Abstract
To determine whether latent Epstein-Barr virus (EBV) modifies DNA damage responses in B lymphocytes, cells were treated with agents either producing DNA cross-links and adducts or generating double-strand breaks. The cyclin-dependent kinase inhibitor p21WAF1 accumulated in mitogen-stimulated primary B cells following exposure to all genotoxins tested. In contrast, when proliferation was EBV driven, p21WAF1 failed to accumulate after treatment with the DNA adduct-producing agents. The tumor suppressor p53 was stabilized and phosphorylated after all treatments, irrespective of whether latent EBV was present. This suggests that regulatory pathways upstream of p53 are unaffected by latent EBV but downstream effectors are altered if DNA adducts or distortions are involved.
Epstein-Barr virus (EBV) is a gammaherpesvirus that produces an asymptomatic infection in the majority of the human population. However, EBV is also associated with a number of human tumors of B-cell origin, including Burkitt's lymphoma, some types of Hodgkin's lymphoma, and immunoblastic B lymphoma in the immunosuppressed. Although the precise contribution that EBV makes to the development of these diseases is not yet known, it has been suggested that interference with cell cycle checkpoints and responses to DNA damage by EBV may play an important role in B lymphomagenesis (reviewed in reference 11).
In vitro, EBV has the ability to infect and transform primary B cells into continuously proliferating lymphoblastoid cell lines (LCLs) that have a pattern of viral gene expression known as latency III. This is characterized by the expression of nine viral proteins: six EBV nuclear antigens (EBNA1, -2, -3A, -3B, -3C, and -LP) and three latent membrane proteins (LMP1, -2A, and -2B). Additional RNA species (the EBV-encoded RNAs and the BamH1A rightward transcripts) are also expressed during latency III (3).
Generally, oncogenic viruses are able to disrupt cell cycle checkpoints that are induced by genotoxic stress (reviewed in reference 11). Although the tumor suppressor p53 is one of the major targets of other oncogenic viruses, several studies indicate that EBV does not specifically target p53 in LCLs (1, 2, 12). Nevertheless, recent studies have demonstrated that although not directly targeting p53, EBV may still interfere with cell cycle checkpoints at both G1/S and G2/M (7, 12, 17). Also, it has been suggested that EBV might target upstream pathways involving Chk2 and may alter the stability of p53 (7, 14).
We previously showed that during cisplatin-induced genotoxic stress in normal B cells, EBV suppresses a checkpoint downstream of p53 by preventing the inactivation of Cdk2 by p21WAF1. This involves the modulation of p21WAF1 stability and can result in the cells undergoing apoptosis (12). However, when normal proliferating B cells respond to gamma irradiation (γ-IR), p21WAF1 protein accumulates and the cells undergo cell cycle arrest irrespective of whether EBV is present (4, 12). Here we explore further this selective facility of latent EBV gene expression to modulate DNA damage responses.
Genotoxic agents producing different types of damage induce different responses in EBV-immortalized LCLs.
Cisplatin is a bifunctional alkylating agent that forms intrastrand and interstrand DNA cross-links, mono-adducts, and DNA-protein adducts, all of which lead to distortions of the DNA double helix (5). In contrast, γ-IR largely disrupts phosphodiester bonds, leading to single-strand breaks and double-strand breaks (DSBs) in DNA (10). In order to test the hypothesis that EBV latency III gene expression modulates responses associated with DNA cross-linking and adduct formation, but not responses to agents that induce other types of damage such as DSBs, a selection of genotoxic agents was investigated.
