Abstract
Though similar to those of herpesvirus saimiri and Epstein-Barr virus (EBV), the Kaposi's sarcoma-associated herpesvirus (KSHV) genome features more splice genes and encodes many genes with bicistronic or polycistronic transcripts. In the present study, the gene structure and expression of KSHV ORF56 (primase), ORF57 (MTA), ORF58 (EBV BMRF2 homologue), and ORF59 (DNA polymerase processivity factor) were analyzed in butyrate-activated KSHV+ JSC-1 cells. ORF56 was expressed at low abundance as a bicistronic ORF56/57 transcript that utilized the same intron, with two alternative branch points, as ORF57 for its RNA splicing. ORF56 was transcribed from two transcription start sites, nucleotides (nt) 78994 (minor) and 79075 (major), but selected the same poly(A) signal as ORF57 for RNA polyadenylation. The majority of ORF56 and ORF57 transcripts were cleaved at nt 83628, although other nearby cleavage sites were selectable. On the opposite strand of the viral genome, colinear ORF58 and ORF59 were transcribed from different transcription start sites, nt 95821 (major) or 95824 (minor) for ORF58 and nt 96790 (minor) or 96794 (major) for ORF59, but shared overlapping poly(A) signals at nt 94492 and 94488. Two cleavage sites, at nt 94477 and nt 94469, could be equally selected for ORF59 polyadenylation, but only the cleavage site at nt 94469 could be selected for ORF58 polyadenylation without disrupting the ORF58 stop codon immediately upstream. ORF58 was expressed in low abundance as a monocistronic transcript, with a long 5′ untranslated region (UTR) but a short 3′ UTR, whereas ORF59 was expressed in high abundance as a bicistronic transcript, with a short 5′ UTR and a long 3′ UTR similar to those of polycistronic ORF60 and ORF62. Both ORF56 and ORF59 are targets of ORF57 and were up-regulated significantly in the presence of ORF57, a posttranscriptional regulator.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a B-lymphocyte-tropic γ-herpesvirus with a genome of ∼165 kb that can encode up to 90 viral proteins (21, 28, 33, 41). Despite its similarity to the organization of other γ-herpesvirus genomes, the KSHV genome is unique in having multiple regions with gene clusters and more split (splice) genes than any other herpesvirus (45). Considerable insight has been gained from recent studies on the structure and expression of individual genes in the KSHV genome, including the identification of ORF50/K8/K8.1 as a cluster gene locus in which all three genes share a common poly(A) signal but utilize alternative promoters and alternative RNA splicing to express each gene (20, 22, 30, 39, 40, 43). ORF73/72/K13 is another cluster gene locus in which all three genes initiate their transcription from the same promoter and polyadenylation from the same poly(A) signal but utilize alternative RNA splicing and translation initiation for the expression of each gene (1, 9, 14, 35, 38). ORF34/35/36/37 was also identified recently as a cluster gene locus that responds to hypoxia for transcription activation, and the hypoxia-induced polycistronic transcripts are presumably polyadenylated from a common poly(A) signal (12, 13).
Several elegant studies have succeeded in mapping and characterizing the transcription start sites of representative genes from the KSHV genome. However, we know little about the 3′ ends of their transcripts except that they are presumed to contain poly(A) signals because such signals are located 3′ to the individual open reading frames (ORFs). Where the transcription of most KSHV genes starts and ends remains largely unknown. In the course of studying KSHV ORF57 gene expression and looking for ORF57 targets, we extensively analyzed the gene structures and expression of four KSHV early genes: ORF56, ORF57, ORF58, and ORF59. KSHV ORF56 encodes a primase protein for viral DNA replication (42) and is positioned immediately upstream of ORF57 on the sense strand of the KSHV genome. KSHV ORF57 has been characterized as a posttranscriptional regulator of viral gene expression (10, 15, 23, 25), as well as a cooperative transcriptional regulator (15, 24). KSHV ORF58 encodes an Epstein-Barr virus BMRF2 homologue of unknown function (18) and is positioned side-by-side with KSHV ORF59 in the reverse direction on the antisense strand of KSHV genome. KSHV ORF59 encodes a viral DNA polymerase processivity factor involved in viral DNA replication (4, 5, 19). Some limited knowledge has been obtained about ORF57, but very little is known about the gene structures and expression of ORF56, ORF58, and ORF59 in the context of the KSHV genome. In this study, we combined all available techniques to precisely map the transcription start sites and polyadenylation sites as well as the expression levels of these genes. We found that KSHV ORF56 and ORF59 are transcribed as bicistronic RNA transcripts and are targets of KSHV ORF57.
MATERIALS AND METHODS
Cells.
The human KSHV-positive, Epstein-Barr virus-positive, PEL-derived B-cell line JSC-1 (3) was a generous gift from Richard Ambinder of The John Hopkins University. Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 10% (vol/vol) fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin in a humidified 5% CO2 atmosphere at 37°C. For induction of the KSHV lytic cycle, JSC-1 cells were treated with sodium butyrate (Aldrich, Milwaukee, WI) at a final concentration of 3 mM for 24 or 48 h.
RT-PCR.
Total RNA extracted from cells using the TRIzol reagent (Invitrogen) was treated with RQ1 DNase I (Promega, Madison, WI) for 20 min at 37°C, followed by heat inactivation, phenol-chloroform extraction, and precipitation. DNase I-treated total RNA (8 μg) was then used in a one-step reverse transcriptase (RT)-PCR containing Superscript II/Platinum Taq (+RT) or Platinum Taq (−RT) (Invitrogen). The gene-specific primers oVM9 (Pr81856; 5′-GGTGAGCGAAGTCACGGTAAC-3′) and oVM11 (Pr82296; 5′-CTCGTCTTCCAGTGTCGGTG-3′) were used to detect spliced ORF56/57 bicistronic transcripts.
RPA.
The radioactive RNA probes were prepared by in vitro transcription in the presence of [α-32P]GTP with Riboprobe System-T7 (Promega), using PCR products with built-in T7 promoter sequences as templates. The following primers were used for DNA template preparation: oVM9 and oVM14 (Pr82297; 5′-TAATACGACTCACTATAGG/GCTCGTCTTCCAGTGTCGGT-3′) for probe a, oVM36 (Pr83438; 5′-TGGAACATCACAGCTTG-3′) and oVM90 (Pr83705; 5′-TAATACGACTCACTATAG/GGCGGTCTGGTGTTGTGTT-3′) for probe b, oVM54 (Pr97374; 5′-CAGGGTGCGTAAGGCGAC-3′) and oVM58 (Pr96704; 5′-TAATACGACTCACTATAGG/GTCCACCCTGACCCCATAGT-3′) for probe c, oVM159 (Pr95717; 5′-TAATACGACTCACTATA/GGGACCAACTGGTGTGAGAGG-3′) and oVM160 (Pr96080; 5′-ACCCGTTTCTGGCTGGGATGGT-3′) for probe d, and oVM91 (Pr94368; 5′-TAATACGACTCACTATAG/GGACAGCACCCAGGCACT-3′) and oVM92 (Pr94768; 5′-ATGCGTGGGCGTATCCG-3′) for probe e. The positions of the individual probes relative to the KSHV genome are shown in Fig. 1 and Fig. 4. The RNase protection assay (RPA) was performed with an RPA III kit (Ambion, Austin, TX) according to the manufacturer's instructions with minor modifications. Briefly, 4 ng of each probe (specific activity, 35,000 cpm/ng) was hybridized overnight at 42°C with 20 to 25 μg of total RNA in hybridization buffer and then digested with an RNase A-T1 mixture for 30 min at 37°C. Five micrograms of yeast RNA was used as a negative control. Protected RNA fragments were separated in a denaturing 8% polyacrylamide gel containing 8 M urea. DNA ladders with 32P end labeling were used as size markers. Autoradiographic data were captured with a Molecular Dynamics PhosphorImager Storm 860 instrument and analyzed with ImageQuant software.
