Abstract
Chloroplast genomes in plants and green algae contain numerous group II introns, large ribozymes that splice via the same chemical steps as spliceosome-mediated splicing in the nucleus. Most chloroplast group II introns are degenerate, requiring interaction with nucleus-encoded proteins to splice in vivo. Genetic approaches in maize (Zea mays) and Chlamydomonas reinhardtii have elucidated distinct sets of proteins that assemble with chloroplast group II introns and facilitate splicing. Little information is available, however, concerning these processes in Arabidopsis (Arabidopsis thaliana). To determine whether the paucity of data concerning chloroplast splicing factors in Arabidopsis reflects a fundamental difference between protein-facilitated group II splicing in monocot and dicot plants, we examined the mutant phenotypes associated with T-DNA insertions in Arabidopsis genes encoding orthologs of the maize chloroplast splicing factors CRS1, CAF1, and CAF2 (AtCRS1, AtCAF1, and AtCAF2). We show that the splicing functions and intron specificities of these proteins are largely conserved between maize and Arabidopsis, indicating that these proteins were recruited to promote the splicing of plastid group II introns prior to the divergence of monocot and dicot plants. We show further that AtCAF1 promotes the splicing of two group II introns, rpoC1 and clpP-intron 1, that are found in Arabidopsis but not in maize; AtCAF1 is the first splicing factor described for these introns. Finally, we show that a strong AtCAF2 allele conditions an embryo-lethal phenotype, adding to the body of data suggesting that cell viability is more sensitive to the loss of plastid translation in Arabidopsis than in maize.
Group II introns are large ribozymes found in bacteria, chloroplasts, and mitochondria that are defined by conserved structural features and patches of sequence similarity (for review, see Michel and Ferat, 1995; Pyle, 1996). Mitochondrial and chloroplast genomes in land plants are particularly intron rich, each encoding approximately 20 group II introns (for review, see Barkan, 2004). The chloroplast genomes of vascular and nonvascular land plants have a similar intron complement, with the small differences attributable to lineage-specific intron loss. In contrast, the chloroplasts of green algae have a distinct intron set, and phylogenetic data imply independent intron acquisition in this lineage (for review, see Barkan, 2004).
Although several group II introns have been shown to splice autocatalytically in vivo, self splicing has not been reported for any of the approximately 40 group II introns in land plant organelles. These introns are believed to be degenerate, having lost elements that are necessary to assemble an RNA-based catalytic center. Intron degeneration has been compensated by the recruitment of protein factors that are obligate participants in the splicing reactions (for review, see Lambowitz and Zimmerly, 2004). Genetic screens in maize (Zea mays) revealed three protein complexes in chloroplasts that are required for the splicing of different intron subsets, and that assemble stably with those introns in vivo (Jenkins et al., 1997; Jenkins and Barkan, 2001; Till et al., 2001; Ostheimer et al., 2003): CRS1 is required specifically for the splicing of the atpF intron, four introns require a CRS2/CAF1 complex, three introns require a CRS2/CAF2 complex, and two introns require both CRS2/CAF1 and CRS2/CAF2 complexes. CRS2 is related to peptidyl-tRNA hydrolase enzymes (Jenkins and Barkan, 2001), whereas CRS1, CAF1, and CAF2 are members of a plant-specific protein family defined by the presence of one or more CRM domains (Till et al., 2001; Ostheimer et al., 2003), a novel RNA binding domain found in prokaryotes and in plants (Barkan et al., 2007). Analogous studies identified three group II intron splicing factors in the chloroplasts of the green algae Chlamydomonas reinhardtii (Perron et al., 1999; Rivier et al., 2001; Merendino et al., 2006), none of which are related to the factors identified in maize.
Despite the intensive use of Arabidopsis (Arabidopsis thaliana) to study chloroplast processes, there is just one report of an Arabidopsis gene that influences the accumulation of a spliced chloroplast RNA: HCF152, a member of the pentatricopeptide repeat family, is required for the accumulation of spliced petB RNA but not for the accumulation of excised petB intron (Meierhoff et al., 2003), suggesting that it promotes exon ligation or stabilization of the spliced mRNA. To determine whether the paucity of chloroplast splicing factors identified in Arabidopsis reflects a fundamental difference between protein-facilitated group II splicing in monocot and dicot plants, we examined the mutant phenotypes associated with T-DNA insertions in the Arabidopsis orthologs of maize crs1, caf1, and caf2 (AtCRS1, AtCAF1, and AtCAF2). We show here that these proteins do promote the splicing of chloroplast group II introns, and that there is generally concordance between the intron specificities of the maize and Arabidopsis orthologs. These results show that these host-derived proteins were recruited to promote the splicing of specific organellar group II introns prior to the divergence of monocot and dicot plants. We also add to the functions established for these proteins by showing that AtCAF1 promotes the splicing of two group II introns that were lost in maize and other monocot grasses, but retained in Arabidopsis. Finally, the fact that a strong Atcaf2 allele conditions an embryo-lethal phenotype adds to the growing body of data indicating that embryonic development and cell viability are more sensitive to the loss of plastid translation in Arabidopsis than in maize and other monocot grasses.