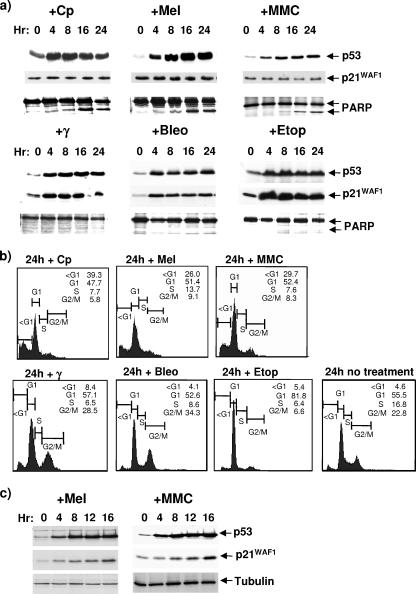
Three newly established LCLs (each produced by EBV infection of primary B cells from mixed donors and cultured for less than 3 months) were treated with mitomycin C and melphalan, two unrelated drugs that, like cisplatin, are bifunctional alkylating agents and generate DNA cross-links and adducts (6, 15). The same LCLs were also treated with the radiomimetic drug bleomycin or the topoisomerase II inhibitor etoposide, which, like γ-IR, can induce DSBs (but not cross-links) in DNA (10). The responses of cells were monitored by Western immunoblotting using antibodies directed against p53, p21WAF1, and poly(ADP-ribose) polymerase (PARP), using techniques described previously (12, 17). The cell cycle distribution was monitored by flow cytometry after staining with propidium iodide, also as described previously (12, 17). Figure 1a and b show the responses of a representative LCL (essentially similar responses were seen in the other two LCLs [data not shown]). Consistent with the hypothesis, melphalan and mitomycin C both produced a response very similar to that induced by cisplatin. That is, they induced the accumulation of p53, but this was not accompanied by an increase in p21WAF1 protein. The outcome was that after 24 h, 25 to 30% of the cells exhibited a sub-G1 cell cycle distribution and proteolytic cleavage of PARP, indicative of caspase activation and apoptosis. In contrast, treatment with γ-IR, bleomycin, or etoposide induced both p53 and p21WAF1, and the cells accumulated in both the G1 and G2 phases of the cell cycle (γ-IR and bleomycin) or only in G1 (etoposide), indicative of cell cycle arrest.
FIG. 1.
Genotoxic agents that produce different types of DNA damage induce different responses in LCLs. (a) Protein was extracted from untreated cells and cells exposed to a variety of genotoxins for the number of hours indicated. After resolution by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the levels of p53, p21WAF1, and PARP were analyzed by Western blotting. The presence of a lower-molecular-weight cleaved fragment of PARP is shown as an indication of caspase activity. (b) The distribution of ED-LCL cells throughout the cell cycle was analyzed by flow cytometry after exposure to the same panel of genotoxins. Samples were stained with propidium iodide to reveal the DNA content. Cells in G1 (with a 2N DNA content) were set at 200 on the FL2-A axis, and those in G2 (with 4N DNA) were set at 400. A DNA content of less than 2N (<G1) indicates cells that are probably in the late stages of apoptosis. (c) Normal peripheral B cells stimulated to proliferate with CD40L/IL-4 were treated with melphalan or mitomycin C and analyzed as for panel a. Gamma-tubulin was used as a loading control. Genotoxic agents used: Cp, cisplatin (Faulding Pharmaceuticals) (10 μg/ml); Mel, melphalan (Glaxo-Wellcome) (10 μg/ml); MMC, mitomycin C (Sigma) (10 μg/ml); γ, gamma irradiation (850rads); Bleo, bleomycin (Kyowa Hakko [United Kingdom] Ltd.) (351U/ml); Eto, etoposide (PCH Pharmachemie) (10 μg/ml). Primary antibodies used: anti-p53 MAb (DO1) and anti-p21WAF1 (SX118) (both gifts from Xin Lu, Ludwig Institute, London, United Kingdom), anti-PARP rabbit polyclonal antibody (Roche), and anti-γ-tubulin MAb (Sigma).
In order to demonstrate that melphalan and mitomycin C can increase p53 and p21WAF1 expression when EBV is absent, primary B cells induced to proliferate by the T-cell-derived mitogens CD40 ligand and interleukin-4 (IL-4) as described previously (12, 13) were treated and analyzed as described above. Figure 1c shows that both drugs induced an accumulation of both p53 and p21WAF1 in EBV-negative proliferating normal B cells.
These results are all consistent with EBV differentially regulating the responses of B cells to DNA damage. They confirm that in normal B cells treated with agents producing DNA strand cross-links, adducts, and distortions, EBV suppresses the accumulation of p21WAF1, but it has no obvious effect in cells responding to DNA strand breaks.
Latent EBV has no effect on the stabilization or phosphorylation of p53 at major serine residues after genotoxic stress.
