Abstract
Geminivirus Rep-interacting kinase 1 (GRIK1) and GRIK2 constitute a small protein kinase family in Arabidopsis (Arabidopsis thaliana). An earlier study showed that a truncated version of GRIK1 binds to the geminivirus replication protein AL1. We show here both full-length GRIK1 and GRIK2 interact with AL1 in yeast two-hybrid studies. Using specific antibodies, we showed that both Arabidopsis kinases are elevated in infected leaves. Immunoblot analysis of healthy plants revealed that GRIK1 and GRIK2 are highest in young leaf and floral tissues and low or undetectable in mature tissues. Immunohistochemical staining showed that the kinases accumulate in the shoot apical meristem, leaf primordium, and emerging petiole. Unlike the protein patterns, GRIK1 and GRIK2 transcript levels only show a small increase during infection and do not change significantly during development. Treating healthy seedlings and infected leaves with the proteasome inhibitor MG132 resulted in higher GRIK1 and GRIK2 protein levels, whereas treatment with the translation inhibitor cycloheximide reduced both kinases, demonstrating that their accumulation is modulated by posttranscriptional processes. Phylogenetic comparisons indicated that GRIK1, GRIK2, and related kinases from Medicago truncatula and rice (Oryza sativa) are most similar to the yeast kinases PAK1, TOS3, and ELM1 and the mammalian kinase CaMKK, which activate the yeast kinase SNF1 and its mammalian homolog AMPK, respectively. Complementation studies using a PAK1/TOS3/ELM1 triple mutant showed that GRIK1 and GRIK2 can functionally replace the yeast kinases, suggesting that the Arabidopsis kinases mediate one or more processes during early plant development and geminivirus infection by activating SNF1-related kinases.
Protein kinases are important components of signal transduction and regulatory pathways in all eukaryotes. In plants, they play critical roles in pathogen recognition and the defense response (for review, see Romeis, 2001; Xing et al., 2002; Pedley and Martin, 2003, 2005). They can also act in concert with pathogen proteins to facilitate infection (Florentino et al., 2006). This partnership is particularly important for viruses that have limited coding capacities. Geminiviruses are small DNA viruses that depend on host machinery to transcribe and replicate their genomes in plant cells (for review, see Hanley-Bowdoin et al., 2004). These important plant pathogens also interact with host factors to move throughout the plant and to overcome host defenses. Host protein kinases have been implicated in these processes through their interactions with geminivirus proteins. Analysis of these interactions will increase our understanding of the mechanisms leading to productive geminivirus infection and provide valuable insight into the roles of these kinases in plant growth and development.
Geminiviruses are a large family of plant-infecting viruses that fall into four genera based on their genome structure, insect vector, and host range (Rojas et al., 2005). Members of the Begomovirus and Curtovirus genera, which infect dicot species, can have single-component or two-component genomes, and some are associated with satellite DNAs (Rojas et al., 2005). These viruses encode three conserved proteins, AL1, AL2, and AL3, on a set of overlapping genes. The AL1 (AC1, C1, or Rep) protein is essential for viral replication and infection (Elmer et al., 1988), while AL3 (C3 or REn) acts as an accessory factor to enhance viral replication (Sunter et al., 1990). AL1 functions as the origin recognition protein (Fontes et al., 1994), catalyzes initiation and termination of viral DNA replication (Laufs et al., 1995; Orozco and Hanley-Bowdoin, 1996), and may act as a DNA helicase during replication (Pant et al., 2001). AL1 also plays a central role in reprogramming mature plant cells to support DNA replication (Kong et al., 2000). The AL2 protein (AC2, C2, or TrAP) activates transcription of viral genes expressed late in infection (Sunter and Bisaro, 1992) and acts as an anti-defense factor (for review, see Bisaro, 2006).
Geminivirus proteins interact with a diverse set of host factors. Many of these interactions have been implicated in the recruitment of plant proteins to participate in essential viral processes. AL1 and AL3 recruit host replication machinery by binding to PCNA and RFC, the clamp and clamp loader of the host DNA polymerase δ (Luque et al., 2002; Castillo et al., 2003; Bagewadi et al., 2004). AL1 binding to a mitotic kinesin and histone H3 may also play a direct role in viral replication (Kong and Hanley-Bowdoin, 2002). The ability of AL3 to enhance viral DNA replication is mediated in part through its interaction with a host NAC1 transcription factor (Selth et al., 2005). The AL1 protein also binds to UBC9, a component of the plant sumoylation pathway (Castillo et al., 2004), but how this interaction contributes to infection is not clear.
Binding of geminivirus proteins can also sequester and/or inhibit the activities of plant proteins to overcome barriers to infection or host defense mechanisms. AL1 and AL3 interact with the retinoblastoma-related protein (pRBR), a negative regulator of cell cycle progression and a differentiation factor (Weinberg, 1995; Ach et al., 1997; Kong et al., 2000). The AL1-pRBR interaction interferes with pRBR binding to the cell cycle-associated E2F transcription factors and leads to the activation of genes encoding proteins required for DNA replication (Egelkrout et al., 2001; Desvoyes et al., 2006). The AL2 protein binds to adenosine kinase (ADK) to inhibit its activity and suppress gene silencing to overcome host defense responses (Wang et al., 2005). Inhibition of the nuclear acetyltransferase AtNSI through its interaction with the geminivirus movement protein BR1 is thought to prevent DNA modifications that interfere with viral DNA replication and movement (Carvalho et al., 2006).
In plants, receptor-like protein kinases (RLKs) are often associated with the plasma membrane and activated by direct interaction with intracellular or extracellular signal molecules (for review, see Morris and Walker, 2003). Signals are also transduced by nonmembrane-associated protein kinase cascades in which upstream kinases activate downstream kinases. In both cases, the terminal kinases phosphorylate substrate proteins to impact their activities, localization, and/or stability. Geminiviruses interact with both types of protein kinase signaling cascades to modulate host processes. The BR1 protein binds to three Leu-rich-repeat RLKs, NIK1, NIK2, and NIK2, to inhibit their kinase and antiviral activities (Fontes et al., 2004). In contrast, interaction and phosphorylation of BR1 by a PERK-like RLK, NsAK, is necessary for efficient infection and full symptom development (Florentino et al., 2006). The AL2 protein inhibits the activity of an SNF1-related kinase (SnRK) through protein interactions, resulting in an enhanced susceptibility phenotype (Hao et al., 2003). In yeast and mammals, the SNF1 and related AMPK kinases are components of a conserved energy-sensing protein kinase cascade (Witters et al., 2006). In an earlier study, we showed that AL1 also binds to a protein kinase, geminivirus Rep-interacting kinase (GRIK), which is up-regulated during infection (Kong and Hanley-Bowdoin, 2002). To gain insight into its function in infection and normal plant processes, we investigated the expression, phylogeny, and likely targets of GRIK and its Arabidopsis (Arabidopsis thaliana) homolog.
RESULTS
A Small Family of Arabidopsis Protein Kinases Interact with a Geminivirus Replication Protein
In an earlier study (Kong and Hanley-Bowdoin, 2002), we identified an Arabidopsis protein kinase that interacts with the AL1 protein of Tomato golden mosaic virus (TGMV) and was designated as GRIK. Comparison with genomic and other cDNA sequence data in GenBank indicated that our GRIK cDNA was truncated at its 5′ end and did not encode the first 11 amino acids of the protein (Kong and Hanley-Bowdoin, 2002). These comparisons also uncovered a second Arabidopsis gene that specifies a closely related protein kinase. We designated the full-length GRIK protein, which is encoded by the At3g45240 gene, as GRIK1 and the related protein, which is specified by the At5g60550 gene, as GRIK2 (Fig. 1).
Figure 1.
Alignment of Arabidopsis GRIK1, GRIK2, and their homologs from M. truncatula and rice. Identical residues are in black blocks and identical or similar residues from at least three proteins are highlighted in gray. The protein kinase catalytic domains are indicated by the regions bordered by the bent arrows. The upper boxed sequences indicate the conserved protein kinase ATP-binding region (PROSITE accession PS00107) and the lower boxed sequences are the Ser/Thr protein kinase catalytic loop (PROSITE accession PS00108). The triangle marks the conserved Lys residue that is required for ATP binding (Hanks and Hunter, 1995) and was altered to Ala in GRIK1(K137A) and GRIK2(K136A). Underlined sequences in GRIK1and GRIK2 correspond to the peptides used for antibody production.
GRIK1 and GRIK2 are 396 and 407 residues in length, respectively, and share 77% amino acid identity and 85% similarity overall (Fig. 1). The GRIK1 kinase domain consists of 262 amino acids, whereas the GRIK2 kinase domain has 264 residues. The domains, which are 88% identical and 93% similar, contain the ATP-binding and catalytic motifs characteristic of Ser/Thr protein kinases (Hanks and Hunter, 1995). The GRIK1 and GRIK2 kinase domains are flanked by N-terminal sequences of 106 and 105 amino acids and C-terminal sequences of 27 and 41 residues, respectively. The N-terminal domains are 57% identical and 71% similar, while the C-terminal sequences are less conserved with 46% identity and 65% similarity.
