Abstract
Primary cell walls are deposited and remodeled during cell division and expansion. Secondary cell walls are deposited in specialized cells after the expansion phase. It is presently unknown whether and how these processes are interrelated. The Arabidopsis (Arabidopsis thaliana) MUR10 gene is required for normal primary cell wall carbohydrate composition in mature leaves as well as for normal plant growth, hypocotyl strength, and fertility. The overall sugar composition of young mur10 seedlings is not significantly altered; however, the relative proportion of pectin side chains is shifted toward an increase in 1 → 5-α-arabinan relative to 1 → 4-β-galactan. mur10 seedlings display reduced fucogalactosylation of tightly cell wall-bound xyloglucan. Expression levels of genes encoding either nucleotide sugar interconversion enzymes or glycosyl transferases, known to be involved in primary and secondary cell wall biosynthesis, are generally unaffected; however, the CesA7 transcript is specifically suppressed in the mur10-1 allele. The MUR10 locus is identical with the CesA7 gene, which encodes a cellulose catalytic subunit previously thought to be specifically involved in secondary cell wall formation. The xylem vessels in young mur10 hypocotyls are collapsed and their birefringence is lost. Moreover, a fucogalactosylated xyloglucan epitope is reduced and a 1 → 5-α-arabinan epitope increased in every cell type in mur10 hypocotyls, including cells that do not deposit secondary walls. mur10 also displays altered distribution of an arabinogalactan-protein epitope previously associated with xylem differentiation and secondary wall thickening. This work indicates the existence of a mechanism that senses secondary cell wall integrity and controls biosynthesis or structural remodeling of primary cell walls and cellular differentiation.
Cell walls are crucial for most aspects of plant life. Primary cell walls form the first intercellular boundary between newly divided cells. During cell expansion, walls have to locally yield to expansive turgor forces and continue to grow by the deposition of new wall material. After a cell has ceased to expand, it differentiates into a mature, specialized cell type. Usually this involves modifications to the cell wall, the most dramatic of which is the deposition of secondary cell wall material resulting in mechanical reinforcement. Several genes involved in primary and secondary cell wall carbohydrate biosynthesis have been identified, initially using forward genetic (Scheible and Pauly, 2004; Seifert, 2004) and, more recently, reverse genetic (Brown et al., 2005; Persson et al., 2005) analyses of Arabidopsis (Arabidopsis thaliana). The products of these genes can be roughly subdivided into four groups: first, nucleotide sugar metabolic enzymes that generate the active precursors for carbohydrate biosynthesis (Seifert, 2004); second, glycosyl transferases that generate specific sugar linkages (Scheible and Pauly, 2004); third, structural proteins, such as arabinogalactan-protein (AGP; Shi et al., 2003; Acosta-Garcia and Vielle-Calzada, 2004; Persson et al., 2005) and extensin-like proteins (Baumberger et al., 2001); and fourth, a number of important genes of unclear biochemical function, such as COBRA, KORRIGAN, or KOBITO1 (Nicol et al., 1998; Schindelman et al., 2001; Pagant et al., 2002). Remodeling of the cell wall involves hydrolytic or oxidative enzymatic and nonenzymatic changes mostly on cell wall matrix components (Rose et al., 2004). Most forward genetic screens, instructive for our understanding of cell wall biosynthesis, were either simple morphological screens (Schiefelbein and Somerville, 1990; Baskin et al., 1992; Benfey et al., 1993; Hauser et al., 1995; Fagard et al., 2000; Favery et al., 2001) and embryo-lethal screens (Lukowitz et al., 2001), or screens for microscopically altered cellular morphology (Turner and Somerville, 1997) and mechanosensory properties (Zhong et al., 1997). A sophisticated screen to identify mutants displaying altered primary cell wall carbohydrate composition was performed in the Somerville lab and has produced the mur (from Latin: murus, wall) mutant series (Reiter et al., 1997). The mur mutants led to the identification of the MUR1 and MUR4 genes, which encode nucleotide sugar interconversion genes (Bonin et al., 1997; Molhoj et al., 2004) and the MUR2 and MUR3 genes, which encode glycosyl transferases (Vanzin et al., 2002; Madson et al., 2003) that are involved in cell wall matrix biosynthesis. The mur1 to mur8 loci are defined by a reduction in one single cell wall sugar, whereas the remaining loci mur9 to mur11 display more complex changes. In addition to their compositional cell wall defects, the mur1, mur9, and mur10 mutants display different morphological phenotypes, such as dwarfism and chlorosis (Reiter et al., 1997). The mur10 locus is defined by two independent mutant alleles that show a reduction in cell wall-bound Xyl and Fuc and a concomitant increase in Ara in 3-week-old leaves. Although mur10 was not further analyzed, the compositional changes suggest an alteration in xyloglucan and pectic arabinan, which are the main primary cell wall carbohydrates containing these sugars. As opposed to more specific changes that are to be expected when individual enzymes are deficient, the complex mur10 phenotype is suggestive of an involvement of MUR10 in the regulation of cell wall biosynthesis or remodeling. To address these possibilities, we decided to further characterize the mur10 phenotype on a biochemical, histological, and molecular genetic level. We describe changes in the primary and secondary cell walls of mur10 and identify mur10-1 and mur10-2 as new mutant alleles of the CesA7/IRX3/FRA5 gene that encodes a secondary cell wall-specific cellulose synthase catalytic subunit. We propose the existence of a quality control mechanism that links secondary cell wall cellulose integrity with primary cell wall deposition or remodeling.