FIG. 1.
KSHV ORF56 is a split gene that produces transcripts of low abundance after lytic induction. (A) Schematic diagrams of the KSHV ORF56 and ORF57 ORFs and their transcripts. The numbers above each ORF are the nucleotide positions of the start and termination codons in the KSHV genome (GenBank accession number U75698) (33). The heavy line with dashes on both ends represents the genomic region encompassing ORF56 and ORF57, with promoters (arrows, designated by their transcription start sites) and 3′-end processing signals [a poly(A) signal and a cleavage site (CS)] indicated. Below the heavy line are bicistronic ORF56/57 and monocistronic ORF57 pre-mRNAs that each contain two exons (boxes 1 and 2) and one intron (dashes between boxes). Primers (heavy arrows) used to characterize KSHV ORF56 and ORF57 transcripts are shown below the pre-mRNAs and are named by the locations of their 5′ ends. Antisense RNA probes a and b for RPA assays and the resulting RNA products protected from each probe are illustrated in the bottom of the diagram, with the sizes (nt) in parentheses. (B) The majority of KSHV ORF56 transcripts are bicistronic ORF56/57 transcripts that are spliced to remove the ORF57 intron. The primers Pr81856 and Pr82296 were used in an RT-PCR on DNase I-treated total RNA (8 μg) isolated from uninduced (0 h) or n-butyrate (NB)-induced (24 and 48 h) JSC-1 cells. Parallel reactions without reverse transcriptase were used as a control. Twenty nanograms of Bac36 DNA (48) was used as a KSHV DNA control. The lower panel shows the splicing junction identified by sequencing of the 333-bp PCR product. (C) RPA analysis of ORF56 and ORF57 transcripts. Twenty micrograms of total RNA from uninduced or butyrate (NB)-induced JSC-1 cells was hybridized with 4 ng of antisense 32P-labeled probe a prepared by in vitro transcription. The protected products were separated along with 100-bp DNA ladders in an 8% denaturing polyacrylamide gel. Yeast total RNA (yRNA) was used as a negative RPA control. (D) Northern blot analysis of ORF56 and ORF57 transcripts. Approximately 1 μg of poly(A)-selected mRNA from uninduced (lane 1) or butyrate-induced (24 h) (lane 2) JSC-1 cells was used in Northern blot analysis with 32P-labeled antisense probe b prepared by in vitro transcription. The membrane was then exposed to X-ray film for various times. ?, unknown transcripts.
FIG.4.
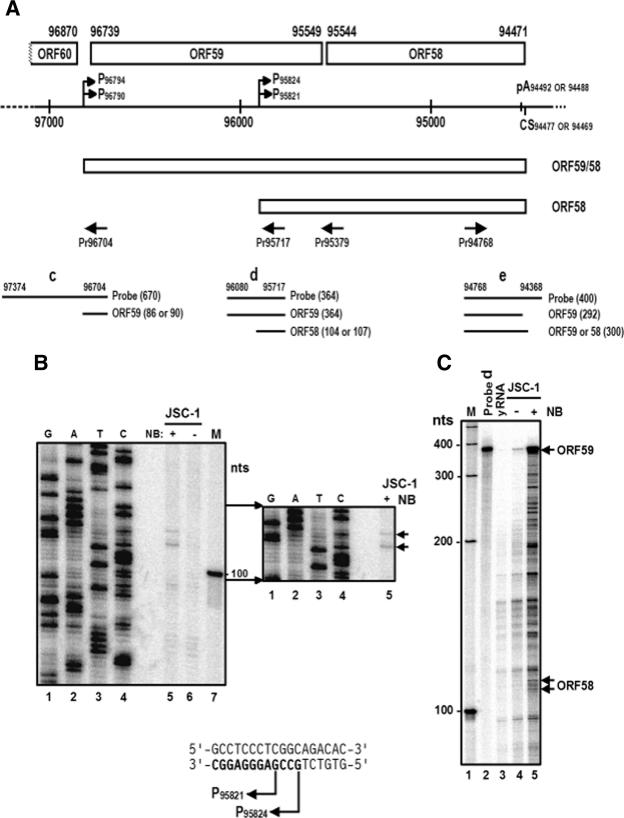
Mapping of the KSHV ORF58 and ORF59 transcription start sites. (A) Schematic diagrams of KSHV ORF58 and ORF59 ORFs and their transcripts. See other details in Fig. 1A. The heavy line below ORF58 and ORF59 with dashes on both ends represents the genomic region encompassing ORF58 and ORF59, with the promoters (arrows, designated by their transcription start sites) and 3′ end processing signals [a poly(A) signal and a cleavage site (CS)] characterized in this study. Below the heavy line are the transcripts of bicistronic ORF58/59 and monocistronic ORF58. Primers (heavy arrows) used for primer extension, sequencing, or RACE are shown below the transcripts. Antisense RNA probes c, d, and e for RPA assays and the resulting RNA products protected from each probe are illustrated in the bottom of the diagram, with the sizes (nt) in parentheses. (B) Mapping of ORF58 transcription start sites by primer extension. Total RNA (20 μg) from uninduced or butyrate-induced JSC-1 cells was used for primer extension with a 32P-labeled primer, Pr95717. The same 32P-labeled primer was used to generate a sequencing ladder from a plasmid containing KSHV genomic DNA from nt 95717 to nt 96080. The extended products were compared with the sequencing ladder in an 8% denaturating polyacrylamide gel, and the corresponding sequence ladders to the extended products are the transcription start sites, as indicated by arrows shown at the bottom of the panel. (C) Mapping of ORF58 transcription start sites by RPA. RPA was performed on 20 μg of total RNA from uninduced or butyrate-induced JSC-1 cells, with 4 ng of 32P-labeled antisense RNA probe d. Arrows indicate the protected ORF58 or ORF59 transcripts. (D) Mapping of ORF58 and ORF59 transcription start sites by 5′-RACE with primer Pr95379. The left panel shows the products of 5′ RACE from regular extension (3 min) cycles. The right panel shows the products of 5′ RACE from short extension (30 s) cycles preferentially amplifying ORF58 transcripts. (E) Mapping of KSHV ORF59 transcription start site by primer extension. A primer extension assay was performed on 20 μg of total RNA from uninduced or butyrate-induced (24 h) JSC-1 cells with a 32P-labeled Pr96704 primer. The same primer was also used to generate a sequencing ladder from a plasmid containing KSHV genomic DNA from nt 97374 to 96704. The extended products were compared with the sequencing ladder in an 8% denaturating polyacrylamide gel, and the corresponding sequence ladders to the extended products are the transcription start sites as indicated by arrows shown at the bottom of the panel. (F) Mapping of ORF59 transcription start sites by RPA. RPA was performed on 25 μg of total RNA from uninduced or butyrate-induced JSC-1 cells with 4 ng of 32P-labeled RNA probe c (see panel A). Arrows indicate RPA products in sizes of 87 and 91 nt, respectively, just above its 1-nt-truncated product.
Primer extension assay.
To map the transcription start site(s) of selected KSHV transcripts or the ORF56/57 branch point(s) for RNA splicing, primer extension assays were carried out using a Primer Extension System-AMV Reverse Transcriptase kit (Promega). Each reaction contained 20 μg of total RNA and one of the following oligonucleotides: oVM158 (Pr95717; 5′-GGGACCAACTGGTGTGAGAGG-3′) for mapping the 5′ end of ORF58 transcripts, oVM73 (Pr96704; 5′-GTCCACCCTGACCCCATAGT-3′) for mapping the 5′ end of ORF59 transcripts, and oVM11 for mapping the ORF56/57 branch point(s).
