 10 min, and finally increased to 95% B for 4 min. The peptides were directly transferred to the NanoElectroSpray source of a Q-TOF Ultima tandem mass spectrometer through a Teflon capillary union and a metal-coated nanoelectrospray tip (Picotip; 50 × 0.36 mm; 10 μm i.d.; Waters). The source temperature was set to 40°C, cone gas flow 50 L/h, and the nanoelectrospray voltage was 1.6 kV. The TOF analyzer was used in reflectron mode. The MS/MS spectra were collected at 0.9-s intervals in the range of 50 to 1,700 m/z. A mixture of 100 fmol/μL human Glu-Fibrinopeptide B and 80 fmol/μL reserpine in 0.1% formic acid/acetonitrile (1:1 v/v) was infused at a flow rate of 0.9 μL/min through the reference NanoLockSpray source every fifth scan to compensate for mass shifts in the MS and MS/MS fragmentation mode due to temperature fluctuations. The data were collected by MassLynx Version 4.0 software. ProteinLynx Global Server Browser Version 2.1.5 (RC7) software (both from Waters) was used for further data processing (deconvolution, baseline subtraction, smoothing), de novo sequence identification, and database searches. The MS/MS data were searched against the NCBI Green Plant Version 1.0 database with the following parameters: peptide tolerance of 20 ppm, fragment tolerance of 0.05 D, estimated calibration error of 0.005 D, one missed cleavage, carbamidomethylation of cysteins, and the possible oxidation of Met. The amino acid sequences of peptides that did not provide conclusive results from the database searches were searched using MS-BLAST Internet (http://www.dove.embl-heidelberg.de) and an in-house-based search engine. Consolidated database (sp_nrdb) containing all nonredundant protein databases (Swiss-Prot, Swiss-ProtNew, SptremblNew, Sptrembl) and PAM30MS matrix with default settings were used for searches (Shevchenko et al., 2001).
RT-PCR Analysis
The total RNA was extracted from local leaves as described in the TRI reagent protocol (Sigma). From all the samples, 1 μg of total RNA was converted to cDNA using a Superscript first-strand synthesis system for RT-PCR according to the manufacturer's instructions (Invitrogen).
Primers were designed from the peptide sequences obtained after LC-MS/MS analysis. Respective sites of the peptides were mapped on nucleotide regions for the respective database entries. Closely related species of N. attenuata were used for primer design when the sequence information of particular genes was unavailable. The primer design scheme is provided in Supplemental Table S1. The location of the peptides within the protein sequence is provided in Supplemental Figure S5.
Two concentrations of cDNA (viz. 1×, 5× or 50×, 250× dilutions of original cDNA derived from 1 μg of RNA) derived from control and induced leaves were used as a template to amplify the respective cDNA fragment with the above primer combinations. PCR conditions were optimized for each primer set. PCR was carried out after denaturing cDNAs at 94°C for 5 min and then 30 cycles of 94°C for 60 s, annealing temperature (see Supplemental Table S1) for 30 s, and extension at 72°C for 60 s. The final extension step in PCR was at 72°C for 10 min. The amplified cDNA fragments were purified from agarose gel using GFX gel elution kit (Amersham) and cloned into PGMT-Easy vector (Promega). Cloned fragments were sequenced and confirmed with the NCBI database and novel sequences were deposited in the database.
Narca Gene Silencing and Analysis
The previously reported N. attenuata 268-nucleotide rca sequence (BU 494545) was used to make the Tobacco rattle virus-based construct for VIGS as described earlier (Saedler and Baldwin, 2004). Three-week-old N. attenuata plants were inoculated with Agrobacterium tumefaciens, transformed with PTV-rca or PTV-phytoene desaturase (pds) or EV (PTV00) constructs (Saedler and Baldwin, 2004; Rayapuram and Baldwin, 2006). Plants were grown in growth chambers at 22°C for 2 to 3 weeks until leaves of pds-transformed plants showed bleaching, a symptom of VIGS (Saedler and Baldwin, 2004). Total RNA from EV and rca-VIGS plants was extracted and analyzed for rca expression using RT-PCR. Induction to +2 leaves with W + OS was carried out with five replicate plants per treatment as described in earlier sections. Protein extraction and 2-DE analyses of the leaf were carried out and compared with EV-transformed plants.