RESULTS
Morphological, Mechanical, and Biochemical Defects in mur10 Seedlings
mur10 plants display a phenotype early during development that is visible on petri dishes in 10-d-old seedlings (Fig. 1). Compared to the wild type, mur10 seedlings show a reduction in growth and dark-green coloration of the aerial parts. Once plants are transferred to soil, the clear differences between mutant and wild-type phenotype are maintained. Both alleles are smaller and darker than the wild type (Fig. 1). As reported previously, mutations in MUR10 cause a dramatic reduction in fertility, typically with fewer than 10 seeds per plant (Reiter et al., 1997). Reciprocal crosses between mur10 and other mutants or wild-type plants were usually successful when mur10 was used as the pollen donor, but failed when mur10 plants were pollinated, indicating reduced female fertility. Mechanical analysis of 4-d-old dark-grown hypocotyls shows significant reduction in the tensile strength of mur10 compared to wild type (Table I). These results indicate that mur10 hypocotyls require less tensile force to break them. In addition, the diameter of mur10 hypocotyls is 10% greater than wild type (Table I).
Figure 1.
Early morphological phenotype of mur10 mutants. Ten-day-old mur10-1 seedlings show early differences on plates compared to the wild type (A). Phenotype on soil of 3-week-old plants, Col-0 (B), mur10-1 (C), and mur10-2 (D).
Table I.
Mechanical properties of 4-d-old hypocotyls
Mechanical parameters were obtained for Col-0 and mur10 hypocotyls. Significant differences were obtained in tensile strength and diameter (average of 20 samples ± sd).
| Col-0 | mur10-1 | Dunnett's Test | |
|---|---|---|---|
| Tensile modulus | 19.54 ± 5.40 MPa | 17.94 ± 3.71 MPa | Not significant |
| Tensile strength | 1.286 ± 0.215 MPa | 1.119 ± 0.197 MPa | P < 0.05 |
| Failure strain | 0.129 ± 0.029 | 0.115 ± 0.031 | Not significant |
| Diameter | 0.253 ± 0.020 mm | 0.284 ± 0.017 mm | P < 0.001 |
Subtle and Complex Alterations of mur10 Primary Cell Walls
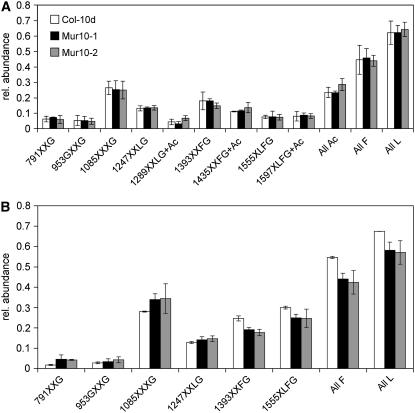
Despite the clear morphological phenotype, cell wall neutral sugar composition does not consistently differ between 10-d-old Columbia (Col-0) and mur10 seedlings. Mutants show a reduction in Fuc, Xyl, Ara, and Man in both alleles (not statistically significant), whereas Rha and Gal differ between alleles (data not shown). Pectin composition of 10-d-old seedlings obtained by polysaccharide analysis using carbohydrate gel electrophoresis (PACE; Barton et al., 2006) shows an increased arabinan-to-galactan ratio in both alleles. However, compared to the wild type, the mur10-1 allele shows a decrease in the relative amount of galactan and arabinan, whereas the mur10-2 allele shows a decrease in galactan, but an increase in arabinan (Table II). Oligosaccharide mass profiling (OLIMP) of cell walls directly treated with a xyloglucan-specific endoglucanase (XEG) highlights the composition of xyloglucan bound relatively loosely between cellulose microfibrils (Lerouxel et al., 2002). Alkali extraction preceding XEG treatment discriminates xyloglucan tightly attached to the surface of cellulose microfibrils (Pauly et al., 1999). OLIMP analysis indicates no differences between wild type and both mutant alleles in loosely bound xyloglucan (Fig. 2A). Analysis of xyloglucan tightly attached to cellulose indicates a relative increase of fucosylated oligosaccharides compared to loosely bound xyloglucan in wild-type cell walls (Table III; Fig. 2B). In both mur10 mutant alleles, however, the amounts of fucosylated oligosaccharides in loosely and tightly bound xyloglucan are equal (Table III; Fig. 2B). As shown below, mur10 displays defective xylem structure. To test whether the difference in fucosylated oligosaccharides observed between wild type and mutant might be due to the delay in development or due to increased water stress, we have analyzed the wild type at four different ages and also in drought conditions. This analysis indicates a steady increase of fucosylation of loosely bound xyloglucan with increasing seedling age (Table III). In all cases, including drought stress, however, fucosylation of tightly bound xyloglucan was considerably higher than in the loosely bound fraction. This indicates that tightly bound xyloglucan at an early developmental stage in both mur10 mutant alleles is specifically underfucosylated.
Table II.
PACE analysis of pectic arabinan and galactan in mur10
Values are relative to wild-type controls ± sd between biological duplicates. In both mutant alleles, pectin is more strongly arabinosylated than galactosylated compared to wild type.
| Genotype | Galactan | Arabinan |
|---|---|---|
| % wild type | % wild type | |
| mur10-1 | 64.0 ± 14.6 | 71.7 ± 0.4 |
| mur10-2 | 88.0 ± 13.6 | 112.5 ± 10.6 |
Figure 2.
Xyloglucan composition after digestion with XEG in Col-0, mur10-1, and mur10-2. Oligosaccharide composition of XEG-treated walls (A). Xyloglucan oligosaccharide composition after subsequent alkali treatment of the residue and XEG digestion of the solubilized xyloglucan (B). Numbers indicate the mass of the oligosaccharides and the corresponding xyloglucan oligosaccharide structure written in the nomenclature by Fry et al. (1993) All Ac, All L, All F, Sum of the abundance of all xyloglucan oligosaccharide ions containing O-acetyl substituents, Gal residues, and/or fucosyl residues, respectively.
Table III.
