Abstract
Mechanisms of cellular transformation associated with human papillomavirus type 5 (HPV5), which is responsible for skin carcinomas in epidermodysplasia verruciformis (EV) patients, are poorly understood. Using a yeast two-hybrid screening and molecular and cellular biology experiments, we found that HPV5 oncoprotein E6 interacts with SMAD3, a key component in the transforming growth factor β1 (TGF-β1) signaling pathway. HPV5 E6 inhibits SMAD3 transactivation by destabilizing the SMAD3/SMAD4 complex and inducing the degradation of both proteins. Interestingly, the E6 protein of nononcogenic EV HPV9 failed to interact with SMAD3, suggesting that downregulation of the TGF-β1 signaling pathway could be a determinant in HPV5 skin carcinogenesis.
Epidermodysplasia verruciformis (EV) is a rare genodermatosis (Online Mendelian Inheritance in Man no. 226400) characterized by abnormal susceptibility to a group of related specific human papillomaviruses (HPVs) (EV HPVs), including oncogenic genotypes HPV5 and HPV8 and nononcogenic subtypes such as HPV9. HPV5 is associated with 80% of skin carcinomas developing in EV patients (13, 18). This has led to the suspicion that EV HPVs has a role in skin carcinogenesis in the general population (20, 24). Numerous studies have focused on the mechanisms exerted by the E6 and E7 proteins of oncogenic genital HPVs in the virus life cycle involving the alteration of specific cellular factors that play a major role in the cell cycle, apoptosis, or some other essential functions that must be activated or inhibited to allow viral replication and transformation (16, 17). In contrast, little is known about the cellular pathways altered by EV HPV oncoproteins, in particular, major oncoprotein E6 of HPV5 (5E6). The E6 protein of oncogenic EV HPVs does not bind the cellular p53 protein, whereas the E7 protein interacts poorly with the retinoblastoma protein pRb (2, 20). It is worth stressing that the E6 oncoprotein of HPV5 and HPV8 abrogates apoptosis by promoting Bak degradation (7, 8).
In order to identify cellular partners of the 5E6 oncoprotein and to get some insight into the cellular pathways altered during HPV5 infection, we performed yeast two-hybrid screening and developed ex vivo assays to better characterize the biological properties of 5E6.
Yeast two-hybrid screening identified SMAD3 as a 5E6-interacting protein.
The 5E6 gene fused to the GAL4 DNA binding domain in pGBKT7 (pGBKT7-5E6 bait plasmid) was used to screen a Matchmaker cDNA library from the human keratinocyte HaCaT cell line (Clontech) (1). This cDNA library was cloned into vector pACT2 (Clontech). The Y187 yeast strain (MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ mel gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ) was transformed with the pACT2-HaCaT cDNA library by standard procedures (4). One aliquot (1.55 × 109 yeast cells) was used for screening by mating with yeast strain AH109 (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ) transformed with the bait plasmid pGBKT7-5E6. Yeast cells were grown on synthetic dextrose medium lacking tryptophan, leucine, and histidine and containing 100 mM 3-aminotriazole, an inhibitor of His3p for the titration of the self-transactivating property of the bait protein. A β-galactosidase overlay assay was performed with 600 His+ colonies to evaluate the second reporter gene. The cDNA inserts of the 200 strongest blue clones were PCR amplified, sequenced, and analyzed with the BLAST computer program. Six clones matched two overlapping cDNA fragments encoding amino acids Y156 to S425 (four clones) and E178 to S425 (two clones) of SMAD3 (accession number P84022), a 48-kDa protein which plays a central role in the transforming growth factor β1 (TGF-β1) signal transduction pathway (23, 27). To further analyze this interaction, full-length viral and cellular genes, as well as specific DNA fragments of SMAD3, were obtained by PCR and inserted into different expression plasmids by GATEWAY Cloning Technology (26).
SMAD3 is specifically targeted by the E6 protein of oncogenic HPV5.
The 5E6 and 5E7 genes were cloned in frame downstream of the glutathione S-transferase gene (GST-5E6 and GST-5E7) into vector pTM1-GST (5). DNA sequences corresponding to the carboxyl-terminal half (residues Y156 to S425) of SMAD3 (YS-SMAD3) were cloned in frame downstream of the triple flag epitope sequence into vector pCI-neo (Promega). BSR cells (21) were transfected with 5E6 or 5E7 and SMAD3 constructs (1 μg) by using polyethylenimine (9), and cytoplasmic extracts were analyzed by GST pull-down assay (12). Only binding of GST-5E6 with YS-SMAD3 was observed, whereas no interaction was detected with GST-5E7 or GST protein (Fig. 1A).