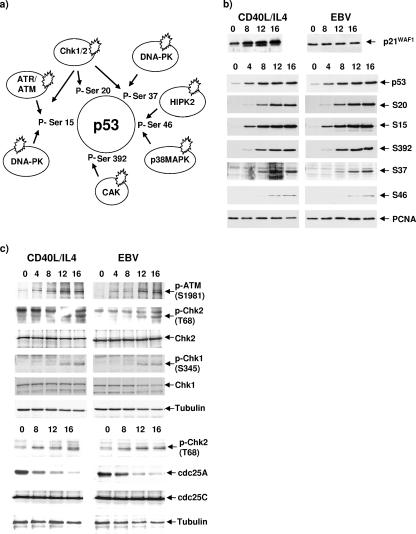
In response to several forms of stress, p53 is phosphorylated at multiple serine residues by a number of kinases (Fig. 2a). Phosphorylation at serines 15, 20, and 37 promotes the stabilization and activation of p53; serine 46 is thought to be involved in the regulation of p53-mediated apoptosis, and modification of serine 392 influences the DNA-binding capacity of p53 (8, 9, 16). It has been proposed that EBNA1 may lower p53 levels during stress, because it can bind to the cellular factor USP7/HAUSP and potentially disrupt its interaction with p53 (14). Furthermore, it has been suggested that EBNA3A might modulate the activity of the checkpoint kinase Chk2, since these proteins can be coimmunoprecipitated (7). We therefore explored events upstream of p53 accumulation.
FIG. 2.
Latent EBV does not alter modifications of p53 or upstream signaling in response to the DNA cross-linking agent cisplatin. (a) Schematic showing the major phosphoserines on p53 and the kinases responsible for their phosphorylation during DNA damage stress responses. (b) EBV-driven (LCL) or CD40L/IL-4-stimulated B cells were treated with 10 μg/ml cisplatin and harvested at 4-hour intervals. Protein extracts were analyzed by Western blotting with antibodies that recognize p21WAF1 (SX118), p53 (DO1), or individual phosphorylated serines (S) on p53. The phospho-specific antibodies against p53 (serines 15, 20, 37, 46, and 392) were all supplied by Cell Signaling Technology. PCNA was used as a loading control (using MAb PC10, a gift from Xin Lu, Ludwig Institute, London, United Kingdom). (c) The ATM/ATR-Chk1/2 pathways are activated in B cells exposed to cisplatin. Following treatment with 10 μg/ml cisplatin, EBV-driven or CD40L/IL-4-driven B cells were harvested at the indicated intervals, and protein was extracted and analyzed by Western blotting. Gamma-tubulin was included as a loading control. S, serine; T, threonine. The phospho-specific antibodies against Chk1 (S345), Chk2 (T68), and ATM (s1981) were all from Cell Signaling Technology. MAbs against cdc25A (F-6) and Chk1 (G-4) and polyclonal antibodies against Chk2 (H-300) and cdc25C (C-20) were from Santa Cruz.
The kinetics of accumulation of total p53 and its main phosphoderivatives were compared in an LCL and in CD40L/IL-4-driven primary B cells driven to proliferate by CD40 ligand and IL-4 following treatment with cisplatin. As predicted, there was an accumulation of p21WAF1 in the mitogen-driven cells but not the LCL cells. However, Western blot analysis of p53 with a pan-specific anti-p53 monoclonal antibody (MAb) and phospho-specific antibodies against various phosphoderivatives revealed no significant differences in the timing of their appearance when CD40/IL-4-driven cells were compared with EBV-driven LCL cells (Fig. 2b). These data strongly suggest that EBV latency III gene expression has little or no effect on the DNA damage response and signaling pathways upstream of p53. To reinforce this, similar Western blots were probed with antibodies specific for components of upstream pathways (Fig. 2c). Levels of activated, phosphorylated ATM, Chk1, and Chk2 kinases increased with similar kinetics in both B-cell populations after exposure to cisplatin. Consistent with these data, the level of cdc25A, which is phosphorylated and thus marked for degradation by active Chk2, decreased in both EBV-positive and -negative populations. We conclude that major DNA damage response pathways functioning upstream of p53 and triggered by cisplatin are not significantly affected by EBV latent gene products.
Similar patterns of p53 phosphorylation are produced by two classes of DNA-damaging agents, and neither is altered by EBV.