Comparison of the relative amount of fucosylated oligosaccharides before and after alkali extraction between samples of different ages and between normal and drought conditions (average values of biological triplicates ± sd)
ND, Not determined.
| Genotype | Age | Ratio of Fucosylated XGO
|
|
|---|---|---|---|
| XEG only | KOH/XEG | ||
| d | |||
| Col-0 | 10 | 0.45 ± 0.094 | 0.55 ± 0.152 |
| mur10-1 | 10 | 0.46 ± 0.182 | 0.44 ± 0.026 |
| mur10-2 | 10 | 0.44 ± 0.036 | 0.42 ± 0.046 |
| Normal condition (wild type) | 6 | 0.36 ± 0.037 | 0.57 ± 0.006 |
| 8 | 0.38 ± 0.014 | 0.58 ± 0.007 | |
| 10 | 0.45 ± 0.094 | 0.55 ± 0.152 | |
| 12 | 0.43 ± 0.019 | ND | |
| 6 | 0.33 ± 0.017 | 0.50 ± 0.031 | |
| Drought condition (wild type) | 8 | 0.37 ± 0.043 | 0.58 ± 0.081 |
| 10 | 0.46 ± 0.005 | 0.55 ± 0.015 | |
| 12 | 0.38 ± 0.093 | 0.59 ± 0.078 | |
Normal Expression of Genes Involved in Cell Wall Biosynthesis
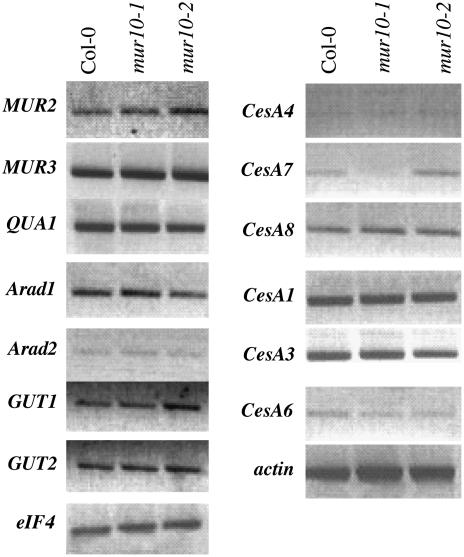
The complex alterations in both structure and amount of different wall polysaccharides prompted us to analyze the transcript levels of genes encoding nucleotide sugar interconversion enzymes involved in the synthesis of Fuc and Xyl and the glycosyl transferase genes involved in primary cell wall polysaccharides, xyloglucan, and pectin. However, neither the transcript levels of genes encoding nucleotide sugar interconversion enzymes (data not shown) nor the glycosyl transferases specifically involved in cell wall matrix polymer biosynthesis (Fig. 3) are affected in mur10 mutants. Genes encoding cellulose synthase catalytic subunits are also unaffected (Fig. 3). The only exception is the CesA7 transcript, which is less abundant in the mur10-1 mutant compared to wild type and the mur10-2 mutant (Fig. 3).
Figure 3.
Expression levels of genes involved in cell wall synthesis. Genes coding for glycosyl transferases involved in the synthesis of xyloglucan and pectin (left). Genes coding for the cellulose synthase catalytic subunit (CesA) involved in primary (CesA1, -3, -6) and secondary (CesA4, -7, -8) cell wall formation (right). Only the CesA7 transcript level is altered in mur10-1. Actin and eIF4, which generally show identical expression, are used as normalization controls.
Identification of MUR10
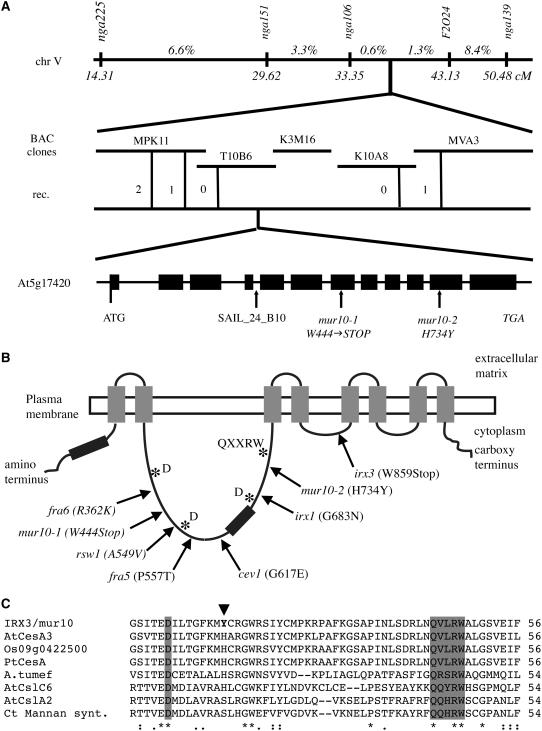
The morphological phenotype segregating in an F2 population mur10-2 (Col-0) × Landsberg erecta (Ler) was mapped to the corresponding genetic locus. Initial linkage analysis, involving 24 mutant plants, locates MUR10 to the top of chromosome V between the simple sequence length polymorphic markers nga106 and nga139. These markers are located at 33.35 and 50.48 cM, respectively, on the recombinant inbred map for Arabidopsis chromosome V (Lister and Dean, 1993). To fine map MUR10, we analyzed a total population of 470 F2 individuals using newly generated markers between nga106 and nga139 markers. This narrows down the position of mur10 to a region of 68 kb, covered by three bacterial artificial chromosome (BAC) clones, T10B6, K3M16, and K10A8 (Fig. 4A). Parallel to the mapping effort, T-DNA insertion lines of different genes, included in these three BACs and possibly involved in cell wall biosynthesis, were phenotypically analyzed. A T-DNA insertion line (Syngenta Arabidopsis Insertion Library [SAIL]_24_B10) in the At5g17420 locus segregates for a phenotype quite similar to mur10 in an approximately 3:1 ratio (20:7). Crosses between herbicide-resistant, phenotypically normal, heterozygous SAIL_24_B10 plants and homozygous mur10 mutants produce approximately 50% mur10-like offspring, confirming allelism. To establish that MUR10 and At5g17420 are identical, the locus was sequenced in both independent mur10 alleles. mur10-1 sequences show a G1332-to-A nucleotide substitution, resulting in the introduction of a stop codon in place of Trp-444 (Fig. 4, A and B); mur10-2 contains a C2200-to-T nucleotide substitution, replacing His-734 with a Tyr residue (Fig. 4, A–C). This His residue is conserved between all cellulose synthase sequences, including primary cell wall-specific Arabidopsis CesA3 (Ellis et al., 2002; Cano-Delgado et al., 2003) and putative cellulose synthase catalytic subunit sequences from rice (Oryza sativa), poplar (Populus tremuloides), and Agrobacterium tumefaciens. It is also conserved in some sequences of the cellulose synthase-like family (Csl), such as AtCslC6, but is replaced with Ser in the 1 → 4-β-mannan synthase from Cyamopsis tetragonoloba (Dhugga et al., 2004) and its putative ortholog in Arabidopsis. The high degree of conservation of this residue is consistent with the strong mutant phenotype of mur10-2 resembling the putative full loss-of-function allele mur10-1. We conclude that MUR10 is identical to At5g17420, which is the gene encoding CesA7/IRX3/FRA5, a cellulose synthase catalytic subunit involved in secondary cell wall biosynthesis (Taylor et al., 1999; Zhong et al., 2003).