BLAST analysis of plant databases uncovered two protein kinase sequences that are related to the Arabidopsis kinases (Fig. 1). The rice (Oryza sativa) Os03g50330 gene (The Institute for Genomic Research nomenclature) encodes an expressed protein that is equally related to GRIK1 (54% identity and 65% similarity) and GRIK2 (52% identity and 64% similarity). The Medicago truncatula bacterial artificial chromosome mth2-22g6 specifies a predicted protein (GenBank ABE79890) that is also equally related to GRIK1 (60% identity and 72% similarity) and GRIK2 (59% identity and 72% similarity). Like the Arabidopsis kinases, the rice and M. truncatula proteins contain highly conserved, central kinase domains flanked by more divergent N- and C-terminal sequences. The M. truncatula protein contains a 15-amino acid sequence in the kinase domain that is absent in the other sequences. The rice protein has two insertions in the N-terminal domain of 21 and six amino acids compared to the dicot proteins. Taken together, these proteins constitute a small protein kinase family in plants.
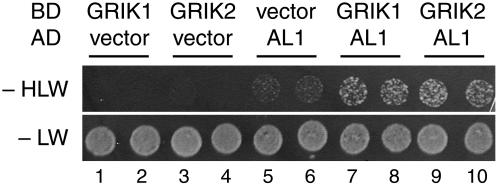
We asked if full-length GRIK1 and GRIK2 bind to TGMV AL1 in yeast two-hybrid assays as shown previously for the truncated GRIK protein (Kong and Hanley-Bowdoin, 2002). Yeast cells were cotransformed with a bait plasmid carrying a GAL4 activation domain-AL1 fusion (Orozco et al., 2000) and a prey plasmid corresponding to GRIK1 or GRIK2 fused to the GAL4 DNA binding domain. Negative controls were the empty DNA binding domain and activation domain vectors. All of the transformants grew on synthetic complete medium lacking Leu and Trp, verifying that they carried the bait and prey plasmids (Fig. 2, spots 1–10). In contrast, yeast cotransformed with the AL1 plasmid and a GRIK1 (Fig. 2, spots 7 and 8) or GRIK2 (spots 9 and 10) plasmid grew more efficiently on synthetic complete medium lacking His, Leu, and Trp than yeast cotransformed with the empty vector controls and the GRIK1 (Fig. 2, spots 1 and 2), GRIK2 (spots 3 and 4), or AL1 (spots 5 and 6) plasmids. These results established that both GRIK1 and GRIK2 bind to TGMV AL1 in yeast, leading to activation of the HIS3 marker.
Figure 2.
Both GRIK proteins interact with a geminivirus replication protein. Full-length GRIK1 (spots 7 and 8) or GRIK2 (spots 9 and 10) fused with the GAL4 DNA binding domain (BD) was assayed for interaction with the TGMV AL1 protein fused with the GAL4 activation domain (AD) in the yeast two-hybrid system. The empty vector controls with GRIK1 (spots 1 and 2), GRIK2 (spots 3 and 4), and TGMV AL1 (spots 5 and 6) are shown. Protein interaction was scored as growth of the yeast transformants expressing the HIS3 reporter gene on the synthetic complete medium without His (−HLW). Equal inoculations are indicated by the growth on medium containing His (−LW). Two independent transformants of each combination are shown.
Peptide Antibodies against GRIK1 and GRIK2
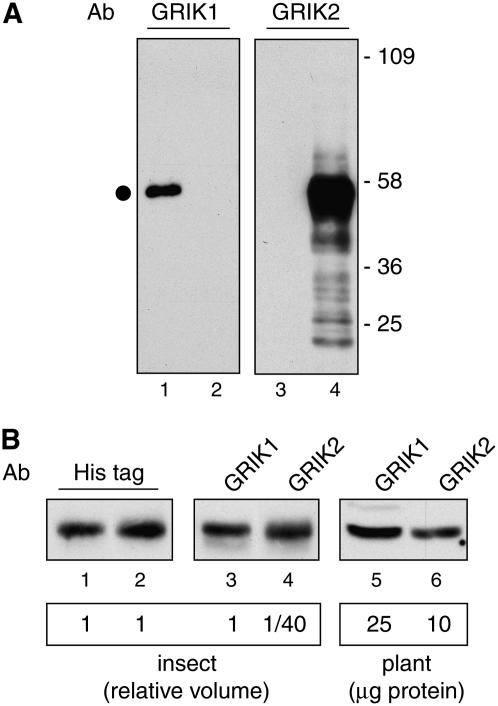
To distinguish between the GRIK1 and GRIK2 proteins, we took advantage of the divergence in their C-terminal sequences to generate unique peptides for antibody production (Fig. 1). The peptide antibodies were used in immunoblotting experiments of total protein extracts from insect cells expressing either His-tagged GRIK1 or GRIK2. The anti-GRIK1 antiserum detected an approximately 50-kD protein in cells expressing recombinant His-GRIK1 (Fig. 3A, lane 1) but not in cells expressing His-GRIK2 (lane 2). Conversely, the anti-GRIK2 antiserum did not cross-react with protein in cells expressing recombinant His-GRIK1 (Fig. 3A, lane 3) and, instead, detected a protein of expected size in cells expressing His-GRIK2 (lane 4). Together, these results show that peptide antibodies are specific for GRIK1 or GRIK2.
Figure 3.
Specificity and relative sensitivity of the GRIK1 and GRIK2 peptide antibodies. A, Total protein extracts from insect cells expressing His-tagged GRIK1 (lanes 1 and 3) or His-tagged GRIK2 (lanes 2 and 4) were resolved by SDS-PAGE followed by immunoblotting with affinity-purified peptide antibodies against GRIK1 (lanes 1 and 2) or GRIK2 (lanes 3 and 4). The dot indicates the GRIK1 or GRIK2 band. Molecular mass markers (in kilodaltons) are indicated. B, Volumes of the insect cell protein extracts were adjusted to contain equal amounts of the His-tagged GRIK1 (lane 1) and His-tagged GRIK2 (lane 2) proteins as determined by immunoblotting with anti-His-tag antibodies. One volume of GRIK1 (lane 3) and 1/40 volume of GRIK2 (lane 4) proteins show similar intensities on immunoblots probed with their corresponding peptide antibodies at 1 μg/mL and 0.1 μg/mL, respectively. The same GRIK1 (lane 5) and GRIK2 (lane 6) antibody dilutions were used to probe a total protein extract (25 and 10 μg, respectively) from young leaves.
The relative sensitivities of the peptide antibodies were compared using the recombinant His-tagged kinases. An anti-His-tag antibody was used to determine the volumes of insect cell protein extracts that gave similar signals for His-GRIK1 and His-GRIK2 on immunoblots (Fig. 3B, compare lanes 1 and 2). Based on this comparison, one volume of the GRIK1 extract and 1/40 volume of the GRIK2 extract produced bands of similar intensity with their respective peptide antibodies (Fig. 3B, compare lanes 3 and 4). These data established that the GRIK2 antibody is approximately 40-fold more sensitive relative to the GRIK1 antibody in our experimental conditions.
Expression of GRIK1 and GRIK2 Is Up-Regulated during Geminivirus Infection
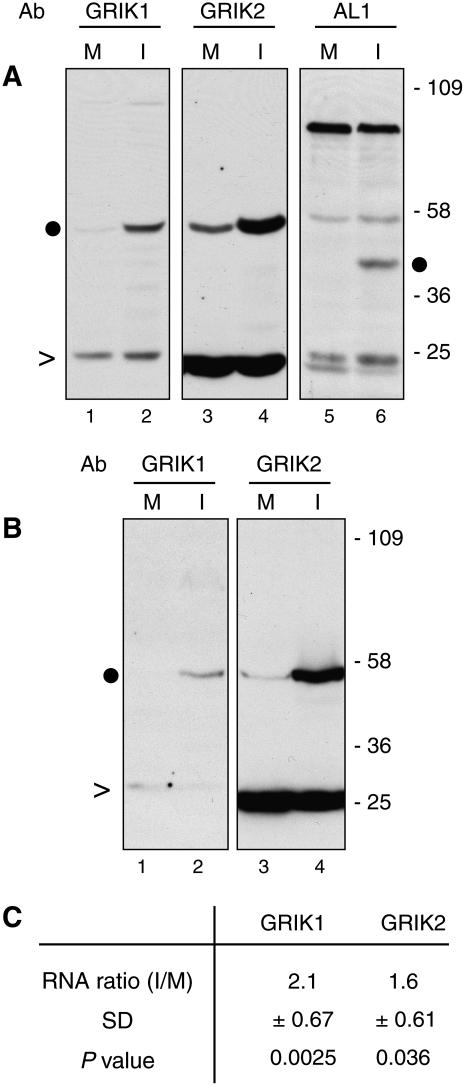
We asked if the affinity-purified peptide antibodies could detect GRIK1 and GRIK2 proteins in total protein extracts from symptomatic Arabidopsis leaves at 12 d postinoculation (dpi) with Cabbage leaf curl virus (CaLCuV), a member of the Begomovirus genus. Bands of the expected size were seen on immunoblots probed with the GRIK1 (Fig. 4A, lane 2) or GRIK2 (lane 4) antibodies. Lower levels of both kinases were detected in extracts from mock-inoculated leaves (Fig. 4A, lanes 1 and 3), demonstrating that their accumulation increases during CaLCuV infection. A polyclonal antibody against the CaLCuV AL1 protein detected the viral replication protein in extracts from infected leaves (Fig. 4A, lane 6) but not from mock-inoculated leaves (lane 5).
Figure 4.