Manual sequencing.
The precise sizes and positions of various RNA fragments obtained by RPA or primer extension assays were determined with sequencing ladders. Templates used for sequencing were prepared by TA cloning of the PCR templates used for probe preparation (probes a, b, c, d, and e) into pCR2.1 vectors using a TA TOPO cloning kit (Invitrogen). Three micrograms of purified plasmid DNA was sequenced with 32P-labeled individual primers using a Sequenase Quick-Denature plasmid sequencing kit (USB, Cleveland, OH) and separated in a denaturing polyacrylamide gel together with RPA and primer extension products.
5′- and 3′-RACE.
5′- and 3′-rapid amplification of cDNA ends (RACE) assays were carried out to amplify the ends of the studied viral transcripts using a BD SMART RACE cDNA amplification kit (BD Biosciences, San Diego, CA), following the manufacturer's instructions. Poly(A)+ RNA (1 μg/reaction) from JSC-1 cells induced with sodium butyrate for 24 h was used as a template. Poly(A)+ RNA was selected with a QuickPrep Micro mRNA purification kit (GE Healthcare, Piscataway, NJ). The primer oVM146 (Pr79608; 5′-AGGGTTACCCTGGGGGCGAGTATCTGGG-3′) was used for ORF56 5′-RACE, oVM67 (Pr83254; 5′-GCATGTAACCTTCTTGGCGAG-3′) for ORF56/57 3′-RACE, oVM47 (Pr95379; 5′-GCGTGACTGGCAGCGACC-3′) for ORF58 and ORF59 5′-RACE, and oVM92 (Pr94768; 5′-ATGCGTGGGCGTATCCG-3′) for ORF58/59 3′-RACE. To amplify the 5′ ends of ORF58 transcripts, the extension time for RACE PCR amplification was shortened to 30 s to favor the amplification of the short ORF58 over the more abundant ORF59 transcripts. Each RACE product was cloned into a pCR2.1 vector (Invitrogen) using TA cloning, and individual colonies were picked up for sequence analysis.
Plasmid construction.
To express the KSHV ORF57-FLAG fusion protein in mammalian cells, a KSHV ORF57 cDNA from nucleotide (nt) 82069 to nt 83541 in the KSHV genome was amplified by RT-PCR from JSC-1 cells using a 5′ sense primer (oVM68; 5′-TACTCAGAATTCACC/ATGGTACAAGCAATGATAGACATGG-3′) containing an EcoRI site in combination with a 3′ antisense primer (oVM69; 5′-ATCGTGGATCC/AGAAAGTGGATAAAAGAATAAACCCTTG-3′) containing a BamHI site. After double digestion with EcoRI and BamHI, the cDNA without an intron was cloned in frame in the mammalian expression vector pFLAG-5.1-CMV (Sigma, St. Louis, MO). The resulting plasmid was named pVM7 and expressed ORF57 with a FLAG tag on its C terminus. A similar strategy was used to construct the KSHV ORF56-FLAG expression vector. ORF56 (nt 79436 to 81964) was amplified with a sense primer, oVM71 (Pr79433; 5′-TACTCAGAATTC/ACCATGGAGACGACATAC CGC-3′), containing an EcoRI site, and an antisense primer, oVM72 (Pr81964; 5′-ATCGTGTCGAC/ACTGGCCAGTCCCACTGGTACCA-3′), containing a SalI site from Bac36 DNA (48). The resulting 2.5-kb PCR product was double digested with EcoRI and SalI and then cloned into the multiple cloning sites of the pFLAG-CMV-5.1 vector (Sigma) using the corresponding restriction sites. The new plasmid was named pVM9 and expresses ORF56 with a FLAG tag on its C terminus.
Cotransfection of 293 cells and sample preparation.
The cotransfection study was performed with 293 cells (5 × 105) in a six-well plate with 1 μg of pVM9 (ORF56-FLAG fusion) together with 0.2 μg of pVM7 (ORF57-FLAG fusion) or empty pFLAG-CMV-5.1 vector using Lipofectamine 2000 (Invitrogen). Protein samples were prepared 24 h after transfection by direct cell lysis in 500 μg of 2× sodium dodecyl sulfate (SDS) protein loading buffer containing 5% (vol/vol) 2-mercaptoethanol. The cytoplasmic and nuclear fractions of total RNA were prepared 24 h after transfection. Briefly, after trypsinization, cells were washed with cold phosphate-buffered saline, transferred into a 1.5-ml tube, and centrifuged at low speed. The cell pellet was resuspended in 200 μl of buffer A (50 mM Tris [pH 8.0], 140 mM NaCl, 1.5 mM MgCl2, 0.2% NP-40, 1 mM dithiothreitol, and 200 U RNAsin), incubated for 5 min on ice, and spun for 2 min at 3,000 × g. The supernatant (cytoplasmic fraction) was transferred into a prechilled tube and used to extract cytoplasmic RNA. The pellet was saved for the extraction of nuclear RNA by the addition of 1 ml of TRIzol reagent (Invitrogen).
Western blot analysis.
Twenty microliters of protein from each sample was separated in a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen) in 1× morpholineethanesulfonic acid running buffer. After transfer, the nitrocellulose membrane was blocked with 5% nonfat milk in Tris-buffered saline (TBS) (10 mM Tris, 150 mM NaCl, pH 7.4) with the addition of Tween 20 at a final concentration of 0.05% (vol/vol) (TTBS) for 1 h at room temperature. After a quick wash with TBS, the membrane was incubated for 1 h at room temperature with primary antibody diluted in TTBS. A monoclonal anti-FLAG M2 antibody (Sigma) diluted 1:5,000 for ORF56-FLAG and ORF57-FLAG detection or an immunoglobulin M (IgM)-type monoclonal anti-β-tubulin antibody (BD Pharmingen, San Jose, CA) diluted 1:1,000 for sample loading controls were used separately in combination with an appropriate horseradish peroxidase-labeled secondary antibody (Sigma) diluted 1:10,000 in 1% nonfat milk in TTBS for 1 h at room temperature. The immunoreactive proteins were detected with enhanced chemiluminescence using the SuperSignal West Pico Western chemiluminescence substrate (Pierce, Rockford, IL). The enhanced chemiluminescence-generated signal was captured on X-ray film or with a charge-coupled-device camera in the Bio-Rad ChemiDoc system and analyzed with Quantity One software (Bio-Rad, Hercules, CA). Before subsequent staining, the membranes were stripped with Restore Western blot stripping buffer (Pierce) according to the manufacturer's instructions.
Northern blot analysis.
Each sample containing 1 μg of poly(A)+ mRNA isolated from butyrate-induced JSC-1 cells was mixed with NorthernMax formaldehyde load dye (Ambion) and denatured at 75°C for 15 min. One microliter of ethidium bromide (1 mg/ml) was added before loading. The samples were then separated in 1% (wt/vol) formaldehyde-agarose gels in 1× morpholinepropanesulfonic acid running buffer, capillary transferred onto a nylon membrane, and immobilized by UV light. After 1 h of prehybridization, hybridization was carried out for 24 h at 65°C or 42°C (for oligonucleotide probes) in SuperHyb hybridization buffer (Sigma) with the addition of shredded salmon sperm DNA (0.1 mg/ml). The 32P-labeled ORF-specific probes (107 cpm) prepared by end labeling (if they were oligonucleotides) or by in vitro transcription (see RPA) were used to detect specific transcripts. The following sense oligonucleotides were used for the hybridizations: oVM45 (Pr94923; 5′-CTGAATAGGTGATGTACTTCCC-3′) for ORF58, oVM73 (Pr96704) for ORF59, oVM169 (Pr97635; 5′-GTCCTTGTCGGCATCGCTGAG-3′) for ORF60, oVM170 (Pr100080; 5′-TGAGAGATTGGGCACACATATCA-3′) for ORF61, and oVM171 (Pr101121;5′-CGGTGAGTTGGTGGAGGTAAA-3′) for ORF62. After hybridization, the membrane was washed once with a 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% SDS solution for 5 min at room temperature and twice with 0.1× SSPE-0.1% SDS for 15 min at 65°C or 42°C (oligonucleotide probes) and exposed to X-ray film at −80°C for various amounts of time.