Figure 4.
Identification of MUR10. MUR10 was mapped to the BAC clone T10B6 on chromosome V. The position of the two mur10 alleles and the T-DNA insertion SAIL_24_B10 are indicated by arrows (A). Summary of various mutations on Arabidopsis CesA isoforms (B). Alignment of selected CesAs (IRX3, AtCesA3/At5g05170, Os09g0422500; poplar/AAM26299; A. tumefaciens/NP_357298); and Csl sequences (AtCslC6 /At3g07330, AtCslA2/At5g22740, C. tetragonoloba 1 → 4-β-mannan synthase/AAR23313) around the mur10-2 mutation (arrowhead). The universally conserved Asp and Gln-Aaa-Aaa-Arg-Trp motifs are boxed gray (C).
Altered Secondary Cell Walls in mur10
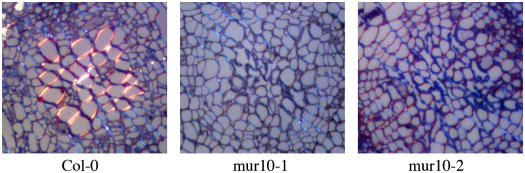
The IRX3 locus was identified in a screen for irregular xylem morphology. The irx mutants show collapse of xylem vessel cells and an almost complete absence of secondary wall cellulose (Turner and Somerville, 1997). To determine whether xylem cells in young mur10 plants are collapsed, we analyzed the hypocotyl histology in toluidine blue-stained semithin sections of 10-d-old plants using polarized light microscopy to additionally highlight the birefringence of thick cellulose deposits. Wild-type hypocotyls contain thick-walled xylem vessels with a relatively round shape, whereas mur10 xylem vessel elements are small and collapsed (Fig. 5). Polarized light microscopy reveals the absence of thick cellulose deposits in both mutant alleles in comparison to the wild type, where secondary cellulose is present in the xylem vessels. On the other hand, collapsed vessels in the mutant show intense blue staining potentially indicating lignin deposition.
Figure 5.
Polarization microscopy of 10-d-old hypocotyl toluidin blue-stained sections of wild-type Col-0, mur10-1, and mur10-2. Birefringence of aligned cellulose is indicated by bright contrast against gray background. The wild-type secondary wall is brightest due to its thickness. Primary walls are less bright and are contrasted equally in wild type and mutants. Dark blue deposits potentially indicate ligin.
The CesA7 Promoter Is Active in Secondary Cell Wall-Forming Tissue
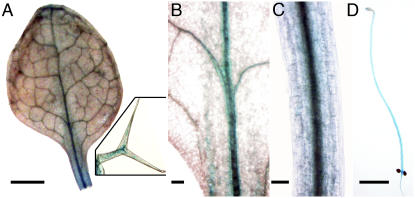
Previous data suggest that IRX3/FRA5/MUR10 expression is strictly associated with secondary cell wall formation (Brown et al., 2005; Persson et al., 2005). The IRX3 promoter:β-glucuronidase (GUS) reporter gene is mainly expressed in vascular tissue of light- and dark-grown hypocotyls and in leaves (Fig. 6). In leaves, expression is also seen in trichomes (Fig. 6A, inset). Expression is higher at the leaf base compared with the more peripheral regions (Fig. 6B). In light-grown hypocotyls, expression is very strong in the vascular cells, but is absent from the epidermis and cortex (Fig. 6C). In dark-germinated seedlings, expression is apparent throughout the length of the elongated hypocotyl (Fig. 6D).
Figure 6.
Expression of the pIRX3:GUS reporter in 10-d-old light-grown seedlings (A–C) and 4-d-old dark-grown seedlings (D). Scale bars = 1 mm and 125 μm for inset (A); 100 μm (B and C); and 3 mm (D).
