Abstract
The mechanisms controlling seed dormancy in Arabidopsis (Arabidopsis thaliana) have been characterized by proteomics using the dormant (D) accession Cvi originating from the Cape Verde Islands. Comparative studies carried out with freshly harvested dormant and after-ripened non-dormant (ND) seeds revealed a specific differential accumulation of 32 proteins. The data suggested that proteins associated with metabolic functions potentially involved in germination can accumulate during after-ripening in the dry state leading to dormancy release. Exogenous application of abscisic acid (ABA) to ND seeds strongly impeded their germination, which physiologically mimicked the behavior of D imbibed seeds. This application resulted in an alteration of the accumulation pattern of 71 proteins. There was a strong down-accumulation of a major part (90%) of these proteins, which were involved mainly in energetic and protein metabolisms. This feature suggested that exogenous ABA triggers proteolytic mechanisms in imbibed seeds. An analysis of de novo protein synthesis by two-dimensional gel electrophoresis in the presence of [35S]-methionine disclosed that exogenous ABA does not impede protein biosynthesis during imbibition. Furthermore, imbibed D seeds proved competent for de novo protein synthesis, demonstrating that impediment of protein translation was not the cause of the observed block of seed germination. However, the two-dimensional protein profiles were markedly different from those obtained with the ND seeds imbibed in ABA. Altogether, the data showed that the mechanisms blocking germination of the ND seeds by ABA application are different from those preventing germination of the D seeds imbibed in basal medium.
After reaching physiological maturity, seeds of many plant species, including Arabidopsis (Arabidopsis thaliana), may enter in a state of deep dormancy during which, upon sowing, the dormant (D) seeds will not germinate or germinate only very slowly compared to the corresponding non-dormant (ND) seeds. Seed dormancy is considered as an adaptive trait that enables seeds to remain quiescent until the conditions for germination become favorable (Finch-Savage and Leubner-Metzger, 2006). In mature seeds, the break of dormancy may either occur gradually in the dry state (after-ripening) or be initiated by imbibition under defined conditions (e.g. cold stratification or chilling at low temperature; Karssen, 1982; Baskin and Baskin, 1998; Koornneef et al., 2002; Donohue et al., 2005; Gubler et al., 2005).
The phytohormone abscisic acid (ABA) has been shown to influence to a large extent both the establishment and the maintenance of dormancy (Wareing and Saunders, 1971; Koornneef et al., 2002). For example, Arabidopsis mutants impaired in ABA biosynthesis or in ABA response during seed development produce seeds that lack dormancy compared to wild type (Koornneef et al., 1982; Léon-Kloosterziel et al., 1996). Also, Okamoto et al. (2006) showed that seeds of the Arabidopsis cyp707a1 and cyp707a2 mutants of the Columbia (Col) accession, which are altered in ABA catabolism, accumulate the phytohormone at higher levels and exhibit enhanced dormancy compared to the wild type. ABA has also been shown to play an essential role in the expression of dormancy in imbibed seeds of several species, as Arabidopsis (Garciarrubio et al., 1997; Jullien et al., 2000; Ali-Rachedi et al., 2004), sunflower (Helianthus annuus; Garello et al., 2000), sweet scented tobacco (Nicotiana plumbaginifolia; Grappin et al., 2000), barley (Hordeum vulgare; Wang et al., 1995; Jacobsen et al., 2002), and lettuce (Lactuca sativa; Toyomasu et al., 1994; Yoshioka et al., 1998). In agreement with this, the application of physiological concentrations of exogenous ABA to ND seeds is well known to inhibit their germination (Milborrow, 1974; Garciarrubio et al., 1997; Gonai et al., 2004), whereas germination of ABA-insensitive Arabidopsis mutants (abi1 to abi5) remains unaffected by such treatment (Koornneef et al., 1984; Finkelstein, 1994). It appears that exogenous ABA can inhibit various hydrolytic enzymes presumed to be involved in weakening of the structures surrounding the embryo, thus facilitating radicle protrusion (Leubner-Metzger et al., 1996; Bewley, 1997). However, exogenous ABA can also inhibit Arabidopsis seed germination by restricting the availability of energy and metabolites, thus limiting the growth potential of the embryo (Garciarrubio et al., 1997). In this context, application of exogenous ABA to turnip (Brassica rapa) seeds was shown to result in reduced activity of isocitrate lyase, a key enzyme of the glyoxylate cycle (Finkelstein and Lynch, 2000). The importance of this cycle, and more generally, of storage lipid mobilization for seedling establishment in Arabidopsis has been well documented (Footitt et al., 2006; Penfield et al., 2006).
Global expression profiling approaches have recently been used to investigate the mechanisms of dormancy release and of germination inhibition in the presence of exogenous ABA. Thus, a cDNA-amplified fragment length polymorphism (AFLP) approach revealed that at least 1,000 cDNA fragments are differentially accumulated in D and ND tobacco seeds and, in particular, that dormancy release entails an up-regulation of numerous genes involved in protein synthesis (Bove et al., 2005). Also, a recent microarray study carried out with the Cape Verde Islands (Cvi) accession of Arabidopsis showed that gene expression associated with protein synthesis is greatly reduced in D seeds (Cadman et al., 2006). For the Arabidopsis Landsberg erecta (Ler) accession, microarray experiments on 4-week-old seedlings disclosed that exogenous ABA regulates the expression of 1,354 genes, among which many encode signal transduction components, ribosomal proteins, and proteins involved in regulated proteolysis (Hoth et al., 2002).
The commonly used Arabidopsis accessions such as Ler and Col only exhibit a very low level of dormancy. However, the Cvi accession collected by Lobin (1983) can display a much deeper dormancy (Alonso Blanco et al., 2003; Ali-Rachedi et al., 2004). In agreement with a role of endogenous ABA in control of dormancy, D Cvi seeds have been shown to contain a higher amount of ABA than corresponding after-ripened ND seeds. Furthermore, during the first hours of imbibition, the ABA content decreased at a much faster rate in ND seeds than in D seeds (Ali-Rachedi et al., 2004).
All these results point to a role of ABA in maintenance of seed dormancy, notably in imbibed seeds following shedding. However, the molecular and biochemical mechanisms involved remain poorly documented. We decided to investigate this question by proteomics. This type of approach brings robust information about the biological functions affected in physiological changes (Pandey and Mann, 2000). In this study, from a comparison of D and ND seeds of the Cvi accession, either under the dry mature state or during imbibition in the presence or absence of ABA, we revealed several processes affected by dormancy and/or ABA. We then determined whether the mechanisms involved in germination inhibition of ND seeds in the presence of exogenous ABA differed from those maintaining dormancy in imbibed D seeds. It appears that inhibition of germination of ND seeds in the presence of exogenous ABA is mediated by regulated proteolysis rather than by inhibition of de novo protein synthesis.
RESULTS
Comparison of Seed Protein Patterns in Cvi and Ler Accessions of Arabidopsis
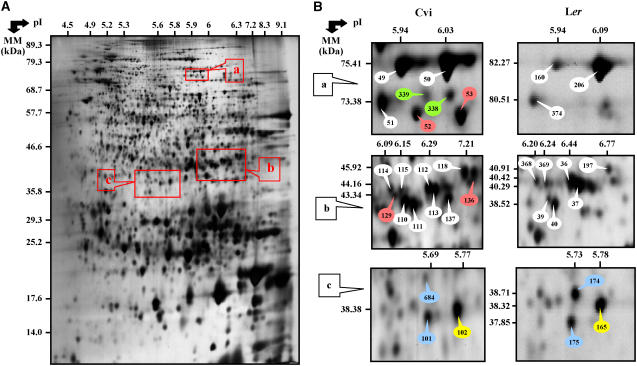
In this study, we used the Cvi accession of Arabidopsis as a model since it exhibits a marked dormancy phenotype (Koornneef et al., 1984; Ali-Rachedi et al., 2004). However, to take advantage of previous proteomic analyses carried out with seeds of the Ler accession (Gallardo et al., 2001, 2002; Job et al., 2005; Rajjou et al., 2006a), we first compared two-dimensional (2D) electrophoretic seed protein patterns from the two accessions. By silver nitrate staining of 2D gels for total soluble protein extracts from the dry mature seeds, 61% of the detected Ler protein spots (704 proteins) exactly matched Cvi protein spots (Fig. 1; Table I). Most importantly, this analysis disclosed a subset of 61 spots that have been independently identified by mass spectrometry (MS) analyses both in Cvi and Ler (see Supplemental Tables S3 and S5). Among them, 54 exhibited the same gene identity in the two accessions (Table I; Supplemental Tables S3 and S5). Furthermore, among the seven proteins assigned to different gene identities in Ler and Cvi, three proteins corresponded to similar functions (Table I; Supplemental Tables S3 and S5). Altogether, these data documented a good conservation in protein sequence from the two genetic backgrounds, a property that was used further in this study to identify proteins in Cvi from those already characterized in Ler (the Ler seed proteome contains 475 identified proteins; Rajjou et al., 2006a).
Figure 1.
Comparison of the seed proteome of the Cvi and Ler accessions. A, A silver nitrate-stained 2D gel of total soluble proteins from dry ND Cvi seeds. The indicated portions of the gel (a, b, and c) are reproduced in B. B, Enlarged windows (a–c) of 2D gels as shown in A for dry Cvi seeds (left) and dry Ler seeds (right). The reference numbers assigned to protein spots for Cvi patterns (left) are reference protein numbers used in this study, while the reference numbers assigned to protein spots for Ler patterns (right) are reference protein numbers from Ler proteome database (Rajjou et al., 2006a; http://seed.proteome.free.fr). The labeled proteins (see Supplemental Tables S3 and S5), numbers 49 (Cvi), 50 (Cvi), 160 (Ler), and 206 (Ler) corresponded to Met synthase; numbers 51 (Cvi) and 374 (Ler) to glycyl tRNA synthetase; numbers 52 and 53 (Cvi) to low temperature induced protein; numbers 338 and 339 (Cvi) to nonidentified proteins; numbers 110 (Cvi), 111 (Cvi), 112 (Cvi), 113 (Cvi), 114 (Cvi), 115 (Cvi), 129 (Cvi), 39 (Ler), 40 (Ler), 368 (Ler), 369 (Ler), 36 (Ler), and 37 (Ler) to cytosolic GAPDH; numbers 118 (Cvi) and 197 (Ler) to 1,6-Fru bisphosphate aldolase; number 119 (Cvi) to Asp aminotransferase; number 136 (Cvi) to hypothetical protein; numbers 101 (Cvi) and 684 (Cvi) to nonidentified proteins; number 102 (Cvi) to malate dehydrogenase; numbers 174 (Ler) and 175 (Ler) to O-acetylserine(thiol) lyase; and number 165 (Ler) to enoyl-[ACP]-carrier protein reductase. The Ler proteins were previously identified by MS (matrix-assisted laser-desorption ionization time of flight [MALDI-TOF] and liquid chromatography coupled with tandem MS) and are listed in Rajjou et al. (2006a; Supplemental Table S3). The Cvi proteins were identified by MALDI-TOF analysis during this study (Supplemental Table S5). The protein spots are labeled in white for matching proteins identified in Ler and Cvi, in red for identified Cvi specific protein spots, in green for unidentified Cvi specific protein spots, in blue for matching spots identified in Ler but not in Cvi, and in yellow for matching spots identified in Cvi and Ler with different genes and functional identity.
Table I.
