Abstract
Small ubiquitin-like modifier (SUMO) conjugation/deconjugation to heat shock transcription factors regulates DNA binding of the peptides and activation of heat shock protein gene expression that modulates thermal adaptation in metazoans. SIZ1 is a SUMO E3 ligase that facilitates SUMO conjugation to substrate target proteins (sumoylation) in Arabidopsis (Arabidopsis thaliana). siz1 T-DNA insertional mutations (siz1-2 and siz1-3; Miura et al., 2005) cause basal, but not acquired, thermosensitivity that occurs in conjunction with hyperaccumulation of salicylic acid (SA). NahG encodes a salicylate hydroxylase, and expression in siz1-2 seedlings reduces endogenous SA accumulation to that of wild-type levels and further increases thermosensitivity. High temperature induces SUMO1/2 conjugation to peptides in wild type but to a substantially lesser degree in siz1 mutants. However, heat shock-induced expression of genes, including heat shock proteins, ascorbate peroxidase 1 and 2, is similar in siz1 and wild-type seedlings. Together, these results indicate that SIZ1 and, by inference, sumoylation facilitate basal thermotolerance through processes that are SA independent.
High temperature stress adversely affects organisms by causing membrane integrity loss, reactive oxygen species (ROS) production, protein inactivation and denaturation, and metabolic and cellular disequilibria, which ultimately lead to cell death (Berry and Björkman, 1980; Quinn, 1988; Lindquist, 1992; Dat et al., 1998b; Los and Murata, 2000; Iba, 2002). Plants have an innate capacity to survive high temperature stress (basal thermotolerance) and can sense and acclimate to high temperatures with metabolic and cellular adjustments that impart a capacity to tolerate heat extremes that were previously lethal (acquired thermotolerance; Vierling, 1991; Alfonso et al., 2001; Clarke et al., 2004; Larkindale et al., 2005). Acquired thermal tolerance responses are coordinated by signaling pathways that regulate heat tolerance determinants to limit stress damage and facilitate reestablishment of cellular homeostasis for survival and growth (Clarke et al., 2004; Larkindale et al., 2005). Thermal adaptation responses include membrane compositional changes necessary for maintenance of functional integrity, activation of oxidative defensive systems through ethylene and salicylic acid (SA), and production of heat shock proteins (HSPs) necessary for cellular protection (Quinn, 1988; Boston et al., 1996; Schöffl et al., 1998; Larkindale and Knight, 2002; Baniwal et al., 2004; Clarke et al., 2004; Larkindale et al., 2005).
Heat shock transcription factor (HSF) activation that facilitates transient production of HSPs is a well-characterized process in acquired thermotolerance (Pirkkala et al., 2001; Larkindale et al., 2005). Vertebrate HSF1, which is the ortholog of yeast (Saccharomyces cerevisiae), Drosophila melanogaster, and Caenorhabditis elegans HSF, exists at ambient temperature as an inactive monomer that is complexed with HSP90 in the cytosol (Zou et al., 1998; Liu and Thiele, 1999; Guo et al., 2001; Hu and Mivechi, 2003). Heat shock causes disruption of the complex, leading to the formation of activated HSF1 homotrimers that migrate to the nucleus (Zandi et al., 1997) and facilitate HSP transactivation through interaction of HSF1 trimers with heat shock elements (5′-AGAAnnTTCT-3′) in the promoters (Pelham, 1982; Westwood and Wu, 1993; Zuo et al., 1994; Fernandes et al., 1995; Liu and Thiele, 1999). HSF1 and HSF3 are the major activators of HSP expression in Arabidopsis (Arabidopsis thaliana; Lohmann et al., 2004). Dysfunctional alleles of either locus independently do not appreciably affect HSP expression; however, hsf1 hsf3 double mutation substantially reduces heat shock-induced HSP expression (Lohmann et al., 2004). hsf1 hsf3 marginally affects thermotolerance, even though high temperature induction of HSP101 expression is substantially less (Lohmann et al., 2004). A tomato (Lycopersicon esculentum) hsfA1 mutation is reported to cause thermosensitivity (Mishra et al., 2002).
HSPs are molecular chaperones that reduce protein denaturation, target denatured proteins for proteasome degradation, facilitate protein folding necessary for proper maturation or renaturation, and regulate the activity of HSFs to control HSP gene expression during thermotolerance acquisition (Johnson and Craig, 1997; Lee and Goldberg, 1998; Lee and Vierling, 2000; Frydman, 2001; Kim et al., 2002). Plant HSPs presumably mediate high temperature stress tolerance, but this is inferred largely because orthologs in other organisms have a thermal adaptive function (Vierling, 1991; Ellis, 2000; Hartl and Hayer-Hartl, 2002). Only the HSP100 family of plant HSPs, which are members of the ClpB chaperone family of ATPases that facilitate disaggregation of denatured proteins, are established functional determinants of acquired thermotolerance (Hong and Vierling, 2000; Queitsch et al., 2000; Lee et al., 2005). Hsa32 (heat shock-associated) protein is necessary for maintenance of acquired thermotolerance in Arabidopsis (Charng et al., 2006). HOT2, HOT3, and HOT4 are genetic loci that facilitate acquired thermotolerance in Arabidopsis but map to positions in the genome that do not encode HSPs (Hong et al., 2003).
Basal thermotolerance is comparatively less understood than acquired thermotolerance (Hong and Vierling, 2000; Larkindale et al., 2005). HSP101 is an essential determinant for basal thermotolerance of seed germination (Hong and Vierling, 2000), and ethylene, SA, and ROS signaling functions in basal thermotolerance at different plant developmental stages (Clarke et al., 2004; Larkindale et al., 2005). The numerous cellular and metabolic processes involved in basal thermotolerance implicate that a signaling network composed of numerous regulators is necessary to exercise concerted control over effector determinants in a developmental context (Larkindale et al., 2005).