Cytoplasmic or nuclear total RNA (∼5 μg) isolated from transfected 293 cells was blotted for ORF56 RNA transcripts with a 32P-labeled antisense oligonucleotide probe (2 × 106 cpm), oVM24 (Pr81676; 5′-AGTGTCGTATGCTTTATCCCAAA-3′), positioned from nt 81676 to 81654 on the antisense strand of the viral genome. Glyceraldehyde-3-dehydrogenase (GAPDH) mRNA and U6 snRNA were probed separately with individual 32P-labeled oligonucleotide probes, oZMZ 270 (5′-TGAGTCCTTCCACGATACCAAA-3′) for GAPDH, and oST 197 (5′-AAATATGGAACGCTTCACGA-3′) for U6 snRNA, as indicators of fractionation efficiency. The nuclear and cytoplasmic fractionation efficiencies were also verified by the presence of 45S and 32S pre-rRNA. The prehybridization and hybridization were performed at 42°C for 20 h, followed by one wash with 2× SSPE-0.1% SDS solution for 5 min at room temperature and two washes with 0.1× SSPE-0.1% SDS for 15 min at 42°C before exposure to a PhosphorImager screen.
RESULTS
KSHV ORF56 is a split gene encoding a bicistronic ORF56/57 mRNA expressed after lytic induction.
KSHV ORF56 and ORF57 physically reside side-by-side in the same orientation on the sense strand of the viral genome and are separated by only 102 nt. A computer analysis of the KSHV ORF56 coding region and its downstream sequences suggests that KSHV ORF56 might share its poly(A) signal with ORF57 for polyadenylation. The poly(A) signal at nt 83608 downstream of the ORF57 coding region had previously been postulated to be used for ORF57 polyadenylation (15). If the two genes share this poly(A) signal, the KSHV ORF56 gene would be a split gene that contains the previously reported ORF57 intron (15). In this case, ORF56 transcripts should be longer than ORF57 transcripts, with a calculated size of more than 4 kb (Fig. 1A). To test this hypothesis, an RT-PCR assay was first conducted with total RNA isolated from butyrate-treated JSC-1 cells to detect ORF56 transcripts. To determine whether ORF56 transcripts contain the ORF57 intron, we performed RT-PCR analyses using a forward primer from the ORF56 coding region approximately 150 nt upstream of the ORF57 transcription start site in combination with a reverse primer downstream of the ORF57 intron. With this strategy, two RT-PCR products of the appropriate sizes were detected, one representing an unspliced, intron-containing ORF56 pre-mRNA and the other representing a spliced ORF56 mRNA (Fig. 1B, top panel). Sequencing of the spliced products (Fig. 1B, bottom panel) revealed the identical splice junction (nt 82117/82226) seen in spliced ORF57 transcripts (15), demonstrating that the ORF56 and ORF57 transcripts contain the same intron. This observation was further confirmed by RPA with probe a. As shown in Fig. 1C, we detected an RPA product of 261 nt, representing spliced ORF56 transcripts, or of 441 nt, representing unspliced ORF56 transcripts, in total RNA isolated from butyrate-induced JSC-1 cells (lane 5). However, the majority of the protected RNA products were spliced ORF57, as indicated by the protection of exon 1 (114 nt) and exon 2 (71 nt; lane 5) at similar levels. Unspliced ORF57 transcripts were less abundant. Consistent with the RPA results described above, Northern blot analysis of poly(A)+ mRNAs using a 32P-labeled antisense probe covering the 3′ end of ORF57 showed that butyrate treatment of JSC-1 cells did induce the cells to produce two major RNA transcripts: an abundant ORF57 of ∼1.6 kb and a much less abundant ORF56 of ∼4.4 kb (Fig. 1D). Together, these data indicate that KSHV ORF56 is a split gene whose 3′ UTR overlaps with ORF57; thus, ORF56 transcripts are bicistronic.
Mapping of the branch point used in the intron splicing of KSHV ORF56 and ORF57 transcripts after lytic induction.
After demonstrating that the KSHV ORF56 and ORF57 transcripts contain the same intron, we used a primer extension assay to map the intron branch point (BP) that mediates the intron recognition by U2 snRNA in spliceosome-mediated splicing of ORF56 or ORF57 RNA. In this strategy, primer extension was performed with total RNA obtained from butyrate-treated JSC-1 cells using the 32P-labeled primer Pr82296, which is complementary to the exon 2 sequences of both the ORF56 and ORF57 transcripts. The same primer was used for both primer extension and dideoxy DNA sequencing reactions. Since primer extension stops one nucleotide before the BP in lariat-containing splicing intermediates, the BP can be mapped by comparing the extended products with the DNA sequencing ladders and showing exactly where the extended products are terminated (46). We detected three extension products from the total RNA obtained from butyrate-activated JSC-1 cells (Fig. 2A, lane 5), one corresponding to spliced ORF57 RNA transcripts with a size of 184 nt and the other two corresponding to extension products of lariat-containing exon 2 RNAs that stopped, respectively, at nt 82186 and nt 82189, with the former being more abundant than the latter. These extension products were not seen when total RNA from uninduced JSC-1 cells was used as a template (Fig. 2A, lane 6). Since primer extension stops one nucleotide before the BP, these results mapped the BP to two adenosine residues on the sense strand at nt 82185 and 82188 (Fig. 2B). Thus, two alternative BPs with overlapping BP sequences, GGCGGAC and GGACCAU (the BP is underlined), are located, respectively, 40 nt and 37 nt upstream of the nt 82226 3′ splice site. Compared with the mammalian BP consensus sequence YNYURAC, the BP sequence GGCGGAC at nt 82185 deviates at two positions, whereas the BP sequence GGACCAU at nt 82188 deviates at four positions. Therefore, the BP at nt 82185 would be predominantly selected for U2 recognition during RNA splicing, as shown in Fig. 2, even though both BP sequences are nonconsensus BP sequences.
FIG. 2.
Splicing of the ORF56/57 intron is mediated through two alternative branch points. Primer extension was performed on 20 μg of total RNA from uninduced or butyrate (NB)-induced JSC-1 cells using a 32P-labeled antisense primer, Pr82296. (A) The sizes of the extended products, whether fully spliced products or splicing intermediates, were compared with the sequencing ladders obtained using the same primer on plasmid pVM1, which contains genomic ORF57 DNA. (B) The identified branch points (arrows pointing to the antisense sequence readout) and the branch point sequences (bolded and boxed) are shown to the right of an enlarged portion of the gel in panel A.
Between the nt 82185 BP and the nt 82226 3′ splice site is a polypyrimidine tract. The polypyrimidine tract contains 10 interspersed purines and is therefore predicted to be suboptimal. Together with the sequence data of the BPs, we conclude that the intron of the ORF56 and ORF57 transcripts is a weak or suboptimal intron.
Mapping of the ORF56 transcription start site and polyadenylation cleavage sites for ORF56 and ORF57 expression in lytic induction.