Comparison between protein patterns of Cvi and Ler accessions
| Cvi | Ler | Common to Both Accessions | |
|---|---|---|---|
| Number of detected protein spots | 978 | 1,142 | 704 |
| Number of identified proteins from the detected protein spots | 124a | 475b | |
| Number of identified proteins from 704 matching protein spots | 92a | 373b | 54ab |
Proteins from Cvi seeds identified by MS analysis in this study (see Supplemental Tables S3 and S5).
Proteins from Ler seeds identified by MS analysis and listed in Rajjou et al. (2006a).
Specific Accumulation of Proteins in D and ND Dry Seeds
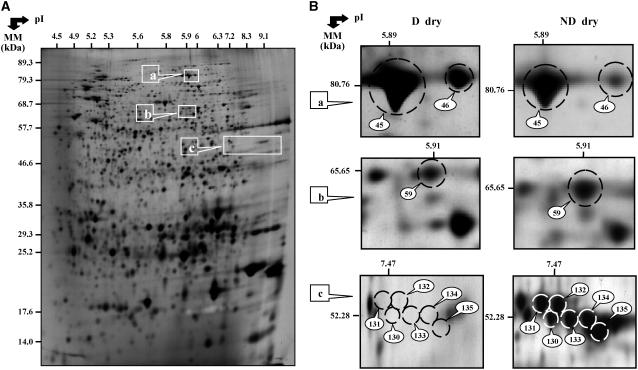
To test for changes in seed protein accumulation levels during after-ripening, we characterized the proteome of the D and ND dry Cvi seeds. This analysis documented differences in the two protein patterns. Thus, from the 978 spots detected in both D and ND seed protein extracts, the accumulation level of 32 proteins (29 identified by MS analysis; see Supplemental Table S5) varied between the two physiological states (Table II). Among them, 22 proteins were up-accumulated in the ND seeds, while seven were down-accumulated in the ND seeds (Table III).
Table II.
Summary of protein variations in dry and imbibed seeds
Data are from 2D gels as shown in Figures 1A and 2A. Numbers of identified proteins are given in parentheses.
| Table No. | Comparisons | Up-Accumulated | Down-Accumulated |
|---|---|---|---|
| III | In dry seeds | 8 | 24 |
| Number of protein changes between 2D protein patterns of D and ND dry seeds (D dry/ND dry) | (7) | (22) | |
| IV | In 1-d imbibed seeds | 15 | 10 |
| Number of protein changes between 2D protein patterns of D and ND seeds imbibed for 1 d in basal medium (D 1 d/ND 1 d) | (15) | (8) | |
| V | Number of protein changes between 2D protein patterns of ND seeds imbibed for 1 d in minimal medium and in the presence of 30 μm ABA (ND 1 d ABA/ND 1 d) | 10 | 61 |
| (7) | (57) |
Table III.
Arabidopsis proteins whose abundance varied significantly between D and ND dry seeds (D dry/ND dry)
AGI, Accession gene identification; Cov., coverage; Exp., Experimental; MM, molecular mass in kD; Theo., theoretical.
| No. | Exp. MM | Exp. pI | Arabidopsis Protein Name and Family | Cov. % | Theo. MM | Theo. pI | AGI No. | Relative Abundanced D Dry/ND Dry |
|---|---|---|---|---|---|---|---|---|
| Storage protein | ||||||||
| 130b | 52.28 | 7.47 | 12S-1 cruciferin precursor | 36 | 50.28 | 7.47 | At5g44120 | ≤0.01 |
| 131b | 52.93 | 7.31 | 12S-1 cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 132c | 52.99 | 7.93 | 12S-1 cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 133c | 52.07 | 8.4 | 12S-1 cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 134c | 53.16 | 8.57 | 12S-1 cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 135c | 52.35 | 9.13 | 12S-1 cruciferin precursor | 42 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 178b | 17.29 | 5.64 | β-Chain of 12S-2 cruciferin | 12 | 20.72 | 6.36 | At1g03880 | ≤0.01 |
| 69b | 61.86 | 6.62 | Similarity to 7S legumin | 24 | 55.06 | 6.64 | At3g22640 | 0.52 ± 0.07 |
| Protease peptidase and hydrolase | ||||||||
| 59b | 65.65 | 5.91 | β-Glucosidase | 26 | 60.02 | 6.2 | At3g21370 | 0.38 ± 0.07 |
| 504a | 45.49 | 5.38 | Mut/nudix family protein (C-terminal portion) | 13 | 45.68 | 5.26 | At1g79690 | 0.50 ± 0.15 |
| Photosynthesis | ||||||||
| 65c | 57.52 | 5.94 | Hypothetical NADP-dependant GAPDH | 11 | 53.06 | 6.23 | At2g24270 | 1.59 ± 0.08 |
| 99c | 45.52 | 5.76 | Chloroplastic GAPDH B-subunit | 20 | 42.80 | 5.6 | At3g55440 | 1.79 ± 0.74 |
| Metabolism and energy | ||||||||
| 854a | 49.11 | 6.02 | 3-Oxoacyl-[acyl-carrier-protein] synthase I, chloroplast | 19 | 50.41 | 8.16 | At5g46290 | ≤0.01 |
| 114b | 43.87 | 6.1 | Cytosolic GAPDH | 28 | 36.91 | 6.62 | At3g04120 | 0.52 ± 0.09 |
| 115c | 43.77 | 6.15 | Cytosolic GAPDH | 27 | 36.91 | 6.62 | At3g04120 | 0.56 ± 0.18 |
| 118b | 45.92 | 7.21 | Probable 1,6-Fru bisphosphate aldolase | 46 | 38.39 | 7.01 | At2g36460 | 0.61 ± 0.29 |
| 123c | 50.37 | 6.26 | Mature 3-oxoacyl-[ACP] synthase I, chloroplastic | 21 | 45.08 | 6.22 | At5g46290 | 0.43 ± 0.36 |
| 76c | 64.44 | 6.69 | Isocitrate lyase | 19 | 64.24 | 6.72 | At3g21720 | 5.56 ± 0.46 |
| Translation protein | ||||||||
| 116c | 42.09 | 6.46 | Initiation factor 3 delta-subunit | 26 | 36.39 | 6.5 | At2g46280 | 0.59 ± 0.14 |
| 45b | 80.76 | 5.89 | Elongation factor EF-2 | 11 | 93.89 | 5.89 | At1g56070 | 1.45 ± 0.74 |
| 46b | 81.11 | 5.89 | Elongation factor EF-2 | 23 | 93.38 | 5.89 | At1g56070 | 2.08 ± 3.34 |
| 417a | 30.09 | 4.48 | Elongation factor 1B alpha-subunit 2 (eF1Balpha2) | 31 | 24.20 | 4.17 | At5g19510 | 1.48 ± 0.47 |
| Cellular division | ||||||||
| 4b | 58.28 | 5.2 | Tubulin alpha-6 chain | 21 | 49.54 | 4.93 | At4g14960 | 1.91 ± 1.36 |
| Chaperone and protein of stress | ||||||||
| 20c | 71.11 | 5.3 | Putative HSP 70 | 24 | 70.91 | 5.3 | At1g16030 | ≤0.01 |
| 41c | 67.90 | 5.7 | Probable seed maturation protein | 13 | 67.20 | 5.78 | At2g42650 | ≤0.01 |
| Mechanism of defense | ||||||||
| 200c | 62.70 | 6.33 | Thioglucosidase | 33 | 60.22 | 6.45 | At3g09620 | ≤0.01 |
| Amino acid metabolism | ||||||||
| 524a | 52.03 | 6.03 | Indole-3-glycerol phosphate synthase | 33 | 44.58 | 6.98 | At2g04400 | ≤0.01 |
| Unclassified proteins | ||||||||
| 60c | 65.32 | 5.86 | 2-Hydroxyphytanol-CoA lyase-like | 17 | 61.47 | 5.74 | At5g17380 | 0.45 ± 0.10 |
| 176c | 17.65 | 5.43 | Probable major latex protein | 27 | 17.62 | 5.91 | At4g23670 | ≤0.01 |
Listed proteins correspond to proteins previously identified in Ler and listed in Rajjou et al. (2006a).
Listed proteins correspond to proteins presently identified in Cvi and previously identified in Ler (Rajjou et al., 2006a).
Proteins specifically found in Cvi. The peptide sequences of proteins identified in Cvi are available in Supplemental Table S5.
Normalized spot volumes in the D dry mature seeds divided by the normalized spot volume in the ND dry mature seeds ± sd: (D dry/ND dry); ≥100 means that the accumulation level of the corresponding protein in the ND dry seeds was close to background; ≤0.01 means that the accumulation level of the corresponding protein in the dry D seeds was close to background. Normalized spot volumes from three different gels and independent extractions. Relative abundance observed was statistically significant.
For the ND dry seeds, up-accumulated proteins corresponded to seed storage proteins, and to proteins involved in energetic and protein metabolisms (Table III). Seed storage proteins were represented by eight cruciferins (six precursor forms of the CRA1 gene [nos. 130–135; Fig. 2] and one subunit form of the CRB gene [no. 178]) and one legumin (no. 69). Metabolism and energy proteins corresponded to a β-glucosidase (no. 59; Fig. 2; Table III) and a thioglucosidase (no. 200; Fig. 4; Table III). The seven down-accumulated proteins in the ND dry seeds corresponded to energetic metabolism and protein metabolism functional classes (Table III). For example, Figure 2 shows the differential accumulation of the translation elongation factor EF-2 (nos. 45 and 46) and Figure 4 depicts the higher accumulation in D dry seeds of the glyoxylate enzyme isocitrate lyase (no. 76).
Figure 2.
Arabidopsis proteins whose abundance varied significantly between D and ND dry seeds. A, 2D gel of total soluble proteins from D dry mature seeds. The indicated portions of the gels (a, b, and c) are reproduced in B. B, Enlarged windows (a–c) of 2D gels as shown in A for the ND dry seeds (right) and the D dry seeds (left). The labeled protein numbers 45 and 46 correspond to elongation factor EF-2, number 59 to β-glucosidase, and numbers 130 to 135 to cruciferin precursors. These proteins were identified by MALDI-TOF analysis (Table III).
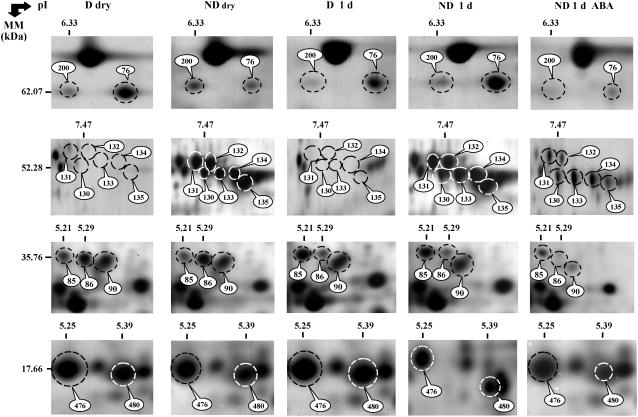
Figure 4.
Comparison of 2D protein patterns in D and ND dry seeds (D dry and ND dry), D and ND seeds imbibed for 1 d in basal medium (D 1 d and ND 1 d), and ND seeds imbibed in 30 μm ABA for 1 d (ND 1 d ABA). The labeled proteins in selected gel windows correspond to thioglucosidase (no. 200), isocitrate lyase (no. 76), cruciferin precursors (nos. 130–135), 60S acidic ribosomal protein POB (nos. 85 and 86), LEA protein (no. 90), and dehydrins (nos. 476 and 480). These proteins are listed in Tables III, IV, V, and Supplemental Tables S1 and S2.