FIG. 1.
5E6 specifically targets SMAD3. (A) Oncoprotein 5E6 binds to truncated SMAD3 as detected by GST pull-down (Pd) assay. BSR cells were cotransfected with plasmid Flag-YS-SMAD3 and with either expression plasmid GST-5E6 or GST-5E7 or the GST expression plasmid as a control. Protein complexes from cleared lysates (L) were assayed by pull-down assay with glutathione Sepharose beads (Amersham Biosciences) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Flag- and GST-tagged proteins were detected by immunoblotting (IB). (B) Full-length SMAD3 interacts with 5E6 as detected by immunoprecipitation experiments. BSR cells were cotransfected with GST-SMAD3 or GST-SMAD4 expression plasmids and Flag-5E6. Cleared lysates were assayed by immunoprecipitation (IP) of complexes with either anti-Flag M2 (Sigma) or anti-GST (Upstate) monoclonal antibodies, and after electrophoresis, complexes were analyzed by immunoblotting. (C) 9E6 or 9E7 does not interact with SMAD3. BSR cells were cotransfected with vectors expressing full-length Flag-SMAD3 and GST-5E6, GST-9E6, or GST-9E7. Cleared lysates were immunoprecipitated and analyzed by immunoblotting.
To determine whether 5E6 was capable of binding full-length SMAD3 or its cellular partner SMAD4 (23), we constructed recombinant plasmids expressing Flag-5E6 and full-length SMAD3 or SMAD4 fused to GST. After transfection of BSR cells, immunoprecipitation experiments indicated that 5E6 interacts with full-length SMAD3 but not with SMAD4 (Fig. 1B). To demonstrate that 5E6 was able to interact with endogenous SMAD3, we established a stable HaCaT cell line ectopically expressing flagged 5E6. After immunoprecipitation of proteins with the appropriate antibodies, the 5E6-SMAD3 complex was clearly identified (see Fig. 4C).
FIG. 4.
The SMAD3/SMAD4 complex is destabilized and degraded by 5E6. (A) Oncoprotein 5E6 interacts with phosphorylated SMAD3. BSR cells were treated with TGF-β1 (2.5 ng/ml) and cotransfected with vectors expressing Flag-SMAD3 or phosphorylation-defective Flag-SMAD3A and GST-5E6. Cells were also transfected with an enhanced green fluorescent protein (EGFP) expression vector as a transfection control. (B) 5E6 destabilizes the SMAD3/SMAD4 complex. BSR cells were transfected with vectors expressing GST-SMAD4 alone (lanes 1 and 2) or cotransfected with Flag-SMAD3 and GST-SMAD4 in the absence (lanes 3 to 6) or presence of 5E6 tagged with Vs epitope from vesicular stomatitis virus (VSV-5E6; lanes 7 to 10). The total amount of DNA transfected (3 μg) was kept constant by adding control plasmid pCI-neo as appropriate. The experiment was carried out in the absence (−) or presence (+) of TGF-β1 (2.5 ng/ml) for 24 h prior lysis. Cell lysates (L) were detected by pull-down (Pd) assay with glutathione Sepharose beads and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting (IB). (C) Ectopically expressed 5E6 associates with endogenous SMAD3. Stable HaCaT cells expressing flagged 5E6 were stimulated for 24 h with TGF-β1 (2.5 ng/ml) and treated with a 20 μM concentration of the proteasome inhibitor MG-132 (Calbiochem) in dimethyl sulfoxide (MG-132 +) or treated with dimethyl sulfoxide alone (MG-132 −) for 6 h prior to lysis. Cleared lysates were assayed by immunoprecipitation of complexes with either anti-Flag M2 or anti-phosphorylated SMAD3 (Santa Cruz Biotechnology) antibodies, and after electrophoresis, complexes were analyzed by immunoblotting. (D) 5E6 induces degradation of SMAD3 and SMAD4. BSR cells were cotransfected with vectors expressing Flag-SMAD3 (1 μg) and GST-SMAD4 (1 μg) and increasing amounts of VSV-5E6 vector (1, 2, 3, 4, and 5 μg). The total amount of transfected DNA (7 μg) was kept constant by adding control plasmid pCI-neo as appropriate. The experiment was carried out in the presence of TGF-β1 (2.5 ng/ml). Cells were treated (+) or not (−) with 20 μM MG-132 for 6 h prior to lysis. The protein concentration in cleared lysates was measured by the Bio-Rad Protein Assay (Bio-Rad). Lysates (80 μg total protein/lane) were electrophoresed and analyzed by immunoblotting. An anti-tubulin monoclonal antibody (Oncogene) was used as a control.