RESULTS
T-DNA Insertions in the Arabidopsis Orthologs of crs1, caf1, and caf2 Cause Embryo-Lethal and/or Albino Seedling Phenotypes
We previously predicted the Arabidopsis orthologs of maize crs1, caf1, and caf2 via phylogenetic analysis (Barkan et al., 2007; summarized in “Materials and Methods”). T-DNA insertion lines for these Arabidopsis genes were obtained from the SALK insertion collection (Table I; Alonso et al., 2003); two insertion lines were obtained for each of AtCRS1, AtCAF1, and AtCAF2. DNA flanking each insertion was amplified and their sequences verified (Supplemental Fig. S1). The position of each insertion is diagrammed in Figure 1A.
Table I.
Summary of Arabidopsis insertion alleles and phenotypes
| AGI Code | Protein | Mutant Allele | Insertion Line | T-DNA Insertion Site | Phenotype |
|---|---|---|---|---|---|
| At5g16180 | AtCRS1 | Atcrs1-1 | SALK_026861 | Exon 1 | Ivory, small |
| Atcrs1-2 | SALK_027605 | Exon 6 | Variegated | ||
| Atcrs1-1/Atcrs1-2 | Noncomplementing progeny | Variegated, ivory | |||
| At2g20020 | AtCAF1 | Atcaf1-1 | SALK_025042 | Intron 1 | Ivory |
| Atcaf1-2 | SALK_021969 | Intron 1 | Ivory | ||
| At1g23400 | AtCAF2 | Atcaf2-1 | SALK_049304 | Exon 2 | Embryo lethal |
| Atcaf2-2 | SALK_008478 | Intron 5 | Pale green, small | ||
| Atcaf2-1/Atcaf2-2 | Noncomplementing progeny | Pale green, small |
Figure 1.
Plant phenotypes associated with T-DNA insertions in AtCRS1 (At5g16180), AtCAF1 (At2g20020), and AtCAF2 (At1g23400). A, Schematic maps of T-DNA insertion sites. Black rectangles are exons and lines are introns. The T-DNA insertion in Atcaf1-2 is within intron 1, 46 nt downstream of that in Atcaf1-1. The sequence flanking each insertion is shown in Supplemental Figure S1. LB, T-DNA left border. B, Seedling phenotypes conditioned by mutant alleles and allele combinations. Where one allele is listed, seedlings are homozygous for the indicated allele. Where two alleles are listed, the plants are the noncomplementing progeny from a cross between plants heterozygous for each allele. Genotypes were confirmed by PCR. Bar=5 mm. C, Segregation of aborted seeds in siliques from a plant heterozygous for Atcaf2-1. The aborted seeds are presumed to be homozygous for the Atcaf2-1 insertion because no homozygous mutants were recovered among the 20 germinating progeny that were genotyped by PCR. The two siliques represent different stages of seed maturation.
The plant phenotypes observed in each line are shown in Figure 1 and summarized in Table I. All of the lines except for Atcaf2-1 segregated chlorophyll-deficient seedlings (Fig. 1B), whereas the Atcaf2-1 line segregated defective seeds (Fig. 1C). In those lines that segregated chlorophyll-deficient mutants, genetic linkage was established between the insertion and the chlorophyll-deficient phenotype (Supplemental Fig. S2). Complementation crosses were performed between the two Atcrs1 alleles and between the two Atcaf2 alleles; these crosses yielded chlorophyll-deficient seedlings (Fig. 1B) harboring both mutant alleles (Supplemental Fig. S3). Taken together, these results show that the chlorophyll-deficient phenotypes are caused by the insertions in the targeted genes.
Although both Atcrs1 alleles condition chlorophyll-deficient seedlings, their phenotypes are distinct: an insertion in exon 1 (Atcrs1-1) yielded albino seedlings, whereas an insertion in exon 6 (Atcrs1-2) yielded variegated seedlings with green and white patches (Fig. 1B). It seems likely that the Atcrs1-1 phenotype (albino) represents the more severe loss of function as the insertion is in an exon near the beginning of the open reading frame (ORF), whereas the Atcrs1-2 insertion disrupts sequences near the carboxyl terminus of the protein and conditions a milder chlorophyll deficiency.
The two insertions in AtCAF2 also condition distinct phenotypes. The siliques resulting from self-pollinating Atcaf2-1/+ plants contained aborted, chlorotic seeds at a frequency of approximately 1:3 aborted to normal seed (Fig. 1C). Furthermore, no homozygous mutant plants were recovered among 20 germinating progeny (data not shown). Together, these results provide strong evidence that Atcaf2-1 conditions an embryo-lethal phenotype. In contrast, Atcaf2-2 conditions slow-growing seedlings with albino cotyledons and pale green leaves (Fig. 1B). The noncomplementing progeny of plants heterozygous for the two alleles (Atcaf2-1/Atcaf2-2) germinated and were similar in appearance to Atcaf2-2 homozygotes. It seems likely that the chlorotic seedling phenotype conditioned by the intronic insertion in Atcaf2-2 represents an incomplete loss of function, whereas the embryo-lethal phenotype conditioned by the Atcaf2-1 insertion, which is in exonic sequences in the middle of the ORF (Fig. 1A), represents the null phenotype.