Geminivirus infection induces the accumulation of GRIK1 and GRIK2 in expanding leaves. A, Total proteins from mock-inoculated (M; lanes 1, 3, and 5) and CaLCuV-infected (I; lanes 2, 4, and 6) leaves (0.5–1.5 cm long) at 12 dpi were resolved by SDS-PAGE followed by immunoblotting with GRIK1 (lanes 1 and 2) and GRIK2 (lanes 3 and 4) peptide antibodies and a polyclonal antiserum against CaLCuV AL1 (lanes 5 and 6). Molecular mass markers (in kilodaltons) are indicated. This experiment was repeated twice with similar results. B, Proteins from mock-inoculated (M; lanes 1 and 3) and BCTV-infected (I; lanes 2 and 4) leaves (0.5–1.5 cm long) at 28 dpi were analyzed similarly using the GRIK1 (lanes 1 and 2) and GRIK2 (lanes 3 and 4) antibodies. In A and B, the dots indicate the GRIK1, GRIK2, or AL1 band, while the carats indicate nonspecific bands as internal controls. C, Real-time quantitative RT-PCR of GRIK1 and GRIK2 mRNAs in CaLCuV-infected leaves (0.5–1.5 cm long). The average ratios of mRNAs from CaLCuV-infected versus mock-inoculated leaves (I/M) were calculated from three independent experiments. The sd of the ratios and the P values from Z tests are given.
We also examined GRIK1 and GRIK2 accumulation in Arabidopsis leaves infected with Beet curly top virus (BCTV Logan), a member of the Curtovirus genus. Because BCTV-inoculated Arabidopsis plants are asymptomatic at 12 dpi, we isolated protein extracts from symptomatic and mock-inoculated leaves at 28 dpi for immunoblot analysis with the peptide antibodies. Higher levels of both kinases were detected in BCTV-infected leaves (Fig. 4B, lanes 2 and 4) than in mock-inoculated leaves (lanes 1 and 3). We were unable to monitor the BCTV replication protein C1 in parallel because our antibodies against begomovirus AL1 proteins do not cross-react with the curtovirus protein. However, the results demonstrated that diverse geminiviruses stimulate the accumulation of the GRIK1 and GRIK2 proteins during infection.
Accumulation of GRIK1 and GRIK2 Is Modulated at Multiple Levels
We investigated if increases in the steady-state levels of GRIK1 and GRIK2 mRNAs during infection are responsible for the higher protein levels in Arabidopsis leaves. Relative mRNA levels were measured in three independent experiments by real-time quantitative reverse transcription (RT)-PCR using gene-specific primers for GRIK1, GRIK2, and the Act2 reference gene. Higher transcript levels were detected for both kinase genes in CaLCuV-infected leaves versus mock-inoculated leaves at 12 dpi, with increases of 2.1-fold and 1.6-fold for GRIK1 and GRIK2 mRNA, respectively (Fig. 4C). Although the increases in GRIK1 and GRIK2 mRNA levels in infected leaves were statistically significant in Z tests (P < 0.05), they were not sufficient to account for the higher protein levels during infection (Fig. 4A).
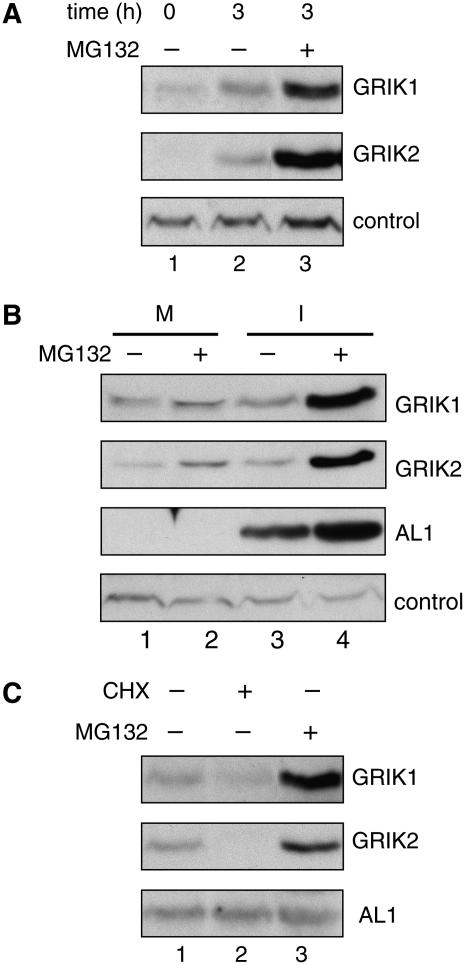
The RNA analysis suggested that the accumulation of GRIK1 and GRIK2 proteins is modulated at least in part by posttranscriptional processes. Protein degradation through the ubiquitin-proteasome pathway is a common mechanism for rapid and effective control of cellular levels of regulatory proteins like protein kinases (Sullivan et al., 2003; Smalle and Vierstra, 2004). Hence, we asked if GRIK1 or GRIK2 are degraded via the proteasome pathway initially in Arabidopsis seedlings and then in infected leaves. GRIK1 and GRIK2 levels were compared in protein extracts from untreated 2-week-old seedlings or in seedlings incubated for 3 h in the presence of the proteasome inhibitor MG132 or solvent alone. Immunoblot analysis revealed that the GRIK1 and GRIK2 proteins are high in MG132-treated seedlings (Fig. 5A, lane 3) but are low or undetectable in untreated (lane 1) or solvent-treated (lane 2) seedlings. These results showed that GRIK1 and GRIK2 are stabilized by MG132 and suggested that intrinsic pathways that maintain GRIK1 and GRIK2 proteins at low levels exist in plant cells.
Figure 5.
GRIK1 and GRIK2 are degraded by the proteasome. A, Two-week-old Arabidopsis seedlings were treated with MG132 (lane 3) or buffer (lane 2) for 3 h and total proteins were subjected to immunoblot analysis using GRIK1 or GRIK2 peptide antibodies. Seedlings collected immediately before treatment (time = 0) were tested in lane 1. A nonspecific band was used as an internal control. B, Mock-inoculated (M; lanes 1 and 2) or CaLCuV-infected (I; lanes 3 and 4) leaves were excised and treated with MG132 (lanes 2 and 4) or buffer only (lanes 1 and 3) for 3 h, and total proteins were analyzed by immunoblotting with antibodies against GRIK1, GRIK2, or CaLCuV AL1. A nonspecific band served as an internal control. C, CaLCuV-infected leaves were excised and treated for 3 h with cycloheximide (CHX; lane 2), MG132 (lane 3), or buffer (lane 1), and total proteins were analyzed by immunoblotting with antibodies against GRIK1, GRIK2, and CaLCuV AL1.
Consistent with the seedling results, there were slight increases in the GRIK1 and GRIK2 levels 3 h after MG132 treatment in mock-inoculated leaves (Fig. 5B, lane 2) compared to the solvent-only control (lane 1). However, MG132 treatment of infected leaves (Fig. 5B, lane 4) resulted in substantial increases in GRIK1 and GRIK2 abundance compared to the solvent treatment (lane 3). The similar levels of GRIK1 and GRIK2 in mock and infected leaves treated with solvent only (Fig. 5B, compare lanes 1 and 3) are consistent with degradation of the kinases during the 3-h treatment in the absence of the inhibitor. Higher levels of the AL1 protein were also observed in the presence of MG132 (Fig. 5B, lanes 3 and 4), but the difference between MG132 and solvent-treated samples was less than seen for the kinases. These results indicated that GRIK1 and GRIK2 are degraded via the ubiquitin-proteasome pathway in CaLCuV-infected plant leaves.
The significantly higher levels of GRIK1 and GRIK2 in CaLCuV-infected leaves versus mock-inoculated leaves in the MG132-treated samples (Fig. 5B, compare lanes 2 and 4) also suggested that the synthesis of both kinases is elevated during infection. We used the protein synthesis inhibitor cycloheximide to confirm that de novo protein synthesis is required to maintain GRIK1 and GRIK2 protein levels during infection. GRIK1 levels were reduced and GRIK2 was not detected in infected leaves incubated with cycloheximide for 3 h prior to protein extraction (Fig. 5C, lane 2) compared to solvent-treated tissue (lane 1). In contrast, AL1 protein levels were not impacted by cycloheximide treatment (Fig. 5C, compare lanes 1 and 2). In parallel assays, high levels of GRIK1 and GRIK2 were detected in the presence of MG132 (Fig. 5C, lane 3). Together, these results indicate that protein synthesis and turnover influence GRIK1 and GRIK2 protein levels during infection.