We next wondered whether SMAD3 was also a target for the E6 or E7 protein from nononcogenic EV HPV9. We constructed pTM1-GST recombinant plasmids expressing 9E6 or 9E7. After immunoprecipitation and immunoblotting analysis, no interaction between SMAD3 and 9E6 or 9E7 was detected (Fig. 1C). This suggests that the interaction between E6 and SMAD3 could be specific for oncogenic HPV5.
5E6 interacts with both the MH1 and MH2 domains of SMAD3.
Initial two-hybrid screening identified an interaction between 5E6 and a truncated SMAD3 protein (YS-SMAD3) comprising part of the linker and the MH2 domain, which is involved in oligomerization and binding of cellular proteins, including SMAD4 (23, 25). This contrasts with HPV16, since the E7 oncoprotein was reported to interact with the MH1 DNA binding domain of SMAD3 (11). According to this, we cloned full-length SMAD3 and deletion mutants SMAD3-MH1 and SMAD3-MH2 (Fig. 2A) fused in frame with the GAL4 DNA binding domain into vector pGBKT7 to perform a two-hybrid test along with the 5E6 gene fused to the GAL4 activation domain into vector pACT2. Interestingly, all three constructs displayed a weak interaction with 5E6 (data not shown). Immunoprecipitation assays were carried out to better define the SMAD3 region interacting with 5E6. We cloned full-length SMAD3 and deletion mutants SMAD3-MH1 and SMAD3-MH2 (Fig. 2A) in frame downstream of the triple flag epitope sequence into vector pCI-neo. Recombinant plasmids expressing GST-16E6 and GST-16E7 were used as controls. 5E6 was found to interact with both the MH1 and MH2 domains (Fig. 2B). Nevertheless, this interaction was clearly weaker than that with the YS-SMAD3 fragment, suggesting that residues Y156 to E228 of SMAD3 could have either a stabilizing effect or directly participate with the interacting domain. As expected, only the SMAD3 MH1 domain interacted with 16E7 (Fig. 2C).
FIG. 2.
Mapping of SMAD3 domains interacting with 5E6 and 16E7. (A) Schematic drawings showing the full-length SMAD3 protein (M1 to S425) and deletion mutant proteins corresponding to part of the linker with the MH2 domain (Y156 to S425, YS-SMAD3), the MH1 domain (M1 to P136), and the MH2 domain (E228 to S425). Oncoprotein 5E6 binds to both the MH1 and MH2 domains of SMAD3 (B), whereas 16E7 interacts only with the MH1 domain (C). BSR cells were cotransfected with vectors expressing the two domains of SMAD3 tagged with Flag and GST-5E6, GST-16E6, or GST-16E7. Cleared lysates (L) were assayed by immunoprecipitation (IP) of complexes and analyzed by immunoblotting (IB).
5E6 oncoprotein inhibits TGF-β1-induced transcriptional activation.
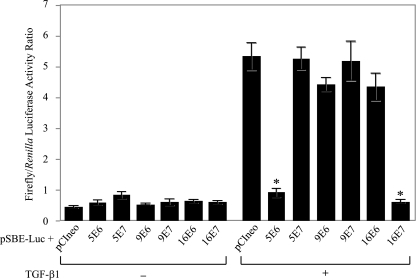
The TGF-β1 signal is known to exert its effect through the activation of R-SMADs (SMAD2 and SMAD3), leading to synthesis of numerous cellular factors such as the inhibitors of cyclin-dependent kinases, among others (6, 14, 15). To investigate the effect of the viral E6 and E7 proteins on the TGF-β1 pathway, a luciferase reporter gene placed downstream of a synthetic SMAD3 regulatory region consisting of six short tandem repeats of the SMAD binding element (SBE) was constructed, as well as mammalian vectors (pCI-neo) expressing E6 and E7 proteins of HPV5, HPV9, and HPV16. As expected, TGF-β1 induction (2.5 ng/ml) markedly increased luciferase activity (maximum activation of 10-fold) in HaCaT cells cotransfected with pSBE/Luc and the pCI-neo control vector (P < 0.001). This SMAD transactivation reporter was specifically down regulated (5- to 10-fold) in the presence of 5E6 or 16E7 in HaCaT cells stimulated with TGF-β1, whereas 5E7, 9E6, 9E7, or 16E6 had no effect on the expression of the reporter plasmid (P < 0.001) (Fig. 3). The experiment was performed four times with the Renilla luciferase expressing vector (hRluc/TK; Promega) as an internal control for transfection efficiency and cellular viability.