Five-week-old plants were used for photosynthesis and nitrate measurements. Net photosynthetic rates and intercellular CO2 concentrations were measured on empty vector and Narca VIGS-silenced plants under saturating light (approximately 1,400 μm m−2 s−1) using a LI-COR 6400 portable photosynthesis system. Net photosynthesis was measured in at least five replicate plants each at six different CO2 concentrations, namely, 800, 600, 400, 200, 100, and 50 μmol/mol. Nitrate was measured spectrophotometrically according to the protocol described by Cataldo et al. (1975). Seven-week-old plants were harvested and dried in the oven at 60°C for 48 h and dry weights of the plants were measured.
Accession Numbers
New sequences of putative partial genes of N. attenuata cloned and characterized in this study were deposited in the public database NCBI. Accession numbers allotted to these genes are as follows (in parentheses): N. attenuata (Na) ATPase, Naatpe (DQ682456); chaperonin, Nacpn (DQ682457); Gln synthase, Nagmps (DQ682458); glyceraldehydes-3-P dehydrogenase, Nagpdh (DQ682459); Gly-rich RNA-binding protein, Nagrp (DQ682460); Mg-protoporphyrin IX chelatase, NaMgpc (DQ682461); oxygen-evolving protein, Naoep (DQ682462); RCA, Narca (DQ682463); S-adenosylmethionine synthetase, Nasmas (DQ682464); translation elongation factor, Natef (DQ682465); PGM, Napgam (DQ682466); ALD, Naald (DQ682467); TK, Natk (DQ682468); mitochondrial formate dehydrogenase, Nafdh (DQ885565); and PME, Napme (DQ885566).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. 2-DE analysis of nuclear proteins.
Supplemental Figure S2. Proteomic maps of two-dimensionally separated control and elicited leaf proteins 6 and 12 h after elicitation.
Supplemental Figure S3. Proteomic maps of two-dimensionally separated control and elicited leaf proteins 48 and 72 h after elicitation.
Supplemental Figure S4. Two-dimensional gel analysis of control and elicited leaf proteins.
Supplemental Figure S5. Location of peptides obtained from protein spots on identified database protein sequence.
Supplemental Table S1. Primers used for RT-PCR analysis.
Acknowledgments
We thank Klaus Gase, Thomas Hahn, and Albrecht Berg for DNA sequencing, providing chemically synthesized FACs, and HPLC analysis; Tamara Krügel and Andreas Weber for growing the plants; and Emily Wheeler for editorial assistance. We also thank Markus Hartl for critically reading the manuscript and Nan Qu for assistance in the preparation of the nuclear extracts and for helpful discussions.
This work was supported by the Max Planck Society. A.P.G. acknowledges the Alexander von Humboldt Foundation, Bonn, for a research fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ian T. Baldwin (baldwin@ice.mpg.de).
The online version of this article contains Web-only data.