GRIK1 and GRIK2 Expression Is Regulated during Plant Development
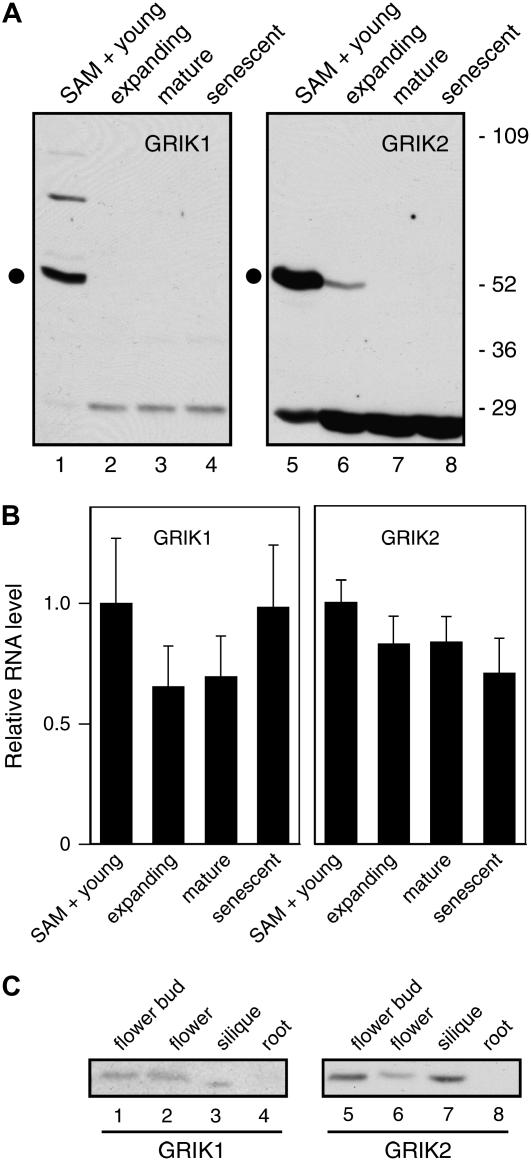
AL1 interacts with the host pRBR to induce the expression of DNA replication machinery and cause mature plant cells to reenter the S phase of cell cycle (Ach et al., 1997; Kong et al., 2000; Nagar et al., 2002). Because GRIK1 and GRIK2 also interact with AL1, we asked if their expression is developmentally regulated, suggestive of intrinsic functions associated with DNA synthesis or differentiation. Leaves from Arabidopsis rosettes provided a good source of tissues at different ages. We divided the leaves of 5-week-old plants grown under a short-day light regime into four fractions: young leaves less than 5 mm in length and the shoot apical meristem (SAM), expanding leaves, fully expanded leaves, and senescent leaves. Only the young leaf sample and, to a lesser extent, the expanding leaf sample contain cells competent for DNA replication (for review, see Traas et al., 1998). Immunoblot analysis of total proteins extracted from these fractions showed that both GRIK1 and GRIK2 are highly expressed in the young leaf fraction (Fig. 6A, lanes 1 and 5). A lesser amount of GRIK2 was detected in expanding leaves (Fig. 5A, lane 6), and a band corresponding to GRIK1 was visible in expanding leaves upon long exposure of the blot (data not shown). Neither GRIK1 nor GRIK2 was detected in mature (Fig. 6A, lanes 3 and 7) or senescent leaves (lanes 4 and 8). These results indicate that both kinases are expressed in young tissues where cells are replicating DNA. Detection of GRIK1 and GRIK2 in expanding leaves, albeit at much reduced levels, excluded the possibility that the kinases accumulate only in the SAM.
Figure 6.
GRIK1 and GRIK2 protein and mRNA accumulate differentially during plant development. A, Leaves from 5-week-old plants were collected in fractions: young leaves (<0.5 cm) with SAM (lanes 1 and 5), expanding leaves (0.5–1.5 cm, lanes 2 and 6), fully expanded leaves (lanes 3 and 7), and senescent leaves (lanes 4 and 8). Total protein extracts were analyzed for GRIK1 (lanes 1–4) and GRIK2 (lanes 5–8) by immunoblotting with their respective peptide antibodies. The dot indicates the GRIK1 or GRIK2 band. Molecular mass markers (in kilodaltons) are indicated. B, Real-time quantitative RT-PCR of GRIK1 and GRIK2 mRNAs in the same leaf tissues. Relative mRNA levels are normalized to the levels in young leaves using Act2 mRNA as reference. The averages and sds of triplicate reactions are shown. This experiment was repeated once with similar results. C, Total proteins from flower buds (lanes 1 and 5), fully opened flowers (lanes 2 and 6), siliques (lanes 3 and 7), and roots (lanes 4 and 8) were analyzed by immunoblotting using antibodies to GRIK1 (lanes 1–4) or GRIK2 (lanes 5–8).
The relative amounts of GRIK1 and GRIK2 in young leaves were compared using different amounts of a total protein extract and their peptide antibodies in Figure 3B. Bands of similar intensities were seen for the two kinases when 25 μg of total plant protein was probed with the GRIK1 antibody and 10 μg of total protein was probed with the GRIK2 antibody (Fig. 3B, compare lanes 5 and 6). Given that the GRIK2 antibody displays approximately 40-fold greater sensitivity than the GRIK1 antibody under these experimental conditions (Fig. 3B, lanes 3 and 4), the amount of GRIK1 protein in young leaves is approximately 10-fold higher than the GRIK2 protein.
The steady-state mRNA levels corresponding to the kinase genes were measured by real-time quantitative RT-PCR in the four leaf samples in two independent experiments. For both GRIK1 and GRIK2, there was no statistically significant difference in relative mRNA abundance between any of the samples (Fig. 6B). These data are consistent with available microarray data showing that GRIK1 and GRIK2 mRNAs levels do not vary in a range of tissue types and developmental stages (http://www.arabidopsis.org) and suggest that their protein levels are posttranscriptionally regulated in developing, mature, and senescent leaves.
GRIK1 and GRIK2 expression in other Arabidopsis organs was examined by immunoblotting with the peptide antibodies. Both kinases were detected in flower buds (Fig. 6C, lanes 1 and 5), fully opened flowers (lanes 2 and 6), and siliques (lanes 3 and 7), but neither was present in roots (lanes 4 and 8). The GRIK2 protein level was higher in flower buds and siliques than in mature flowers (Fig. 6C, lanes 5 and 7 with lane 6), further supporting the idea that the GRIK proteins are associated with young plant tissues that contain dividing and endoreduplicating cells.
GRIK1 and GRIK2 Display Different Immunolocalization Patterns
We compared the cellular and subcellular distributions of GRIK1 and GRIK2 in 5-week-old Arabidopsis plants using the peptide antibodies for immunolocalization. These studies focused on the SAM, leaf primordia, and emerging petioles of leaves immediately flanking the primordium, because the kinases are the predominant immunoreactive proteins in these tissues (Fig. 6A, lanes 1 and 5). In addition, these tissues contain few or no plastids, which were also a source of potential background. Both peptide antibodies stained chloroplasts in mature leaves (not shown) even though neither kinase protein was detected in this tissue by immunoblotting (Fig. 6A, lanes 3 and 7).
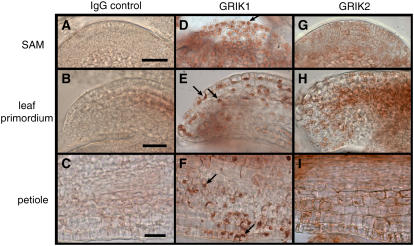
The GRIK1 antibody stained a variety of cells in the SAM (Fig. 7D), leaf primordium (Fig. 7E), and petiole (Fig. 7F). The stained cells included cells in the SAM central zone, epidermal and emerging mesophyll cells in the leaf primordium, and epidermal cells in the petiole. The strongest GRIK1 staining colocalized with 4′,6-diamidino-2-phenylindole staining (data not shown), indicating that the kinase is in the nucleus. Cytoplasmic GRIK1 staining was also visible in the SAM (Fig. 7D) but was not readily apparent in the leaf primordium or petiole sections when compared to the corresponding normal rabbit antibody controls (compare Fig. 7, B and E, with C and F). These data are consistent with earlier results using a GRIK polyclonal antibody showing that the kinase is in nuclei of developing leaf cells of Nicotiana benthamiana plants (Kong and Hanley-Bowdoin, 2002). Cells positive for GRIK1 could be found adjacent to unstained cells in the petiole sections (Fig. 7F). The spotty pattern may reflect differential GRIK1 accumulation in adjacent cells, loss of nuclei resulting from sectioning, or constraints on antibody access as cells become larger and less likely to lie in the sectioning plane.
Figure 7.
Immunolocalization of GRIK1 and GRIK2 in the SAM and young leaves. Sections from 5-week-old Arabidopsis SAM (A, D, and G), leaf primordium (B, E, and H), and emerging petiole (C, F, and I) were incubated with the GRIK1 (D, E, and F) or GRIK2 (G, H, and I) antibodies followed by visualization using a secondary antibody HRP-conjugate and the AEC substrate. Normal rabbit IgG (A–C) was used as a primary antibody control. Arrows indicate some of the stained nuclei. The bars indicate 100 nm.
In contrast, GRIK2 antibody staining was weak in all three tissues (Fig. 7, G–I). This was unexpected because of the greater sensitivity of the antibody on immunoblots. The weak staining could not be attributed to the 20-kD cross-reacting band detected on immunoblots because the same band was observed in young and mature leaf extracts (Fig. 6A, compare lanes 5 and 7), but mature leaves did not show staining in parallel assays (data not shown). Instead, the weak staining may reflect the distribution of GRIK2 throughout the cell, making it more difficult to detect. Consistent with this idea, there appears to be more general GRIK2 staining in all three tissues compared to the normal rabbit antibody controls (compare Fig. 7, A and G; B and H; and C and I). However, a few stained nuclei can be seen in the leaf primordium and petiole sections (Fig. 7, H and I), indicating that GRIK2 is not excluded from the nucleus. Another possibility is that the GRIK2 C terminus is masked and unavailable for antibody binding in the native protein in the fixed tissue sections. In either case, the distinct staining patterns for GRIK1 and GRIK2 are indicative of differences in their local concentrations and/or differential localization.