Proteomics of D and ND Seeds during Imbibition
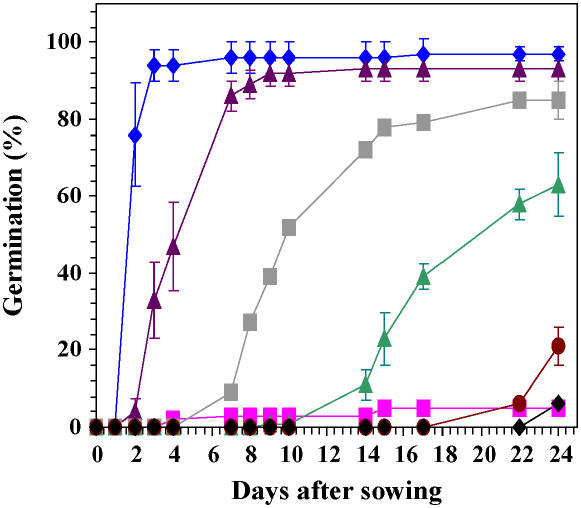
ND seeds started to germinate after 2 d of imbibition on basal medium and completed germination at day 4 (Fig. 3). In agreement with previous data (Ali-Rachedi et al., 2004), the D seeds did not germinate under these experimental conditions (Fig. 3). To characterize proteins possibly involved in the maintenance of dormancy during imbibition of the D seeds in basal medium, we compared the 2D protein profiles of the D and ND seeds at 1 d of imbibition (Table IV; Supplemental Tables S1 and S2), a time point that is prior to radicle emergence of the ND seeds (see Fig. 3). Therefore, variations in the proteome were strictly restricted to the phase of germination sensu stricto and could not simply result from different developmental stages achieved by the two seed samples following imbibition. Under these conditions, 2D gel analysis disclosed 25 proteins (23 proteins identified; Table II; see Supplemental Table S5) that exhibit differential accumulation in the two physiological states (Tables I and IV; Fig. 4). Among them, 15 proteins displayed higher accumulation in the D imbibed seeds than in the ND seeds, which corresponded to storage proteins, chaperones, and proteins associated with amino acid metabolism and translation functional classes (Table IV; Fig. 4). The eight identified proteins that displayed higher accumulation in ND imbibed seeds were assigned to cruciferin precursors, HSP70 and isocitrate lyase (Table IV; Fig. 4). It is noted that among the 23 proteins differentially accumulated between D and ND imbibed seeds, three cruciferin forms (nos. 66, 178, and 188), the isocitrate lyase enzyme (no. 76), the HSP70 protein (no. 20), the chloroplastic ATP synthase (no. 9), the 60S acidic ribosomal protein PO-B (no. 86), the mitochondrial Asp aminotransferase (no. 119), and the major latex protein (no. 176), were also subject to significant differential accumulation in the transition from dry to imbibed states (see Table IV; Supplemental Tables S1 and S2).
Figure 3.
Germination kinetics of D Cvi seeds imbibed on basal medium (pink squares), and ND Cvi seeds imbibed on basal medium (blue diamonds) or on ABA at a concentration of 1 μm (violet triangles), 3 μm (gray squares), 10 μm (green triangles), 30 μm (red circles), and 100 μm (black diamonds).
Table IV.
Arabidopsis proteins whose abundance varied significantly between D and ND seeds imbibed for 1 d in basal medium (D 1 d/ND 1 d)
AGI, Accession gene identification; Cov., coverage; Exp., experimental; MM, molecular mass in kD; Theo., theoretical.
| No. | Exp. MM | Exp. pI | Arabidopsis Protein Name and Family | Cov. % | Theo. MM | Theo. pI | AGI No. | Relative Abundanced D 1 d/ND 1 d |
|---|---|---|---|---|---|---|---|---|
| Storage protein | ||||||||
| 68b | 61.86 | 6.62 | Similarity to 7S legumin | 24 | 55.06 | 6.64 | At3g22640 | ≥100 |
| 69b | 60.20 | 6.6 | Similarity to 7S legumin | 30 | 55.06 | 6.64 | At3g22640 | ≥100 |
| 66b | 57.58 | 6.12 | 12S-3 cruciferin precursor | 47 | 58.24 | 6.53 | At4g28520 | 4.46 ± 0.22 |
| 131b | 52.93 | 7.31 | 12S-1 cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 132c | 52.99 | 7.93 | 12S-1 cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 133c | 52.07 | 8.4 | 12S-1 cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 134c | 53.16 | 8.57 | 12S-1 cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 135c | 52.35 | 9.13 | 12S-1 cruciferin precursor | 42 | 52.60 | 7.68 | At5g44120 | ≤0.01 |
| 188b | 24.94 | 8.23 | α-Chain of 12S-2 cruciferin | 29 | 27.24 | 6.34 | At1g03880 | ≥100 |
| 178b | 17.29 | 5.64 | β-Chain of 12S-2 cruciferin | 12 | 20.24 | 6.36 | At1g03880 | 0.42 ± 0.1 |
| Metabolism and energy | ||||||||
| 76c | 64.44 | 6.69 | Isocitrate lyase | 19 | 64.24 | 6.72 | At3g21720 | 0.4 ± 0.1 |
| Chaperone and protein of stress | ||||||||
| 20c | 71.11 | 5.3 | Putative HSP 70 | 24 | 70.91 | 5.3 | At1g16030 | ≤0.01 |
| 27c | 64.21 | 5.67 | Chaperonin β precursor | 29 | 64.19 | 6.21 | At1g55490 | 2.30 ± 1.40 |
| 562a | 17.94 | 5.50 | HSP17.7 | 42 | 17.83 | 5.22 | At5g12030 | 1.93 ± 0.3 |
| 906a | 17 | 7.78 | Peptidyl-prolyl cis-trans isomerase | 41 | 18.60 | 7.83 | At4g38740 | ≥100 |
| 476a | 17.66 | 5.25 | Dehydrin | 6 | 18.44 | 7.95 | At5g66400 | 2.11 ± 0.14 |
| 480a | 17.42 | 5.39 | Dehydrin | 6 | 18.44 | 7.95 | At5g66400 | 2.74 ± 0.25 |
| Photosynthesis | ||||||||
| 9c | 60.07 | 5.29 | Chloroplastic ATP synthase alpha chain | 22 | 55.33 | 5.19 | AtCg00120 | 3.4 ± 0.6 |
| Amino acid metabolism | ||||||||
| 119c | 44.83 | 7.37 | Mitochondrial aspartate aminotransferase | 30 | 47.76 | 8.36 | At2g30970 | 2.71 ± 0.45 |
| Translation protein | ||||||||
| 86b | 35.48 | 5.29 | 60S acidic ribosomal protein PO-B | 33 | 34.14 | 4.99 | At3g09200 | 3.94 ± 2.02 |
| Unclassified protein | ||||||||
| 929a | 29.91 | 5.68 | Dormancy related protein | 22 | 59.8 | 12.5 | At1g54870 | ≥100 |
| 176c | 17.65 | 5.43 | Probable major latex protein | 27 | 17.62 | 5.91 | At4g23670 | ≥100 |
| 928a | 13.95 | 6.33 | Major latex protein | 25 | 17.86 | 7.18 | At1g14940 | ≥100 |
Listed proteins correspond to proteins previously identified in Ler and listed in Rajjou et al. (2006a).
Listed proteins correspond to proteins presently identified in Cvi and previously identified in Ler (Rajjou et al., 2006a).
Proteins specifically found in Cvi. The peptide sequences of proteins identified in Cvi are available in Supplemental Table S5.
Normalized spot volumes in the D imbibed seeds divided by the normalized spot volume in the ND imbibed seeds ± sd: (D 1 d/ND 1 d); ≥100 means that the accumulation level of the corresponding protein in the ND imbibed seeds was close to background; ≤0.01 means that the accumulation level of the corresponding protein in the D imbibed seeds was close to background. Normalized spot volumes from three different gels and independent extractions. Relative abundance observed was statistically significant.
Proteomic Analysis of the Effect of Exogenous ABA Application during Imbibition of ND Cvi Seeds
As Figure 3 shows, ABA inhibited the germination of ND seeds. For an ABA concentration of 30 μm, the germination behavior of these seeds was very similar to that of the D seeds imbibed on basal medium alone. From these data, the possibility that an exogenous application of ABA to ND seeds can mimic maintenance of dormancy in D seeds imbibed on basal medium was assessed by proteomics.
The application of ABA to the ND seeds revealed 71 proteins (64 proteins identified; see Supplemental Table S5) whose accumulation levels varied compared to the ND seeds incubated under the same conditions but in the absence of the phytohormone (Table II). Among these 64 identified proteins, 57 proteins were down-accumulated (e.g. isocitrate lyase no. 76 in Fig. 4) and seven proteins were up-accumulated (Table V). Down-accumulated proteins corresponded to seed storage protein precursors, β-glucosidase, proteases, enzymes involved in energetic and protein metabolisms, and proteins involved in plant response to abiotic and biotic stress (Fig. 4; Table V). Up-accumulated proteins corresponded to 7S legumins (nos. 68 and 69), 12S-3 cruciferin precursor (no. 209), 1,6-Fru bisphosphate aldolase (no. 103), two isoforms of cytosolic glyceraldehyde-3-P dehydrogenase (GAPDH; nos. 114 and 115), and chloroplastic 3-oxoacyl-[ACP] synthase I (no. 123; Table V).
Table V.
Arabidopsis proteins whose abundance significantly varied in ND seeds imbibed for 1 d in 30 μm ABA (ND 1 d ABA) or in basal medium (ND 1 d)
AGI, Accession gene identification; Cov., coverage; Exp., experimental; MM, molecular mass in kD; Theo., theoretical.