Sumoylation is a posttranslational modification process that conjugates the small ubiquitin-like modifier (SUMO) peptide to the K residue in the Ψ-K-X-D/E (Ψ, large hydrophobic residue; X, any residue) target motif of protein substrates (Bernier-Villamor et al., 2002; Melchior et al., 2003; Schmidt and Müller, 2003; Johnson, 2004). SUMO conjugation of substrates occurs in a series of biochemical steps that are mediated by E1-activating, E2-conjugating, and E3-ligation enzymes. SUMO has been linked in fungi and metazoans to processes such as innate immunity, cell cycle progression, thermal adaptation, DNA repair, nucleocytoplasmic trafficking, subnuclear targeting, ubiquitination antagonism, and transcriptional regulation (Mao et al., 2000; Saitoh and Hinchey, 2000; Freiman and Tjian, 2003; Bohren et al., 2004; Dohmen, 2004; Johnson, 2004; Gill, 2005; Hay, 2005; Shuai and Liu, 2005; Zhao and Blobel, 2005; Hietakangas et al., 2006). Conjugation of SUMO to human (h) HSF1, hHSF2, and hHSF4b and Xenopus HSF2 regulates DNA binding and HSP expression (Goodson et al., 2001; Hong et al., 2001; Hietakangas et al., 2003, 2006; Hilgarth et al., 2003, 2004). In plants, high temperature induces SUMO1/2 conjugation to peptides, inferring that sumoylation may be involved in responses to high temperatures (Kurepa et al., 2003; Miura et al., 2005).
Arabidopsis SIZ1 is an ortholog of mammalian PIAS (protein inhibitor of activated signal transducer and activator of transcription) and yeast Siz family SUMO E3 ligases that facilitate sumoylation of transcription factors (Gill, 2005; Hay, 2005; Miura et al., 2005). Loss-of-function analyses described herein establish that the independent dysfunctional T-DNA insertion alleles siz1-2 or siz1-3 (Miura et al., 2005) cause thermosensitivity. Experimental results indicate that SIZ1 is a positive regulator of processes that are necessary for basal thermotolerance through functions that are independent of SA. However, SIZ1, dependent or independent of sumoylation function, does not regulate acquired thermotolerance as it does in fungi and metazoans (Goodson et al., 2001; Hong et al., 2001; Hietakangas et al., 2003; Hilgarth et al., 2003, 2004). Apparently, SUMO conjugation/deconjugation facilitates high temperature tolerance in plants through processes that have yet to be identified in other organisms.
RESULTS
The SUMO E3 Ligase SIZ1 Facilitates High Temperature Tolerance
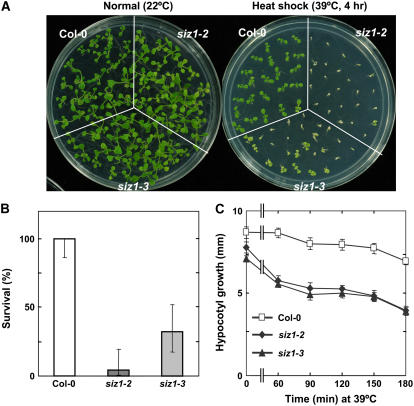
Ten-day-old siz1-2 and siz1-3 seedlings exhibited substantial thermosensitivity, as determined by using a heat shock survival assay (Fig. 1, A and B). siz1 seedlings rapidly developed severe heat shock symptoms in response to high temperature that were not evident in wild type. Substantial reduction in leaf surface area (shrinkage) was visible immediately after treatment, followed by severe chlorosis 24 h later. siz1 seedlings that developed these extreme symptoms did not survive (Fig. 1, A and B). The maximal heat shock temperature (4-h treatment) that wild-type and siz1 seedlings could survive was 43°C and 40°C, respectively (data not shown).
Figure 1.
siz1-2 and siz1-3 seedlings are thermosensitive. A and B, Ten-day-old wild-type (Col-0), siz1-2, and siz1-3 seedlings were subjected to a heat shock treatment of 39°C for 4 h in the dark at 60% relative humidity. Untreated wild-type, siz1-2, and siz1-3 seedlings (normal) were maintained in light at 22°C for the 4-h period, and all remained viable (100% survival) throughout the duration of the experiment. A, Photograph of representative seedlings 4 d after completion of heat shock treatment and B, seedling survival determined for the same experiment as in A 4 d after treatment, mean with 95% confidence intervals, n = 24 to 25. C, Wild-type, siz1-2, and siz1-3 seedlings were subjected to a heat shock treatment of 39°C in the dark for the time indicated and returned to the dark at 22°C/18°C (16 h/8 h). Hypocotyl growth was determined 2.5 d after treatment, mean ± se and n ≥ 18.
Thermosensitivity of siz1-2 and siz1-3 seedlings was evident also in a hypocotyl elongation assay (Fig. 1C). High soil temperatures early in seedling development can restrict hypocotyl elongation, which may prevent or delay shoot emergence from the soil (Lin et al., 1984; Hong and Vierling, 2000). Hypocotyl elongation of siz1 seedlings was sensitive to even a brief exposure to 39°C (Fig. 1C). siz1 seedlings also exhibited a reduction in hypocotyl elongation at 37°C and 38°C, but sensitivity relative to wild type was less than at 39°C. Similar results in heat shock-sensitive phenotypes caused by independent siz1 alleles (Fig. 1) are indicative that SIZ1 functions in thermotolerance.