References
- Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422: 198–207 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Thatcher LF, Singh KB (2005) Plant defence responses: conservation between models and crops. Funct Plant Biol 32: 21–34 [DOI] [PubMed] [Google Scholar]
- Baldwin IT (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol 127: 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Snyder M (2005) Prospects and challenges in proteomics. Plant Physiol 138: 560–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Schaller A, Bieri F, Kessler F, Amrhein N, Schmid J (2001) Molecular characterization of tomato 3-dehydroquinate dehydratase-shikimate:NADP oxidoreductase. Plant Physiol 125: 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43: 545–580 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Botha FC, Mani S, Hiten FN, Rigden DJ, Verbruggen N (2005) Characterization and functional investigation of an Arabidopsis cDNA encoding a homologue to the d-PGMase superfamily. J Exp Bot 56: 1129–1142 [DOI] [PubMed] [Google Scholar]
- Campo S, Carrascal M, Coca M, Abian J, San Segundo B (2004) The defense response of germinating maize embryos against fungal infection: a proteomics approach. Proteomics 4: 383–396 [DOI] [PubMed] [Google Scholar]
- Cataldo DA, Haroon M, Schrader LE, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant-tissue by nitration of salicylic-acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102: 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittoor JM, Leach JE, White FF (1997) Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 10: 861–871 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Simon WJ, Yu XL, Yalpani N, Slabas AR (2005) Pathogen elicitor-induced changes in the maize extracellular matrix proteome. Proteomics 5: 4894–4904 [DOI] [PubMed] [Google Scholar]
- Franceschetti M, Fornale S, Tassoni A, Zuccherelli K, Mayer MJ, Bagni N (2004) Effects of spermidine synthase overexpression on polyamine biosynthetic pathway in tobacco plants. J Plant Physiol 161: 989–1001 [DOI] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI (1999) A sampling of the yeast proteome. Mol Cell Biol 19: 7357–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinski M, Weckwerth W (2006) The role of mass spectrometry in plant systems biology. Mass Spectrom Rev 25: 173–214 [DOI] [PubMed] [Google Scholar]
- Greenshields DL, Liu GS, Selvaraj G, Wei YD (2005) Differential regulation of wheat quinone reductases in response to powdery mildew infection. Planta 222: 867–875 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Bednarek P, Ciolkowski I, Hamberger B, Heise A, Liedgens H, Logemann E, Nurnberger T, Schmelzer E, Somssich IE, et al (2003) Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc Natl Acad Sci USA 100: 14569–14576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Gase K, Hui DQ, Schmidt DD, Baldwin IT (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VI. Microarray analysis reveals that most herbivore-specific transcriptional changes are mediated by fatty acid-amino acid conjugates. Plant Physiol 131: 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125: 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ (2005) Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43: 828–835 [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Hayashi Y, Nito K, Kato A, Hayashi M, Hara-Nishimura I, Nishimura M (2002) Molecular characterization of an Arabidopsis acyl-coenzyme A synthetase localized on glyoxysomal membranes. Plant Physiol 130: 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol 125: 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DQ, Iqbal J, Lehmann K, Gase K, Saluz HP, Baldwin IT (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. V. Microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol 131: 1877–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Merrill AR, Andersen GR (2006) The life and death of translation elongation factor 2. Biochem Soc Trans 34: 1–6 [DOI] [PubMed] [Google Scholar]
- Kang J, Wang L, Giri AP, Baldwin IT (2006) Silencing threonine deaminase and the JAR1 homologue in Nicotiana attenuata impairs JA-isoleucine-mediated defenses against the specialist herbivore, Manduca sexta. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53: 299–328 [DOI] [PubMed] [Google Scholar]
- Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, Foyer CH (2003) Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana. Antioxid Redox Signal 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4: 3569–3578 [DOI] [PubMed] [Google Scholar]
- Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol 57: 315–332 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Chung HS, Kobayashi Y, Howe GA (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2005) Comprehensive post-genomic data analysis approaches integrating biochemical pathway maps. Phytochemistry 66: 413–451 [DOI] [PubMed] [Google Scholar]