| No. | Exp. MM | Exp. pI | Arabidopsis Protein Name and Family | Cov. % | Theo. MM | Theo. pI | AGI No. | Relative Abundanced ND 1 d ABA/ND 1 d |
|---|---|---|---|---|---|---|---|---|
| Storage protein | ||||||||
| 68b | 61.86 | 6.62 | Similarity to 7S legumin | 24 | 55.06 | 6.64 | At3g22640 | ≥100 |
| 69b | 60.20 | 6.6 | Similarity to 7S legumin | 30 | 55.06 | 6.64 | At3g22640 | ≥100 |
| 131b | 52.93 | 7.31 | 12S-1 Cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | 0.26 ± 0.01 |
| 132c | 52.99 | 7.93 | 12S-1 Cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | 0.29 ± 0.02 |
| 133c | 52.07 | 8.4 | 12S-1 Cruciferin precursor | 48 | 52.60 | 7.68 | At5g44120 | 0.50 ± 0.02 |
| 134c | 53.16 | 8.57 | 12S-1 Cruciferin precursor | 51 | 52.60 | 7.68 | At5g44120 | 0.39 ± 0.04 |
| 135c | 52.35 | 9.13 | 12S-1 Cruciferin precursor | 42 | 52.60 | 7.68 | At5g44120 | 0.39 ± 0.10 |
| 163b | 21.05 | 7.07 | β-Chain of 12S-3 cruciferin | 16 | 21.20 | 6.19 | At4g28520 | 0.39 ± 0.25 |
| 209c | 58.54 | 6.53 | 12S-3 Cruciferin precursor | 30 | 31.18 | 6.35 | At4g28520 | 2.00 ± 0.08 |
| 107b | 48.22 | 5.87 | 12S-2 Cruciferin precursor | 29 | 50.56 | 6.53 | At1g03880 | 0.43 ± 0.15 |
| 160c | 25.47 | 6.94 | α-Chain of 12S-2 cruciferin | 21 | 27.24 | 6.34 | At1g03880 | 0.27 ± 0.05 |
| 178b | 17.29 | 5.64 | β-Chain of 12S-2 cruciferin | 12 | 20.24 | 6.36 | At1g03880 | 0.29 ± 0.01 |
| Protease, peptidase, and hydrolase | ||||||||
| 32b | 56.28 | 5.57 | Cytosolic leucyl aminopeptidase | 24 | 54.41 | 5.66 | At2g24200 | 0.42 ± 0.03 |
| 57b | 65.39 | 6.02 | β-Glucosidase | 32 | 60.02 | 6.2 | At3g21370 | 0.68 ± 0.09 |
| 58b | 64.06 | 6.02 | β-Glucosidase | 31 | 60.02 | 6.2 | At3g21370 | 0.30 ± 0.01 |
| 59b | 65.65 | 5.91 | β-Glucosidase | 26 | 60.02 | 6.2 | At3g21370 | 0.40 ± 0.19 |
| 61b | 62.95 | 5.97 | β-Galactosidase | 13 | 94.78 | 6.81 | At4g36360 | 0.28 ± 0.08 |
| 74b | 73.69 | 6.38 | Subtilisin-like proteinase, nodule specific | 15 | 78.21 | 5.81 | At5g67360 | 0.55 ± 0.12 |
| Photosynthesis | ||||||||
| 37b | 57.08 | 5.74 | Ribulose bisphosphate carboxylase large chain | 18 | 47.62 | 6.13 | AtCg00490 | 0.23 ± 0.17 |
| 72b | 66.90 | 6.35 | Malic enzyme | 21 | 64.28 | 6.32 | At2g19900 | 0.27 ± 0.01 |
| 117c | 43.54 | 6.7 | Chloroplastic GAPDH A-subunit | 30 | 36.28 | 6.67 | At3g26650 | 0.43 ± 0.23 |
| Metabolism and energy | ||||||||
| 11b | 66.25 | 5.29 | Vacuolar ATP synthase subunit α | 12 | 68.81 | 5.11 | At1g78900 | 0.16 ± 0.10 |
| 25c | 79.28 | 5.37 | Pyruvate orthophosphate dikinase | 10 | 104.79 | 5.85 | At4g15530 | 0.30 ± 0.02 |
| 26c | 81.87 | 5.37 | Aconitase | 13 | 98.77 | 5.98 | At4g35830 | 0.04 ± 0.14 |
| 44c | 81.11 | 5.86 | Aconitase | 15 | 98.15 | 5.98 | At4g35830 | 0.21 ± 0.09 |
| 31c | 54.23 | 5.54 | Xyl isomerase | 22 | 53.15 | 5.74 | At5g57655 | 0.47 ± 0.15 |
| 62b | 60.66 | 5.88 | Mitochondrial ATP synthase α-chain | 32 | 55.05 | 6.23 | At2g07698 | 0.27 ± 0.07 |
| 63b | 61.46 | 6.03 | Cytosolic Glu-6 phosphate isomerase | 18 | 61.72 | 6.19 | At5g42750 | 0.54 ± 0.00 |
| 75c | 70.70 | 6.73 | Phosphoenolpyruvate carboxykinase | 15 | 73.41 | 6.61 | At4g37870 | 0.34 ± 0.04 |
| 76c | 64.44 | 6.69 | Isocitrate lyase | 19 | 64.24 | 6.72 | At3g21720 | 0.17 ± 0.10 |
| 100b | 45.08 | 5.8 | Alcohol dehydrogenase | 35 | 41.21 | 5.83 | At1g77120 | 0.56 ± 0.21 |
| 102c | 38.38 | 5.77 | Malate dehydrogenase | 47 | 34.01 | 5.68 | At3g47520 | 0.36 ± 0.05 |
| 103c | 40.21 | 5.46 | Putative 1,6-Fru bisphosphate aldolase | 20 | 42.94 | 6.79 | At4g38970 | 5.8 ± 2.7 |
| 108c | 49.88 | 5.87 | Similarity to isocitrate dehydrogenase | 22 | 45.47 | 6.13 | At1g65930 | 0.42 ± 0.08 |
| 111b | 42.88 | 6.22 | Cytosolic GAPDH | 46 | 36.91 | 6.62 | At3g04120 | 0.52 ± 0.10 |
| 112b | 44.11 | 6.29 | Cytosolic GAPDH | 34 | 36.91 | 6.62 | At3g04120 | 0.13 ± 0.03 |
| 113b | 44.16 | 6.34 | Cytosolic GAPDH | 45 | 36.91 | 6.62 | At3g04120 | 0.32 ± 0.01 |
| 114c | 43.87 | 6.1 | Cytosolic GAPDH | 28 | 36.91 | 6.62 | At3g04120 | 3.38 ± 0.01 |
| 115c | 43.77 | 6.15 | Cytosolic GAPDH | 27 | 36.91 | 6.62 | At3g04120 | 2.67 ± 0.05 |
| 129c | 43.34 | 6.09 | Cytosolic GAPDH | 19 | 36.91 | 6.62 | At3g04120 | 0.48 ± 0.23 |
| 118b | 45.92 | 7.21 | Probable Fru bisphosphate aldolase | 46 | 38.39 | 7.01 | At2g36460 | 0.33 ± 0.03 |
| 123c | 50.37 | 6.26 | Mature 3-oxoacyl-[ACP] synthase I, chloroplastic | 21 | 45.08 | 6.22 | At5g46290 | 5.78 ± 0.05 |
| 144b | 25.94 | 5.43 | Cytosolic triose phosphate isomerase | 34 | 27.17 | 5.39 | At3g55440 | 0.27 ± 0.02 |
| Amino acid metabolism | ||||||||
| 49b | 75.41 | 5.94 | Methionine synthase | 15 | 84.36 | 6.09 | At5g17920 | 0.44 ± 0.44 |
| 50b | 75.24 | 6.03 | Methionine synthase | 26 | 84.36 | 6.09 | At5g17920 | 0.20 ± 0.05 |
| 122b | 48.12 | 6.62 | Cytoplasmic aspartate aminotransferase | 48 | 44.27 | 6.8 | At5g19550 | 0.41 ± 0.03 |
| Translation protein | ||||||||
| 45b | 80.76 | 5.89 | Elongation factor EF-2 | 11 | 93.89 | 5.89 | At1g56070 | 0.25 ± 0.09 |
| 85c | 35.76 | 5.21 | Putative 60S acidic ribosomal protein PO-B | 26 | 34.13 | 4.99 | At3g09200 | 0.38 ± 0.03 |
| 95b | 49.77 | 5.42 | Initiation factor EIF-4A1 | 36 | 41.85 | 7.63 | At3g13920 | 0.31 ± 0.10 |
| 97b | 51.87 | 5.45 | DEAD box RNA helicase RH 15 | 25 | 48.38 | 5.49 | At5g11200 | 0.30 ± 0.24 |
| Cellular division | ||||||||
| 79c | 46.62 | 5.31 | Actin 8 | 36 | 41.89 | 5.37 | At1g49240 | 0.27 ± 0.11 |
| Chaperone and protein of stress | ||||||||
| 562a | 17.94 | 5.50 | HSP 17.7 | 42 | 17.83 | 5.22 | At5g12030 | ≤0.01 |
| 8b | 60.15 | 5.20 | Late embryogenesis abundant protein | 39 | 52.05 | 5.29 | At3g53040 | 0.30 ± 0.15 |
| 20c | 71.11 | 5.3 | Putative HSP 70 | 24 | 70.91 | 5.3 | At1g16030 | 0.24 ± 0.17 |
| 27c | 64.21 | 5.67 | Chaperonin β precursor | 29 | 64.19 | 6.21 | At1g55490 | 0.21 ± 0.14 |
| 64c | 59.42 | 5.95 | Chloroplastic gluthatione disulfide reductase | 25 | 52.70 | 5.89 | At3g54660 | 0.38 ± 0.04 |
| 137c | 29.33 | 5.25 | Putative lactoylglutathione lyase | 33 | 31.96 | 5.11 | At1g11840 | 0.50 ± 0.11 |
| 480a | 17.42 | 5.39 | Dehydrin | 6 | 18.45 | 7.95 | At5g66400 | ≤0.01 |
| 476a | 17.66 | 5.25 | Dehydrin | 6 | 18.45 | 7.95 | At5g66400 | 0.52 ± 0.16 |
| Mechanism of defense | ||||||||
| 33b | 54.35 | 5.59 | Probable myrosinase binding protein | 38 | 51.21 | 5.5 | At2g33070 | 0.47 ± 0.44 |
| 70c | 58.34 | 6.96 | Catalase | 28 | 56.93 | 6.63 | At1g20630 | 0.17 ± 0.04 |
| 124c | 39.56 | 5.86 | Probable NADPH 2 quinone reductase P | 23 | 38.13 | 5.81 | At5g16970 | 0.48 ± 0.01 |
| Unclassified protein | ||||||||
| 39c | 60.47 | 5.8 | Dehydrogenase | 14 | 61.67 | 6.26 | At5g62530 | 0.42 ± 0.09 |
| 183b | 14.06 | 6.15 | Major latex protein 1 | 52 | 17.89 | 6.37 | At1g14950 | 0.22 ± 0.07 |
Listed proteins correspond to proteins previously identified in Ler and listed in Rajjou et al. (2006a).
Listed proteins correspond to proteins presently identified in Cvi and previously identified in Ler (Rajjou et al., 2006a).
Proteins specifically found in Cvi. The peptide sequences of proteins identified in Cvi are available in Supplemental Table S5.
Normalized spot volumes in the ND seeds imbibed for 1 d in the presence of 30 μm ABA divided by the normalized spot volume in the ND seeds imbibed for 1 d in basal medium ± sd: (ND 1 d ABA/ND 1 d); ≥100 means that the accumulation level of the corresponding protein in the ND seeds imbibed for 1 d in basal medium was close to background; ≤0.01 means that the accumulation level of the corresponding protein in the ND seeds imbibed for 1 d in the presence of 30 μm ABA was close to background. Normalized spot volumes from three different gels and independent extractions. Relative abundance observed was statistically significant.