Cell Wall Alterations in mur10 Xylem and Nonxylem Cells
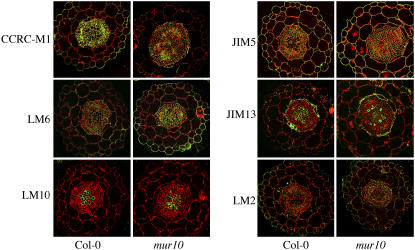
A series of antibodies directed against primary and secondary cell wall polysaccharide epitopes decorate wild-type and mur10 hypocotyl sections (Fig. 7). Labeling reveals subtle differences between sections of mur10 and wild type in their cross-reactivity with CCRC-M1 and LM6 antibodies, which bind to components of primary cell wall, fucogalactosylated xyloglucan (Freshour et al., 1996), and pectic (1 → 5) α-l-arabinan (Willats et al., 1998), respectively. On the one hand, there is a marked reduction of CCRC-M1 labeling in mur10 compared to wild type, affecting all tissues, including epidermal and ground cells, which largely have primary cell walls, which is consistent with the OLIMP data. On the other hand, LM6 labeling is stronger in mur10 than in wild type, which is consistent with PACE data. The LM10 monoclonal antibody that binds to xylan (McCartney et al., 2005), a hemicellulose enriched in secondary cell walls, labels both wild type and mur10 with comparable intensity, but clearly highlights the collapse of the xylem vessels in mur10. JIM13, a monoclonal antibody cross-reacting with a carbohydrate epitope present on AGP, has previously been shown to bind to root cells destined to differentiate into xylem (Dolan et al., 1995). In sections of wild-type hypocotyls, JIM13 labels endodermis cells and large xylem vessels in the center of the section. In mur10 sections, JIM13 also labels the endodermis, but the collapsed xylem vessels are unlabeled. Instead, JIM13 epitopes are highly abundant in the walls of small cells (probably xylem parenchyma) in the proximity of the xylem vessels. Labeling with JIM5, which binds to predominantly deesterified homogalacturonan (Clausen et al., 2003), and LM2, which recognizes a d-glucuronyl-containing AGP epitope (Yates et al., 1996), does not vary between mur10 and wild type.
Figure 7.
Immunoreflection labeling with antibodies against cell wall polysaccharide epitopes in medial transverse sections of 10-d-old light-grown hypocotyls. Antibody distribution is shown in green; tissue autofluorescence is shown in red.
DISCUSSION
MUR10 and IRX3/FRA5 Are Identical
In this study, we show that the MUR10 gene, which has previously been shown to be required for wild-type neutral sugar composition in primary cell walls, is identical to the IRX3/FRA5 locus that is necessary for cellulose deposition in secondary walls. This is supported by mapping of the morphological mur10 phenotype to the IRX3/FRA5 region, identification of single-base-pair substitutions in both independent mur10 alleles, observation that T-DNA insertions in IRX3/FRA5 in the Col-0 background display the mur10 morphological phenotype, and, by a similar, but less pronounced, neutral sugar phenotype in the irx3-1 mutant.
Both mur10 mutations occur between two different highly conserved regions important for substrate binding and catalysis (Fig. 4B) and probably represent complete or near-complete loss-of-function alleles. Whereas the missense mur10-2 mutation affects a His residue located between a conserved Asp residue and a conserved QxxRW motif, which is conserved among all CesA genes from various organisms but is not totally conserved in the CesA/Csl superfamily (Fig. 4C), the mur10-1 mutation causes a premature termination of translation in place of the residue Trp-444. Although there might be some residual function in mur10-2, the growth phenotype and all other phenotypic aspects were very similar to mur10-1, which presumably represents a full loss-of-function allele. This is probably also the case with the irx3-1 allele that results in premature termination of translation at amino acid residue Trp-859 (Taylor et al., 1999). The much more dramatic effect of IRX3 loss of function on growth observed with mur10 and irx3-4 (Brown et al., 2005), both in the Col-0 background, compared to irx3 in the Ler background, is intriguing because it might point to genetic differences in the way these two accessions respond to aberrant secondary cell walls.
Comparative reverse transcription-PCR analysis of transcript levels of CesA genes in wild type and two mur10 alleles indicates a specific reduction of the CesA7 transcript in mur10-1, whereas all other CesA genes have equal relative abundance in both mutant alleles and wild type. A reduction in CesA7 transcript abundance in mur10-1 might be related to the nature of the mutation in the CesA7 gene because premature stop codons are known to reduce transcript abundance (Carol et al., 2005). Initially, IRX3 was defined by the irx3 allele in the Ler background (Turner and Somerville, 1997). Neutral sugar analysis indicated no significant differences between wild type and irx3 or two other irx mutants (Turner and Somerville, 1997; Ha et al., 2002). However, their cell wall material was obtained from mature hypocotyls and, in a later study, isolated sclerenchyma cells and showed a composition like the xylan-rich secondary cell walls of xylem. In the screen for the mur mutants, on the other hand, leaves that are rich in primary cell walls were used (Reiter et al., 1997). This explains why alterations in primary cell walls might have been overlooked in the original irx3 analysis. However, in both studies, the relative content of Ara was increased in irx3 compared to the wild type, which is also a feature of mur10. Another difference between irx3 and mur10 is the growth and sterility phenotype. It was found that irx1 and irx3 mutants grew normally except being smaller than wild type and that irx3 displayed a tendency to recline (Turner and Somerville, 1997). In a later reverse genetic study, T-DNA insertion lines in IRX3 in the Col-0 background were found to cause the same morphological phenotype as mur10, including very small, dark green, and infertile plants (Brown et al., 2005). Therefore, the differences between irx3 and mur10 are, at least partially, due to a different effect of mutations in IRX3 in the Col-0 and Ler genetic backgrounds.