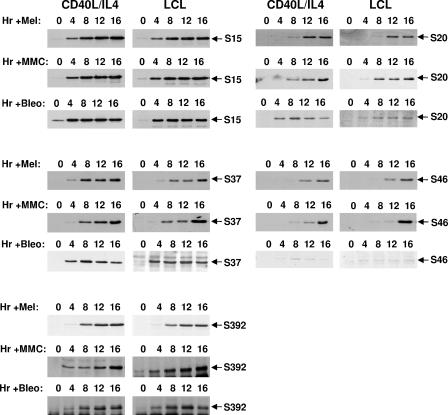
The responses of p53 in EBV-positive and EBV-negative normal B cells to the cross-linking agents melphalan and mitomycin C were compared with those in similar cells responding to the DSB-inducing agent bleomycin (Fig. 3). Stabilization and phosphorylation of p53 were apparently unaffected by EBV gene expression. The only residue that was not consistently phosphorylated under all circumstances was serine 46. This was not detectably phosphorylated in either LCLs or mitogen-stimulated B cells treated with bleomycin for 16 h. This serine was, however, phosphorylated in cells exposed to melphalan and mitomycin C for a similar period. Since phosphorylation of serine 46 is specifically associated with transactivation of p53AIP1 and the induction of apoptosis (9), this result is consistent with the bleomycin-treated cells arresting rather than dying (Fig. 1 and data not shown).
FIG. 3.
Phosphorylation on p53 is largely unaltered by the type of DNA damage and unaffected by EBV gene expression. LCL or CD40L/IL-4-stimulated primary B cells were exposed to either melphalan (Mel), mitomycin C (MMC), or bleomycin (Bleo) as described in the legend to Fig. 2. Protein extracts were Western blotted and probed with antibodies specific to individual phosphoserines on p53.
p53 target genes are activated to similar extents in EBV-positive and EBV-negative B cells responding to cisplatin.
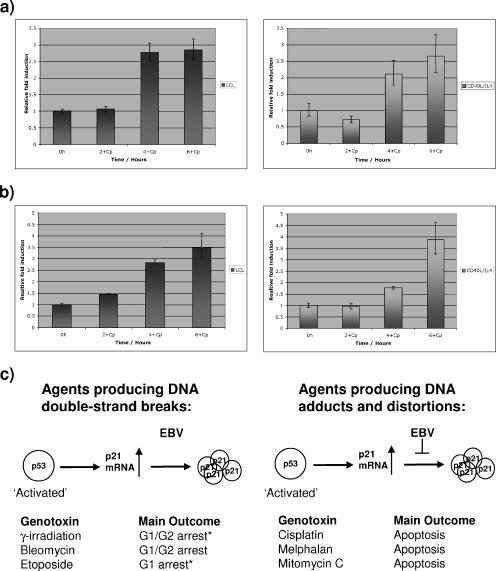
Finally, to show that p53 transactivates target genes as efficiently in the presence of EBV as in its absence, the levels of p21WAF1 and GADD45B mRNAs extracted from B cells treated with cisplatin were determined by quantitative real-time PCR. The results (Fig. 4a and b) indicate that EBV has no significant effect on p53-mediated transactivation of the p21WAF1 or GADD45B gene. While these data do not rule out the possibility that up-regulation might involve factors other than p53 (e.g., p73), they confirm that the EBV-mediated deregulation of p21WAF1 is posttranscriptional and are consistent with previous data showing that deregulation is at the level of p21WAF1 protein turnover (12). Taken together with our previous demonstration that latent EBV does not prevent the induction of hMDM2 after p53 activation (2, 12), these results indicate that EBV does not impair the ability of activated p53 to enhance transcription of target genes or nonspecifically block the accumulation of p53-induced proteins.
FIG. 4.
Quantitative real-time PCR analysis of mRNA corresponding to (a) p21WAF1 and (b) GADD45B extracted from LCL or CD40L/IL-4-stimulated B cells at the times indicated after exposure to cisplatin. (Error bars indicate standard deviations.) There are no significant differences between the increases in transcription of these p53-responsive genes with or without latent EBV. (c) Summary of responses described in this study. Genotoxins that produce DSBs induce p21WAF1 accumulation in B cells, regardless of latent EBV; in contrast, genotoxins that induce cross-links and DNA distortions induce p21WAF1 only in the absence of EBV. Generally, when there was an increase in p21WAF1, the cells underwent cell cycle arrest, but sometimes (*) there was also some apoptosis detected.