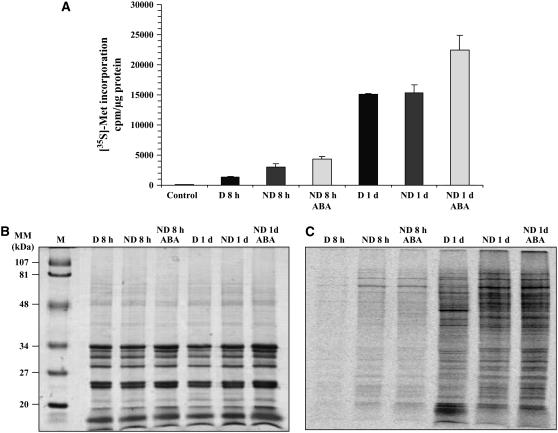
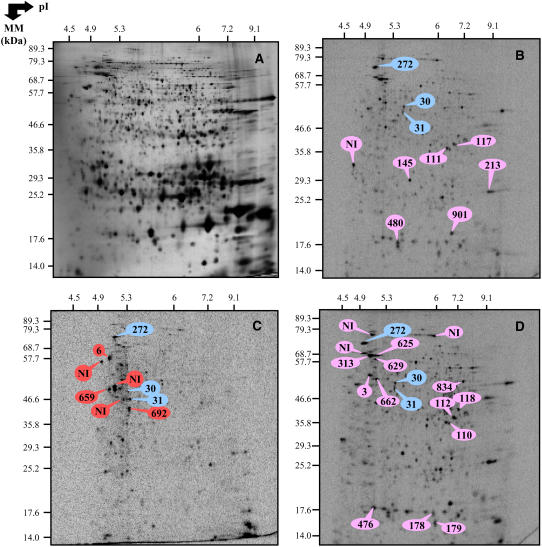
Since exogenous ABA entailed a decrease in the accumulation level of several proteins involved in protein biosynthesis (Table V), we assessed the possibility that impediment of germination of the ND seeds brought about by the phytohormone was due to an inhibition of de novo protein synthesis. In parallel, we investigated whether this hypothesis could also account for impediment of germination of the D seeds incubated on basal medium. Proteins synthesized de novo were labeled by adding [35S]-Met in the incubation medium, and total incorporation of the labeled precursor into proteins was quantitated after TCA precipitation, as described in “Materials and Methods.” The extent of [35S]-Met incorporation into protein was similar for the D and ND seed samples imbibed on basal medium and was even substantially higher for the ND seeds incubated in the presence of ABA (Fig. 5A). This finding was reinforced by qualitative analysis of de novo protein synthesis using one-dimensional (1D) and 2D gel electrophoresis (Figs. 5 and 6). As Figure 6C shows, some proteins were specifically synthesized de novo in the D imbibed seeds, as was, for example, the case for a 26S protease regulatory subunit 6A (no. 659), a hydrolase α/β-family protein (no. 692), and a vacuolar ATP synthase a-chain (no. 6; Supplemental Table S4). On the other hand, the de novo protein patterns of the ND seeds imbibed on basal medium or in the presence of 30 μm ABA were very similar (Fig. 6, B and D; Supplemental Table S4), as inferred from the accumulation of proteins involved in defense mechanism (jasmonate-inducible protein [no. 625]), energetic metabolism (cytosolic GAPDH [nos. 110, 111, and 112] and 1,6-Fru bisphosphate aldolase [no. 118]), cellular division (tubulin a-chains [nos. 3 and 662]), seed storage proteins (β-chain of 12S-2 cruciferin [nos. 145, 178, 179, 213, and 901] and cruciferin precursor [no. 834]), photosynthesis (chloroplastic GAPDH β-subunit [no. 117]), and several proteins associated with stress response (HSP no. 313 and dehydrins nos. 476 and 480; spots labeled in pink color in Fig. 6D). Finally, three identified proteins were found to be synthesized de novo both in D and ND seeds imbibed on basal medium and in ND seeds incubated in the presence of ABA (spots labeled in blue color in Fig. 6B). These proteins corresponded to enolase (no. 30), Xyl isomerase (no. 31), and a MUT/nudix family protein (no. 272; Fig. 6B).
Figure 5.
Incorporation of [35S]-Met into proteins synthesized de novo from D and ND seeds imbibed on basal medium or ABA. A, Quantification of [35S]-Met incorporation into proteins synthesized de novo from D seeds imbibed on basal medium for 8 and 24 h, and from ND seeds imbibed on basal medium or on 30 μm ABA for 8 and 24 h, following TCA precipitation as described in “Materials and Methods.” B, Coomassie blue staining of 1D gel of total soluble proteins from the seed samples shown in A. C, Autoradiogram of the corresponding 1D gel shown in B. M stands for size markers.
Figure 6.
2D patterns of de novo protein synthesis in D and ND seeds imbibed for 1 d on basal medium and in ND seeds imbibed for 1 d in the presence of 30 μm ABA. Radiolabeling of proteins was effected by adding [35S]-Met in the germination assays. Total soluble proteins were extracted from seed samples shown in Figure 5A, submitted to 2D gel electrophoresis and the radiolabeled proteins revealed by Phosphorimager analysis as described under “Materials and Methods.” A, Silver-stained 2D gel of total soluble proteins from the ND seeds imbibed for 1 d in basal medium. B, Autoradiogram of the 2 D gel for proteins extracted from the ND seeds imbibed in basal medium for 1 d. C, Autoradiogram of the 2 D gel for proteins extracted from the D seeds imbibed in basal medium for 1 d. D, Autoradiogram of the 2 D gel for proteins extracted from the ND seeds imbibed for 1 d in the presence of 30 μm ABA. The labeled protein spots were identified either by MS or by comparison with Arabidopsis seed protein reference maps from Ler (Rajjou et al., 2006a; http://seed.proteome.free.fr; see Supplemental Tables S3 and S5). They are listed in Supplemental Table S4. The proteins labeled in blue color correspond to proteins synthesized de novo in the D and ND seeds imbibed in basal medium for 1 d and in the ND seeds imbibed for 1 d in the presence of 30 μm ABA (no. 30, enolase; no. 31, Xyl isomerase; no. 272, MUT/nudix family protein). The proteins labeled in red color correspond to proteins synthesized de novo in the D seeds imbibed for 1 d in basal medium (no. 6, vacuolar ATP synthase α-chain; no. 692, hydrolase alpha β-chain; no. 659, 26S protease regulatory family). The proteins labeled in pink color correspond to proteins synthesized de novo in the ND seeds imbibed for 1 d in basal medium or in the presence of 30 μm ABA.
DISCUSSION
Comparison of 2D Protein Patterns from Ler and Cvi Mature Seeds
A comparison of 2D gels obtained from protein extracts of dry seeds of Ler (Rajjou et al., 2006a, 2006b) and Cvi (this study) accessions revealed high similarities between the two protein patterns (Table I; Supplemental Table S3). Out of the 704 common spots that were evidenced in the two genetic backgrounds, 92 were presently identified in Cvi (Supplemental Table S4) and 373 were previously identified in Ler (Rajjou et al., 2006a). For 61 of these matching spots independently identified by MS analysis in the two accessions, most of them identified the same gene (Supplemental Table S3). This enlightened both the reliability of the proteomic approach and a strong conservation of the seed proteome in the two accessions, a property that was used in this work to help characterize the proteome of Cvi seeds. Chevalier et al. (2004) investigated the root proteome of eight Arabidopsis accessions and demonstrated that for 250 protein spots detected for each accession, one-quarter was found in all the accessions, and one-half of proteins identified in Col-0 were shared by at least five other accessions. When the comparison was restricted to two or three accessions, the number of common spots increased. This result is therefore in accordance with our finding on the conservation of the seed proteome in Arabidopsis accessions. Altogether, these data pointed out the usefulness of proteomics for comparative genomic approaches using core collections of Arabidopsis accessions (Koornneef et al., 2004; McKhann et al., 2004; Mitchell-Olds and Schmitt, 2006).
Accumulation of Storage Proteins in ND Dry Seeds
The 2D gels from the ND dry seeds specifically displayed a predominant train of six proteins (protein nos. 130–135) at 52 kD (Fig. 2). The proteins in this train were identified as the precursor form of the CRA1 gene (Table III; Supplemental Table S5). Since this specific train of proteins was absent in the protein profile of the D dry seeds (Fig. 2), one possibility could be that it accumulated by de novo synthesis during after-ripening. Storage proteins are accumulated in seeds during the late stages of seed development (Bewley and Black, 1994; Harada, 1997) and are used by the germinating seedlings as a source of amino acids and nitrogen (Spencer, 1984; Lambert and Yarwood, 1992). Cruciferins, which are the major seed storage proteins in Arabidopsis (Gruis et al., 2003), are classified as globulins according to their solubility in saline solutions (Osborne, 1924; Shewry et al., 1995). Cruciferins are synthesized in developing embryos in a precursor form consisting of a protein chain of about 60 kD. This precursor form is then cleaved during seed maturation so that the mature form of cruciferins consists of six subunit pairs interacting noncovalently, each of these pairs being constituted of an acidic α-subunit of Mr equals approximately 40,000 and a basic β-subunit of Mr equals approximately 20,000 that are linked via disulfide bridges (Gruis et al., 2003). In agreement with our data, the presence of cruciferin precursors was also detected in the dry mature seeds of the ND Ler accession (Gallardo et al., 2001). Therefore, our present results revealed that the accumulation of cruciferin precursors can still proceed in dry mature seeds during after-ripening leading to dormancy breakage. This implies, quite remarkably, that the maturation program can continue after seed dispersal. In this context, it is noted that Leubner-Metzger (2005) and Bove et al. (2005) identified mRNA accumulation during after-ripening of tobacco seeds, although seeds kept a constant moisture content (8%–13%) during the after-ripening process. These observations raise the question of the mechanisms responsible for gene expression under the very low hydration conditions prevailing in orthodox seeds after desiccation on the mother plant. Recent 1H-magnetic nuclear resonance microimaging studies identified regions of local higher water content in after-ripened seeds that could explain occurrence of such a metabolic activity in dry seeds (Leubner-Metzger, 2005).
The train of cruciferin precursor spots accumulated in the ND dry seeds decreased sharply upon imbibition (Fig. 4; Supplemental Table S1). This result was in accordance with previous data obtained with Ler seeds (Gallardo et al., 2001; Rajjou et al., 2004).
An Accumulation of Energetic Metabolism and Hydrolytic Enzymes Is Triggered in ND Dry and Germinating Seeds and Repressed by ABA
Compared to the D dry seeds, the ND dry seeds up-accumulated proteins involved in energetic metabolism (Table III), as was the case for the neoglucogenesis enzymes 1,6-Fru bisphosphate aldolase (no. 118) and cytosolic isoforms of GAPDH (nos. 114 and 115). Thus, the ND dry seeds seemed well equipped to express neoglucogenesis activity during early germination, possibly to provide energy from stored lipids required for seedling establishment (Penfield et al., 2005, 2006; Cernac et al., 2006; Footitt et al., 2006). In addition, the level of isocitrate lyase (no. 76), an enzyme from the glyoxylate cycle that is involved in storage lipid mobilization in germinating seeds, sharply increased upon imbibition of ND seeds (Table IV; Fig. 4). In marked contrast, this level remained constant (and lower than that seen for the ND imbibed seeds) during imbibition of D seeds (Fig. 4; Supplemental Table S2). Isocitrate lyase catalyzes the reversible cleavage of isocitrate to glyoxylate and succinate and plays a major role in enabling gluconeogenesis to occur from stored lipids in germinating seeds (Eastmond et al., 2000). Therefore, these data point out two important mechanisms to allow resumption of metabolism from quiescent D seeds. The one leads to an accumulation of a subset of important (controlling) metabolic enzymes during after-ripening. The other allows an accumulation of another, complementary, subset of such controlling enzymes during imbibition, but only in the ND seeds. Gallardo et al. (2002) described an accumulation of isocitrate lyase in germinating Arabidopsis seeds of the Ler accession, occurring at the moment of radicle protrusion. Since no such accumulation was detected during imbibition of seeds of the Ler ga1 GA-deficient mutant, these authors concluded that the accumulation of isocitrate lyase during seed germination was dependent on GA action, in agreement with the data of Marriott and Northcote (1975) showing that exogenous GAs can stimulate the induction of isocitrate lyase activity during germination of castor bean seeds. It is noted that application of exogenous ABA to ND seeds strongly repressed the accumulation of isocitrate lyase (Table V; Fig. 4). Altogether, these observations suggest the occurrence of antagonistic interaction of ABA and GA in the control of the seed germination process. Strikingly, several enzymes involved in energetic metabolism were repressed by ABA in ND imbibed seeds (Table V). It is thus likely that mounting metabolic pathways in imbibed seeds is indispensable for germination and seedling establishment.