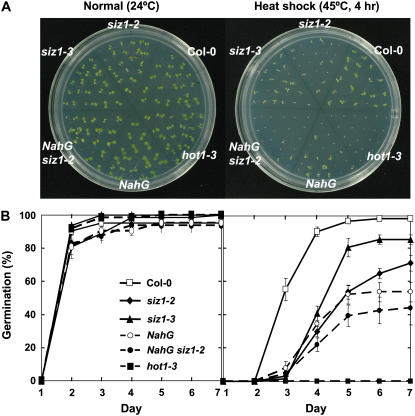
During latter stages of seed maturation, HSP101 accumulates and facilitates basal thermotolerance during germination (Hong and Vierling, 2001). siz1 seeds were sensitive to heat shock administered after imbibition and stratification, inhibiting both germination rate and seedling development (Fig. 2; Columbia [Col-0] and siz1-2, P < 0.01; Col-0 and siz1-3, P < 0.05). siz1-2 caused greater seed germination sensitivity than siz1-3 (Fig. 2; siz1-2 and siz1-3, P < 0.05). Heat treatment did not significantly alter seedling viability after germination (data not shown) but impaired development (Fig. 2A) and increased leaf chlorosis of siz1-2 and siz1-3 seedlings relative to wild type (data not shown).
Figure 2.
siz1 increases thermosensitivity during seed germination. Stratified wild-type, siz1-2, siz1-3, NahG siz1-2, NahG, and hot1-3 seeds (3 d in the dark at 4°C) were immediately subjected to heat shock treatment of 45°C for 4 h or incubated at 24°C (normal), sown onto plates, and then maintained under a 16-h daily photoperiod at 24°C. Germination was assessed at the indicated intervals. A, Illustration of representative seeds/seedlings 6 d after heat treatment and B, seed germination data from three independent experiments, mean ± se, n = 21.
SIZ1 Mediates Basal But Not Acquired Thermotolerance
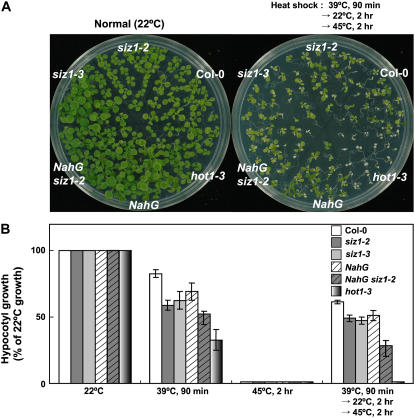
Plants, like most other organisms, exhibit both basal (innate) and acquired thermotolerance (Larkindale et al., 2005). The latter phenomenon is an acclimation that occurs in response to brief exposure to high temperature or longer-term exposure to increasing temperature and facilitates survival of thermal extremes that previously were lethal (Hong and Vierling, 2000; Hong et al., 2003). siz1 mutations cause heat sensitivity in assays that assess basal thermotolerance (Fig. 3B; Col-0 and siz1-2, P < 0.01; Col-0 and siz1-3, P < 0.05); consequently, their effects on acquired thermotolerance were examined. siz1-2 and siz1-3 seedlings exhibited a similar capacity for acquired thermotolerance as wild type, as indicated from viability and hypocotyl elongation assays (Fig. 3, A and B). An exposure to 39°C for 90 min is a sublethal heat shock treatment for all the genotypes compared in this experiment. hot1-3 seedlings did not acclimate in response to the sublethal temperature pretreatment stress, because the mutation abrogates capacity for acquired thermotolerance (Hong and Vierling, 2001). By inference, SIZ1 function in high temperature adaptation seems restricted to basal thermotolerance.
Figure 3.
siz1-2 and siz1-3 seedlings exhibit reduced basal thermotolerance but not acquired thermotolerance. A, Ten-day-old wild-type, siz1 (siz1-2 and siz1-3), NahG siz1-2, NahG, and hot1-3 seedlings were not acclimated (normal, 22°C) or high-temperature acclimated by exposure to 39°C for 90 min. After a recovery period of 2 h at 22°C, seedlings were exposed to a heat shock of 45°C for 2 h under the conditions described in the Figure 1 legend. Illustrated are representative seedlings 4 d after heat shock treatment. B, Stratified seeds of genotypes described in A were incubated in the dark at 22°C/18°C (16 h/8 h). Three days thereafter, seedlings were high-temperature acclimated by exposure to 39°C for 90 min. After a recovery period of 2 h at 22°C, seedlings were exposed to a heat shock of 45°C for 2 h under conditions described in the Figure 1 legend and then grown for 2.5 d at 22°C/18°C (16 h/8 h). Illustrated are relative growth determinations (100% indicates the growth of genotypes at 22°C) from three independent experiments, mean ± se, n ≥ 18 seedlings/experiment.
SIZ1 Mediates Basal Thermotolerance through a SA-Independent Process(es)
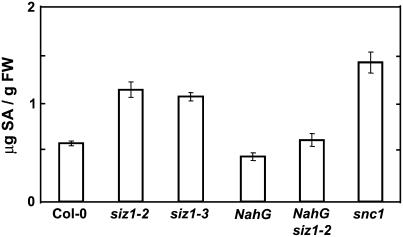
SA signaling is implicated to effect basal thermotolerance (Clarke et al., 2004; Larkindale et al., 2005). siz1 seedlings and plants hyperaccumulate SA to a greater extent than wild type (Fig. 4; Lee et al., 2006b; Col-0 and siz1-2, P < 0.05; Col-0 and siz1-3, P < 0.05) yet are more thermosensitive (Figs. 1–3). The interaction between SIZ1 and SA was assessed by genetic analysis of heat shock effects on siz1-2 and NahG siz1-2 seed germination and seedling growth. NahG transgenic plants express the Pseudomonas putida salicylate hydroxylase that catabolizes SA and effectively prevents accumulation (Delaney et al., 1994). NahG expression in wild type caused thermosensitivity (Figs. 2 and 3), which is consistent with data of others and supports the notion that SA facilitates basal high temperature tolerance (Clarke et al., 2004; Larkindale et al., 2005). NahG expression in siz1-2 caused additive hypocotyl elongation thermosensitivity (Fig. 3B; NahG siz1-2 and siz1-2, P < 0.05; NahG siz1-2 and siz1-3, P < 0.05; NahG siz1-2 and NahG, P < 0.01), and this correlated with SA levels that are comparable to wild type (Fig. 4; Lee et al., 2006b). NahG siz1-2 seedlings exhibited impaired development after heat shock treatment during seed imbibition (Fig. 2A). NahG siz1-2 also resulted in hyperthermosensitivity during germination (Fig. 2B; NahG siz1-2 and siz1-2, P < 0.01; NahG siz1-2 and siz1-3, P < 0.01; NahG siz1-2 and NahG, P = 0.12). Catechol is a product of SA degradation by NahG and itself causes a loss of nonhost pathogen resistance in Arabidopsis (van Wees and Glazebrook, 2003). However, catechol treatment does not affect thermotolerance in Arabidopsis (Clarke et al., 2004), suggesting that thermosensitivity caused by NahG is due specifically to decreased SA levels. Together, these results indicate that SIZ1-mediated sumoylation positively affects basal thermotolerance independent of SA. SIZ1 also negatively regulates SA accumulation (Fig. 4; Lee et al., 2006b), yet the positive affect of the SUMO E3 ligase on high temperature adaptation supercedes that of SA.