GRIK1 and GRIK2 Are Related to the Yeast and Animal SNF1/AMPK-Activating Kinases
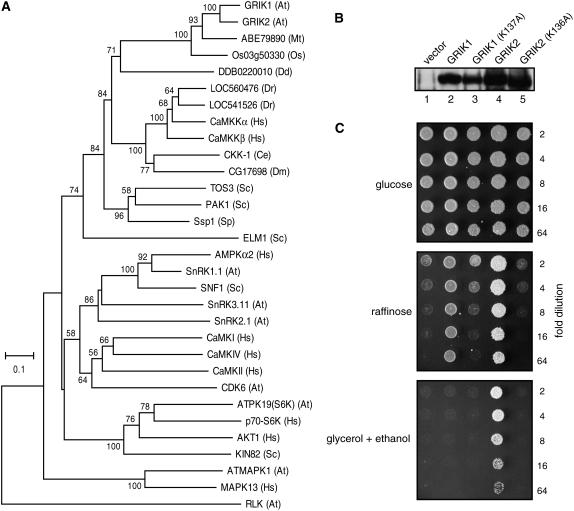
GRIK1 and GRIK2 form a small protein kinase family in Arabidopsis and are distinct from their next most similar protein kinase, AtKIN10, a SnRK1 family protein kinase. BLAST searches of the Saccharomyces cerevisiae and human proteomes revealed that GRIK1 and GRIK2 are most similar to a family of protein kinases that activate the yeast SNF1 kinase or its mammalian homolog AMPK (for review, see Witters et al., 2006). When their catalytic domains are compared, the Arabidopsis kinases are 41% to 53% similar to yeast PAK1, TOS3, and ELM1 and display 52% to 55% similarity to human CaMKKα and CaMKKβ. We constructed a phylogenetic tree by aligning the catalytic domains of GRIK1 and GRIK2, their homologs in rice and M. truncatula, the yeast PAK1/TOS3/ELM1, the human CaMKKα/β, and homologs from other species with available draft genome sequences (Fig. 8A). The tree also includes AtKIN10, SNF1, and AMPK, as the most similar kinases to GRIK1/2, PAK1/TOS3/ELM1, and CaMKKα/β in their respective species. Arabidopsis GRIK1/2, yeast PAK1/TOS3/ELM1, human CaMKKα/β, and similar proteins from other species cluster in a single clade distinct from their most similar proteins within their own species (Fig. 8A), indicative of a common origin. Within the clade, the Arabidopsis, M. truncatula, and rice proteins are more closely related to each other than to the fungal and animal kinases, which in turn form discrete subclusters with the exception of yeast ELM1.
Figure 8.
GRIK1 and GRIK2 are related to activating kinases for the yeast SNF1 and mammalian AMPK. A, A phylogenetic tree containing GRIK1, GRIK2, their plant homologs, and related protein kinases from nonplant species. The protein kinase catalytic domains were aligned and used for building the tree with the neighbor-joining method. The numbers at branches indicate the percentage of bootstrap support (only those greater than 50% are shown). The two letter abbreviations in the brackets indicate the species sources of the kinases: At, Arabidopsis; Mt, M. truncatula; Os, rice; Dd, D. discoideum; Dr, zebrafish; Hs, human; Ce, C. elegans; Dm, D. melanogaster; Sc, S. cerevisiae; and Sp, S. pombe. An Arabidopsis RLK (At4g22730) was used as an outgroup control. B, Protein expression of GRIK1, GRIK2, and their mutant forms GRIK1(K137A) and GRIK2(K136A) in the S. cerevisiae pak1/tos3/elm1 mutant MCY5138. Total protein from yeast cells containing the expression plasmids or the empty vector were analyzed by immunoblotting with a mixture of GRIK1 and GRIK2 antibodies. C, Complementation of the S. cerevisiae pak1, tos3, and elm1 mutations by GRIK1, GRIK1(K137A), GRIK2, and GRIK2(K136A). Equal volumes of a 2-fold dilution series of the transformants containing the expression plasmids were spotted onto synthetic complete media containing either Glc, or raffinose, or glycerol and ethanol as carbon sources for further growth. Transformants carrying the empty expression vector was used as control.
In yeast, the PAK1/TOS3/ELM1 kinases phosphorylate and activate the downstream kinase SNF1, which facilitates utilization of non-Glc carbon sources (Hong et al., 2003). Human CaMKKα/β activate their substrate AMPK, which is homologous to yeast SNF1 (Hawley et al., 2005; Hong et al., 2005; Hurley et al., 2005). Both CaMKKα and CaMKKβ can complement a yeast triple mutant disrupted in the PAK1/TOS3/ELM1 genes, which regains the capacity to grow on medium containing non-Glc carbon sources in the presence of the human kinases (Hong et al., 2005). We asked if Arabidopsis GRIK1 or GRIK2 is also able to functionally replace the yeast PAK1/TOS3/ELM1 kinases. The triple mutant strain MCY5138 (Hong et al., 2003) was transformed with expression cassettes corresponding to GRIK1, GRIK2, or their mutant forms, GRIK1(K136A) and GRIK2(K137A) (Fig. 1), under the control of the yeast ADH1 gene promoter. Immunoblotting of protein extracts from transformants grown in Glc-containing medium indicated that GRIK1 (Fig. 8B, lane 2), GRIK1(K136A) (lane 3), GRIK2 (lane 4), and GRIK2(K137A) (lane 5) were expressed in the pak1/tos3/elm1 mutant. We then plated a dilution series for each transformant on synthetic complete media containing Glc, raffinose, or glycerol-ethanol as the carbon source. MCY5138 cells transformed with an empty expression cassette served as the negative control. All of the transformants showed similar growth on Glc medium (Fig. 8C, top). On raffinose medium, weak growth was seen only at lower dilutions for the transformants carrying the empty vector, the GRIK1(K136A) cassette, or the GRIK2(K137A), while both GRIK1 and GRIK2 transformants grew well at all the dilutions tested (Fig. 8C, middle). On glycerol-ethanol medium, growth was only detected for the GRIK2 transformant, which grew at all dilutions tested (Fig. 8C, bottom). These data demonstrated that GRIK1 or GRIK2 can complement the pak1/tos3/elm1 mutations in yeast and that complementation depends on their kinase activities. They also suggest that Arabidopsis GRIK1 and GRIK2 mediate similar biochemical functions to yeast PAK1/TOS3/ELM1 and mammalian CaMKKα/β. However, GRIK2 complementation was stronger than GRIK1, indicating that the Arabidopsis kinases may not be functionally equivalent.
DISCUSSION
Geminiviruses have coevolved with their plant hosts to use endogenous cellular processes to replicate, express, and transport their DNA genomes and to overcome resistance mechanisms (Rojas et al., 2005). This is achieved in part by modulating existing signal transduction pathways through the interaction of viral proteins and host protein kinases. Geminivirus proteins bind to RLKs that are part of the plant pathogen surveillance system and interfere with the ability of the host to mount a resistance response (Fontes et al., 2004). Susceptibility to geminivirus infection is also enhanced by interaction with an SnRK1, implicating this evolutionarily conserved, energy-sensing protein kinase in the plant defense response (Hao et al., 2003). In this article, we provide another connection between SnRK1 and geminivirus infection by showing that the AL1 partners GRIK1 and GRIK2 can act as SNF1-activating kinases in yeast. Data showing that the GRIK1 and GRIK2 proteins accumulate preferentially during infection and in young Arabidopsis tissues suggest that the SnRK1 cascade regulates a diverse range of functions in higher plants.
The Arabidopsis genome encodes two related protein kinases, GRIK1 and GRIK2, which are the sole members of the kinase family 4.2.7 as defined by the PlantsP Kinase Classification Scheme (http://plantsp.genomics.purdue.edu). Related kinases are encoded by the rice and M. truncatula genomes (Fig. 1), demonstrating that the 4.2.7 family is conserved in both monocot and dicot plant species. The GRIK1 and GRIK2 coding sequences display 82% nucleotide identity, and their genes are arranged similarly with respect to exon-intron location and exon size, characteristic of a gene duplication event. GRIK1 and GRIK2 also have many attributes in common, including their expression patterns, posttranslational regulation, interaction with a geminivirus replication protein, and complementation of the yeast pak1/tos3/elm1 triple mutant. These similarities suggest that GRIK1 and GRIK2 are functionally redundant, but two observations argue against this conclusion. Immunolocalization studies suggested that they are in different cellular compartments or associated with different protein complexes (Fig. 7). In addition, GRIK2 displayed a stronger complementation phenotype in the yeast triple mutant (Fig. 8C). This was most striking when yeast was grown in the presence of glycerol and ethanol, with only GRIK2 showing complementation.
GRIK1 and GRIK2 accumulation is associated with young leaf tissues (Fig. 6A) and induced in older leaves by geminivirus infection (Fig. 4, A and B). Immunolocalization studies showed that the kinases are abundant in all cell types of the SAM, leaf primordium, and emerging petiole. Cells at these early developmental stages and geminivirus-infected cells share the capacity to replicate DNA either as part of a mitotic cell cycle or an endocycle (Traas et al., 1998; Hanley-Bowdoin et al., 2004). GRIK2 is also highly abundant in developing flower buds (Fig. 6C), which contain replication competent cells. The uniform GRIK1 and GRIK2 mRNA levels during leaf development (Fig. 6B) indicate that accumulation of the kinases is modulated by posttranscriptional mechanisms. The proteasome inhibitor MG132 stabilized both kinases in Arabidopsis seedlings and CaLCuV-infected leaves (Fig. 5). Hence, GRIK1 and GRIK2 accumulation is regulated, at least in part, by protein degradation via the proteasome pathway, which controls the levels of many proteins associated with the cell cycle and DNA replication (del Pozo et al., 2002; Planchais et al., 2004; Yamamoto et al., 2004). However, the uniform GRIK1 expression pattern in the SAM (Fig. 7D) is not consistent with modulation of protein levels in a cyclic manner. Instead, the declining proportion of GRIK1 positive cells in the emerging petiole suggests that its turnover is activated during differentiation. The absence of detectable GRIK1 and GRIK2 in mature tissues is consistent with efficient degradation in differentiated cells.