The hydrolytic enzyme, β-glucosidase (no. 59), was found to be most abundant in ND dry seeds (Table III; Fig. 2). In germinating barley seeds, this enzyme was shown to be implied in sugar liberation from β-glucans derived from cell wall polysaccharides, thus providing carbohydrates to the plantlet (Leah et al., 1995). The complete hydrolysis of β-glucans is facilitated by several enzymes, including endo-(1–3)-β-glucanase (Leah et al., 1995). Recent studies in tobacco seeds revealed that an up-accumulation of β-1,3 glucanase correlated with the release of seed-coat imposed dormancy during after-ripening (Leubner-Metzger, 2002, 2005). Here, Leubner-Metzger et al. (2005) proposed that hydrolysis of β-glucans contributes to release the seed-coat barrier to facilitate radicle protrusion. Break of birch (Betula pubescens) bud dormancy by β-1,3 glucanase has also been described (Rinne et al., 2001). In agreement with these findings, we observed that application of ABA to ND seeds reduced the level of this β-glucosidase (spot no. 59 in Table V).
For the ND imbibed Cvi seeds, exogenous ABA led to a decrease in the accumulation levels of several peptidases, proteases, and hydrolases (nos. 32, 57, 58, 59, and 74; Table V). The accumulation level of another hydrolase, β-galactosidase (no. 61), also decreased under these conditions (Table V). This enzyme has been implicated in a number of biological processes including plant growth and fruit ripening. β-Galactosidase has been identified in mung bean (Vigna radiata) seedlings and its activity was shown to increase to a high level between days 4 and 6 of germination (Li et al., 2001). It has been suggested that this enzyme is synthesized or activated to mobilize or modify cell wall components during seed germination (Edwards et al., 1985; Konno and Tsumuki, 1993; Li et al., 2001).
Genes Responsive to Biotic and Abiotic Stresses Are Induced by Dormancy Release
ND dry seeds specifically accumulated a thioglucosidase (myrosinase; no. 200; Table III; Fig. 4) that is known to catalyze the hydrolysis of glucosinolates during germination (Chew, 1988; McGregor, 1988). Although intact glucosinolates are relatively nontoxic, their breakdown products have important biological effects, as for the goitrogenic species that perturb thyroid function or the very reactive isothiocyanates that present antibacterial and antifungal properties (Rask et al., 2000). Our results indicate, therefore, that dormancy release is associated with the mounting of specific defense mechanisms to protect germinating seeds against insects and/or pathogenic microorganisms.
Proteins involved in the protection against cellular damage induced by abiotic stress also appeared to be regulated in germinating Cvi seeds. Dehydrins (nos. 476 and 480; Table IV) encoded by the RAB18 gene were more abundant in D imbibed seeds than in ND seeds imbibed either in basal medium or ABA (Fig. 4; Tables IV and V). RAB18 belongs to the group 2 LEA gene family and encodes an ABA-inducible protein that is expressed in Arabidopsis developing seeds (Lång et al., 1994) and believed to be important for desiccation tolerance or in response to hyperosmotic stress (Allagulova et al., 2003). In Arabidopsis, dehydrins can also accumulate in early stages of seed germination, during a time window in which the germinating seed can recapitulate late phases of the seed maturation program, most presumably to mount adaptive reactions in response to fluctuating water levels in the soil (Lopez-Molina et al., 2002). Our present data showing an active de novo synthesis of dehydrins in ND germinating Cvi seeds (Fig. 6, B and D; Supplemental Table S4) are thus in good agreement with data obtained with Ler seeds (Rajjou et al., 2004, 2006a). This de novo protein synthesis was a specific feature of the ND seeds since it was not observed with the D imbibed seeds (Fig. 6). Even if the level of dehydrins is lower in 1-d imbibed ND seeds than in D seeds, this biosynthesis activity in the ND seeds should confer competence response immediately to water stress. Interestingly, the accumulation level of a dehydrin has been shown to decrease during cold or warm stratification of seeds of Taiwan flowering cherry (Prunus campanulata) but, on the contrary, to increase upon a sequential application of both treatments (Lee et al., 2006). Given that dormancy breaking in this woody species requires a warm plus a cold stratification, these data are consistent with our results in Arabidopsis and suggest that a specific mechanism of de novo dehydrin synthesis in the ND germinating seeds could be generalized for a wide range of species. We also conclude that the time window during which germinating seeds can recapitulate the late maturation program (Lopez-Molina et al., 2002) can only occur after the break of dormancy, implying, therefore, that this developmental time window is an integral part of the germination process. In the context of the Cvi seeds, it is also noted that the ABA treatment did not affect de novo dehydrin synthesis in the ND seeds (Fig. 6; Supplemental Table S4). Since the D seeds have been shown to accumulate higher ABA amounts during imbibition than the ND seeds (Ali-Rachedi et al., 2004), it appears that endogenous ABA is not able to elicit de novo synthesis of dehydrins in the D imbibed seeds. Our data therefore suggest an ABA-independent regulation of dehydrin biosynthesis in imbibed seeds, in contrast to the situation that holds during the seed maturation phase (Han et al., 1997).
An HSP70 (no. 20) was present at high level in ND dry seeds (Table III). HSPs are synthesized in all cells in response to elevated temperature (Spremulli, 2000) but are also induced by other environmental stresses (Noven et al., 1992). They participate in diverse cellular processes by acting as molecular chaperones (Hong and Vierling, 2001). They are also described as being developmentally regulated, being abundant in dry mature seeds, and disappearing during germination (Wehmeyer et al., 1996). It is noted that a proteomic study showed an accumulation of a 17.6-kD HSP after 3 weeks of cold stratification of seeds of the weed species waterhemp (Amaranthus rudis; Leon et al., 2006). Altogether, these data suggest that HSPs are required during dormancy release to maintain the proper folding of other proteins.
Protein Synthesis and Stability Are Key Components Controlling Seed Germination
Transcriptome analyses provided evidence for an accumulation of several mRNAs encoding translation initiation factors in after-ripened Arabidopsis seeds of the Cvi accession (Cadman et al., 2006), suggesting that protein synthesis is repressed in D imbibed seeds. Also, the germination of Arabidopsis seeds has been shown to be completely halted by cycloheximide, an inhibitor of cytosolic translation (Osherov and May, 2000), indicating the importance of de novo protein synthesis in the control of seed germination (Rajjou et al., 2004). This finding has been recently confirmed by transcriptome analysis of the Col-0 accession of Arabidopsis during germination (Nakabayashi et al., 2005). Our data obtained from quantification of [35S]-Met incorporation in D and ND imbibed seeds (Fig. 5A) clearly indicated that the failure of the D imbibed seeds to germinate was not due to an incapability of these seeds to support de novo protein synthesis. On the contrary, we found that de novo protein synthesis reached rather similar levels for both the D and ND imbibed seeds (Fig. 5A). Consistent with this, the proteomic data obtained by silver staining of 2D gels showed that three EF-2 elongation factors (nos. 45, 46, and 417; Fig. 2; Table III) are up accumulated in the D dry seeds. Moreover, protein synthesis activity even substantially increased for the ND seeds incubated in ABA (Fig. 5A). Higgins et al. (1982) have reported that ABA controls protein synthesis both negatively and positively in barley aleurone cells by regulating the level of mRNA and possibly also by changing the efficiency of translation. However, the obtained 2D profiles for de novo synthesized proteins differed markedly for the ND and D imbibed seeds (Fig. 6, B and C; Supplemental Table S4). Thus, it appears that only the seeds that successfully break dormancy can acquire the capacity to reprogram the pattern of protein biosynthesis during imbibition, allowing the completion of germination. This suggests that during imbibition, ND seeds are committed to translate a specific set of genes, from mRNA pools stored during maturation and/or synthesized de novo, both of them being required for germination and seedling establishment (Rajjou et al., 2004; Nakabayashi et al., 2005). Among the proteins identified on the autoradiograms of Figure 6 (as listed in Supplemental Table S4), it is noted a specific labeling of tubulin α-chains (nos. 3 and 662) in the ND imbibed seeds. Interestingly, a GA treatment induced both seed germination of the Ler ga1 GA-deficient mutant and a concomitant accumulation of tubulin proteins (Gallardo et al., 2002). Furthermore, accumulation of β-tubulin in tomato (Lycopersicon esculentum) seed germination and priming has been well documented (Groot et al., 1988; de Castro et al., 2000). Altogether, these data point out that de novo synthesis and accumulation of tubulin proteins in ND germinating seeds is an important determinant of completion of germination and growth initiation.
The 2D patterns for protein accumulation (Fig. 4; Tables IV and V) and de novo protein synthesis (Fig. 6; Supplemental Table S4) were very different in D seeds imbibed in basal medium and in ND seeds imbibed in ABA. Thus, out of the 64 identified proteins whose accumulation levels were regulated by exogenous ABA in ND imbibed seeds (Table V), only 10 proteins appeared to be subject to a similar differential accumulation when comparing the D and ND imbibed seeds (Table IV). It is striking that the application of ABA to the ND seeds induced a down-accumulation of a large part (90%) of the 64 differentially accumulated proteins (Table V). There are at least two possibilities to account for this feature. In the first, down-accumulation of seed proteins would result from inhibition of their de novo synthesis. However, the observed patterns of de novo protein synthesis with the ND seeds imbibed in ABA or in basal medium are not in favor of this hypothesis (Fig. 6, D and B). An alternative possibility would be that the observed prominent down-accumulation of proteins originates from proteolytic mechanisms induced by ABA in the ND imbibed seeds, notably observing that some of the down-accumulated proteins (Table V) are actively synthesized de novo (e.g. spot nos. 111 and 112; Fig. 6; Supplemental Table S4). In support of this hypothesis, Hoth et al. (2002) reported that genes encoding proteins involved in regulated proteolysis are up-regulated at the RNA level in Arabidopsis seedlings subjected to an ABA treatment. Also, genetic studies in Arabidopsis have provided evidence for a role of the ubiquitin/26S proteasome pathway in ABA responses, notably during germination (Smalle et al., 2003).