Figure 4.
siz1 hyperaccumulates high levels of SA. Ten-day-old wild-type, siz1 (siz1-2 and siz1-3), NahG siz1-2, NahG, and snc1 seedlings grown at 24°C on medium were harvested. SA content was determined by HPLC analysis. Illustrated are data from three independent experiments, mean ± se (micrograms of SA per gram fresh weight). Experiments were repeated four times in the same condition. snc1 seedlings result hyperaccumulation of SA (Zhang et al., 2003).
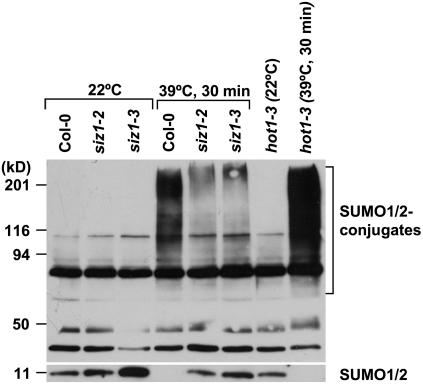
SIZ1 Controls Heat Shock-Induced SUMO Conjugation
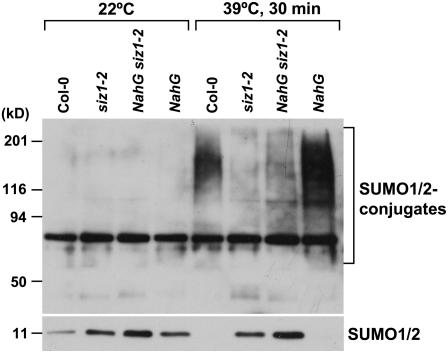
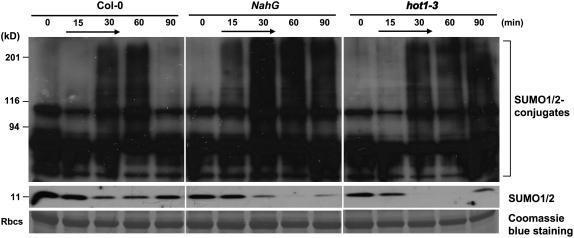
Heat shock induced an increase in conjugation of SUMO1/SUMO2 to substrate proteins at 34°C to 43°C (Figs. 5 and 6; Kurepa et al., 2003; Miura et al., 2005). A 39°C heat shock treatment induced sumoylation in wild type but to a lesser extent in siz1 seedlings (Figs. 5 and 6). Immunoblots were probed with anti-SUMO1 that detects both SUMO1 and SUMO2, and heat shock does not induce SUMO3 conjugation in Arabidopsis (Kurepa et al., 2003). Increased SUMO conjugation corresponded with a decrease in free SUMO1/2, indicating that accumulation is facilitated by increased sumoylation and not by reduced desumoylation. Together, these results indicate that SIZ1 mediates high temperature-induced accumulation of SUMO conjugation products. SUMO1/2 conjugation was substantially greater in NahG and hot1-3 seedlings compared to wild type after 30 min of heat shock exposure (Figs. 5 and 6). Interestingly, SUMO conjugation and deconjugation rates were different in NahG and hot1-3 seedlings (Fig. 7). NahG seedlings exhibited more rapid SUMO conjugation in response to heat shock and delayed SUMO deconjugation after return to ambient temperature (Fig. 7, center). SUMO deconjugation of hot1-3 seedlings was impaired during the recovery period. siz1-2 suppressed induction of sumoylation that is associated with NahG expression (Fig. 6). This is indicative that SIZ1-mediated SUMO conjugation may be a biochemical process by which the E3 ligase regulates thermotolerance responses independently of SA.
Figure 5.
Heat shock-induced SUMO1/2 conjugation is suppressed by siz1-2 and siz1-3. Ten-day-old wild-type, siz1-2, siz1-3, and hot1-3 seedlings were exposed to a 30-min heat shock treatment (39°C, dark, 60% relative humidity). Total protein was extracted from untreated or heat shock-treated seedlings. Twenty micrograms of protein were loaded onto an SDS-PAGE, and the immunoblot was probed with anti-SUMO1, which detects both SUMO1 and SUMO2 (Kurepa et al., 2003).
Figure 6.
Heat shock-induced SUMO1/2 conjugation in NahG seedlings requires SIZ1. Ten-day-old wild-type, siz1-2, NahG siz1-2, and NahG seedlings were exposed to a 30-min heat shock at 39°C in the dark. Total protein was extracted from seedlings as described in Figure 5. Ten micrograms of protein were separated by SDS-PAGE and the immunoblot was probed with anti-SUMO1.
Figure 7.