Defects in IRX3/FRA5/MUR10 Cause Alterations in Xyloglucan and Pectin Remodeling throughout the Plant
We started our initial characterization of mur10 based on previously published information that suggested a role for MUR10 in primary cell wall composition or structure. The previously observed increase in Ara and decrease in Xyl and Fuc indicated complex changes in pectin and xyloglucan. In fact, it has previously been demonstrated that less xyloglucan is solubilized from mur10 compared to wild type (Lerouxel et al., 2002). We show a clear morphological defect in both mur10 alleles in 10-d-old seedlings and, because the previous studies were performed in relatively mature plants grown on soil, we decided to reproduce the analysis at an earlier stage in seedlings grown at 100% humidity on petri dishes. On the one hand, we failed to find pronounced alteration in neutral sugar composition. This could indicate that crude primary cell wall alterations only become manifest at later stages of growth. On the other hand, we show an alteration in both types of pectin side chains as well as in fucosylation of xyloglucan tightly bound to cellulose. Although the content of xyloglucan fucosylation in wild-type seedlings increases with age between 6 and 10 d, fucosylation in the tightly bound fraction is always significantly higher than in the loose fraction. Drought stress does not change this relationship. The specific relative reduction of fucosylation in the tightly bound fraction in mur10 is therefore unlikely to be a consequence of slower development or of increased drought stress, but it could be a more specific consequence of the mutation. It appears that the alteration in xyloglucan fucosylation affects cell types not undergoing substantial secondary wall thickening as indicated by immunolabeling with CCRC-M1 antibody. We only observe a gradual decrease of labeling throughout the section consistent with the partial reduction of fucosylation in all cell types. We also find an increased proportion of pectic arabinan versus pectic galactan in both mur10 alleles compared to wild type. There are, however, differences between the two mur10 alleles in relation to the wild-type value, indicating a relative increase of arabinan and a decrease of galactan in mur10-2 compared to wild type and a relative decrease of both arabinan and galactan in mur10-1. Consistent with an increase in pectic arabinan, we find more intense labeling of mur10 with the LM6 antibody that recognizes the 1 → 5 α-arabinan epitope. As with CCRC-M1, differential labeling is not restricted to any particular cell type. This, together with other previously published data (Reiter et al., 1997; Lerouxel et al., 2002), indicates that the alterations of secondary cell walls lead to alterations of primary cell wall composition throughout the plant. Alterations of the primary cell wall as a response to secondary cell wall defects might seem counterintuitive because the formation of primary cell walls has normally already stopped when secondary walls are being deposited. However, cell wall carbohydrates are remodeled in muro by various enzymes such as glycosyl hydrolases and carbohydrate esterases after their deposition. It is entirely possible that the physiological responses triggered by defective secondary cell walls in the mur10 mutant lead to an alteration of xyloglucan and pectin remodeling, causing a decrease of fucosylation specifically in tightly bound xyloglucan, whereas newly deposited xyloglucan is unchanged. Another possibility to explain the alterations of primary cell walls in mur10 is that the stress responses triggered by defective secondary cell walls that are deposited early in development lead to a gradual change in plant physiology that would also affect primary cell wall deposition in tissue that forms at a later stage. A subset of genes, thought to be specifically involved in cell wall polymer biosynthesis, does not show apparent alterations in transcript abundance; however, many more unknown genes are involved in the biosynthesis and remodeling of primary cell walls. This question could be addressed with genome-wide transcript analysis.
We also observe that immunoreactivity with the monoclonal antibody JIM13, which binds to a specific subset of AGP, is subtly altered between wild type and mur10. Whereas the wild type is labeled in its xylem, the mutant is not and, instead, has strongly labeled cells at the periphery of the collapsed xylem. JIM13 has recently been associated with xylem differentiation in roots (Dolan et al., 1995) and has been shown to bind to xylogen, an AGP-like glycoprotein that acts as an inductive signal in vascular development (Motose et al., 2004). The strong expression of the JIM13 reactive epitope in cells in the zone surrounding the collapsed mur10 xylem might indicate enhanced differentiation activity in response to the xylem defects. Increased cambial divisions might also explain the increase in hypocotyl thickness in mur10.
We find a small, but significant, decrease in the tensile strength of 4-d-old dark-grown hypocotyls in mur10. In much more mature stem tissue, which contains a higher proportion of secondary cell walls, the bending modulus and maximal bending stress of wild type increased between 34- and 42-d-old plants, but were much lower and failed to increase with age in irx3 stems (Turner and Somerville, 1997). The reduced tensile strength in young hypocotyls is probably a result of the reduction of cellulose in the xylem. However, because xyloglucan composition in this organ is known to affect their tensile properties (Ryden et al., 2003), the reduction of strength could also be an indirect consequence of the alterations in primary cell wall matrix polymers.
Alterations of Cell Wall Components Have Complex Effects on Plant Physiology
It has become clear that the cell wall is an extremely complex reservoir of chemical and mechanical information (Somerville et al., 2004). Numerous studies indicate that defective cell walls can trigger dramatic responses. The most prominent examples come from the field of plant pathology, where several disease resistance pathways become activated in cell wall-defective mutants (for review, see Pilling and Hofte, 2003; Schulze-Lefert, 2004; Vorwerk et al., 2004). Mutations in the CesA3 gene, which is involved in primary cell wall biosynthesis, trigger ectopic lignification, constitutive ethylene and jasmonic acid signaling, and powdery mildew resistance (Cano-Delgado et al., 2000; Ellis and Turner, 2001; Ellis et al., 2002). The disruption of COBRA, encoding a glycosylphosphotidylinositol-anchored protein required for normal cellulose microfibril orientation in elongating cells (Roudier et al., 2005), leads to constitutive production of jasmonic acid and increased expression of defense-related genes (Ko et al., 2006). Mutation in callose synthase and a pectate lyase were isolated as powdery mildew resistance loci (Vogel et al., 2002; Nishimura et al., 2003). Mutations in the AtXTH27 gene, coding for a cell wall-remodeling enzyme, cause a lesion-mimic phenotype (Matsui et al., 2005). RNA interference with pectin biosynthesis, pharmacological interference with cellulose biosynthesis, and AGP damage in suspension cell function all triggered programmed cell death (Guan and Nothnagel, 2004; Duval et al., 2005; Ahn et al., 2006). The above examples indicate that defective primary cell walls affect signaling events leading to local and systemic stress responses. Our finding suggests that secondary cell wall changes are sensed by younger growing tissues and also lead to complex alterations in primary cell wall composition throughout the plant.