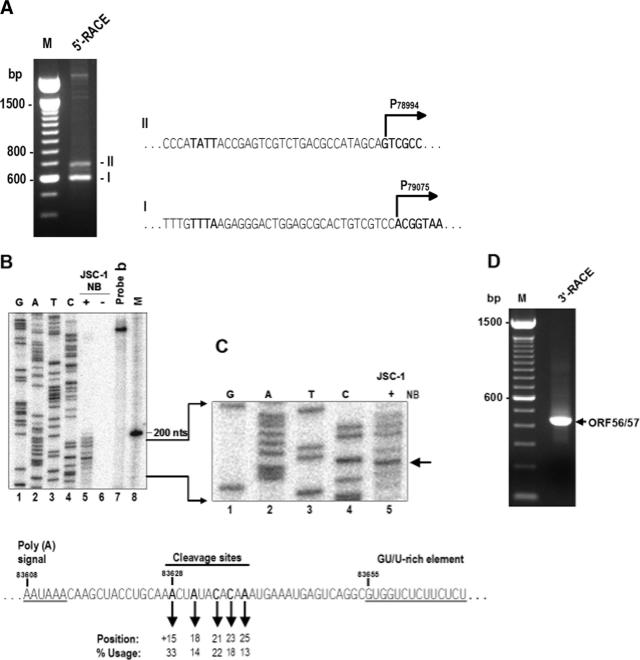
As described above, KSHV ORF56 transcripts are much less abundant than ORF57 transcripts in activated KSHV-positive cells. Initially, it was difficult to map the transcription start site of ORF56 with conventional RPA assays, which had been used for mapping the ORF57 (15) and K8.1 (40) transcription start sites. To locate the transcriptional start site of ORF56, we performed a 5′-RACE assay on poly(A)-selected mRNAs obtained from butyrate-treated JSC-1 cells by using an ORF56-specific primer, Pr79608 (see primer position in Fig. 1A). As shown in Fig. 3A, two RACE products of ∼600 bp (product I) and ∼700 bp (product II) were obtained, cloned, and sequenced to determine the 5′ end of each transcript. Among the 12 clones sequenced, 7 showed an ORF56 transcription start site at nt 79075, 4 at nt 78994, and 1 at nt 78993. The mapped ORF56 transcription start site at nt 79075 corresponds to the major 5′ RACE product I, and the mapped ORF56 transcription start sites at nt 78994 or 78993 correspond to the minor 5′ RACE product II, demonstrating that KSHV ORF56 utilizes at least two alternative transcription start sites (one major and one minor) to initiate its transcription during viral lytic infection. The major transcription start site A, at nt 79075, and the minor transcription start site G, at nt 78994, are positioned, respectively, 361 nt and 442 nt upstream of the first AUG at nt 79436. Analysis of the region 5′ to each start site revealed TATA-like boxes, TTTA, 26 bp upstream of nt 79075, and TATT, 24 bp upstream of nt 78994.
FIG. 3.
Mapping of the ORF56 transcription start site and cleavage sites for ORF56 and ORF57 polyadenylation. (A) Mapping of the ORF56 transcription start site. The 5′-RACE was conducted on total poly(A)+ mRNA isolated from butyrate-induced JSC-1 cells using an ORF56-specific primer, Pr79608. The sequences of RACE products I and II are shown to the right of the gel, where the start sites are indicated. (B) An RNase protection assay was performed on 20 μg of total RNA from uninduced or butyrate-induced JSC-1 cells with 4 ng of 32P-labeled antisense RNA probe b (see Fig. 1A) covering KSHV genome nt 83438 to 83705. The same PCR product was also cloned into the pCR2.1 vector (Invitrogen) and served as a template for sequencing with the 32P-labeled Pr83438 primer. The protected products were separated along with sequencing ladders and a 100-bp DNA ladder on an 8% denaturing polyacrylamide gel. (C) A portion of the gel in panel B, enlarged to show the protected products and the sequencing ladders. The arrow indicates a major protected product from butyrate-induced JSC-1 cells. The diagram shown below B and C contains the calculations of alternative cleavage site usage; regulatory elements upstream [a poly(A) signal] or downstream (a GU/U-rich element) of the cleavage site are also shown. (D) Mapping of ORF56/57 cleavage sites by 3′-RACE by using a primer, Pr83254 (see its position in Fig. 1A). An RACE product with the predicted size of ∼390 bp was detected, cloned, and sequenced.
A highly conserved hexanucleotide signal, AAUAAA, at nt 83608 downstream of the ORF57 coding region, could potentially be used as a poly(A) signal for cleavage to generate the 3′ ends of nascent ORF56 or ORF57 transcripts for polyadenylation. To map the cleavage sites, a 32P-labeled antisense RNA probe from nt 83438 to 83705 that includes the putative 3′ ends of the transcripts was prepared for RPA analysis of total RNA isolated from butyrate-treated JSC-1 cells. The primer Pr83438, used to generate a DNA template for preparation of the antisense RNA probe, was also used for sequencing after end labeling with 32P. The cleavage sites were mapped by comparing the RPA products with the DNA sequencing ladders. As shown in Fig. 3B and C, the protected products detected in the total RNA of butyrate-induced JSC-1 cells displayed considerable heterogeneity, with multiple (at least five) RPA products with a 2- or 3-nt difference in size, but no products were detected in uninduced JSC-1 cells. By comparing the protection pattern with the DNA sequence ladders, five major RPA products were mapped to individual corresponding cleavage sites at least 15 nt downstream of the nt 83608 poly(A) signal (Fig. 3C, see the arrow to the left), with the predominant cleavage site (over 33%) at nt 83628 for ORF56 and ORF57 polyadenylation. Four other minor cleavage sites were also mapped downstream of the nt 83628 cleavage site just one or two nucleotides apart (Fig. 3, see diagram below the gels). The heterogeneity and precise positions of the ORF56 and ORF57 cleavage sites mapped by RPA were further confirmed by 3′-RACE analysis of poly(A)+ mRNA from butyrate-treated JSC-1 cells (Fig. 3D) by which the Pr83254 primer was used, and a RACE product with the predicted size of ∼390 bp was detected, cloned, and sequenced. The sequencing results confirmed the heterogeneity of the cleavage sites mapped by RPA.
Sixteen nucleotides downstream of the last cleavage site of ORF56 and ORF57 is a suboptimal GU/U-rich element. This element downstream of a cleavage site is often a recognition site for cleavage stimulation factor, part of the RNA polyadenylation machinery. Together, our data indicate that the KSHV ORF56/57 bicistronic RNA and ORF57 monocistronic mRNA are polyadenylated from the same poly(A) site during lytic KSHV infection.
Mapping of transcription start sites for KSHV ORF58 and ORF59 expression in lytic KSHV induction.