Heat stress-induced SUMO1/2 conjugation/deconjugation is impaired in NahG and hot1-3 seedlings. Ten-day-old wild-type, NahG, and hot1-3 seedlings were exposed to a 39°C heat stress for 15 or 30 min, returned to 24°C, and collected at indicated time points. Black arrow indicates the serial of heat shock. Total protein was extracted as described in Figure 5. Ten micrograms of protein were separated by SDS-PAGE, and the immunoblot was probed with anti-SUMO1.
SIZ1-Mediated Thermotolerance Is Independent of HSF Regulon Expression
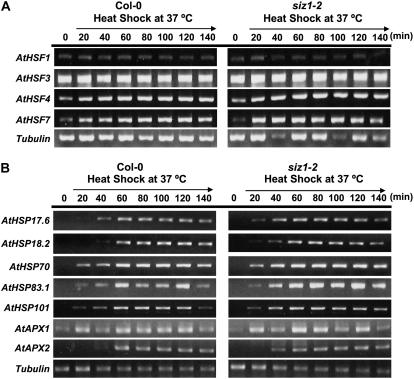
HSF complexity in plants is predicted to be substantially greater than in other organisms. Yeast, Drosophila, and C. elegans have one HSF and vertebrates have four (Morimoto, 1998; Nakai, 1999; Nover et al., 2001; Baniwal et al., 2004), while bioinformatic analyses predict 21, 18, and 23 HSFs in Arabidopsis, tomato, and rice (Oryza sativa), respectively, based on sequence similarity (Baniwal et al., 2004). Transcript abundance of HSF1, 3, 4, and 7 was similar in wild-type and siz1-2 seedlings, indicating that SIZ1-dependent sumoylation does not regulate expression of these genes (Fig. 8A). HSF1 and HSF3 are major regulators of high temperature-induced HSP expression (Lohmann et al., 2004). Expression of heat shock-induced genes was also similar in wild-type and siz1-2 seedlings (Fig. 8B). Included in the survey were genes that encode ascorbate peroxidase (APX)1 and APX2, and HSPs, all of which are regulated by HSF1 and HSF3 and implicated to facilitate thermotolerance (Storozhenko et al., 1998; Hong and Vierling, 2000; Panchuk et al., 2002; Lohmann et al., 2004). APX1 and APX2 function as antioxidant enzymes that detoxify hydrogen peroxide, which is produced in response to heat stress and is presumed to be a major effector of cellular dysfunction (Panchuk et al., 2002). HSFs, APXs, and HSPs expression patterns were similar in siz1-2 and wild-type seedlings after heat shock treatment at 39°C (data not shown). Furthermore, heat shock-induced expression of these genes in NahG siz1-2 was similar to siz1-2 seedlings (data not shown), indicating that SIZ1 functions in thermal adaptation independent of the HSF-controlled regulon.
Figure 8.
HSF and heat shock-induced gene expression is similar in wild-type and siz1 seedlings. Ten-day-old wild-type and siz1-2 seedlings were exposed to 37°C for the times indicated. Transcript abundance was determined by semiquantitative reverse transcription-PCR for four major HSFs (HSF1, 3, 4, and 7), and five different HSPs (HSP17.6A [class II sHSP], HSP18.2 [class I sHSP], HSP70, HSP83.1, HSP101) and APX1 and APX2. Black arrows indicate the period of heat shock.
DISCUSSION
The results of this study implicate a function for SIZ1 SUMO E3 ligase in basal thermotolerance of Arabidopsis. siz1-2 and siz1-3 cause thermal hypersensitivity (Figs. 1–3) that is manifested during seed germination, hypocotyl elongation, and seedling survival, inferring that SIZ1-mediated sumoylation is necessary for heat shock tolerance at numerous developmental stages. SUMO conjugation/deconjugation, in other organisms, is necessary for both ambient and high temperature-dependent HSF interactions with heat shock elements that regulate HSP expression (Goodson et al., 2001; Hong et al., 2001; Hilgarth et al., 2003, 2004; Hietakangas et al., 2006). However, the biological role of sumoylation in heat stress responses is unresolved, because it is not evident if SUMO conjugation to HSFs positively or negatively regulates thermotolerance (Goodson et al., 2001; Hong et al., 2001; Hietakangas et al., 2006). SUMO1/2 conjugates accumulation is an early plant response (within minutes) to high temperature stress, implicating a function for sumoylation in thermotolerance (Kurepa et al., 2003; Miura et al., 2005). The evidence herein confirms that high temperature-induced SUMO1/2 conjugation is attributable to SIZ1 SUMO E3 ligase function (Fig. 5; Miura et al., 2005). However, these data do not resolve if overall increased SUMO1/2 conjugation is a phenomenon that is linked causally to thermotolerance. Together, these results indicate that SIZ1 controls basal, but not acquired, thermotolerance, and infer that sumoylation/desumoylation of specific substrates is necessary.
AtSIZ1 is, by domain composition and functional data, a member of the PIAS family of SUMO E3 ligases (Kurepa et al., 2003; Miura et al., 2005). AtSIZ1 regulates gene expression and root architecture responses that are caused by phosphate deprivation (Miura et al., 2005). The MYB transcription factor PHR1 that is a controller of phosphate starvation-induced gene expression is a sumoylation substrate of SIZ1 (Miura et al., 2005). SIZ1 is implicated also in both thermal adaptation (herein) and pathogen defense (Lee et al., 2006b). Together, these results indicate that sumoylation/desumoylation is a key control process in the regulation of signal networks in plants. It is still to be resolved in any organism how sumoylation/desumoylation regulates diverse biological processes. However, emerging evidence implicates a function in gene expression regulator that involves chromatin remodeling (Gill, 2005; Hay, 2005).