FIG. 3.
Regulation of TGF-β1-induced transcriptional activity by proteins E6 and E7 of HPV5, HPV9, and HPV16. HaCaT cells grown in 24-well culture plates were cotransfected with the pSBE/Luc firefly luciferase-expressing vector in the presence of the empty pCI-neo control vector or the pCI-neo vector expressing protein E6 or E7 of HPV5, HPV9, and HPV16, as shown. The Renilla luciferase-expressing vector (hRluc/TK; Promega) was added to each sample as a control for transfection efficiency. Cells were left untreated (−) or treated (+) for 24 h with TGF-β1 (2.5 ng/ml) and harvested into 100 μl of passive lysis buffer (Dual-Luciferase Reporter Assay system; Promega) per well. The luciferase activities from both firefly and Renilla luciferase constructs were quantified by using the reagents and protocol provided by the manufacturer. The results were scored as the ratio of firefly luciferase activity normalized to Renilla luciferase activity per well. Values given are means ± standard deviations of four separate transfections. *, P < 0.001.
5E6 participates in SMAD3 degradation through the proteasome.
TGF-β1 induces the phosphorylation of SMAD3 at S422, S423, and S425, and the resulting conformational change allows the formation of homodimers and heterotrimers with SMAD4. These complexes are translocated to the nucleus, inducing the synthesis of numerous cell factors (6, 23, 25). We wondered whether 5E6 interacts with the phosphorylated SMAD3 protein or interferes with the formation and stability of SMAD3/SMAD4 complexes. To address these questions, a SMAD3 phosphorylation-defective mutant was generated with S422, S423, and S425 changed to alanine (SMAD3A). 5E6 failed to interact with SMAD3A, suggesting the involvement of SMAD3 phosphorylation in the formation of the 5E6/SMAD3 complex (Fig. 4A). We show that the 5E6 oncoprotein alters the formation and stability of the SMAD3/SMAD4 complex in cells cotransfected with 5E6, SMAD3, and SMAD4 (Fig. 4B). We also demonstrate that ectopically expressed flagged 5E6 in HaCaT cells can associate with endogenously expressed SMAD3 in a phosphorylated state since immunocomplexes were only captured and detected with an anti-P-SMAD3 antibody. Moreover, immunocomplexes were only observed in the presence of proteasome inhibitor MG-132, suggesting that 5E6 could induce the proteasomal degradation of SMAD3 (Fig. 4C). Finally, comparative experiments done with or without the proteasome inhibitor MG-132 and increasing amounts of 5E6 reinforce the hypothesis by which 5E6 leads to the degradation of both SMAD3 and SMAD4 via the proteasome (Fig. 4D).
Our data demonstrate that the E6 protein of oncogenic EV HPV5 interacts with SMAD3 in the TGF-β1 signaling pathway. Similarly, binding of HPV16 E7 to SMAD3 has been reported (11). In contrast, this interaction does not occur for the E6 or E7 protein of nononcogenic EV HPV9. Recent studies have shown that the interaction between SMAD3 and proteins from viruses (human T-cell lymphotropic virus type 1, hepatitis C virus, and Kaposi's sarcoma-associated herpesvirus) is implicated in cell transformation (3, 10, 19, 22). This indicates that the TGF-β1 signaling pathway is a common target for several oncogenic viruses. It is worth stressing that the TGF-β1 pathway leads to the synthesis of inhibitors (p16, p17, p21, p27, etc.) of the cyclin-dependent kinases that play a crucial role in the cell cycle (6, 15, 23, 25). It can be speculated that the interaction of oncoprotein 5E6 or 16E7 with the SMAD3 protein could negatively regulate the inhibitors of the cell cycle and favor cell transition from the G1 to the S phase to allow viral DNA vegetative replication and cell transformation.