The primer extension, RPA, and 5′-RACE strategies were also used to map the KSHV ORF58 and ORF59 transcription start sites. Because ORF58 and ORF59 are located side-by-side on the opposite strand of the viral genome relative to ORF56 and ORF57, a sense probe for ORF58 (probe d) and a sense probe for ORF59 (probe c; see their relative positions in Fig. 4A) were synthesized for RPA by in vitro transcription from individual PCR templates. The same sense primers, Pr95717 for ORF58 and Pr96704 for ORF59, were also labeled with 32P for primer extension and DNA sequencing to map where the extension products would stop. As shown in Fig. 4B, we detected two weak extension products for ORF58 in total cell RNA obtained from butyrate-induced JSC-1 cells but did not detect these products in uninduced JSC-1 total RNA (compare lane 5 to lane 6). The major extension product for ORF58 was 105 nt in size, corresponding to a nucleotide C position (lane 4) at nt 95821 of the viral genome sense strand, and the minor extension product was 108 nt in size, also corresponding to a nucleotide C position (lane 4) at nt 95824 of the viral genome sense strand. Since ORF58 is positioned on the opposite strand, the two alternative transcription start sites of ORF58 were thus mapped by primer extension to a G at nt 95821 (major) and a G at nt 95824 (minor). RPA analysis of the total cell RNA from butyrate-induced JSC-1 cells using sense probe d confirmed the presence of two RPA products with sizes similar to those of the extension products (Fig. 4C, compare lane 5 to lane 4), but their expression levels were much lower than the level of ORF59 detected by probe d (Fig. 4C, lane 5), in spite of some RNA degradation in the reaction. The two ORF58 transcription start sites were also verified by 5′-RACE (Fig. 4D), cDNA cloning, and sequencing analysis of butyrate-induced JSC-1 poly(A)+ mRNA, showing that the majority of 5′-RACE products had a 5′ end at nt 95821 (six of nine clones), with a minority at nt 95824 (three of nine clones). Analysis of the region 5′ to each transcription start site revealed a TATA-like box, TTTA, 25 bp upstream of nt 95821 and 22 bp upstream of nt 95824.
Figure 4E shows three primer extension products, using primer Pr96704 for ORF59 from total cell RNA isolated from butyrate-induced JSC-1 cells, which were 87, 91, and 92 nt in size according to the sequencing ladders generated with the same primer (compare lane 5 to lanes 1 to 4). The three products correspond, respectively, to a T at nt 96790, a T at nt 96794, and a G at nt 96795 on the sense strand of the viral genome. Since ORF59 is also positioned on the opposite strand, the transcription start sites for ORF59 mapped by primer extension should be an A at nt 96790 and 96794 and a C at nt 96795. Products with similar sizes were also detected in RPA assays using probe c on total RNA from butyrate-induced, but not from uninduced, JSC-1 cells (Fig. 4F, compare lane 4 to lane 3). The 5′ ends of the ORF59 transcripts were further verified at nt 96790 (two of seven clones) and 96794 (five of seven clones) by sequencing analysis of cloned 5′-RACE products (Fig. 4D), but none of the seven cDNA clones sequenced showed a 5′ end at nt 96795. Altogether, the ORF59 transcription start sites were mapped by the three means to nt 96790 (minor) and nt 96794 (major), each starting with an A on the opposite strand. By scanning the region 5′ to each transcription start site, we found a potential TATA-like box, TATTTA, 23 nt upstream from nt 96794 and 27 nt upstream from nt 96790.
KSHV ORF58 and ORF59 transcripts utilize two overlapping poly(A) sites and two alternative cleavage sites for mRNA polyadenylation.
After mapping the transcription start sites for both ORF58 and ORF59, we further mapped the 3′ ends of each transcript. A search for a poly(A) signal 3′ to the ORF58 and ORF59 coding regions showed two overlapping AAUAAA hexanucleotides at nt 94492 and nt 94488, respectively, of the viral genome, a region that resides in the 3′ end of the ORF58 coding region. Although another AAUAAA hexanucleotide was identified in the middle of the ORF59 coding region at nt 96557, the lack of a GU/U-rich element downstream suggests that this is a cryptic poly(A) signal. This presumption was further supported by various assays in which we never found a truncated form of ORF59 (∼0.24 kb) derived from usage of this poly(A) signal (data not shown). The unique organization of the two overlapping poly(A) signals near the 3′ end of the ORF58 coding region implies that KSHV ORF59 mRNAs must be bicistronic.
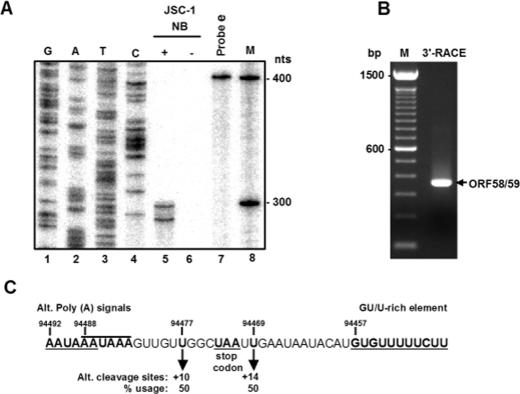
To determine whether the two putative overlapping poly(A) signals can actually be used for both ORF58 and ORF59 expression, we designed a sense probe, probe e, from nt 94368 to 94768 to cover the two putative overlapping poly(A) sites, as well as their downstream and upstream sequences. To map the selection of each poly(A) signal-mediated cleavage site for the addition of a poly(A) tail to individual transcripts, we performed an RPA analysis using this probe on total cell RNA isolated 24 h after butyrate induction of JSC-1 cells. The results in Fig. 5A show that two RNA fragments of 291 and 299 nt were protected by the probe from RNase digestion in butyrate-induced JSC-1-cell total RNA (lane 5). In contrast, no RPA products were detected in total RNA from uninduced JSC-1 cells (lane 6). By comparing the RPA products with the DNA sequencing ladder generated by the 32P-labeled antisense primer Pr94768, the two protected RNA products were determined to be at nt 94477 and nt 94469 of the viral genome, respectively. The bands were of equal density, and each product had a U residue at its 3′ end (compare lane 5 to lane 3). Thus, two alternative cleavage sites were determined to be at positions +10 and +14, respectively, downstream of the two alternative poly(A) signals. The two cleavage sites determined by RPA were further verified by 3′-RACE and cDNA sequence analysis (Fig. 5B). Consistent with the role of these cleavage sites in polyadenylation, we found an optimal GU/U-rich element, which facilitates cleavage, 12 nt downstream of the cleavage sites (Fig. 5C). Since the ORF58 stop codon at nt 94473 is positioned between the two alternative cleavage sites, the only cleavage site that could provide ORF58 transcripts for polyadenylation without losing the stop codon would be the cleavage site at nt 94469. However, either cleavage site could be selected for ORF59 expression, because the ORF58 coding region only serves as a 3′ UTR for ORF59.
FIG. 5.
Mapping of the KSHV ORF58 and ORF59 polyadenylation cleavage sites by RPA and 3′ RACE. (A) RPA was performed on 20 μg of total RNA from uninduced or butyrate-induced (24 h) JSC-1 cells with 4 ng of 32P-labeled RNA probe e (see Fig. 4A). The protected RNA products were then compared with a sequence ladder that was generated by the antisense primer Pr94768 from a plasmid containing KSHV genomic DNA from nt 94368 to 94768. The protected products were separated along with the sequencing ladders and a 100-bp DNA ladder in an 8% denaturating polyacrylamide gel. (B) Mapping of ORF58 and ORF59 cleavage sites by 3′ RACE by using a primer, Pr94768 (see its position in Fig. 4A). An RACE product with the predicted size of ∼320 bp was detected, cloned, and sequenced. (C) The sequence reading showing two alternative cleavage sites from the mappings, along with two alternative overlapping poly(A) signals upstream and one GU/U-rich element downstream of the cleavage site. The +10 position is relative to the first poly(A) signal, and the +14 position is relative to the second.
Expression of KSHV ORF58 as a monocistronic transcript and ORF59 as a bicistronic transcript in KSHV lytic induction.