PIAS family members are implicated to function as transcriptional regulators independent of the SP-RING domain that facilitates E3 ligase activity (Lee et al., 2006a; Sharrocks, 2006). The SAP domain of PIAS proteins is associated with transcriptional regulation resulting from chromatin remodeling (Shuai and Liu, 2005; Sharrocks, 2006). Consequently, there is a foundation to support the notion that SIZ1 may regulate basal thermotolerance through a sumoylation-independent process. Alternatively, it is also possible that SIZ1 is a determinant of thermotolerance through sumoylation-dependent and -independent processes.
SIZ1 Controls Basal Thermotolerance through a SA-Independent Process(es)
A genetic analysis of basal and acquired thermotolerance in Arabidopsis implicated the involvement of numerous heat shock-response pathways that do not involve a HSF regulon (Larkindale et al., 2005). Evidence implicates SA, ethylene, and ROS as possible intermediary signal molecules (Dat et al., 1998a; Cronjé and Bornman, 1999; Larkindale and Knight, 2002; Clarke et al., 2004; Larkindale et al., 2005). Understanding of the biological role of SA in thermal adaptation is rudimentary, but SA regulates HSP17.6 expression in Arabidopsis and HSP70 in tomato (Cronjé and Bornman, 1999; Clarke et al., 2004). However, HSP expression (particularly that of HSP101 and Hsa32) is presumed to be associated with acquired thermotolerance, and SA is not considered to be a principal regulator of this process (Hong and Vierling, 2000, 2001; Clarke et al., 2004; Larkindale et al., 2005; Charng et al., 2006). Our data further illustrate that NahG expression affects basal but not acquired thermotolerance and that SA does not regulate expression of numerous heat shock-regulated genes (Fig. 3; data not shown). siz1 mutations cause SA accumulation in seedlings and plants, yet seed germination and seedling growth and survival are adversely affected by high temperature. NahG expression causes thermal sensitivity of Arabidopsis (Clarke et al., 2004; Larkindale et al., 2005; Figs. 2 and 3) and increases hypersensitivity resulting from siz1 mutations (i.e. NahG siz1-2), indicating that SA has some thermal protective function that is precluded by processes controlled by SIZ1. That is, SIZ1 negatively regulates SA biosynthesis and/or catabolism but positively regulates basal thermotolerance through a SA-independent process(es).
Heat shock-induced SUMO conjugation/deconjugation is impaired in NahG and hot1-3 seedlings. Thiobarbituric acid reactive substances are products of lipid oxidation and thiobarbituric acid reactive substance levels are induced by heat stress in NahG and hot1-3 seedlings (Larkindale et al., 2005). ROS are reported to increase SUMO conjugation (Saitoh and Hinchey, 2000; Kurepa et al., 2003), even though low concentrations of ROS cause desumoylation of most substrates (Bossis and Melchior, 2006). High concentrations of ROS cause inactivation of the SUMO isopeptidase SENP-1 that leads to accumulation of SUMO conjugation products (Bossis and Melchior, 2006). A paradigm is emerging that ROS species and/or levels constitute a regulatory system that controls sumoylation/desumoylation of protein substrates (Kurepa et al., 2003; Bossis and Melchior, 2006). These results support the notion that ROS may regulate differences in heat shock-induced sumoylation and desumoylation evident in NahG and hot1-3 seedlings that correlate with increased thermal sensitivity, although it is not possible to link SUMO conjugation/deconjugation of specific substrates based on the results presented herein.
SIZ1-Mediated Thermotolerance Is Not Due to HSF Regulon Expression
Although SIZ1 is apparently not essential (Miura et al., 2005), it is evident that the SUMO E3 ligase is necessary for sumoylation that is associated with plant stress responses, as are PIAS and Siz orthologs in yeast, Drosophila, C. elegans, and vertebrates (Schmidt and Müller, 2002; García-Estrada et al., 2003; Takahashi and Kikuchi, 2005). HSF-controlled gene expression is critical for high temperature tolerance in these organisms, and sumoylation of Xenopus and human HSFs regulates transactivation of HSP expression (Hong et al., 2001; Hilgarth et al., 2004). Although sumoylation of hHSF1, hHSF2, and hHSF4b likely result in transcriptional repression, it is postulated that SUMO conjugation and deconjugation are dynamic regulatory processes that are necessary for fine tuning regulation of basal and acquired thermotolerance (Anckar et al., 2006; Hietakangas et al., 2006).
At present, there is no evidence that Arabidopsis HSF family members are substrates for SUMO conjugation or that sumoylation/desumoylation of HSF is necessary for regulation of high temperature-induced HSP expression or thermotolerance in plants. It is important to note that redundant regulatory effect of AtHSF1 and AtHSF3 on HSP expression does not markedly influence thermotolerance (Lohmann et al., 2004) as does LeHSF1 in tomato (Mishra et al., 2002). Also, we detected no difference in high temperature-induced mRNA expression patterns of HSPs in siz1 and wild-type seedlings (Fig. 8), indicating that SIZ1-dependent sumoylation does not regulate acquired thermotolerance in plants (Fig. 3). Attempts at in vitro sumoylation of AtHSF1 or AtHSF3 were inconclusive, although the assay does mediate SUMO conjugation to the transcription factor PHR1 (Miura et al., 2005).
Together, our results establish that SIZ1 facilitates basal thermotolerance in Arabidopsis through a SA-independent process(es). SIZ1, independent or dependent of sumoylation function, does not regulate acquired thermal adaptation responses in plants, unlike in other organisms as diverse as human, yeast, and Xenopus. The protein targets of SIZ1-dependent sumoylation that mediate thermotolerance remain to be identified as do the processes that control basal thermotolerance. Determinants that control high temperature signaling that regulate sumoylation/desumoylation and control thermal adaptation await identification. However, the results herein indicate that SUMO conjugation is a necessary process for basal thermotolerance at different plant developmental stages.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 ecotype genetic resources for this research were siz1-2, siz1-3 (Miura et al., 2005), hot1-3 (kindly provided by Dr. Elizabeth Vierling, University of Arizona, Tucson), NahG (Delaney et al., 1994), NahG siz1-2 (Lee et al., 2006b), and snc1 (kindly provided by Dr. Xin Li, University of British Columbia, Vancouver). Seeds were stratified for 3 d at 4°C and then sown onto a medium in petri plates containing 1× Murashige and Skoog basal salt mixture, 2% Suc, 2.5 mm MES, pH 5.7, and 0.8% agar. Seeds and seedlings, unless otherwise noted, were incubated under a 16-h-light (100 μmol m−2 s−1)/8-h-dark photoperiod at 22°C/18°C. For the hypocotyl elongation assay, seeds were sown onto agar (1.2%) medium and plates were placed in a vertical position in the dark under conditions as described above.