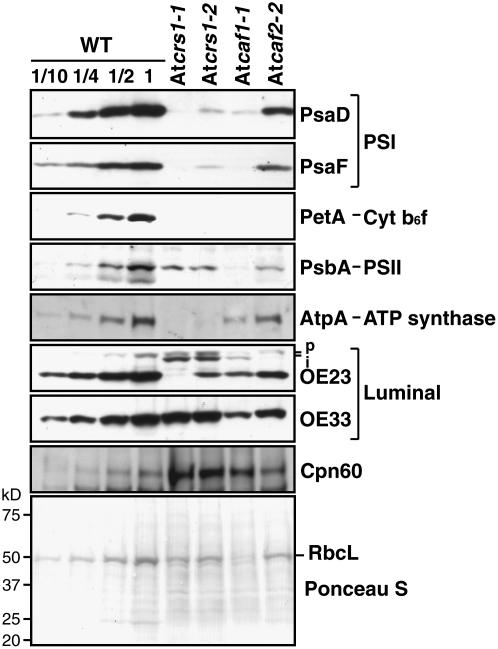
Immunoblots were performed to assess the accumulation of the major photosynthetic enzyme complexes in the leaves of those mutants that germinate (Fig. 2). Representative subunits of each complex were assayed whose abundance reflects that of other closely associated subunits of the same complex: PsaD and PsaF for PSI, PetA for the cytochrome b6f complex, PsbA for PSII, AtpA for the ATP synthase, and RbcL for Rubisco. Whereas the nucleus-encoded chaperone protein Cpn60 accumulated to increased levels in all of the mutants, the levels of all of the assayed core subunits of PSI, PSII, the cytochrome b6f complex, the ATP synthase, and Rubisco were reduced. These protein deficiencies are consistent with underlying global defects in chloroplast gene expression, as each of the affected complexes includes at least one plastid-encoded subunit. The AtCRS1 mutants differ from the AtCAF1 and AtCAF2 mutants in that AtpA is reduced considerably more than the other proteins assayed. This is analogous to what is observed in maize crs1 mutants, which lack the ATP synthase complex due to the failure to splice the atpF RNA (Jenkins et al., 1997), and which have a less severe loss of other plastid-encoded proteins due to a global reduction in chloroplast translation (Till et al., 2001). Interestingly, AtCRS1 mutants accumulate increased levels of the stromal intermediate of the luminal protein OE23 (iOE23), indicating a defect in its translocation across the thylakoid membrane. OE23 is translocated to the thylakoid lumen via the TAT translocation pathway, which is driven by a trans-thylakoid ΔpH. It seems likely that the severe ATPase deficiency in AtCRS1 mutants disrupts this targeting pathway by preventing the generation of the necessary pH gradient.
Figure 2.
Accumulation of representative subunits of photosynthetic enzyme complexes in AtCRS1, AtCAF1, and AtCAF2 mutants. Duplicate immunoblots of total leaf extract (5 μg protein or the indicated dilution of the wild-type sample) were probed with antibodies to the indicated proteins. The mutant plants were homozygous for the indicated allele. The bottom section shows the filter that had been probed for PsbA stained with Ponceau S, to demonstrate similar protein loading, and to detect the large subunit of Rubisco (RbcL). OE23 and OE33 are luminal proteins found in the oxygen-evolving complex associated with PSII. Bands corresponding to the putative precursor (p) and stromal intermediate (i) forms of OE23 are marked.
Chloroplast Splicing Defects in AtCRS1, AtCAF1, and AtCAF2 Mutants Mirror Those in Maize
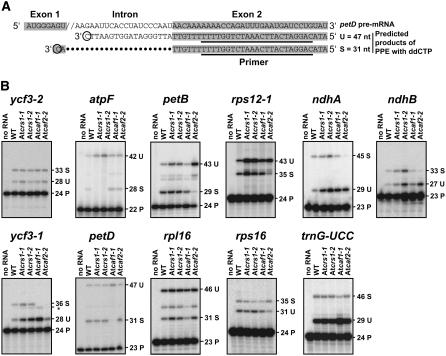
To determine whether AtCRS1, AtCAF1, and AtCAF2 function similarly to their maize orthologs in chloroplast group II intron splicing, the metabolism of intron-containing plastid transcripts was analyzed for those mutant alleles that germinated on Suc-containing medium. A poisoned-primer extension (PPE) assay was used for this purpose: an oligonucleotide complementary to exon sequences near a 3′ splice junction was used to prime reverse transcription in the presence of a dideoxynucleotide that terminates after different distances on spliced and unspliced RNA templates (see diagram in Fig. 3A). With this assay, all transcripts for which the intron is spliced are detected as one band, and all those in which the intron is retained are detected as a second band.
Figure 3.
Ratio of spliced to unspliced transcripts from Arabidopsis plastid genes with orthologs in maize. A, Diagram of the PPE assay. The petD intron is used as an example of the strategy. Shaded nts represent exon sequences, and the primer is underlined. Dideoxy CTP is included in the primer extension reaction, such that primer extension terminates at the first guanidine residue encountered in the template RNA (see circled residues). The lengths of the extension products differ for spliced (S) and unspliced (U) transcripts. B, Results of PPE assays for introns whose maize orthologs are influenced by mutations in maize crs1, caf1, or caf2. Mutants were homozygous for the indicated allele. The predicted sizes of the PPE products for spliced (S) and unspliced (U) transcripts are stated in nts, next to each product. Bands corresponding to the radiolabeled primers (P) are marked. Ten micrograms of seedling leaf RNA was analyzed, except in the first lane in each section, which show reactions lacking leaf RNA. The size of the product marked with an asterisk in the ycf3-1 section is consistent with that predicted for chain termination at the branchpoint adenosine. The sizes of the unmarked bands in the petB and rpl16 assays do not correlate with those predicted for termination at the branchpoint adenosines; the origin of these products is unknown.