By mapping and identifying the KSHV ORF58 and ORF59 transcription start sites and polyadenylation sites, we revealed that the two transcripts share the same poly(A) signal for polyadenylation despite each gene having its own transcription start site. To test whether ORF58 mRNA is monocistronic and ORF59 mRNA is bicistronic, probe e was further used for a Northern blot analysis of poly(A)+ RNA isolated from uninduced or butyrate-induced JSC-1 cells. Transcripts of ∼1.5 kb, corresponding to monocistronic ORF58 [1.35 kb plus a poly(A) tail], were approximately 18-fold less abundant than transcripts of ∼2.5 kb, corresponding to bicistronic ORF59 [2.3 kb plus a poly(A) tail] in JSC-1 cells receiving butyrate treatment (Fig. 6A and B). In contrast, no products were detected in uninduced JSC-1 cells, suggesting that the products are specific to lytic induction. Surprisingly, several additional transcripts, including an abundant RNA product of ∼6.7 kb and two less-abundant products of ∼5.7 kb and ∼3.3 kb, were detected by Northern blot analysis from butyrate-induced JSC-1 cells. Although a search for other poly(A) signals 3′ to ORF60 (small subunit of ribonucleotide reductase), ORF61 (large subunit of ribonucleotide reductase), and ORF62 (assembly and DNA maturation protein) on the antisense strand of the KSHV genome localized an AAUAAA hexamer at nt 98298 in the ORF61 coding region, none of the detected products had a size nearly matching a possible product from the usage of this hexamer for polyadenylation in our Northern blot analysis. We therefore hypothesized that these three genes might share the same poly(A) signal with ORF58 and ORF59. Accordingly, the predicted ORF60, ORF61, and ORF62 (Fig. 6A) matched the detected products in size (Fig. 6B), indicating that the three genes are all transcribed as polycistronic RNA transcripts, with ORF62/61/60/59/58 being most abundant during lytic viral induction. This observation was further confirmed by Northern blot analysis using probes from all five ORFs (Fig. 6C). Except for monocistronic ORF58, all other RNA transcripts from individual ORFs were detectable by two or more ORF-specific probes, e.g., ORF59 transcripts by two, ORF60 by three, and ORF62 by all five. In all cases, ORF61 transcripts were under the detectable level for butyrate-activated JSC-1 cells.
FIG. 6.
Transcription profiling of KSHV ORF58, ORF59, ORF60, ORF61, and ORF62 by Northern blotting. (A) Schematic diagram of predicted positions and transcription orientations of the ORFs from nt 94471 to 101194 in the KSHV genome. Dashes below the 6.7-kb transcript are individual ORF-specific probes used in this study. (B and C) Northern blot analysis. Approximate 1 μg of poly(A)+ mRNAs were isolated from uninduced (lane 1 in panel B and lanes 1, 3, 5, 7, and 9 in panel C) or butyrate-induced (lane 2 in panel B and lanes 2, 4, 6, 8, and 10 in panel C) JSC-1 cells, separated in a 1% formaldehyde-morpholinepropanesulfonic acid agarose gel, transferred onto a nylon membrane, fixed with UV light, and probed with 32P-labeled antisense probe e prepared by in vitro transcription (B) (see Fig. 4A) or a 32P-labeled oligonucleotide prepared by end labeling (C).
Enhancement of ORF56 and ORF59 expression by ORF57.
Previous studies in our laboratory and others showed that ORF57 expression promotes the expression of several viral genes, including KSHV ORF59 (a viral DNA polymerase processivity factor) (15, 23) and PAN RNA (nut-1 RNA; polyadenylated nuclear RNA with unknown function) (15), presumably at the posttranscriptional level. We examined whether this regulation is limited to ORF59 or whether other KSHV genes are targets for regulation by ORF57. In this experiment, the ORF56 ORF was cloned downstream of a cytomegalovirus immediate-early promoter and fused in frame with a C-terminal FLAG tag. Since ORF57 does not up-regulate the cytomegalovirus immediate-early promoter (15), cotransfection of ORF56 with ORF57 expression vectors allowed us to examine the posttranscriptional effect of ORF57 on ORF56 transcripts. As shown in Fig. 7A, cytoplasmic and nuclear RNAs were separated by cell fractionation and RNA isolation after cotransfection of 293 cells, as indicated by the presence of precursor rRNAs (45S), immature rRNAs (32S), and U6 only in the nuclear total RNA. When probed with an ORF56-specific probe, the amounts of ORF56 RNA in the cytoplasmic and nuclear total RNAs obtained from 293 cells cotransfected with wild-type ORF57 were roughly 14- and 19-fold, respectively, higher than the corresponding levels seen in the absence of ORF57 (compare lanes 4 and 5 to lanes 2 and 3 or lanes 6 and 7) after normalization of the signal to GAPDH RNA or U6 to the control for sample loading. The data indicate that KSHV ORF56 is subject to ORF57 regulation in vivo. Further experimental approaches by Western blotting showed that ORF57-mediated accumulation of the nuclear and cytoplasmic ORF56 RNAs led to a significant level of ORF56 protein expression (compare lanes 1 and 3 to lane 2 in Fig. 7B).
FIG. 7.
Enhancement of ORF56 expression by ORF57. (A) Detection of ORF56 transcripts by Northern blotting. Fractionated cytoplasmic (C) or nuclear (N) total RNA from 293 cells transfected with 1 μg of pVM9 (ORF56-FLAG) in the presence or absence of 0.2 μg of pVM7 (ORF57-FLAG) or pFLAG-CMV-5.1 control vector was prepared 24 h after transfection. Five micrograms of total RNA was used for Northern blot analysis with a 32P-labeled ORF56-specific probe. The same membrane was reprobed separately with a 32P-labeled GAPDH-specific probe and a U6-specific probe for sample loading and fractionation efficiency. The nuclear and cytoplasmic fractionation efficiencies were also verified by the presence or absence of 45S and 32S pre-rRNA. T, total RNA from unfractionated 293 cells. (B) Detection of ORF56-FLAG fusion protein by Western blot analysis. Protein samples were prepared after 24 h of transfection and blotted with anti-FLAG antibody to detect ORF56- and ORF57-FLAG fusions. The membrane was reprobed with anti-β-tubulin antibody for sample loading.
Using a similar strategy, we tried several times to determine whether ORF58 is also subject to ORF57 regulation but failed due to an extremely low level of ORF58 expression (below detection) even after cotransfection with ORF57.
DISCUSSION
KSHV infection proceeds through two phases. Latent KSHV infection in B-cell lines and Kaposi's sarcoma tissues is limited to the expression of only a few viral latent genes (7). Lytic KSHV infection features the production of infectious virus particles from infected cells and is inducible from the viral latent stage by the viral transactivator ORF50 (RTA) (22, 37) or by chemicals, such as n-butyrate (27), phorbol esters (for example, 12-O-tetradecanoylphorbol-13-acetate) (31), or valproic acid (17). Although the mechanism of conversion from the viral latent stage to the lytic stage remains largely unknown, it has been explored by analyzing various viral promoters and their activities under different conditions that drive viral latent gene expression during latent viral infection and viral lytic gene expression after viral induction. The initial approach has been to locate individual promoters through mapping of the transcription start sites of individual genes. Today, the transcription start sites of more than 10 KSHV genes have been mapped, including those of ORF50 (22), K8 (20), K8.1 (39), ORF57 (15), ORF73/72/K13 (34), K14/ORF74 (16), ORF34 (13), ORF35 (13), PAN (36, 47), K3 (32), and K5 (11). These mapping analyses have provided fundamental insights into the mechanisms by which the expression and transcriptional regulation of KSHV genes occur during KSHV infection and reactivation. In contrast, the transcripts of many fewer genes have been mapped to define their 3′ ends for polyadenylation, an important RNA processing event in the posttranscriptional regulation of gene expression.