Thermotolerance Assays
Seedlings were subjected to heat shock in the dark at 60% relative humidity in a plant growth chamber (E-30B, Percival Scientific) with the capacity to control temperature fluctuation control by ±1°C. These conditions were used to minimize photooxidative and high humidity stresses (Larkindale and Knight, 2002; Zhou et al., 2004; Larkindale et al., 2005). Survival was monitored daily beginning 4 d after heat shock treatment. Stratified seeds were subjected to heat shock treatment in a temperature-controlled water bath (ISOTEMP 210, Fisher Scientific) and then sown onto medium. Seed germination was monitored every 24 h. The hypocotyl elongation assay was carried out as described by Hong and Vierling (2000). Briefly, 3-d-old etiolated seedlings were incubated at 22°C or heat shock-treated at 39°C or 45°C in dark for the indicated time. Plumule position of each seedling was recorded by marking the plate, and the plate was rewrapped and incubated at 22°C/18°C (16 h/8 h) in dark. Hypocotyl growth after heat shock treatment was measured after 2.5 d.
Total RNA Isolation and Semiquantitative Reverse Transcription-PCR Analysis
Total RNA from 10-d-old seedlings grown at 22°C or heat shock treated for the indicated time was isolated by using PureLink Micro-to-Midi Total RNA Purification system (no. 12183–018, Invitrogen). Two micrograms of total RNA were used as template for first-strand cDNA synthesis with ThermoScript reverse transcription-PCR system (no. 11146–016, Invitrogen) and an oligo(dT)20 primer. Gene-specific primers were used to amplify PCR products of approximately 500 bp in length (Supplemental Table S1).
In Vivo Analysis of Sumoylation Profiles
Total protein was extracted from 10-d-old seedlings grown on medium under conditions described above. Plant tissues (0.2 g) were extracted with a mortar and pestle in grinding buffer (100 mm Na-MOPS, pH 7.5, 10 mm NaCl, 1 mm EDTA, pH 8.0, 10% Suc, 5% β-mercaptoethanol, and 4% SDS) at room temperature. Protein concentration was measured by using Bio-Rad Protein Assay (no. 500–0006, Bio-Rad), and protein was separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane (no. 162–0177, Bio-Rad), probed with anti-SUMO1 antibody (ab5316, Abcam), and detected by using ECL plus Western Blotting Detection system (Amersham Biosciences).
SA Quantification
Shoots of 10-d-old seedlings that were grown on medium under conditions described above were harvested and frozen in liquid nitrogen. Tissue (0.2 g fresh weight, without roots) was extracted in 4 mL of methanol for 24 h at 4°C and then in a solution of 2.4 mL of water plus 2 mL of chloroform with 40 μL of 5 mm 3,4,5-trimethoxy-trans-cinamic acid (internal standard) for 24 h at 4°C. Supernatants were dried by speed vacuum. The residue was resuspended in 0.4 mL of water:methanol (1:1, v/v), and SA was quantified by HPLC as described by Freeman et al. (2005).
Statistical Analysis
All data except germination rates were analyzed by Student's t test for pairwise comparison. Germination was compared at different time points using method of logistic regression with repeated measurements using SAS PHREG software and P values are indicated.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Gene-specific primer sequences used to detect heat shock-related genes by RT-PCR.
Acknowledgments
We thank Dr. Yun Joo Yoo (Department of Biostatistics, University of Alabama at Birmingham) for the statistical analyses.