Several lines of evidence indicated that GRIK1 and GRIK2 expression is regulated by multiple mechanisms during geminivirus infection. GRIK1 and GRIK2 mRNAs are elevated approximately 2-fold in infected leaves (Fig. 4C), but this change is not sufficient to account for the much larger rise in protein levels (Fig. 4A), further indicating that posttranscriptional mechanisms contribute to the accumulation of the kinases. Consistent with this idea, MG132 treatment of CaLCuV-infected leaves established that GRIK1 and GRIK2 are degraded via the proteasome pathway and uncovered a large increase in de novo protein synthesis during infection (Fig. 5, B and C). The net accumulation of GRIK1 and GRIK2 most likely reflects a balance between these two processes. Interestingly, the 5′-untranslated regions of both GRIK1 and GRIK2 mRNAs contain multiple upstream small open reading frames (data not shown), which have been associated with translational regulation of other mRNAs (Morris and Geballe, 2000; Wiese et al., 2004; Hanfrey et al., 2005). AL1 binding to GRIK1 and GRIK2 may protect them from degradation. It is also possible that viral infection negatively impacts the expression of a specific E3 ubiquitin ligase that targets GRIK1 and GRIK2 for turnover.
Phylogenetic analysis showed that GRIK1 and GRIK2 are related to the S. cerevisiae kinases PAK1, TOS3, and ELM1 and to human CaMKKs. This analysis also uncovered related kinases in Schizosaccharomyces pombe, Dictyostelium discoideum, Caenorhabditis elegans, zebrafish (Danio rerio), and Drosophila melanogaster, indicating that GRIK1 and GRIK2 belong to a kinase family that is conserved across eukaryotes. This family branch in the phylogenetic tree is supported by a bootstrap value of 74%, which increases to 84% if the more diverse ELM1 is not considered (Fig. 8A). These data agree with an earlier study comparing protein kinases from Arabidopsis and budding yeast that did not conclusively identify GRIK1 and GRIK2 as homologs of PAK1/TOS3/ELM1 (Wang et al., 2003a). However, several observations support the idea that these Arabidopsis and yeast kinases are related. First, a bootstrap value of 84% is also seen for the branch containing only plant GRIKs and mammalian CaMKKs, indicating that the plant and animal kinases show similar divergence from the yeast kinases. Second, the functional relationship between the CaMKKs and the yeast kinases has been established in complementation assays using a yeast pak1/tos3/elm1 triple mutant (Hong et al., 2005). We showed that GRIK1 and GRIK2 expression also restores the ability of the same yeast mutant to use non-Glc carbon sources (Fig. 8C). Last, the most similar kinases to PAK1/TOS3/ELM1, CaMKKs and GRIKs, e.g. the yeast kinase SNF1, the animal AMPK, and the plant SnRK1, constitute a homologous family of protein kinases involved in sugar metabolism (Wang et al., 2003a; Halford et al., 2004). This conservation is underscored by the ability of CaMKK to phosphorylate the Arabidopsis SnRK1 AKIN10 in vitro (Sugden et al., 1999). These data strongly suggest that like the kinases PAK1/TOS3/ELM1 and CaMKKs, the substrates of GRIK1 and GRIK2 are members of the SNF1 family.
The Arabidopsis SnRK family has 38 members constituting three subfamilies (Hrabak et al., 2003). The SnRK1 subfamily contains three members that are orthologs of the yeast SNF1 and mammalian AMPKs. This subfamily has been shown to play roles in regulating and coordinating plant carbon and nitrogen metabolism (for review, see Halford et al., 2003, 2004; Francis and Halford, 2006). Two additional subfamilies, SnRK2 and SnRK3, have 10 and 25 members, respectively. As a result, plant SnRKs are more diverse and thought to play roles in a variety of cellular processes in addition to nutrient and energy metabolism (Hrabak et al., 2003; Halford et al., 2004). Because of the size and diversity of the Arabidopsis SnRK family, it is likely that their activities are regulated through multiple mechanisms (Chikano et al., 2001). In animals, AMPK can be activated by multiple upstream protein kinases, including the CaMKKs (Hawley et al., 2005; Hurley et al., 2005; Woods et al., 2005) and the AMP-stimulated LKB1 protein kinase (Woods et al., 2003). It is possible that a yet-to-be-discovered SnRK-activating kinase regulates nutrient metabolism in mature plant tissues, while GRIK1 and GRIK2 function as upstream activators of one or more SnRKs to coordinate growth and energy supplies in young tissues (Thelander et al., 2004) and sugar control of cell cycle progression (Riou-Khamlichi et al., 2000). This idea is supported by the specific expression of GRIK1 and GRIK2 in young tissues and provides a mechanism for plants to address the unique energy requirements and high biosynthetic activity of sink tissues. Alternatively, GRIK1 and GRIK2 might activate an SnRK that regulates a process specifically associated with early development and unrelated to energy balance.
Geminiviruses modify differentiation and cell cycle controls in mature leaf cells to induce replication of host chromosomal DNA as well as their own genomes (Nagar et al., 2002). They also dramatically change the transcriptome (J. T. Ascencio-Ibañez and L. Hanley-Bowdoin, unpublished data) and presumably the proteome of infected tissues. Synthesis of DNA, RNA, and protein depends on adequate precursor pools and high ATP levels, and it is likely that geminiviruses use existing host mechanisms to control nutrient availability and energy balance. Thus, the accumulation of GRIK1 and GRIK2 may reflect the recruitment of an SnRK cascade that normally is active only in young tissues to support the high metabolic requirements of infected cells. The cell autonomous nature of GRIK accumulation is consistent with activation of the cascade only in virus-positive cells and not in adjacent uninfected cells (Kong and Hanley-Bowdoin, 2002), thereby providing a mechanism to selectively ensure that potentially limiting compounds and energy stores are available for viral processes.
GRIK1 and GRIK2 might also contribute to the host defense response by up-regulating the SnRK1 pathway and conferring reduced susceptibility to geminivirus infection (Hao et al., 2003). The ability of the SnRK1 pathway to interfere with infection is inhibited by the CaLCuV protein AL2, which interacts with and inactivates SnRK1 (Hao et al., 2003). AL2 also binds to and inhibits ADK, which catalyzes the synthesis of AMP from adenosine and may control SnRK1 activation mediated by AMP-dependent inhibition on SnRK1 dephosphorylation in plants (Sugden et al., 1999; Wang et al., 2003b). AL2-mediated inhibition of ADK would further down-regulate SnRK1 activity in infected cells. Up-regulation of GRIK1 and GRIK2 in infected cells would provide a mechanism to combat the negative effect of AL2 on SnRK1 activity and raise the barrier for establishment of a successful infection.
The potential involvement of GRIK1 and GRIK2 in the defense response seems counter to the hypothesis that the kinases facilitate infection by activating SnRK-regulated nutrient metabolism. However, these possibilities may not be mutually exclusive in a dynamic, ordered process in which AL1 and AL3 expression and GRIK accumulation precedes AL2 expression and inhibition of a SnRK cascade. Early in the infection process, the viral and host replication components are put into place, with GRIK1 and GRIK2 activation of the SnRK cascade ensuring that the requisite precursors and energy sources are available for virus propagation. Later, production of the AL2 protein counters the capacity of activated SnRK1 to also interfere with the infection process. This fine temporal control could have evolved during the long evolutionary relationship between geminiviruses and their hosts as a way to ensure viral propagation and plant survival (Rojas et al., 2005). Alternatively, different SnRK cascades may be involved in precursor and energy regulation and in the host defense response. In this case, GRIK1 and GRIK2 would control the metabolic pathways, whereas a different upstream activator would induce SnRK1 in the defense pathway. It is also possible that GRIK1 and GRIK2 act solely in the SnRK1 defense cascade and that geminiviruses have developed two mechanisms to inhibit this pathway through AL2 binding to SnRK1 and AL1 binding to GRIK1 and GRIK2. Future experiments that identify and characterize the SnRK substrates of GRIK1 and GRIK2 during development and geminivirus infection and determine the impact of AL1 interaction on their activities will distinguish between these possibilities.
MATERIALS AND METHODS
Plant Growth and Treatments
Arabidopsis (Arabidopsis thaliana) Columbia-0 plants were grown at 20°C in a Percival reach-in chamber with 8/16-h light/dark cycles under a light intensity of 15,000 Lux. Leaves from 5-week-old rosette plants or flower buds, fully open flowers, and siliques from bolted plants grown in soil were collected for protein and RNA extractions. Young seedlings were grown on petri dishes containing 1× Murashige and Skoog salts, 1× Gamborg's B5 vitamins, 1% Suc, and 0.7% agar for 2 weeks. The seedling roots were collected for protein extraction, or whole seedlings were removed and subjected to MG132 treatment before extraction.
Five-week-old, soil-grown plants were infected in the apex by syringe inoculation of Agrobacterium tumefaciens carrying pNSB1090 and pNSB1091, which contain partial tandem copies of CaLCuV A and B DNA, respectively (Egelkrout et al., 2002). Alternatively, they were inoculated with A. tumefaciens containing a plasmid (D.M. Bisaro, unpublished data) with a partial tandem copy of the single-component BCTV Logan genome (Stenger et al., 1991). A. tumefaciens strains carrying corresponding empty cloning vectors were used for the mock-inoculation controls. Leaves (0.5–1.5 cm long) were collected for protein and RNA extraction or for treatment with MG132 or cycloheximide prior to protein extraction from CaLCuV-infected plants at 12 dpi and from BCTV-infected plants at 28 dpi.