To define the structure and expression of four KSHV early genes, ORF56, ORF57, ORF58, and ORF59, we first mapped the transcription start sites of ORF56, ORF58, and ORF59; the ORF57 transcription start site had been mapped previously in another laboratory (15). Subsequently, we mapped all cleavage sites for the addition of a poly(A) tail in the individual transcripts of ORF56, ORF57, ORF58, and ORF59. Our analysis demonstrated that the four genes are derived from two gene clusters on separate strands of the viral genome: ORF56/ORF57 on the sense strand and ORF59/ORF58 on the antisense strand. The genes in each cluster have their own promoters but share a common poly(A) signal for polyadenylation of each transcript. Because of this clustered gene organization, ORF56 and ORF59 are transcribed, respectively, as bicistronic ORF56/57 and ORF59/58 transcripts, whereas the ORF57 and ORF58 transcripts are monocistronic. Except for the ORF57 transcription start site, which was previously mapped to a T nucleotide (15), all transcription start sites mapped for ORF56, ORF58, and ORF59 in our study were initiated from a purine (A or G), which is common for the initiation of eukaryotic gene transcription (2). A TATA-like box, TTTA, positioned 22 to 26 bp upstream of the individual start site, is another common feature among the three genes. In metazoans, the TATA box is typically located about 25 to 30 nt upstream of the transcription start site (2). Interestingly, each of the four mapped genes could initiate its transcription from two alternative transcription start sites.
RNA polyadenylation plays an important role in the control of viral and eukaryotic gene expression and involves cleavage of the nascent transcript and addition of a poly(A) tail with 150 to 200 adenylate residues. Cleavage and polyadenylation are tightly coupled events that are triggered through recognition of three RNA signals by the cellular polyadenylation machinery: a highly conserved AAUAAA hexamer, a cleavage site generally positioned 10 to 30 nt downstream of AAUAAA, and a GU-rich element that is ≤30 nt further downstream of the cleavage site (8). In this study, our extensive 3′ end mapping revealed KSHV ORF56 and ORF57 transcripts with heterogeneous 3′ ends derived from five tandem cleavage sites starting 15 nt downstream of the canonical AAUAAA sequence. A major cleavage site for the ORF56 and ORF57 transcripts was mapped to nt 83628. Four minor cleavage sites, including one at nt 83636 that was reported previously (15), were mapped downstream of the major site, with only one or two nucleotides separating them. The heterogeneity of the cleavage site usage in ORF56 and ORF57 transcripts can presumably be ascribed to the weak content of the GU/U-rich element downstream and may simply reflect selection flexibility, as is common in mammals (29). Recognition of the GU/U-rich element by cleavage stimulation factor in the cellular polyadenylation machinery will stabilize the binding of cleavage and polyadenylation specificity factor to AAUAAA (6, 44).
The maturation of the 3′ end of the KSHV ORF58 and ORF59 transcripts involves cleavage of the nascent transcripts at two cleavage sites that are separated by 7 nt. Since the ORF58 stop codon resides between the two cleavage sites, the only one that can be used for ORF58 polyadenylation is the one immediately downstream of the stop codon, because selection of the cleavage site upstream of the stop codon would disrupt ORF58 translation termination and restrict ORF58 expression. However, either cleavage site could be efficiently selected for ORF59 polyadenylation, because the ORF58 coding region is part of the 3′ UTR of the bicistronic ORF59 transcript, giving a selection advantage for ORF59 expression. This was further supported by the presence of bicistronic ORF59 transcripts in much greater abundance than monocistronic ORF58 in butyrate-induced JSC-1 cells. In addition, our expression analysis demonstrated that the polyadenylation sites mapped for ORF58 and ORF59 expression may also be used for ORF60, ORF61, and ORF62 expression, resulting in production of multiple polycistronic transcripts from this region. The similar sizes of the transcripts from this region were reported from another study (34). The polycistronic nature of the transcripts from this region was taken into account when the transcription start sites of ORF58 and ORF59 were determined.
The finding that both ORF56 and ORF59, but not ORF58, are targets of ORF57 implies involvement of ORF57 in KSHV DNA replication during lytic viral infection. However, exactly how ORF57 selectively enhances expression of both genes at the posttranscriptional level remains to be understood. Previous studies in other laboratories have indicated that ORF57 promotes viral RNA export from the nucleus to the cytoplasm through interactions with the cellular export factor REF (25, 26). However, our recent study showed that the binding of ORF57 to REF is not essential for promoting viral RNA export, and ORF57 acts to enhance accumulation of ORF59 RNA in the nucleus (23). Consistent with this, similar ORF57 activity was observed in promoting ORF56 expression in the nucleus, suggesting that ORF57 might play an important role in nuclear viral RNA processing.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, S. R., and B. Chandran. 2000. Characterization of human herpesvirus 8 ORF59 protein (PF-8) and mapping of the processivity and viral DNA polymerase-interacting domains. J. Virol. 74:10920-10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., M. Ciustea, and R. P. Ricciardi. 2005. Processivity factor of KSHV contains a nuclear localization signal and binding domains for transporting viral DNA polymerase into the nucleus. Virology 340:183-191. [DOI] [PubMed] [Google Scholar]
- 6.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 7.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilmartin, G. M. 2005. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev. 19:2517-2521. [DOI] [PubMed] [Google Scholar]
- 9.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque, M., D. A. Davis, V. Wang, I. Widmer, and R. Yarchoan. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: relevance to lytic induction by hypoxia. J. Virol. 77:6761-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque, M., V. Wang, D. A. Davis, Z. M. Zheng, and R. Yarchoan. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated herpesvirus ORF34-37 gene cluster. J. Virol. 80:7037-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirshner, J. R., K. Staskus, A. Haase, M. Lagunoff, and D. Ganem. 1999. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klass, C. M., L. T. Krug, V. P. Pozharskaya, and M. K. Offermann. 2005. The targeting of primary effusion lymphoma cells for apoptosis by inducing lytic replication of human herpesvirus 8 while blocking virus production. Blood 105:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagenaur, L. A., and J. M. Palefsky. 1999. Regulation of Epstein-Barr virus promoters in oral epithelial cells and lymphocytes. J. Virol. 73:6566-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, K., C. Y. Dai, and R. P. Ricciardi. 1998. Cloning and functional analysis of Kaposi's sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J. Virol. 72:6228-6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majerciak, V., K. Yamanegi, S. H. Nie, and Z. M. Zheng. 2006. Structural and functional analyses of Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365-28378. [DOI] [PubMed] [Google Scholar]
- 24.Malik, P., D. J. Blackbourn, M. F. Cheng, G. S. Hayward, and J. B. Clements. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155-2166. [DOI] [PubMed] [Google Scholar]
- 25.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 26.Malik, P., and J. B. Clements. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32:5553-5569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 28.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauws, E., A. H. van Kampen, S. A. van de Graaf, J. J. de Vijlder, and C. Ris-Stalpers. 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids. Res. 29:1690-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 32.Rimessi, P., A. Bonaccorsi, M. Sturzl, M. Fabris, E. Brocca-Cofano, A. Caputo, G. Melucci-Vigo, M. Falchi, A. Cafaro, E. Cassai, B. Ensoli, and P. Monini. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 39.Tang, S., K. Yamanegi, and Z. M. Zheng. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J. Virol. 78:2609-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 277:14547-14556. [DOI] [PubMed] [Google Scholar]
- 41.Taylor, J. L., H. N. Bennett, B. A. Snyder, P. S. Moore, and Y. Chang. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol. 79:15099-15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamanegi, K., S. Tang, and Z. M. Zheng. 2005. Kaposi's sarcoma-associated herpesvirus K8β is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8α (K-bZIP) to induce p21 and p53 and blocks K8α-CDK2 interaction. J. Virol. 79:14207-14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, Z. M. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng, Z. M., P. He, and C. C. Baker. 1996. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J. Virol. 70:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]