CONCLUSION
In summary, we have demonstrated that mur10 mutants show complex alterations in their primary cell wall structure that are most likely the consequence of altered cell wall remodeling triggered by the disruption of secondary cell wall structure caused by a defect in the CesA7 locus. Because primary and secondary cell wall formation are temporally and spatially separated, this suggests the involvement of a systemic signaling mechanism that transports information about the integrity of the secondary cell wall in specific cells to the machinery that makes and remodels the primary cell wall throughout the plant.
MATERIALS AND METHODS
Mutant Lines, Growth Conditions, and Mapping
The mutant Col-0 lines mur10-1 and mur10-2 were kindly provided by Wolf-Dieter Reiter. Seeds were surface sterilized in 10% sodium hypochlorite for 10 min and rinsed three times in sterile deionized water. Seeds were then plated on Murashige and Skoog medium (pH 5.8; Duchefa), supplemented with 1% Suc, and 0.5% Phytagel. Seeds were placed at 4°C for 48 h in darkness and then grown in continuous light (80 μmol m−2 s−1) at 25°C in vertically oriented petri dishes. For drought stress induction, seedlings were transferred to new petri dishes with filter paper instead of Murashige and Skoog medium. Filter paper was watered with 1 mL water and afterward filter paper was watered every 2 d, reducing the amount of water from 1 mL to 250 μL. This treatment resulted in gradual leaf darkening and inhibition of growth. Prolonged exposure led to complete dehydration of the plants.
The mur10 locus was mapped using plants with a mutant phenotype as described below in the F2 from a cross between mur10-2 (Col-0) and Ler. Initial mapping was performed using simple sequence length polymorphic markers from The Arabidopsis Information Resource (http://www.arabidopsis.org). Additional markers for fine mapping were generated based on the information available by CEREON (Jander et al., 2002). DNA was amplified in 20-μL reactions as described by Bell and Ecker (1994). PCR was performed using Taq polymerase (Invitrogen Life Technologies) in a Peltier thermal cycler (MJ Research). PCR conditions were as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 10 s, 50°C to 60°C for 10 s, and 72°C for 20 s.
Reverse Transcription-PCR
RNA was isolated from 10-d-old seedlings using an RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 10 μL of total RNA using oligo(dT)12 (Invitrogen) and 200 units reverse transcriptase (Invitrogen) and incubated in the following program, 42°C for 60 min, 50°C for 20 min, and 92°C for 2 min. cDNA was diluted with water. For comparative PCR, different cDNAs were normalized using eukaryotic translation initiation factor 4E (eIF4E, AT4g18040) and actin 8 (At1g49240) specific primers. Glycosyl transferase genes analyzed were MUR2, At2g03220; MUR3, At2g20370; QUA1, At3g25140; Arad1, At2g35100, Arad2, At4g44930; GUT1, At5g61840; GUT2, At1g27440; CesA1, At4g32410; CesA3, At5g05170; CesA6, At5g64740; CesA4, At5g44030; CesA7, At5g17420; CesA8, At4g18780.
Promoter-GUS Fusions
Primers CAGAATTCAAGTAGCTGCCCA and GCGTCGACAGGGACGGCCGGAGATTAGCT were used with Pfu DNA polymerase (Promega) to amplify an 849-bp IRX3 promoter fragment from cosmid L6 (Taylor et al., 1999). This PCR product was digested with EcoRI and SalI and ligated to similarly digested pBluescript KSII (Stratagene) to give plasmid pP8, which was sequenced and digested with SalI and EcoRI, to ligate the 850-bp insert to pCAMBIA1381Z yielding plasmid pIRX3:GUS. Ler plants were transformed by Agrobacterium tumefaciens (GV3101) carrying pIRX3:GUS according to Clough and Bent (1998). Transformants were selected on hygromycin. A number of lines showing a common staining pattern were selected for further study and grown to homozygosity. Seedlings were stained for GUS activity in 1 mm X-Gluc in 50 mm NaHPO4 (pH 7.2), 0.5% Triton X-100 for 24 h at 37°C. After clearing in three changes of 70% ethanol, seedlings were mounted in water for imaging. Whole leaves and dark-grown hypocotyls were photographed using the Macro function on a Nikon Coolpix 4500 and the higher magnification leaf and hypocotyls were photographed using the same camera attached to a Nikon Eclipse E-600 microscope.
Immunohistochemistry
Reflection microscopy of silver-enhanced immunogold-labeled resin-embedded tissue sections and electron microscopy was performed as previously described (Bush and McCann, 1999). All treatments of wild type and mutants, including image acquisition and processing, were performed in parallel.
OLIMP
Xyloglucan oligosaccharides were analyzed on cell wall material of seedlings using the protocol described by Lerouxel et al. (2002). After XEG digestion, an alkali extraction with 4 m KOH was performed to analyze the xyloglucan tightly bound to cellulose microfibrils. For alkali treatment, 600 μL of KOH were added to the cell wall material present in the remaining pellet after XEG digestion. After incubation with KOH for 16 h, supernatants were neutralized with acetic acid and dialyzed against water. Supernatants were added to Microcon columns and centrifuged for 70 min at maximal speed. Columns were washed three times with 500 μL deionized water and centrifuged for 45 min after each wash. Finally, columns were transferred to a new tube and centrifuged at 1,000 relative centrifugal force for 3 min. The alkali-solubilized xyloglucan polymers were digested with XEG by incubating the samples o/n at 37°C in 500 μL of XEG buffer and the corresponding amount of enzyme. OLIMP analysis was performed on the supernatant.
PACE Analysis of Galactan and Arabinan
Fresh 10-d-old seedlings were harvested and transferred to 2-mL screw-cap microtubes. Samples were frozen in liquid nitrogen and freeze dried to keep them at room temperature. Cell wall was extracted using a modification of the protocol described by Barton et al. (2006). Plant materials were incubated for 10 min in 65% ethanol at 65°C to inactivate enzymes. They were ground in a ball Mixer Mill MM200 (Glen Creston). The homogenate was centrifuged at 16,000g for 10 min. The pellet was washed with 65% ethanol centrifuged at 16,000g for 10 min, washed with methanol:chloroform (2:3 [v/v]; two times), centrifuged at 16,000g for 10 min, and washed with 100% acetone (two times). The remaining cell wall pellet was dried in a Speed-Vac concentrator SVC100H (Savant). PACE analysis of (1 → 4)-β-galactan, (1 → 5)-α-arabinan was performed as previously described (Barton et al., 2006).