For treatment with MG132, plant materials were submerged for 3 h in 1× Murashige and Skoog salts solution containing 50 μm MG132 previously dissolved in dimethyl sulfoxide. The equivalent volume of dimethyl sulfoxide in 1× Murashige and Skoog salts was used as the control treatment. Plant tissues were treated similarly with 100 μm cycloheximide except that ethanol was used as the solvent.
cDNA Cloning and Recombinant Baculoviruses
To obtain the full-length cDNA for GRIK1, Arabidopsis leaf RNAs were reverse transcribed using the primer 5′-TATGAGCATGAAGGTACATGAG-3′. The cDNA was amplified using the same primer and the second primer, 5′-GGCTCGAGGATCCGATGTTTTGTGATAGT-3′. The DNA fragment corresponding to the 5′-end of the GRIK1 coding sequence was digested with BamHI and DraIII and used to replace the BamHI-DraIII fragment of pNSB1016, a yeast two-hybrid prey plasmid that specifies GRIK1 residues 12 to 396 (Kong and Hanley-Bowdoin, 2002). The resulting plasmid, pNSB1185, contains the full-length GRIK1 coding region.
The cDNA for full-length GRIK2 was cloned by RT-PCR using the primer 5′-GGAGCTCGAATTCTTAGTTAGGATCTGAGGTTTC-3′ for both reactions and the primer 5′-GGAACGCCATATGTTTCGTGATAGTTTTTTGTTTGC-3′ for PCR. The amplification product was digested with NdeI and EcoRI and subcloned into the same sites of pNSB910 (Castillo et al., 2004) to give pNSB1184, which contains a His-tag fused to the 5′-end of the GRIK2 coding region in the pMON27025 background (Luckow et al., 1993).
The insect transfer vector pNSB1184 was used to make a recombinant baculovirus corresponding to His-tagged GRIK2. The transfer vector for His-tagged GRIK1(12–396) was described previously (Kong and Hanley-Bowdoin, 2002). Recombinant baculovirus generation by Tn7-mediated transposition with the bacmid plasmid bMON14242, Spodoptera frugiperda Sf9 cell transfection, virus amplification, and protein production in monolayer cells were carried out according to Kong and Hanley-Bowdoin (2002).
Antibodies
Two peptides, CKRKAEEEEDQNHS and SKIEEGEANGISETS, located at the C termini of GRIK1 and GRIK2, respectively, were synthesized and conjugated to keyhole limpet hemocyanin for immunizing rabbits (Washington Biotechnology). Specific antibodies were affinity purified with the GRIK1 and GRIK2 peptides coupled to the SulfoLink Coupling Gel (Pierce) and Affi-Gel 102 (Bio-Rad) resins, respectively, according to the manufacturers' instructions. The antibodies were purified further by passage through keyhole limpet hemocyanin-agarose (Sigma). Antiserum against CaLCuV AL1 was prepared by immunizing rabbits with His-tagged recombinant protein produced in Escherichia coli (J.T. Ascencio-Ibañez and L. Hanley-Bowdoin, unpublished data). Monoclonal antibodies against the His tag were from CLONTECH.
Protein Extraction and Immunoblotting
Proteins used in this study were from total cell lysates. Plant tissues were collected and ground into fine powders in liquid nitrogen, suspended in 1 volume of 2× SDS-PAGE sample buffer, boiled for 15 min, and centrifuged at 16,000g for 10 min to remove cell debris. Attached insect cells expressing recombinant proteins were solubilized with 1× SDS-PAGE sample buffer, boiled, and centrifuged in the same way. Yeast (Saccharomyces cerevisiae) cells from a 5-mL overnight culture in liquid synthetic medium were collected by centrifugation and suspended in 100 μL of 2× SDS-PAGE sample buffer. The cell suspension was vortexed vigorously for 1 min in the presence of 20 μL glass beads, frozen in liquid nitrogen for 5 min, thawed at 37°C for 5 min, and cleared by centrifugation for 10 min. Total soluble protein in 0.5 μL was quantified using the Bio-Rad protein assay reagent.
Fifty micrograms of plant or yeast proteins or the indicated volumes of insect cell proteins were separated in 10% (w/v) gels by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% (w/v) nonfat milk in 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl and probed with primary antibody in Tris-buffered saline plus 0.1% Tween 20 (TBST) for 1 h at 22°C. Typically, the primary antibodies to GRIK1 and GRIK2 were used at 1 μg/mL and 0.5 μg/mL, respectively. The antiserum to AL1 was diluted 1:500, and the monoclonal His-tag antibody was diluted 1:2,000. After washing in TBST, the blots were probed with horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare) diluted 1:2,500 in TBST for 1 h, washed in TBST, and incubated with Pierce's SuperSignal West Pico chemiluminescent substrate. The signals were detected by exposing x-ray films with the membranes.
Immunohistochemical Staining
The central, aboveground part of 5-week-old, soil-grown Arabidopsis rosette seedlings was fixed overnight at 4°C in 4% (w/v) formaldehyde, 15 mm PIPES, pH 6.8, 80 mm KCl, 20 mm NaCl, 0.5 mm EDTA, 2 mm EGTA, 0.15 mm spermine, 0.5 mm spermidine, 1 mm dithiothreitol, and 0.01 mm sodium acetate. Vertical cross sections were made by embedding the plant material in 5% (w/v) agarose in phosphate-buffered saline (PBS; 20 mm sodium phosphate, pH 7.5, and 130 mm NaCl) and slicing into 50-μm-thick pieces using a Vibratome sectioning system (Technical Products International). Endogenous hydrogen peroxidases were quenched by incubating the sections in 3% (v/v) hydrogen peroxide in methanol for 30 min at 22°C. The sections were then treated with PBS, pH 7.5, containing 1.5% (v/v) normal horse serum and 3% (w/v) bovine serum albumin for 1 h. Primary antibodies at 1 μg/mL in the blocking solution containing 0.1% Tween 20 were incubated with the sections for 1 h. Normal rabbit IgG (Sigma) was used as a negative control. After washing in PBS with 0.1% Tween 20, the sections were incubated in biotinylated horse anti-rabbit IgG antibodies (Vector Laboratories) followed by the Vectastain Elite ABC kit (Vector Laboratories) containing avidin and biotinylated HRP as instructed. The AEC kit (Vector Laboratories) with the substrate 3-amino-9-ethyl carbazole was applied for 10 min, and the sections were then washed in PBS, counter-stained with 1 μg/mL 4′,6-diamidino-2-phenylindole for 10 min, and mounted onto a glass slide in 90% (v/v) glycerol in PBS. Immunostained sections were observed with a Nikon Eclipse E800 microscope.
RNA Extraction and Real-Time Quantitative RT-PCR
The NucleoSpin RNA Plant kit (CLONTECH) was used to extract RNAs from 100 mg of plant tissue powders. RNA (5 μg) was transcribed to cDNA using PowerScript reverse transcriptase (CLONTECH) and oligo(dT)15 as primer. The cDNAs were diluted to 100 μL with water and 2 μL was subjected to real-time quantitative PCR with gene-specific primers and DyNAmo SYBR Green qPCR reagents (Finnzymes) in Stratagene Mx3000P Real-Time PCR system. All the reactions were performed in triplicate, and mRNA corresponding to the Arabidopsis Act2 gene (At3g18780) was used as reference. Amplification efficiencies for the genes were estimated according to the slopes of their amplification curves (Ramakers et al., 2003). The GRIK1 primers are 5′-GTGATACACTTCAAGATACTTA-3′ and 5′-TATCAAGAGTCTTCTCAATACA-3′; the primers for GRIK2 are 5′-CGGGATATTGTTACTGGACT-3′ and 5′-ATGCTCCGACACATTCTTCA-3′; and those for Act2 are 5′-AGGTCGTTGCACCACCTGAA-3′ and 5′-AGGTCGTTGCACCACCTGAA-3′.
Yeast Two-Hybrid and Complementation Assays
The GRIK1 coding region was PCR amplified from pNSB1185 with the primers 5′-CCGGAATTCATGTTTTGTGATAGT-3′ and 5′-CGAAGCTCGAGTCAGCTATGGTTTTG-3′; and the coding region of GRIK2 was obtained from pNSB1184 with the primers 5′-CCGGAATTCATGTTTCGTGATAGT-3′ and 5′-CGAAGCTCGAGTTAGTTAGGATCTGA-3′. The DNA fragments were digested with EcoRI and XhoI and fused with the GAL4 DNA binding domain by ligation into the vector pGBKT7 (TRP1; CLONTECH) also digested with EcoRI and SalI. The resulting plasmids, pNSB1398 (GRIK1) and pNSB1399 (GRIK2), were used as baits in yeast strain AH109 (MATa, leu2, trp1, ura3, his3, gal4Δ, gal180Δ, LYS2:GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3:MEL1UAS-MEL1TATA-lacZ) cotransformed with the prey plasmid pNSB809 (LEU2) carrying a GAL4 activation domain fused to TGMV AL1 (Orozco et al., 2000). Liquid cultures of the transformants were grown at 30°C for 24 h in synthetic complete medium lacking Leu and Trp to similar densities and diluted 1:16 in water. The dilutions (4 μL) were spotted onto complete medium plates either minus Leu and Trp or His, Leu, and Trp, and the yeast was grown at 30°C for 4 d. The host HIS3 reporter gene was used for scoring protein-protein interaction according to growth of the transformants on medium lacking His, Leu, and Trp. The empty vectors were used as negative controls.