In summary, EBV, by suppressing the accumulation of the cyclin-dependent kinase inhibitor p21WAF1, modulates a checkpoint induced by agents that cause DNA cross-links and the formation of adducts. However, EBV appears to have no effect if other forms of DNA damage are involved (summarized in Fig. 4c). A discussion of which EBV latency gene products might be involved in these phenotypes has been presented elsewhere (11). Irrespective of the nature of the insult, the phosphorylation, stabilization, and activation of p53 appear to be unaffected by EBV in LCLs. We can only assume that the data presented here differ from the report suggesting that EBNA1 might affect p53 turnover (14) because we have examined responses in B cells expressing EBNA1 from latent EBV. In contrast, the results of Saridakis and colleagues (14) were obtained when EBNA1 was (over)expressed from plasmids in carcinoma or osteosarcoma cells and therefore removed from the context of EBV latency.
Acknowledgments
We thank the Wellcome Trust for their financial support of this research.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Allday, M. J., G. J. Inman, D. H. Crawford, and P. J. Farrell. 1995. DNA damage in human B cells can induce apoptosis, proceeding from G1/S when p53 is transactivation competent and G2/M when it is transactivation defective. EMBO J. 14:4994-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allday, M. J., A. Sinclair, G. Parker, D. H. Crawford, and P. J. Farrell. 1995. Epstein-Barr virus efficiently immortalizes human B cells without neutralizing the function of p53. EMBO J. 14:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. London B 356:437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannell, E. J., P. J. Farrell, and A. J. Sinclair. 1998. Cell cycle arrest following exposure of EBV-immortalised B-cells to gamma irradiation correlates with inhibition of cdk2 activity. FEBS Lett. 439:297-301. [DOI] [PubMed] [Google Scholar]
- 5.Eastman, A. 1987. The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol. Ther. 34:155-166. [DOI] [PubMed] [Google Scholar]
- 6.Frank, A. J., S. J. Proctor, and M. J. Tilby. 1996. Detection and quantification of melphalan-DNA adducts at the single cell level in hematopoietic tumor cells. Blood 88:977-984. [PubMed] [Google Scholar]
- 7.Krauer, K. G., A. Burgess, M. Buck, J. Flanagan, T. B. Sculley, and B. Gabrielli. 2004. The EBNA-3 gene family proteins disrupt the G2/M checkpoint. Oncogene 23:1342-1353. [DOI] [PubMed] [Google Scholar]
- 8.Meek, D. W. 2004. The p53 response to DNA damage. DNA Repair (Amsterdam) 3:1049-1056. [DOI] [PubMed] [Google Scholar]
- 9.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 10.Olive, P. L., and J. P. Banath. 1993. Detection of DNA double-strand breaks through the cell cycle after exposure to X-rays, bleomycin, etoposide and 125IdUrd. Int. J. Radiat. Biol. 64:349-358. [DOI] [PubMed] [Google Scholar]
- 11.O'Nions, J., and M. J. Allday. 2004. Deregulation of the cell cycle by the Epstein-Barr virus. Adv. Cancer Res. 92:119-186. [DOI] [PubMed] [Google Scholar]
- 12.O'Nions, J., and M. J. Allday. 2003. Epstein-Barr virus can inhibit genotoxin-induced G1 arrest downstream of p53 by preventing the inactivation of CDK2. Oncogene 22:7181-7191. [DOI] [PubMed] [Google Scholar]
- 13.O'Nions, J., and M. J. Allday. 2004. Proliferation and differentiation in isogenic populations of peripheral B cells activated by Epstein-Barr virus or T cell-derived mitogens. J. Gen. Virol. 85:881-895. [DOI] [PubMed] [Google Scholar]
- 14.Saridakis, V., Y. Sheng, F. Sarkari, M. N. Holowaty, K. Shire, T. Nguyen, R. G. Zhang, J. Liao, W. Lee, A. M. Edwards, C. H. Arrowsmith, and L. Frappier. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18:25-36. [DOI] [PubMed] [Google Scholar]
- 15.Schlade-Bartusiak, K., A. Stembalska-Kozlowska, M. Bernady, M. Kudyba, and M. Sasiadek. 2002. Analysis of adaptive response to bleomycin and mitomycin C. Mutat. Res. 513:75-81. [DOI] [PubMed] [Google Scholar]
- 16.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 17.Wade, M., and M. J. Allday. 2000. Epstein-Barr virus suppresses a G2/M checkpoint activated by genotoxins. Mol. Cell. Biol. 20:1344-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]