Acknowledgments
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM 2004 00 2308) and the Ligue Nationale contre le Cancer (contract R05/75-129). J.-A.M. is supported by a long-term fellowship from the Fundación Gran Mariscal de Ayacucho and the Universidad de Los Andes, Venezuela.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boxman, I. L. A., L. H. Mulder, F. Noya, V. de Waard, S. Gibbs, T. R. Broker, F. ten Kate, L. T. Chow, and J. ter Schegget. 2001. Transduction of the E6 and E7 genes of epidermodysplasia-verruciformis-associated human papillomaviruses alters human keratinocyte growth and differentiation in organotypic cultures. J. Investig. Dermatol. 117:1397-1404. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, P. L., M. H. Chang, C. H. Chao, and Y. H. Wu Lee. 2004. Hepatitis C viral proteins interact with Smad3 and differentially regulate TGF-β/Smad3-mediated transcriptional activation. Oncogene 23:7821-7838. [DOI] [PubMed] [Google Scholar]
- 4.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horikami, S. M., R. E. Hector, S. Smallwood, and S. Moyer. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261-270. [DOI] [PubMed] [Google Scholar]
- 6.Itoh, S., F. Itoh, M. J. Goumans, and P. ten Dijke. 2000. Signaling of transforming growth factor-β family members through Smad proteins. Eur. J. Biochem. 267:6954-6967. [DOI] [PubMed] [Google Scholar]
- 7.Jackson, S., C. Harwood, M. Thomas, L. Banks, and A. Storey. 2000. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson, S., and A. Storey. 2000. E6 proteins from diverse cutaneous HPV types inhibit apoptosis in response to UV damage. Oncogene 19:592-598. [DOI] [PubMed] [Google Scholar]
- 9.Jacob, Y., E. Real, and N. Tordo. 2001. Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J. Virol. 75:9613-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, D. K., B.-C. Kim, J. N. Brady, K.-T. Jeang, and S.-J. Kim. 2002. Human T-cell lymphotropic virus type 1 tax inhibits transforming growth factor-beta signaling by blocking the association of Smad proteins with Smad-binding element. J. Biol. Chem. 277:33766-33775. [DOI] [PubMed] [Google Scholar]
- 11.Lee, D. K., B.-C. Kim, Y. K. Isaac, E-A. Cho, D. J. Satterwhite, and S.-J. Kim. 2002. The human papilloma virus E7 oncoprotein inhibits transforming growth factor-signaling by blocking binding of the Smad complex to its target sequence. J. Biol. Chem. 277:38557-38564. [DOI] [PubMed] [Google Scholar]
- 12.Le May, N., S. Dubaele, L. Proietti de Santis, A. Billecocq, M. Bouloy, and J. M. Egly. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541-550. [DOI] [PubMed] [Google Scholar]
- 13.Majewski, S., S. Jablonska, and G. Orth. 1997. Epidermodysplasia verruciformis. Immunological and nonimmunological surveillance mechanisms: role in tumor progression. Clin. Dermatol. 15:321-334. [DOI] [PubMed] [Google Scholar]
- 14.Major, M. B., and D. A. Jones. 2004. Identification of a gadd45beta 3′ enhancer that mediates SMAD3- and SMAD4-dependent transcriptional induction by transforming growth factor. J. Biol. Chem. 279:5278-5287. [DOI] [PubMed] [Google Scholar]
- 15.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 16.Münger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 21:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 18.Orth, G. 1987. Epidermodysplasia verruciformis, p. 199-243. In P. M. Howley and N. P. Salzman (ed.), The papillomaviruses. Plenum Press, New York, N.Y.
- 19.Pavio, N., S. Battaglia, D. Boucreux, B. Arnulf, R. Sobesky, O. Hermine, and C. Brechot. 2005. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-β pathway. Oncogene 24:6119-6132. [DOI] [PubMed] [Google Scholar]
- 20.Pfister, H. 2003. Human papillomavirus and skin cancer. J. Natl. Cancer Inst. Monogr. 31:52-56. [DOI] [PubMed] [Google Scholar]
- 21.Sato, M., N. Maida, H. Yoshida, M. Urade, S. Saito, T. Miyazaki, T. Shibata, and M. Watanabe. 1977. Plaque formation of herpes virus hominidis type 2 and rubella virus in variants isolated from the colonies of BHK21/WI-2 cells formed in soft agar. Arch. Virol. 53:269-273. [DOI] [PubMed] [Google Scholar]
- 22.Seo, T., J. Park, and J. Choe. 2005. Kaposi's sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res. 65:1738-1747. [DOI] [PubMed] [Google Scholar]
- 23.Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 24.Sterling, J. C. 2005. Human papillomaviruses and skin cancer. J. Clin. Virol. 32S:S67-S71. [DOI] [PubMed] [Google Scholar]
- 25.ten Dijke, P., and C. S. Hill. 2004. New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 29:265-273. [DOI] [PubMed] [Google Scholar]
- 26.Walhout, A. J., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple, M. A. Brasch, N. Thierry-Mieg, and M. Vidal. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287:116-122. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y., X. Feng, R. We, and R. Derynck. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383:168-172. [DOI] [PubMed] [Google Scholar]