Determination of Tensile Strength of Hypocotyls
Tensile strength was determined in basal regions of 4-d-old dark-grown hypocotyls grown on horizontally oriented petri dishes as previously described (Ryden et al., 2003).
Acknowledgments
We gratefully acknowledge the technical help of David Rico Arcos for the neutral sugar composition analysis and the provision of infrastructure for PACE by Paul Dupree. Kim Findlay and Sue Bunnewell helped with histological preparations. We are grateful for general technical help by Chris Barber. Michael Hahn and Paul Knox provided monoclonal antibodies and Wolf-Dieter Reiter provided mur10 seeds. The Arabidopsis Biological Resource Center provided T-DNA lines. S.B. identified the MUR10 locus, performed most experiments, and cowrote the manuscript; L.N. and M.P. planned and performed OLIMP and neutral sugar analyses; C.J.B. performed pectin PACE. P.R. provided mechanical analyses; N.G.T. analyzed IRX3:GUS expression; and G.J.S. initiated and supervised the project in the K.R. lab and cowrote the manuscript.
This work was supported by a Marie Curie student grant (grant no. MEST–CT–2004–504273 to S.B.), by the Biotechnology and Biological Science Research Council (grant no. 208/D10332 to G.J.S.) and the European Union (grant no. QLK5–CT–2001–00443 [EDEN] to G.J.S.), by a Royal Society University Research fellowship (to N.G.T.), and by the Biotechnology and Biological Science Research Council (grants to C.J.B. and P.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Georg J. Seifert (georg.seifert@boku.ac.at).
Open Access articles can be viewed online without a subscription.
References
- Acosta-Garcia G, Vielle-Calzada JP (2004) A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JW, Verma R, Kim M, Lee JY, Kim YK, Bang JW, Reiter WD, Pai HS (2006) Depletion of UDP-D-apiose/UDP-D-xylose synthases results in rhamnogalacturonan-II deficiency, cell wall thickening, and cell death in higher plants. J Biol Chem 281: 13708–13716 [DOI] [PubMed] [Google Scholar]
- Barton CJ, Tailford LE, Welchman H, Zhang Z, Gilbert HJ, Dupree P, Goubet F (2006) Enzymatic fingerprinting of Arabidopsis pectic polysaccharides using polysaccharide analysis by carbohydrate gel electrophoresis (PACE). Planta 224: 163–174 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Betzner AS, Hoggart R, Cork A, Williamson RE (1992) Root morphology mutants in Arabidopsis thaliana. Austr J Plant Physiol 19: 427–437 [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci USA 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MS, McCann MC (1999) Pectic epitopes are differentially distributed in the cell walls of potato (Solanum tuberosum) tubers. Physiol Plant 107: 201–213 [Google Scholar]
- Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado AI, Metzlaff K, Bevan MW (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Clausen MH, Willats WG, Knox JP (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al (2004) Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Dolan L, Linstead P, Roberts K (1995) An AGP epitope distinguishes a central metaxylem initial from other vascular initials in the Arabidopsis root. Protoplasma 189: 149–155 [Google Scholar]
- Duval I, Brochu V, Simard M, Beaulieu C, Beaudoin N (2005) Thaxtomin A induces programmed cell death in Arabidopsis thaliana suspension-cultured cells. Planta 222: 820–831 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Guan Y, Nothnagel EA (2004) Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol 135: 1346–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha MA, MacKinnon IM, Sturcova A, Apperley DC, McCann MC, Turner SR, Jarvis MC (2002) Structure of cellulose-deficient secondary cell walls from the irx3 mutant of Arabidopsis thaliana. Phytochemistry 61: 7–14 [DOI] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN (1995) Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Kim JH, Jayanty SS, Howe GA, Han KH (2006) Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J Exp Bot 57: 2923–2936 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Seveno M, Usadel B, Faye L, Lerouge P, Pauly M (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C (1993) Recombinant inbred lines for mapping rflp and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42: 525–534 [DOI] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Molhoj M, Verma R, Reiter WD (2004) The biosynthesis of d-galacturonate in plants: functional cloning and characterization of a membrane-anchored UDP-d-glucuronate 4-epimerase from Arabidopsis. Plant Physiol 135: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878 [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H (1998) A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Hofte H (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS (1999) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling E, Hofte H (2003) Feedback from the wall. Curr Opin Plant Biol 6: 1–6 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Rose JK, Saladie M, Catala C (2004) The plot thickens: new perspectives of primary cell wall modification. Curr Opin Plant Biol 7: 296–301 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden P, Sugimoto-Shirasu K, Smith AC, Findlay K, Reiter WD, McCann MC (2003) Tensile properties of Arabidopsis cell walls depend on both a xyloglucan cross-linked microfibrillar network and rhamnogalacturonan II-borate complexes. Plant Physiol 132: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Pauly M (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7: 285–295 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P (2004) Knocking on heaven's wall: pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr Opin Plant Biol 7: 377–383 [DOI] [PubMed] [Google Scholar]
- Seifert GJ (2004) Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol 7: 277–284 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Willats WG, Marcus SE, Knox JP (1998) Generation of monoclonal antibody specific to (1 → 5)-alpha-l-arabinan. Carbohydr Res 308: 149–152 [DOI] [PubMed] [Google Scholar]
- Yates EA, Valdor JF, Haslam SM, Morris HR, Dell A, Mackie W, Knox JP (1996) Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6: 131–139 [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison III WH, Freshour GD, Hahn MG, Ye ZH (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH (1997) Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]