Eleven of the 17 group II introns found in both maize and Arabidopsis plastids were examined, including orthologs of all of the introns that require maize CRS1, CAF1, or CAF2. The results show that the splicing of orthologous introns is generally promoted by orthologous splicing factors (Fig. 3B; summarized in Table II). For example, the ycf3-2 intron was unaffected by mutations in any of these Arabidopsis genes, as in maize. AtCRS1, like maize CRS1, is required for the splicing of the atpF intron but not for the other introns examined. AtCAF2, like maize CAF2, is required for the splicing of the petB, rps12-1, ndhA, ndhB, and ycf3-1 introns, but not for the splicing of the other introns examined. Like its maize counterpart, reduction of AtCAF1 function caused a strong defect in the splicing of the petD and ycf3-1 introns and a distinct defect in rpl16 splicing, but had no impact on the petB, rps12-1, atpF, or ycf3-2 introns. Residual splicing of affected introns in Atcaf1-1 and Atcaf2-2 mutants could be due to the incomplete disruption of gene function by the intron insertions in these alleles.
Table II.
Intron specificities of CRS1, CAF1, and CAF2 in maize and Arabidopsis
| Introna | Required in Arabidopsis
|
Required in Maize
|
||||
|---|---|---|---|---|---|---|
| AtCRS1 | AtCAF1 | AtCAF2 | CRS1 | CAF1 | CAF2 | |
| atpF | Yes | No | No | Yes | No | No |
| petD | No | Yes | No | No | Yes | No |
| rpl16 | No | Yes | No | No | Yes | No |
| rps16 | No | Reducedb | No | No | Yes | No |
| trnG-UCC | No | Reducedb | No | No | Yes | No |
| ndhB | No | Reducedb | Yes | No | Reducedb | Yes |
| petB | No | No | Yes | No | No | Yes |
| rps12-1 (trans) | No | No | Yes | No | No | Yes |
| ndhA | No | Reducedb | Yes | No | Reducedb | Yes |
| ycf3-1 | No | Yes | Yes | No | Yes | Yes |
| ycf3-2 | No | No | No | No | No | No |
| clpP-1 | No | Yes | No | Introns not present in maize | ||
| clpP-2 | No | No | No | |||
| rpoC1 | No | Yes | No | |||
Introns whose splicing has been assayed in all three mutant lines in maize and Arabidopsis are listed. Maize data are from Jenkins et al. (1997) and Ostheimer et al. (2003).
Splicing is reduced in the mutant background but considerable spliced RNA accumulates. Incomplete disruption of splicing in the Arabidopsis mutants could be due to incomplete disruption of gene function by the intron-localized T-DNA insertions.
The role of AtCAF1 in the splicing of the rps16, trnG-UCC, ndhB, and ndhA introns is less clear cut from these data, in that these all show mild splicing defects in Atcaf1-1 mutants. The results for ndhA and ndhB are similar to those in maize, where these introns are strongly dependent on CAF2 and weakly dependent on CAF1 (Ostheimer et al., 2003; G. Ostheimer and A. Barkan, unpublished data). The mild ndhA splicing defect in maize caf1 mutants may be an indirect effect, as this intron did not strongly coimmunoprecipitate with CAF1 from chloroplast extract (Schmitz-Linneweber et al., 2005). A more notable difference from the results in maize concerns the rps16 and trnG-UCC introns: in maize, the splicing of these introns is strongly CAF1 dependent and these introns strongly coimmunoprecipitate with CAF1 from stromal extract (Ostheimer et al., 2003; Schmitz-Linneweber et al., 2005). The small effect of the Atcaf1-1 mutation on these introns may reflect a true difference in the intron specificities of maize and Arabidopsis CAF1, redundancy between AtCAF1 and AtCAF2 function with regard to these introns, or simply an incomplete disruption of AtCAF1 gene function by the intron-localized insertion.
PPE assays of the ycf3-1 intron in Atcaf1-1 mutants yielded increased amounts of an extension product that is approximately 1 nucleotide (nt) shorter than the product from spliced RNA (marked with an asterisk in Fig. 3). Chain termination at this position corresponds to the predicted branchpoint adenosine, whose 2′OH group forms a branched structure after the first of the two splicing steps; this structure could impede the progress of reverse transcriptase. Thus, AtCAF1 may be necessary specifically for the second of the two trans-esterification reactions involved in group II intron splicing.