Differential Protein Accumulation and de Novo Protein Synthesis Do Not Result from a Control of Gene Expression at the RNA Level in D and ND Imbibed Seeds
Cadman et al. (2006) recently analyzed the transcriptome of 1-d imbibed D and ND (after-ripened) Arabidopsis Cvi seeds using Affymetrix technology. It was therefore of interest to compare their data with our present proteomic results, which might help decipher at which level (mRNA and/or protein) gene expression is regulated during germination and dormancy release. Table IV lists 23 proteins, corresponding to 16 different genes, whose accumulation levels differed in D and ND 1-d imbibed seeds, as revealed by silver staining of 2D gels. Out of them, only the two genes At5g12030 (protein no. 562) and At2g30970 (protein no. 119), encoding the HSP17.7 and the mitochondrial Asp aminotransferase, respectively, were identified as being differentially accumulated at the RNA level between D and ND states (supplemental table S1 in Cadman et al., 2006). Moreover, the accumulation of the mitochondrial Asp aminotransferase (At2g30970) was higher in D than in ND imbibed seeds at the protein level (Table IV), while the converse was observed at the RNA level (supplemental table S1 in Cadman et al., 2006). Also, none of the 15 genes encoding the 21 identified proteins that are specifically synthesized de novo in ND and in D imbibed seeds (Fig. 6, B–D; Supplemental Table S4) are characterized as being differentially expressed by the transcriptome analysis (supplemental tables in Cadman et al., 2006). From these observations, it appears that there exists only very limited correlation between the proteomic (this study) and transcriptomic (Cadman et al., 2006) data. This feature could originate from slight differences in the experimental procedure used to conduct seed imbibition. Thus, Cadman et al. (2006) have compared D seeds imbibed for 24 h in the dark with ND seeds that have been imbibed for 24 h either in the dark or for 20 h in the dark and then for 4 h in red light conditions. In our study, the proteome analysis has been conducted with D and ND seeds that had been imbibed under a 16-h photoperiod (see “Materials and Methods”). An alternative possibility to account for this apparent lack of correlation between the transcriptome and proteome data could result from the occurrence of translational and/or posttranslational mechanisms that are well known to occur during early stages of seed germination. Thus, as mentioned above, the importance of protein synthesis for seed germination has been stressed by Nakabayashi et al. (2005) from transcriptomic analysis of Arabidopsis Col-0 seed germinating. Moreover, a higher transcript accumulation of genes involved in protein synthesis machinery has been shown to be associated to dormancy release in sweet scented tobacco (Bove et al., 2005) and in the Cvi accession of Arabidopsis (Cadman et al., 2006). Cadman et al. (2006) raised the hypothesis that translation runs at a lower rate in imbibed D Arabidopsis seeds than in corresponding ND seeds. Our data on de novo protein synthesis (Figs. 5 and 6) are not in agreement with such a hypothesis. We show here that imbibed D seeds are competent for protein synthesis but that their pattern of de novo protein synthesis is different from that seen with the ND seeds incubated in minimal medium or in ABA (Fig. 6). Perhaps, this may arise from differential regulation of translation activity from either the stored or the neosynthesized mRNAs in D and ND imbibed seeds. Both classes of mRNAs have been shown to be required for seed germination (Bewley, 1997; Rajjou et al., 2004). It is also noted that 14 of the 23 proteins differentially accumulated between D and ND imbibed seeds were already accumulated in the dry seeds (Table IV; Supplemental Tables S1 and S2). Therefore, it is possible that their differential accumulation was due to posttranslational modifications rather than from transcriptional control. Consistent with this hypothesis, protein phosphorylation is considered as a major mechanism controlling seed germination. Thus, Gallie et al. (1997) and Le et al. (1998) have shown that the phosphorylation state of several translation initiation factors (eIF4A, eIF4B, and eIF2) varies during wheat (Triticum aestivum) seed germination and, most importantly, that this posttranslational modification correlates with resumption of protein synthesis in germinating seeds. Altogether, these findings support the proposal that the D and ND imbibed seeds can display comparable protein synthesis activity but that they are subjected to differential control of protein synthesis from different pools of mRNA and of posttranslational protein modifications from stored and/or de novo synthesized proteins. In addition to transcription regulation (Cadman et al., 2006), this may provide fine tuning of seed germination under the best environmental conditions ensuring successful establishment of vigorous seedlings.
CONCLUSION
This data documented that reference protein maps established for the seeds of a given Arabidopsis accession (e.g. Ler) can be used to identify proteins in other accessions as exemplified here for Cvi. They also illustrated the robustness of the proteomic approach based on the use of 2D gels, as a number of matching spots from Ler and Cvi seed protein extracts showed the same gene identity following independent protein identification by MS analyses.
A specific feature of seed dormancy release during after-ripening concerned the accumulation of several protein spots in ND seeds that were assigned to cruciferin precursors. This raises the question of the mechanisms responsible for such an accumulation despite the very low hydration level of dry mature embryos during after-ripening. Further work is in progress to address this question.
An analysis of de novo synthesis protein patterns demonstrated that (1) the nature of proteins synthesized de novo, but not the global level of protein synthesis activity, is determinant for germination, and (2) exogenous ABA application to ND seeds does not mimic the behavior of D seeds imbibed in basal medium. Since imbibed D seeds accumulate ABA to higher levels than imbibed ND seeds (Ali-Rachedi et al., 2004), these results suggest that ABA targets different genes during imbibition of D and ND seeds. Moreover, this work suggests that germination inhibition by exogenous ABA of ND seeds may be mediated by regulated proteolysis, a level of posttranslational control that has not been identified in imbibed D seeds.
MATERIALS AND METHODS
Plant Material
D and ND seeds of Arabidopsis (Arabidopsis thaliana) L. Heynh, ecotype Cvi, were used for all experiments. Seeds were supplied by Dr. Maarten Koornneef (Wageningen, The Netherlands). Plants were grown in a growth chamber at 19°C/20°C under a 16-h photoperiod of artificial light (Orsam L58/31830 luminux plus Wanton Wan White tubes, 45 μm m−2 s−1) and 70% of relative humidity (RH). After full maturation of the seeds, plants were no more watered, kept dry during 3 weeks, and seeds harvested. The collected D seeds were stored at 7°C, 40% RH in darkness to maintain the D state. To obtain the release of dormancy, D seeds were maintained at room temperature (21°C–24°C) in dry conditions (35%–40% RH), which was fully achieved after 6 to 12 months of such a dry storage.
Germination Assays
Seed samples were subjected to germination assays. For each test, 100 seeds were aseptically sown on a basal medium composed of distilled water buffered with MES (3 mm, pH 5.7) and gelified with agar (7 g/L Noble agar, Difco). Germination assays were conducted in controlled culture room under 16-h photoperiod (Philips TRM HOW/33 RS tubes, 170 μm m−2 s−1), at 25°C (light period)/20°C (dark period) and a constant 70% RH.
Preparation of Total Soluble Protein Extracts
Total soluble protein extracts were prepared from mature dry and imbibed seeds at different times of imbibition. Seeds (200 mg) were homogenized in liquid nitrogen and reduced to powder using mortar and pestle. The powder was suspended in 1.2 mL of a thiourea/urea lysis buffer (Harder et al., 1999) containing 7 m urea (Amersham Pharmacia Biotech), 2 m thiourea (Merck), 4% (w/v) CHAPS (Amersham Pharmacia Biotech), and 1% (v/v) Pharmalyte, pH 3 to 10, carrier ampholytes (Amersham Pharmacia Biotech). This extraction buffer also contained 18 mm Tris-HCl (Trizma HCl; Sigma), 14 mm Trizma Base (Sigma), 215 μL of protease inhibitor cocktail complete Mini (Roche Diagnostics) from one tablet dissolved in 1.5 mL of sterilized water, 53 units mL−1 Dnase I (Roche Diagnostics), 4.9 Kunitz units mL−1 Rnase A (Sigma), and 0.2% (v/v) Triton X-100. After 10 min at 4°C, protein extracts were centrifuged (35,000g for 10 min) at 4°C. The supernatants, containing the total soluble protein extracts, were submitted to a second clarifying centrifugation as above and stored at −20°C. Protein contents in the extracts were measured according to Bradford (1976), using bovine serum albumin as a standard.
2D Gel Electrophoresis
Isoelectric focusing (IEF) of 200 μg protein in reswelling buffer (8 m urea, 2% [w/v] CHAPS, 0.5% [v/v] immobilized pH gradient buffer 4–7, 20 mm dithiothreitol [DTT], and 0.01% [w/v] bromphenol blue) was run using 18-cm immobilized pH gradient (3–10 nonlinear) immobilized pH gradient strips on a Multiphor II system (Amersham-Pharmacia Biotech). Strips were rehydrated for 14 h at 22°C with the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100, 20 mm DTT, and the protein extracts. IEF was performed at 22°C for 1 h at 300 V and for 7 h at 3,500 V. After IEF, proteins were separated according to size. IEF strips were equilibrated for 2 × 20 min in 2 × 100 mL of equilibration solution containing 6 m urea, 30% (v/v) glycerol, 2.5% (w/v) SDS, 0.15 m bis-Tris, and 0.1 m HCl (Görg et al., 1987; Harder et al., 1999). DTT (50 mm) was added to the first equilibration solution and iodoacetamide (4% [w/v]) was added to the second (Harder et al., 1999). Equilibrated gel strips were placed on top of vertical polyacrylamide gels (10% [v/v] acrylamide, 0.33% [w/v] piperazine diacrylamide, 0.18 m Trizma base, 0.166 m HCl, 0.07% [w/v] ammonium persulfate, and 0.035% [v/v] TEMED). A denaturing solution (1% [w/v] low-melting agarose, 0.4% [w/v] SDS, 0.15 m bis-Tris, and 0.1 m HCl) was loaded on gel strips. After agarose solidification, electrophoresis was performed at 10°C in a buffer (pH 8.3) containing 25 mm Trizma Base, 200 mm taurine, and 0.1% (w/v) SDS, for 1 h at 35 V and 110 V overnight. Ten gels were run in parallel (Isodalt system from Amersham Pharmacia Biotech). For each condition analyzed 2D gels were made in triplicate.
Protein Staining and Gel Analyses
2D gels were stained with silver nitrate as described (Blum et al., 1987) using the Hoefer Automated Gel stainer apparatus from Amersham Pharmacia biotech. Silver-stained gels were scanned with the Sharp JX-330 scanner equipped with the Labscan version 3.00 from Amersham Pharmacia Biotech. After spot detection and background subtraction (mode: average on boundary), 2D gels were compared, matched, and the quantitative determination of the spot volumes was performed (mode: total spot volume normalization). For each analysis, statistical data showed a high level of reproducibility between normalized spot volumes of gels produced in triplicate. Data were subjected to statistical analysis by one-way analysis of variance. Where F values indicated significance (P < 0.05), individual means were compared using Student's t test (α = 0.05). Proteins listed in Tables reporting the various comparisons investigated were found to be significantly different.
Protein Identification by Matrix-Assisted Laser-Desorption Ionization Time of Flight MS
Spots of interest were excised from GelCode-stained 2D gels with a sterile syringe and placed in Eppendorf tubes. Pieces of gel were then washed using a Multiprobe II robot (Perkin Elmer) in several steps with water, 25 mm ammonium carbonate, and acetonitrile. Protein identification was done by the laboratory of Dr. Michel Rossignol (Institut National de la Recherche Agronomique, Montpellier). Proteins were digested with trypsin (12.5 μg/mL in 25 mm ammonium carbonate). Supernatants were mixed with equal volumes of matrix solution (α-cyano-4-hydroxycinnamic acid) and spotted onto targets. Peptide mass fingerprints were acquired using a Biflex III mass spectrometer (Bruker Daltonics). Spectra were calibrated internally and annotated automatically. The MASCOT (http://www.matrixscience.com) search engine software (Matrix Science) was used to search NCBInr database. The following parameters were used for database search: mass tolerance of 100 ppm, a minimum of four matched peptides and one miscleavage allowed.
De Novo Protein Synthesis
Labeled proteins were synthesized in vivo by Cvi seeds imbibed on basal medium and 30 μm ABA for 8 and 24 h in the presence of [35S]-Met (1.85 MBq; ICN Biomedicals SARL). Following incubation, protein extracts were prepared and protein synthesis was measured by TCA precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after eight washing steps in cold 5% TCA and 0.04 m sodium pyrophosphate and two washing steps in absolute methanol, filters were dried and counted for radioactivity in a liquid scintillation counter (Dietrich et al., 1985). Protein extracts were also submitted to 2D gel electrophoresis as described above. Proteins on the 2D gels were stained by silver nitrate. Then, stained 2D gels were dried for 2 d at room temperature in a sandwich composed of, from bottom to top: one sheet of cellophane model Gel Dryer (Bio-Rad), 2D gel, one sheet of Saran wrap (VWR international SAS), and one sheet of cellophane model Gel Dryer. After drying, the upper sheet of cellophane and the Saran sheep were peeled and gels were submitted to Phosphorimager analysis (Molecular Dynamics Storm 840 phosphorimager, Amersham Biosciences). Labeled 2D protein patterns were scanned as described above for the silver nitrate-stained gels. Relative protein neosynthesis levels were quantitated by densitometric analyses of the spots on the autoradiograms as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Proteins differentially accumulated in dry and 1-d imbibed ND seeds.