This work was supported by the National Science Foundation Plant Genome Award (DBI–98–13360), by the Basic Science Project of Korea Science and Engineering Foundation (grant no. RO1–2006–000–10123–0 and postdoctoral fellowship to H.C.P.), by the Environmental Biotechnology National Core Research Center Project of Korea Science and Engineering Foundation (grant no. R15–2003–012–01002–00), and by the Biogreen 21 Program of the Rural Development Administration, Korea. This work is Purdue University Agricultural Research Program Paper 2006–17918.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paul M. Hasegawa (paul.m.hasegawa.1@purdue.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alfonso M, Yruela I, Almárcegui S, Torrado E, Pérez MA, Picorel R (2001) Unusual tolerance to high temperatures in a new herbicide-resistant D1 mutant from Glycine max (L.) Merr. cell cultures deficient in fatty acid desaturation. Planta 212: 573–582 [DOI] [PubMed] [Google Scholar]
- Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L (2006) Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol 26: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29: 471–487 [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Berry J, Björkman O (1980) Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol 31: 491–543 [Google Scholar]
- Bohren KM, Nadkarni V, Song JH, Gabbay KJ, Owerbach D (2004) A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem 279: 27233–27238 [DOI] [PubMed] [Google Scholar]
- Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21: 349–357 [DOI] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E (1996) Molecular chaperones and protein folding in plants. Plant Mol Biol 32: 191–222 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Mur LA, Wood JE, Scott IM (2004) Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J 38: 432–447 [DOI] [PubMed] [Google Scholar]
- Cronjé MJ, Bornman L (1999) Salicylic acid influences Hsp70/Hsc70 expression in Lycopersicon esculentum: dose- and time-dependent induction or potentiation. Biochem Biophys Res Commun 265: 422–427 [DOI] [PubMed] [Google Scholar]
- Dat JF, Foyer CH, Scott IM (1998. a) Changes in salicylic acid and antioxidants during induced thermotolerance in mustard seedlings. Plant Physiol 118: 1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat JF, Lopez-Delgado H, Foyer CH, Scott IM (1998. b) Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol 116: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ (2004) SUMO protein modification. Biochim Biophys Acta 1695: 113–131 [DOI] [PubMed] [Google Scholar]
- Ellis RJ (2000) Chaperone substrates inside the cell. Trends Biochem Sci 25: 210–212 [DOI] [PubMed] [Google Scholar]
- Fernandes M, Xiao H, Lis JT (1995) Binding of heat shock factor to and transcriptional activation of heat shock genes in Drosophila. Nucleic Acids Res 23: 4799–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Garcia D, Kim D, Hopf A, Salt DE (2005) Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol 137: 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Tjian R (2003) Regulating the regulators: lysine modifications make their mark. Cell 112: 11–17 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- García-Estrada C, Reguera RM, Villa H, Requena JM, Müller S, Pérez-Pertejo Y, Balaña-Fouce R, Ordóñez D (2003) Identification of a gene in Leishmania infantum encoding a protein that contains a SP-RING/MIZ zinc finger domain. Biochim Biophys Acta 1629: 44–52 [DOI] [PubMed] [Google Scholar]
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 276: 18513–18518 [DOI] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R (2001) Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem 276: 45791–45799 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L (2003) Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol 23: 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgarth RS, Hong Y, Park-Sarge OK, Sarge KD (2003) Insights into the regulation of heat shock transcription factor 1 SUMO-1 modification. Biochem Biophys Res Commun 303: 196–200 [DOI] [PubMed] [Google Scholar]
- Hilgarth RS, Murphy LA, O'Connor CM, Clark JA, Park-Sarge OK, Sarge KD (2004) Identification of Xenopus heat shock transcription factor-2: conserved role of sumoylation in regulating deoxyribonucleic acid-binding activity of heat shock transcription factor-2 proteins. Cell Stress Chaperones 9: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Lee U, Vierling E (2003) Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol 132: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276: 40263–40267 [DOI] [PubMed] [Google Scholar]
- Hu Y, Mivechi NF (2003) HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J Biol Chem 278: 17299–17306 [DOI] [PubMed] [Google Scholar]
- Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53: 225–245 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson JL, Craig EA (1997) Protein folding in vivo: unraveling complex pathways. Cell 90: 201–204 [DOI] [PubMed] [Google Scholar]
- Kim SY, Sharma S, Hoskins JR, Wickner S (2002) Interaction of the DnaK and DnaJ chaperone system with a native substrate, P1 RepA. J Biol Chem 277: 44778–44783 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: accumulation of SUMO 1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Verling E (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL (1998) Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol 18: 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Quinn JC, Prasanth KV, Swiss VA, Economides KD, Camacho MM, Spector DL, Abate-Shen C (2006. a) PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev 20: 784–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al (2006. b) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J (in press) [DOI] [PubMed]
- Lee U, Wie C, Escobar M, Williams B, Hong SW, Vierling E (2005) Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell 17: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Roberts JK, Key JL (1984) Acquisition of thermotolerance in soybean seedlings: synthesis and accumulation of heat shock proteins and their cellular localization. Plant Physiol 74: 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S (1992) Heat-shock proteins and stress tolerance in microorganisms. Curr Opin Genet Dev 2: 748–755 [DOI] [PubMed] [Google Scholar]
- Liu PC, Thiele DJ (1999) Modulation of human heat shock factor trimerization by the linker domain. J Biol Chem 274: 17219–17225 [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Los DA, Murata N (2000) Regulation of enzymatic activity and gene expression by membrane fluidity. Sci STKE 2000: pe1. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Liu LF (2000) SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem 275: 26066–26073 [DOI] [PubMed] [Google Scholar]
- Melchior F, Schergaut M, Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28: 612–618 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Kood YD, Jin JB, Bressan RA, et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nakai A (1999) New aspects in the vertebrate heat shock factor system: Hsf3 and Hsf4. Cell Stress Chaperones 4: 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR (1982) A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30: 517–528 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PJ (1988) Effects of temperature on cell membranes. Symp Soc Exp Biol 42: 237–258 [PubMed] [Google Scholar]
- Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275: 6252–6258 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Müller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Müller S (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci 60: 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A (1998) Regulation of the heat-shock response. Plant Physiol 117: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD (2006) PIAS proteins and transcriptional regulation-more than just SUMO E3 ligase? Genes Dev 20: 754–758 [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B (2005) Regulation of gene-activation pathways by PIAS proteins in the immune system. Nature Rev Immunol 5: 593–605 [DOI] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inzé D, Kushnir S (1998) The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol 118: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi Y (2005) Yeast PIAS-type Ull/Siz1 is composed of SUMO ligase and regulatory domains. J Biol Chem 280: 35822–35828 [DOI] [PubMed] [Google Scholar]
- van Wees SC, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33: 733–742 [DOI] [PubMed] [Google Scholar]
- Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 570–620 [Google Scholar]
- Westwood JT, Wu C (1993) Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol 13: 3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi E, Tran TN, Chamberlain W, Parker CS (1997) Nuclear entry, oligomerization, and DNA binding of the Drosophila heat shock transcription factor are regulated by a unique nuclear localization sequence. Genes Dev 11: 1299–1314 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1–1 constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Menke FL, Yoshioka K, Moder W, Shirano Y, Klessig DF (2004) High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and gene expression. Plant J 39: 920–932 [DOI] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94: 471–480 [DOI] [PubMed] [Google Scholar]
- Zuo J, Baler R, Dahl G, Voellmy R (1994) Activation of the DNA-bind ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol 14: 7557–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]