AtCAF1 Is Required for the Splicing of Two Arabidopsis Plastid Introns That Lack Maize Orthologs
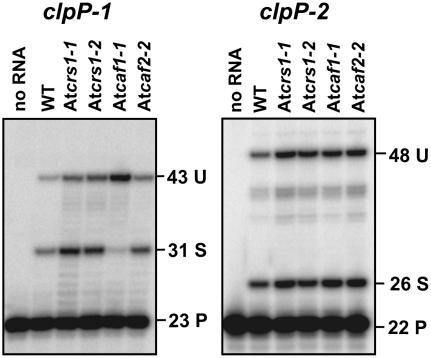
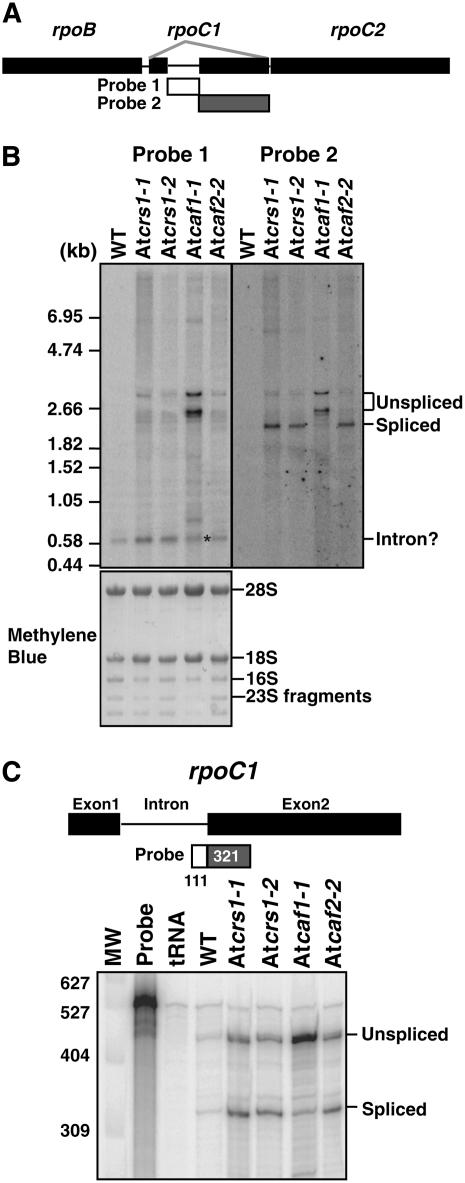
The Arabidopsis plastid genome encodes three group II introns that are absent in maize: two in the clpP gene and one in the rpoC1 gene. To determine whether the splicing of these introns is dependent on AtCRS1, AtCAF1, or AtCAF2, their splicing was monitored in the corresponding mutant backgrounds. PPE assays showed that the splicing of the clpP-1 intron requires AtCAF1 but not AtCRS1 or AtCAF2, whereas clpP-2 intron splicing was not disrupted in any of these mutant backgrounds (Fig. 4; Supplemental Fig. S4A). Splicing of rpoC1 was monitored by RNA gel-blot hybridizations and a ribonuclease protection assay (Fig. 5). The RNA gel blots (Fig. 5B) were hybridized with intron-specific (probe 1) and exon-specific (probe 2) probes; the results showed a dramatic increase in the abundance of two unspliced precursors and the near absence of the spliced rpoC1 mRNA in Atcaf1-1 mutants. A low Mr transcript that hybridized specifically to the intron probe (marked with an asterisk in Fig. 5B) may represent excised intron; the ratio of this transcript to the unspliced precursors is also reduced in Atcaf1-1 mutants. The ribonuclease protection assay (Fig. 5C; Supplemental Fig. S4B) likewise revealed a substantial increase in the ratio of unspliced-to-spliced rpoC1 RNAs in Atcaf1 mutants, but not in the other mutants. These results show that AtCAF1, but not AtCRS1 or AtCAF2, is required for the splicing of both the clpP-1 and rpoC1 introns. AtCAF1 is the first protein to be identified that functions in the splicing of these introns.
Figure 4.
PPE assay of clpP intron splicing. The Arabidopsis plastid clpP gene contains two group II introns, clpP-1 and clpP-2. PPE assays were performed as described in Figure 3. U, Unspliced; S, spliced; P, primer. The Atcaf1-2 allele gave results similar to those seen with Atcaf1-1 (Supplemental Fig. S4A).
Figure 5.
Analysis of rpoC1 splicing in AtCRS1, AtCAF1, and AtCAF2 mutants. A, Map of the Arabidopsis plastid rpo locus and the probes used for RNA gel-blot hybridizations. B, RNA gel-blot hybridizations showing rpoC1 transcripts in each mutant line. Three micrograms of seedling leaf RNA was analyzed in each lane. Duplicate blots were probed with intron-specific (probe 1) and exon-specific (probe 2) probes. The asterisk marks a band that may correspond to the excised intron. The positions of RNA size markers are shown to the left. The blot used with probe 1 was stained with methylene blue, and is shown below to illustrate the rRNAs in each sample. C, RNAse protection assay of rpoC1 splicing. The radiolabeled probe spanned the 3′-splice junction, with the indicated number of nts corresponding to intron (111 nt) and exon (321 nt) sequences. Protection of the probe by unspliced or spliced RNAs is predicted to yield 432 nt and 321 nt products, respectively. Three micrograms of total leaf RNA was analyzed. The lane labeled tRNA shows the results when yeast (Saccharomyces cerevisiae) tRNA was substituted for leaf RNA. The positions of DNA size markers are shown to the left. The Atcaf1-2 allele gave results similar to those seen with Atcaf1-1 (Supplemental Fig. S4B).
DISCUSSION
Proteins that facilitate the splicing of group II introns fall into two general classes: conserved maturases that are usually encoded within the intron whose splicing they facilitate, and proteins of diverse origin that were coopted from the host genome (for review, see Lambowitz and Zimmerly, 2004). Little is known about the relative timing of intron acquisition and the recruitment of host-encoded splicing factors, or the coevolution of these proteins with their cognate introns once the relationship had been established. Phylogenetic analyses indicate that most of the group II introns in land plants were acquired prior to the divergence of the charophyte algae, whereas group II introns were acquired independently in more distantly related algal lineages such as the chlorophytes (which include Chlamydomonas; Turmel et al., 2006; for review, see Barkan, 2004). Given the distinct history of intron acquisition in maize and Chlamydomonas plastids, it is perhaps not surprising that there is no similarity among the plastid group II intron splicing factors identified to date in these organisms. Results presented here show that the splicing functions and intron specificities of three group II intron splicing factors in maize are generally conserved with their Arabidopsis orthologs, indicating that these proteins were recruited to function in group II intron splicing prior to the divergence of monocot and dicot plants. The Arabidopsis CAF1 ortholog has an additional role in the splicing of two introns that are found in Arabidopsis but not in maize (rpoC1 and clpP-1), and, to our knowledge, is the first splicing factor to be discovered for these introns. A summary of the intron specificities of CAF1, CAF2, and CRS1 in maize and Arabidopsis is presented in Table II.