The same EcoRI-XhoI-digested DNA fragments were ligated into the yeast expression vector pWS93 (URA3) digested with EcoRI and SalI (Song and Carlson, 1998). The resulting plasmids, pNSB1404 and pNSB1406, specified hemagglutinin-tagged GRIK1 and GRIK2, respectively, under the control of the yeast ADH1 gene promoter. The mutant forms GRIK1(K137A) and GRIK2(K136A) were generated by Stratagene QuikChange site-directed mutagenesis system. The yeast expression cassette (pNSB1405) for GRIK1(K137A) was made using pNSB1404 as template and the primers 5′-AAGCATTATGCTATTgcGGCTTTTCACAAGTC-3′ and 5′-GACTTGTGAAAAGCCgcAATAGCATAATGCTT-3′. The yeast expression cassette (pNSB1407) for GRIK2(K136A) was constructed using pNSB1406 as template and the primers 5′-CAGTATTATGCTATCgcGGCATTTCACAAGCT-3′ and 5′-GACTTGTGAAATGCCgcGATAGCATAATACTG-3′ (the altered nucleotides are shown in lowercase type).
In the yeast triple mutant strain MCY5138 (MATα, ura3, trp1, ade2, his3, can1, leu2, pak1Δ∷kanMX4, tos3Δ∷kanMX4, elm1Δ∷ADE2), the genes for the kinases PAK1, TOS3, and ELM1 are disrupted (Hong et al., 2003). The mutant was transformed with the wild type and mutant GRIK1 and GRIK2 expression plasmids, and transformants were maintained on synthetic complete medium without uracil but containing 2% (w/v) Glc. Complementation was assayed by growing 4-μL aliquots of 2-fold dilutions of overnight liquid cultures on synthetic complete medium containing 2% (w/v) raffinose for 2 d, or 2% (v/v) glycerol and 3% (v/v) ethanol for 3 d, as compared to the growth on medium containing Glc for 2 d.
Phylogenetic Analysis
Proteins with similar amino acid sequences were retrieved from GenBank by performing protein-protein BLAST searches. Sequence alignment of GRIK1, GRIK2, and their plant homologs were done with ClustalX 1.83 and adjusted manually. The kinase domains of the proteins from Arabidopsis, Medicago truncatula, rice (Oryza sativa), Dictyostelium discoideum, zebrafish (Danio rerio), human, Caenorhabditis elegans, Drosophila melanogaster, S. cerevisiae, and Schizosaccharomyces pombe were also aligned and used to build a phylogenetic tree by the neighbor joining method with MEGA 3.1 (Kumar et al., 2004). Bootstrap support for the tree branches was performed by comparing 1,000 random replicate trees. The proteins included in the phylogenetic tree are (database accession numbers in parentheses): GRIK1 (AAL07158), GRIK2 (AAM20697), ABE79890, Os03g50330 (AAK18832), DDB0220010 (EAL67851), LOC560476 (XM_683879), LOC541526 (AAH91900), CaMKKα (AAH43487), CaMKKβ (AAK91829), CKK-1 (AAA19242), CG17698 (EAL24539), TOS3 (NP_011336), PAK1 (AAC03227), Ssp1 (NP_588360), ELM1 (NP_012876), AMPKα2 (AAH69680), SnRK1.1 (AAQ56829), SNF1 (AAB64904), SnRK3.11 (AAM20472), SnRK2.1 (ABD85157), CaMKI (AAI06756), CaMKIV (AAH16695), CaMKII (NP_751911), CDK6 (AAM98149), ATPK19 (AAK17162), p70-S6K (AAA36411), AKT1 (AAA55732), KIN82 (NP_010015), ATMAPK (AAN15381), MAPK13 (AAH00433), and AtRLK (AAR23717).
Acknowledgments
We thank Drs. David M. Bisaro (The Ohio State University, Columbus, OH), Luisa Lopez-Ochoa, Sharon B. Settlage, Dominique Robertson, and Ralph E. Dewey (North Carolina State University, Raleigh, NC) for their critical comments on this manuscript; Dr. Marian Carlson (Columbia University, New York) for providing the yeast mutant MCY5138 and the plasmid pWS93; David Bisaro for the BCTV strain; Jose T. Ascencio-Ibañez and Luisa Lopez-Ochoa for the protein samples from BCTV-infected Arabidopsis leaves; and Dr. Gerardas Dambrauskas for cloning the full-length GRIK1 and GRIK2 cDNAs and preparing the insect cell expressed GRIK1 and GRIK2 proteins.
This work was supported by the National Science Foundation (grant no. IBN–0235251).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Linda Hanley-Bowdoin (linda_hanley-bowdoin@ncsu.edu).
References
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagewadi B, Chen S, Lal SK, Choudhury NR, Mukherjee SK (2004) PCNA interacts with Indian mung bean yellow mosaic virus rep and downregulates Rep activity. J Virol 78: 11890–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344: 158–168 [DOI] [PubMed] [Google Scholar]
- Carvalho MF, Turgeon R, Lazarowitz SG (2006) The geminivirus nuclear shuttle protein NSP inhibits the activity of AtNSI, a vascular-expressed Arabidopsis acetyltransferase regulated with the sink-to-source transition. Plant Physiol 140: 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AG, Collinet D, Deret S, Kashoggi A, Bejarano ER (2003) Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312: 381–394 [DOI] [PubMed] [Google Scholar]
- Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER (2004) Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol 78: 2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H (2001) Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet 264: 674–681 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L (2002) Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Robertson D, Hanley-Bowdoin L (2001) Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13: 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer JS, Brand L, Sunter G, Gardiner WE, Bisaro DM, Rogers SG (1988) Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res 16: 7043–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP (2006) A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J Virol 80: 6648–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EP, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L (1994) Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J Biol Chem 269: 8459–8465 [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Halford NG (2006) Nutrient sensing in plant meristems. Plant Mol Biol 60: 981–993 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul MJ (2004) Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J Exp Bot 55: 35–42 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ (2005) A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem 280: 39229–39237 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Robertson D (2004) Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol Plant Pathol 5: 149–156 [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG (2005) Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2: 9–19 [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Momcilovic M, Carlson M (2005) Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J Biol Chem 280: 21804–21809 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280: 29060–29066 [DOI] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, et al (2000) A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Laufs J, Traut W, Heyraud F, Matzeit V, Rogers SG, Schell J, Gronenborn B (1995) In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci USA 92: 3879–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow VA, Lee SC, Barry GF, Olins PO (1993) Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol 67: 4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque A, Sanz-Burgos AP, Ramirez-Parra E, Castellano MM, Gutierrez C (2002) Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302: 83–94 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ER, Walker JC (2003) Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol 6: 339–342 [DOI] [PubMed] [Google Scholar]
- Nagar S, Hanley-Bowdoin L, Robertson D (2002) Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14: 2995−3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Hanley-Bowdoin L (1996) A DNA structure is required for geminivirus replication origin function. J Virol 70: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Kong LJ, Batts LA, Elledge S, Hanley-Bowdoin L (2000) The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J Biol Chem 275: 6114–6122 [DOI] [PubMed] [Google Scholar]
- Pant V, Gupta D, Choudhury NR, Malathi VG, Varma A, Mukherjee SK (2001) Molecular characterization of the Rep protein of the blackgram isolate of Indian mungbean yellow mosaic virus. J Gen Virol 82: 2559–2567 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Planchais S, Samland AK, Murray JA (2004) Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J 38: 616–625 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43: 361–394 [DOI] [PubMed] [Google Scholar]
- Romeis T (2001) Protein kinases in the plant defence response. Curr Opin Plant Biol 4: 407–414 [DOI] [PubMed] [Google Scholar]
- Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA (2005) A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17: 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Song W, Carlson M (1998) Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J 17: 5757–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger DC, Revington GN, Stevenson MC, Bisaro DM (1991) Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc Natl Acad Sci USA 88: 8029–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG (1999) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19: 433–439 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4: 948–958 [DOI] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM (1992) Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G, Hartitz MD, Hormuzdi SG, Brough CL, Bisaro DM (1990) Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179: 69–77 [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H (2004) Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J 23: 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J, Hulskamp M, Gendreau E, Hofte H (1998) Endoreduplication and development: rule without dividing? Curr Opin Plant Biol 1: 498–503 [DOI] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79: 7410–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harper JF, Gribskov M (2003. a) Systematic trans-genomic comparison of protein kinases between Arabidopsis and Saccharomyces cerevisiae. Plant Physiol 132: 2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao L, Shung CY, Sunter G, Bisaro DM (2003. b) Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witters LA, Kemp BE, Means AR (2006) Chutes and ladders: the search for protein kinases that act on AMPK. Trends Biochem Sci 31: 13–16 [DOI] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D (2005) Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab 2: 21–33 [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D (2003) LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13: 2004–2008 [DOI] [PubMed] [Google Scholar]
- Xing T, Ouellet T, Miki BL (2002) Towards genomic and proteomic studies of protein phosphorylation in plant-pathogen interactions. Trends Plant Sci 7: 224–230 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kimura S, Mori Y, Oka M, Ishibashi T, Yanagawa Y, Nara T, Nakagawa H, Hashimoto J, Sakaguchi K (2004) Degradation of proliferating cell nuclear antigen by 26S proteasome in rice (Oryza sativa L.). Planta 218: 640–664 [DOI] [PubMed] [Google Scholar]