Supplemental Table S2. Proteins differentially accumulated in dry and 1-d imbibed D seeds.
Supplemental Table S3. Protein identification of corresponding matching spots in Ler and Cvi accessions.
Supplemental Table S4. Identity of neosynthesized proteins in D, ND 1-d imbibed seeds, and in ND treated with ABA.
Supplemental Table S5. Peptide sequences identified by MALDI-TOF-MS in Cvi.
This work was supported by the French Ministry of Agriculture. The Ph.D. thesis of Kamel Chibani was supported by the Algerian and French Ministries of Education.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Philippe Grappin (grappin@versailles.inra.fr).
The online version of this article contains Web-only data.
References
- Ali-Rachedi S, Bouinot D, Wagner M-H, Bonnet M, Sotta B, Grappin P, Jullien M (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488 [DOI] [PubMed] [Google Scholar]
- Allagulova CHR, Gimalov FR, Shakirova FM, Vakhitov VA (2003) The plant dehydrins: structure and putative functions. Biochemistry (Mosc) 68: 945–951 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Baskin CC (1998) Seeds: Ecology, Biogeography and Evolution of Dormancy and Germination. Academic Press, San Diego
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination. Plenum Press, New York
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Bove J, Lucas P, Godin B, Ogé L, Jullien M, Grappin P (2005) Gene expression analysis by cDNA-AFLP highlights a set of new signalling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia. Plant Mol Biol 57: 593–612 [DOI] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46: 805–822 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141: 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Martin O, Rofidal V, Devauchelle AD, Barteau S, Sommerer N, Rossignol M (2004) Proteomic investigation of natural variation between Arabidopsis ecotypes. Proteomics 4: 1372–1381 [DOI] [PubMed] [Google Scholar]
- Chew FS (1988) Searching for defensive chemistry in the Cruciferae, or do glucosinolates always control interactions of cruciferae with their potential herbivores and symbionts? No! In KC Spencer, ed, Chemical Mediation of Coevolution. Academic Press, San Diego, pp 81–112
- de Castro RD, van Lammeren AAM, Groot SPC, Bino RJ, Hilhorst HWM (2000) Cell division and subsequent radicle protrusion in tomato seeds are inhibited by osmotic stress but DNA synthesis and formation of microtubular cytoskeleton are not. Plant Physiol 122: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Teissère M, Job C, Job D (1985) Poly(dAT) dependent trinucleotide synthesis catalyzed by wheat-germ RNA polymerase II: effects of nucleotide substrates and cordycepin triphosphate. Nucleic Acids Res 13: 6155–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J (2005) Niche construction through germination cueing: life-history responses to timing of germination in Arabidopsis thaliana. Evolution Int J Org Evolution 59: 771–785 [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Dea ICM, Bulpin PV, Reid JSG (1985) Xylogucan (amyloid) mobilization in the cotyledon of Trapeolum majus L. seeds following germination. Planta 163: 133–140 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA responsive loci are similar to abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) Abscisic acid inhibition of radicle emergence but not seedlings growth is suppressed by sugars. Plant Physiol 122: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Marquez J, Schmuths HY, Baker A, Theodoulou FL, Holdsworth M (2006) Analysis on the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J Exp Bot 57: 2805–2814 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Le H, Caldwell C, Tanguay RL, Hoang NX, Browning KS (1997) The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J Biol Chem 272: 1046–1053 [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203: 182–187 [DOI] [PubMed] [Google Scholar]
- Garello G, Barthe P, Bonelli M, Bianco-Trinchant J, Bianco J, Le Page-Degivry MT (2000) Abscisic acid-regulated responses of dormant and non-dormant embryos of Heliantus annuus: role of ABA-inducible proteins. Plant Physiol Biochem 6: 473–482 [Google Scholar]
- Gonai T, Kawahara S, Tougou M, Satoh S, Hashiba T, Hirai N, Kawaide H, Kamiya Y, Yoshioka T (2004) Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J Exp Bot 55: 111–118 [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L (1987) Elimination of point streaking on silver-stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis 8: 122–124 [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210: 279–285 [DOI] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM (1988) Gibberellin-deficient tomato seeds prior to radicle protrusion. Planta 174: 500–504 [DOI] [PubMed] [Google Scholar]
- Gruis D, Schulze J, Jung R (2003) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8: 183–187 [DOI] [PubMed] [Google Scholar]
- Han B, Hughes WD, Galau GA, Bewley DJ, Kermode AR (1997) Changes in late-embryogenesis-abundant (LEA) messenger RNAs and dehydrins during maturation and premature drying of Ricinus communis L. seeds. Planta 201: 27–35 [DOI] [PubMed] [Google Scholar]
- Harada JJ (1997) Seed maturation and control of germination. In BA Larkins, IK Vasil, eds, Advances in Cellular and Molecular Biology of Seed Development. Kluwer Academic Press, Dordrecht, The Netherlands, pp 545–592
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A (1999) Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis 20: 826–829 [DOI] [PubMed] [Google Scholar]
- Higgins TJV, Jacobsen JV, Zwar JA (1982) Gibberellic acid and abscisic acid modulate protein synthesis and mRNA levels in barley aleurone layers. Plant Mol Biol 1: 191–215 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4899 [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D (2005) Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol 138: 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien M, Bouinot D, Ali-Rachedi S, Sotta B, Grappin P (2000) Abscisic acid control of seed dormancy expression in Nicotiana plumbaginifolia and Arabidopsis thaliana. In J Crabbé, JD Viemont, eds, Dormancy in Plants: From Whole Plant Behaviour to Cellular Control. Second International Symposium on Plant Dormancy, Angers. CAB International, Wallingford, UK, pp 195–210
- Karssen CM (1982) Seasonal patterns of dormancy in weed seeds. In Kahn AA, ed, The Physiology and Biochemistry of Seed Development, Dormancy and Germination. Elsevier Biomedical, Amsterdam, pp 243–270
- Konno H, Tsumuki H (1993) Purification of a β-galactosidase from rice shoots and its involvement in hydrolysis of the natural substrate in cell walls. Physiol Plant 89: 40–47 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 6: 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid–insensitive mutants of Arabidopsis thaliana. Plant Physiol 61: 377–383 [Google Scholar]
- Lambert N, Yarwood JN (1992) Engineering legume seed storage proteins. In PR Shewry, S Guteridge, eds, Plant Protein Engineering. Cambridge University Press, Cambridge, UK, pp 167–187
- Lång V, Mäntylä E, Welin B, Sundberg B, Palva ET (1994) Alterations in water status, endogenous abscisic acid content and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol 104: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Browning KS, Gallie DR (1998) The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J Biol Chem 273: 20084–20089 [DOI] [PubMed] [Google Scholar]
- Leah R, Kigel J, Svendesen I, Mundy J (1995) Biochemical and molecular characterization of a barley seed beta-glucosidase. J Biol Chem 270: 15789–15797 [DOI] [PubMed] [Google Scholar]
- Lee CS, Chien CT, Lin CH, Chiu YY, Yang YS (2006) Protein changes between dormant and dormancy-broken seeds of Prunus campanulata Maxim. Proteomics 26: 4147–4154 [DOI] [PubMed] [Google Scholar]
- Leon RG, Basshaman DC, Owen MDK (2006) Germination and proteome analyses reveal intraspecific variation in seed dormancy regulation in common waterhemp (Amaranthus tuberculatus). Weed Sci 54: 305–316 [Google Scholar]
- Léon-Kloosterziel KM, Alvarez-Gil M, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G (2002) Seed after-ripening and over-expression of class I beta-1,3-glucanase confer maternal effects on tobacco testa rupture and dormancy release. Planta 215: 958–968 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G (2005) Beta 1,3-glucanase gene expression in low hydrated seeds as a mechanism for dormancy release during tobacco-after-ripening. Plant J 41: 133–145 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Frundt C, Meins FJR (1996) Effects of gibberellins, darkness and osmotica on endosperm rupture and class I β-1,3-glucanase induction in tobacco seed germination. Planta 199: 282–288 [Google Scholar]
- Li SC, Han JW, Chen KC, Chen KC (2001) Purification and characterization of isoforms of β-galactosidases in mung bean seedlings. Phytochemistry 57: 349–359 [DOI] [PubMed] [Google Scholar]
- Lobin W (1983) The occurrence of Arabidopsis thaliana in the Cape Verde Islands. Arab Inf Serv 20: 119–123 [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua N-H (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Marriott KM, Northcote DH (1975) The induction of enzyme activity in the endosperm of germinating castor-bean seeds. Biochem J 152: 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor DI (1988) Glucosinolate content of developing rapessed (Brassica napus L. Midas) seedlings. Can J Plant Sci 68: 367–380 [Google Scholar]
- McKhann HI, Camilleri C, Berard A, Bataillon T, David JL, Reboud X, LeCorre V, Calloustian C, Gut IG, Brunel D (2004) Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J 38: 193–202 [DOI] [PubMed] [Google Scholar]
- Milborrow BV (1974) Biosynthesis of abscisic acid in a cell-free system. Phytochemistry 13: 131–136 [Google Scholar]
- Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 22: 947–952 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Noven LG, Haskell DW, Guy CL, Denslow N, Klein PA, Green LG, Silverman A (1992) Association of 70-kilodalton heat-shock cognate proteins with acclimation to cold. Plant Physiol 99: 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwarhara A, Seo M, Kushiro T, Asami T, Hirai N, Kamya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode ABA 8′-hydroxylases are indispensable for a proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 14: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TB (1924) The Vegetable Proteins. Longmans, Green, London
- Osherov N, May G (2000) Conidial germination in Aspergillus nidulans requires RAS signalling and protein synthesis. Genetics 155: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Mann M (2000) Proteomics to study genes and genomes. Nature 405: 837–846 [DOI] [PubMed] [Google Scholar]
- Penfield S, Graham S, Graham IA (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D (2006. a) Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol 141: 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Job C, Job D (2006. b) Proteome analysis for the study of developmental process in plants. In C Finnie, ed, Plant Proteomics. Blackwell Publishing, Oxford, pp 151–184
- Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Rinne PLH, Kaikuranta PM, van der Schoot C (2001) The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J 26: 249–264 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D (1984) The physiological role of storage proteins in seeds. Philos Trans R Soc Lond B Biol Sci 304: 275–785 [Google Scholar]
- Spremulli L (2000) Protein synthesis, assembly, and degradation. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry & Molecular Biology of Plants. American Society of Plant Physiologists, Rockeville, MD, pp 412–454
- Toyomasu T, Yamane H, Murofushi N, Inoue Y (1994) Effects of exogenously applied gibberellin and red light on the endogenous levels of abscisic in photoblastic lettuce seeds. Plant Cell Physiol 35: 127–129 [Google Scholar]
- Wang M, Heimovaara-Dijkstra S, Van Duijin B (1995) Modulation of germination of embryos isolated from dormant and non-dormant barley grains by manipulation of endogenous abscisic acid. Planta 195: 586–592 [Google Scholar]
- Wareing PF, Saunders PF (1971) Hormones and dormancy. Annu Rev Plant Physiol 22: 261–288 [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E (1996) Synthesis of small heat-shock protein is part of the developmental program of late seed maturation. Plant Physiol 112: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Endo T, Satoh S (1998) Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol 39: 307–312 [Google Scholar]