We previously identified two high-affinity binding sites for CRS1 within its atpF intron substrate, in intron domains 1 and 4 (Ostersetzer et al., 2005). The sequences of these binding sites are not highly conserved in Arabidopsis, and yet the genetic data presented here imply that AtCRS1 associates with the Arabidopsis atpF intron. It is possible that maize and Arabidopsis CRS1 recognize similar RNA structures in analogous intron regions, or that distinct RNA/protein interactions evolved subsequent to the divergence of maize and Arabidopsis. In either case, sets of orthologous splicing factor/intron pairs like this provide natural variants that can be used to deduce the determinants of sequence-specific RNA recognition by these proteins.
Relatively little information is available concerning plastid splicing factors in Arabidopsis, with just one candidate described previously: the pentatricopeptide repeat protein HCF152 is required for the accumulation of spliced petB RNA but not for the accumulation of the excised intron (Meierhoff et al., 2003). The genetic identification of plastid splicing factors in Arabidopsis might be hindered by the maintenance in the Arabidopsis plastid genome of several conserved ORFs whose tobacco (Nicotiana tabacum) orthologs have been shown to be essential for cellular viability: ycf1, ycf2, accD, and clpP (Drescher et al., 2000; Kuroda and Maliga, 2003; Kode et al., 2005). The failure to splice any of the group II introns in plastid pre-tRNAs, ribosomal protein pre-mRNAs, or in clpP is therefore predicted to cause embryo lethality in Arabidopsis by preventing the synthesis of one or more of these essential proteins. We show here that the Atcaf2-1 allele, which is predicted to be a null allele, conditions an embryo-lethal phenotype; embryo lethality can be accounted for by the role of AtCAF2 in promoting the splicing of the rps12 mRNA, whose Escherichia coli ortholog encodes an essential ribosomal protein (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp). In contrast, a null allele of maize caf2 conditions germinating seedlings with an albino phenotype (Ostheimer et al., 2003). This difference between the maize and Arabidopsis null phenotypes (albino seedling versus embryo lethality) correlates with a difference in plastid gene content between the two species: the maize plastid genome lacks the essential plastid genes ycf1, ycf2, and accD (Maier et al., 1995) and the plastid-encoded ClpP in maize is not essential for cellular viability (Cahoon et al., 2003). Thus, there are no ORFs in the maize plastid genome that are known to be essential for cellular viability. This difference may account for the fact that severe defects in the splicing of plastid tRNAs and ribosomal protein mRNAs in maize result in albino seedlings whose molecular phenotypes can be analyzed. The role of AtCAF1 in promoting clpP mRNA splicing suggests that a complete loss of AtCAF1 function is likely to cause embryo lethality; the Atcaf1 mutant alleles analyzed here are likely to be weak alleles as the T-DNA insertions reside within an intron and are distant from both splice junctions.
Maize CAF1 and CAF2 facilitate splicing in complexes with a protein called CRS2, which is related to peptidyl-tRNA hydrolase enzymes and that is necessary for the splicing of CAF-dependent group II introns (Ostheimer et al., 2003, 2005, 2006). There are two predicted CRS2 orthologs in Arabidopsis (Ostheimer et al., 2005), but just one in rice (Oryza sativa; R. Williams-Carrier and A. Barkan, unpublished data), suggesting a gene duplication event in the Arabidopsis lineage. We have attempted to determine the role of each AtCRS2 coortholog by analysis of T-DNA insertions (R. Williams-Carrier and A. Barkan, unpublished data); however, an exonic insertion in one of these did not cause a phenotype, whereas no insertions are available in the other gene. In light of the findings presented here, it seems likely that one or both of these CRS2 coorthologs function in concert with AtCAF1 and AtCAF2 to promote the splicing of plastid group II introns. The confounding effects of species-specific duplicate loci and embryo lethality on genetic screens for chloroplast regulatory factors emphasize the importance of using multiple model organisms to study these processes.
MATERIALS AND METHODS
Ortholog Prediction
Arabidopsis (Arabidopsis thaliana) orthologs of CRS1, CAF1, and CAF2 were identified by generating multiple sequence alignments of all members of the CRM domain family in rice (Oryza sativa; 14 proteins) and Arabidopsis (16 proteins), together with maize (Zea mays) CRS1, CAF1, and CAF2. The alignments were used to generate neighbor-joining and parsimony trees; the two methods yielded trees with identical topology and high bootstrap values (Barkan et al., 2007), with orthologs among the three species clustering in the trees.