- Leitner M, Boland W, Mithofer A (2005) Direct and indirect defences induced by piercing-sucking and chewing herbivores in Medicago truncatula. New Phytol 167: 597–606 [DOI] [PubMed] [Google Scholar]
- Li ZY, Zhang JS, Chen SY (1999) Molecular clotting, expression analysis and chromosomal mapping of salt-responsive cDNAs in rice (Oryza sativa L.). Sci China C Life Sci 42: 506–516 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manter DK, Kerrigan J (2004) A/Ci curve analysis across a range of woody plant species: influence of regression analysis parameters and mesophyll conductance. J Exp Bot 55: 2581–2588 [DOI] [PubMed] [Google Scholar]
- Matt P, Krapp A, Haake V, Mock HP, Stitt M (2002) Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J 30: 663–677 [DOI] [PubMed] [Google Scholar]
- Mazarei M, Lennon KA, Puthoff DP, Rodermel SR, Baum TJ (2003) Expression of an Arabidopsis phosphoglycerate mutase homologue is localized to apical meristems, regulated by hormones, and induced by sedentary plant-parasitic nematodes. Plant Mol Biol 53: 513–530 [DOI] [PubMed] [Google Scholar]
- Peck SC (2005) Update on proteomics in Arabidopsis. Where do we go from here? Plant Physiol 138: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis AR (1995) The regulation of Rubisco by Rubisco activase. J Exp Bot 46: 1285–1291 [DOI] [PubMed] [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al (2006. a) Genomics of hybrid poplar (Populus trichocarpa x deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Ralph SG, Yueh H, Friedmann M, Aeschliman D, Zeznik JA, Nelson CC, Butterfield YSN, Kirkpatrick R, Liu J, Jones SJM, et al (2006. b) Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell Environ 29: 1545–1570 [DOI] [PubMed] [Google Scholar]
- Rayapuram C, Baldwin IT (2006) Using nutritional indices to study LOX3-dependent insect resistance. Plant Cell Environ 29: 1585–1594 [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A, Halitschke R, Steppuhn A, Baldwin IT (2004) Individual variability in herbivore-specific elicitors from the plant's perspective. Mol Ecol 13: 2421–2433 [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55: 151–157 [DOI] [PubMed] [Google Scholar]
- Saravanan RS, Rose JKC (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4: 2522–2532 [DOI] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol 125: 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt DD, Voelckel C, Hartl M, Schmidt S, Baldwin IT (2005) Specificity in ecological interactions. Attack from the same lepidopteran herbivore results in species-specific transcriptional responses in two solanaceous host plants. Plant Physiol 138: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster AM, Davies E (1983) Ribonucleic acid and protein metabolism in pea epicotyls. I. The aging process. Plant Physiol 73: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Sunyaev S, Loboda A, Shevehenko A, Bork P, Ens W, Standing KG (2001) Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time of flight mass spectrometry and BLAST homology searching. Anal Chem 73: 1917–1926 [DOI] [PubMed] [Google Scholar]
- Stenzel O, Teuber M, Drager B (2006) Putrescine N-methyltransferase in Solanum tuberosum L., a calystegine-forming plant. Planta 223: 200–212 [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT (2004) Nicotine's defensive function in nature. PLoS Biol 2: 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57: 755–766 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K (2000) Two-component systems in plant signal transduction. Trends Plant Sci 5: 67–74 [DOI] [PubMed] [Google Scholar]
- Vargas-Suarez M, Ayala-Ochoa A, Lozano-Franco J, Garcia-Torres I, Diaz-Quinonez A, Ortiz-Navarrete VF, Sanchez-de-Jimenez E (2004) Rubisco activase chaperone activity is regulated by a post-translational mechanism in maize leaves. J Exp Bot 55: 2533–2539 [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Havecker M, Schlogl R, Baldwin IT (2006) Caterpillar-elicited methanol emission: a new signal in plant-herbivore interactions? Plant J 46: 948–960 [DOI] [PubMed] [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19: 195–216 [DOI] [PubMed] [Google Scholar]
- Yamada S, Komori T, Hashimoto A, Kuwata S, Imaseki H, Kubo T (2000) Differential expression of plastidic aldolase genes in Nicotiana plants under salt stress. Plant Sci 154: 61–69 [DOI] [PubMed] [Google Scholar]
- Yan S-P, Zhang Q-Y, Tang Z-C, Su W-A, Sun W-N (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5: 484–496 [DOI] [PubMed] [Google Scholar]
- Zangerl AR, Hamilton JG, Miller TJ, Crofts AR, Oxborough K, Berenbaum MR, de Lucia EH (2002) Impact of folivory on photosynthesis is greater than the sum of its holes. Proc Natl Acad Sci USA 99: 1088–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT (2004) Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol 134: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]