Plant Material and Growth Conditions
Seeds from the following T-DNA insertion lines were obtained from the Salk Institute Genomic Analysis Laboratory: SALK_026861 (Atcrs1-1), SALK_027605 (Atcrs1-2), SALK_025042 (Atcaf1-1), SALK_021969 (Atcaf1-2), SALK_049304 (Atcaf2-1), and SALK_08478 (Atcaf2-2). Phenotypically normal seedlings were used as the source of wild-type tissue for phenotypic analyses. Seeds were surface sterilized with 5% (v/v) bleach and 0.1% Tween 20 for 5 min, washed three times in sterilized water, and then sowed on Murashige and Skoog plates (0.3% [w/v] Phytagel [Sigma], 1× Murashige and Skoog salts [Sigma], 1× Gamborg's B5 vitamin [Sigma], and 2% [w/v] Suc, pH 5.8). Plants were grown in a growth chamber at 22°C, 14-h-light/10-h-dark cycles with an irradiance of approximately 200 μE m−2s−1. Plants used for protein and RNA analysis were between 2 and 3 weeks old. Homozygous mutants were seedling or embryo lethal, so mutations were propagated by transferring heterozygous plants to vermiculite 3 weeks after germination, and feeding with a nutrient solution (Liquid Fruit and Bloom, 2-5-1, Down to Earth) until seeds were produced.
DNA Extraction and PCR Amplification
Seedlings were frozen in liquid nitrogen and ground to a powder with a toothpick in a 1.5 mL microfuge tube. TPS buffer (100 mm Tris-HCl, pH 9.5, 1 m KCl, and 10 mm EDTA; Thomson and Henry, 1995; 20 μL for mutant seedlings and 50 μL for wild-type seedlings) was added and mixed thoroughly. Samples were boiled at 95°C for 10 min, and debris was removed by centrifugation at 21,000g for 10 min. Supernatants were diluted 10-fold with distilled water.
T-DNA insertions were confirmed by PCR amplification of either the mutant or wild-type alleles, using the T-DNA left border primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and gene-specific primers as follows. Atcrs1-1: At-crs1-1, 5′-GTCCAAAGCTGGTCCACATC-3′; At-crs1-2, 5′-GCAAGTCAGCATCAACGGGA-3′; or atCRS15 (1405), 5′-CTCTCCCATGGCATTTTCCC-3′. Atcrs1-2: At-crs1-3, 5′-AAACCATCGGAGGGAGATGA-3′ and At-crs1-4, 5′-TTGGACCCAATTTTGCGTAGC-3′. Atcaf1-1 and Atcaf1-2: AtCAF1-1, 5′-CCTCTGCTCGTTCGATTCCGGGTC-3′ and AtCAF1-2, 5′-TAACTGCTCGCAAACATTATCCAT-3′. Atcaf2-1: Atcaf2-1, 5′-GATTCCGGCGTCTCGTATCA-3′ and Atcaf2-2, 5′-GGTTCAAGCCCTGGGCAGTC-3′. Atcaf2-2: 1g234-3′, 5′-TTGCAATCACTTAAGGTCAATCTCAAATG-3′ and 1g234-5′, 5′-GTGTTCTGCAGCCAGTGAAGTGCCTT-3′.
Protein Analysis
Protein was extracted from the leaves of 2- to 3-week-old seedlings grown on agar medium, as described previously for maize (Barkan, 1998). Antibody to Cpn60 was generously provided by Harry Roy. The other antibodies were described in Roy and Barkan (1998).
RNA Analysis
RNA was extracted from the leaf tissue of 2- to 3-week-old seedlings grown on agar medium, by using TriZol reagent (GIBCO/BRL) according to the manufacturer's instructions. Phenotypically normal siblings from the same plates were used as the source of wild-type RNA. RNA gel-blot hybridizations and RNAse-protection assays were performed as described previously for maize (Jenkins et al., 1997). RNA quantities are indicated in the figure legends. The probes for RNA gel-blot assays of rpoC1 RNA were generated by PCR from total Arabidopsis DNA using the following primers: Probe 1 (Intron), YA at rpoC1 F3 (5′-TGTGATTTGATCGAAAT-3′) and YA at rpoC1 R3 (5′-ATTGGGATAGGGTCCACT-3′); Probe 2 (Exon), YA at rpoC1 F4 (5′-TTTTCTTTTGCTAGGCCCATAACT-3′) and YA at rpoC1 R4 (5′-TTAGGTATCATATGAACAGG-3′). The probes were radiolabeled by the random-priming method. The probe for the ribonuclease-protection assay of rpoC1 splicing was generated by cloning the PCR product obtained from total Arabidopsis DNA with the primers YA At rpoC1-F2 (5′-AATTTGAAAGCGAGACCCCT-3′) and YA At rpoC1-R2 (5′-AGCTAATTCCATACGTCTAACTA-3′) into pGEM-T (Promega), digesting the clone with NcoI and generating radiolabeled RNA by in vitro transcription with SP6 RNA polymerase.
Poisoned primer extension reactions were performed with 10 μg of leaf RNA. Antisense primers were designed to obtain extension productions of different sizes from unspliced and spliced RNA templates when performed in the presence of a specific dideoxynucleotide. The primers and dideoxynucleotide used for each intron were as follows: ycf3-1, primer YAAtycf3-R3 5′-AATTTCCTTCAGATTGAGCCGACA-3′ ddGTP; ndhA, primer YAAtNdhAR2 5′-ATCAACTGTACTTAAACTGTTAG-3′, ddCTP; ndhB, primer YAAtNdhBR2, 5′-AAGCAACGACTGGAGTGGGAGA-3′, ddATP; ycf3-2, primer YAAtycf3-1R2, 5′-CTTGTTGAATGGCCTGTTCTCCAC-3′, ddCTP; petB, primer YAAtpetB-R2, 5′-ACGTTCTTCGAACCAATCATAAAC-3′, ddCTP; petD, primer YAAtpetD-R, 5′-CAGGATCATTCAAATCTGGTTTT-3′, ddCTP; atpF, primer YAAtatpF-2, 5′-CTTTCGGTTATCTAATAAATCA-3′, ddCTP; clpP-1, primer YAAtclpP-1-R, 5′-ATCTTTCTCGATAAAGTCGGTTG-3′, ddCTP; clpP-2, primer YAAtclpP-2-R, 5′-GCGGGTTGATGGATCATTACCC-3′, ddCTP; trnG(UCC), primer YAAttrnG[UCC]-R 5′-ATCGTTAGCTTGGAAGGCTAAGGG-3′, ddGTP; rpl16, primer YAAtrpl16-R 5′-TGTTGTTTACGAAATCTGGTTCT-3′, ddCTP; rps16, primer YAAtrps16-R 5′-TTGCAAGGATTCGATAAACGGCTC-3′, ddATP; rps12-1, primer YAAtrps12-1 5′-AAGCAGAGTTTGGTTTTTTGGGGG-3′, ddCTP. Primers were 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The RNA was suspended in 4 μL 10 mm Tris-HCl, pH 8.5, 100 mm KCl, combined with 1 μL of labeled primer (50,000–100,000 cpm), heated briefly to 95°C and cooled to 48°C by placing in a 48°C heat block for 10 min. Primer extension was initiated by the addition of 5 μL of a solution containing 5 units AMV reverse transcriptase (Promega) and 10 units of RNAsin Ribonuclease Inhibitor (Promega) in 50 mm Tris-HCl, pH 8.5, 20 mm MgCl2, 20 mm dithiothreitol, 0.5 mm of the appropriate ddNTP, and 0.5 mm of the other three dNTPs. Reactions were incubated at 45°C for 30 min and then stopped by the addition of 12 μL 90% formamide, 20 mm Tris-HCl, pH 8.0, 20 mm EDTA, 0.04% bromphenol blue, and 0.04% xylene cyanole and boiling for 2 min. Ten microliters of each reaction was applied to a 15% polyacrylamide gel containing 7 m urea and 1× Tris-borate/EDTA, and electrophoresis was performed at 27 mA with 1× Tris-borate/EDTA buffer. Gels were imaged with a Storm PhosphorImager (Molecular Dynamics).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. T-DNA insertion sites in AtCRS1, AtCAF1, and AtCAF2.
Supplemental Figure S2. Linkage between chlorophyll-deficient phenotypes and T-DNA insertions.
Supplemental Figure S3. Presence of both mutant alleles in noncomplementing progeny derived from complementation crosses.
Supplemental Figure S4. Defects in clpP-1 and rpoC1 splicing in Atcaf1-2 mutants.
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and Harry Roy for providing antibody to Cpn60. We also are grateful to Shingo Kikuchi, Athea Vichas, Jana Prikryl, Mark Carrier, Kenny Watkins, Nigel Walker, and Rosalind Williams-Carrier for technical advice, and to Susan Belcher and Oren Ostersetzer for contributing to the early stages of the AtCRS1 and AtCAF2 work.
This work was supported by the National Science Foundation (grant no. DBI–0421799).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alice Barkan (abarkan@molbio.uoregon.edu).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Barkan A (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Barkan A (2004) Intron splicing in plant organelles. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 281–308
- Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP (2007) The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA (in press) [DOI] [PMC free article] [PubMed]
- Cahoon A, Cunningham K, Stern D (2003) The plastid clpP gene may not be essential for plant cell viability. Plant Cell Physiol 44: 93–95 [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Jenkins B, Barkan A (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J 20: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B, Kulhanek D, Barkan A (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S (2004) Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Koessel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol 251: 614–628 [DOI] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Perron K, Rahire M, Howald I, Rochaix JD, Goldschmidt-Clermont M (2006) A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res 34: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Ferat J-L (1995) Structure and activities of group II introns. Annu Rev Biochem 64: 435–461 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Watkins K, Cooke A, Barkan A (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17: 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer G, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer GJ, Hadjivassiliou H, Kloer DP, Barkan A, Matthews BW (2005) Structural analysis of the group II intron splicing factor CRS2 yields insights into its protein and RNA interaction surfaces. J Mol Biol 345: 51–68 [DOI] [PubMed] [Google Scholar]
- Ostheimer GJ, Rojas M, Hadjivassiliou H, Barkan A (2006) Formation of the CRS2-CAF2 group ll intron splicing complex is mediated by a 22 amino acid motif in the C-terminal region of CAF2. J Biol Chem 281: 4732–4738 [DOI] [PubMed] [Google Scholar]
- Perron K, Goldschmidt-Clermont M, Rochaix J-D (1999) A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J 18: 6481–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A (1996) Catalytic reaction mechanisms and structural features of group II intron ribozymes. In F Eckstein, DMJ Lilley, eds, Nucleic Acids and Molecular Biology, Vol 10. Springer-Verlag, Berlin, pp 75–108
- Rivier C, Goldschmidt-Clermont M, Rochaix J (2001) Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J 20: 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy LM, Barkan A (1998) A secY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J Cell Biol 141: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D, Henry R (1995) Single-step protocol for preparation of plant tissue for analysis by PCR. Biotechniques 19: 394–397, 400 [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C (2006) The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23: 1324–1338 [DOI] [PubMed] [Google Scholar]