Abstract
The herpesvirus tegument is a layer of viral and cellular proteins located between the capsid and envelope of the virion. The VP1/2 tegument protein is critical for the propagation of all herpesviruses examined. Using an infectious clone of the alphaherpesvirus pseudorabies virus, we have made a collection of truncation and in-frame deletion mutations within the VP1/2 gene (UL36) and examined the resulting viruses for spread between cells. We found that the majority of the VP1/2 protein either was essential for virus propagation or did not tolerate large deletions. A recently described amino-terminal deubiquitinase-encoding domain was dispensable for alphaherpesvirus propagation, but the rate of propagation in an epithelial cell line and the frequency of transport in axons of primary sensory neurons were both reduced. We mapped one essential domain to a conserved sequence at the VP1/2 carboxy terminus and demonstrated that this domain sufficient to redirect the green fluorescent protein to capsid assemblons in nuclei of infected cells.
All herpesviruses share a common structure that consists of an icosahedral capsid surrounded by a layer of additional proteins, collectively called the tegument. The capsid and tegument are enclosed in an envelope consisting of a lipid bilayer and membrane-associated proteins. Although the capsid and envelope are similar to components found in many virus families, the herpesvirus tegument is rather unique.
The tegument is composed of both virally encoded proteins and proteins derived from the host cell (7, 21, 30, 34, 36). Herpesviruses encode a dozen or more tegument proteins, and a fraction of these are conserved in the alpha-, beta-, and gammaherpesvirus subfamilies (these are sometimes referred to as the “ancient” tegument proteins) (5, 6). Among the conserved tegument proteins is a large protein of >300 kDa. In the alphaherpesviruses herpes simplex virus (HSV) and pseudorabies virus (PRV), the large tegument protein is referred to as VP1/2 (although the names ICP1, ICP1/2, and VP1-3 are also used) and is encoded by the UL36 gene (20).
VP1/2 is essential for alphaherpesvirus propagation and is suggested to participate in a diverse array of functions (16, 24). Based on studies done with PRV in epithelial cells and sensory neurons, after entering a cell, capsids shed the majority of the tegument layer but notably retain the VP1/2 protein during translocation from the plasma membrane to the nucleus (12, 17). Studies using HSV in epithelial cells demonstrate that once capsids are docked at nuclear pores, viruses encoding a temperature-sensitive form of VP1/2 fail to deliver the viral DNA into the nucleus at nonpermissive temperatures (2). HSV VP1/2 isolated from infected epithelial cell nuclear extracts binds to viral DNA probes and is suggested to participate in viral genome cleavage and packaging, and nuclear capsids in PRV-infected epithelial cells have a reduced ability to exit the nucleus in the absence of VP1/2 (4, 18). In the cytoplasm, VP1/2 is required for the transport of capsids along microtubules in PRV-infected epithelial cells (18). Finally, VP1/2 is required for the envelopment of cytosolic HSV and PRV capsids into mature virions (8, 11).
The specific activities that VP1/2 performs in the above-described pathways are poorly understood. To date, VP1/2 is recognized to have two activities. First, the N terminus of the protein is a cysteine protease that can function as a deubiquitinase, and this activity is conserved throughout the herpesvirus family (13, 22, 32). Second, VP1/2 binds at least two other tegument proteins: UL37 (shown with HSV and PRV) and VP16 (shown with HSV) (14, 31). UL37 and VP16, in turn, appear to recruit additional tegument proteins onto capsid surfaces, which leads to efficient cytoplasmic capsid envelopment (10, 15). However, VP1/2 binding to UL37 is not essential for propagation of PRV (11).
Using an infectious clone of PRV, we have made a collection of viruses harboring truncation and in-frame deletion mutations within the UL36 gene. The majority of mutants failed to propagate; however, the N terminus of VP1/2 showed decreased growth kinetics, indicating that the deubiquitinase is an important but nonessential activity in the alphaherpesviruses. Finally, we mapped one essential domain to a small conserved 62-amino-acid (aa) sequence in the C terminus of the VP1/2 protein and showed that this domain is targeted to nuclear sites of capsid assembly, referred to as assemblons (33).
MATERIALS AND METHODS
Viruses.
Viruses encoding either the green fluorescent protein (GFP) or monomeric red fluorescence protein (mRFP1) fused to the capsid VP26 protein (PRV-GS443 and PRV-GS847) and their respective infectious clones (pGS443 and pGS847) were previously described (26, 27). Truncations of the UL36 gene were made using a RED-GAM/Flp-recombinase protocol that we have previously described (18). RED-GAM mutagenesis was used to insert first a stop codon and then a kanamycin resistance cassette flanked by Flp recombination target (FRT) sites at the desired location within the UL36 coding sequence. The kanamycin resistance cassette was then excised by Flp-mediated recombination, leaving behind a 37-nucleotide (nt) insertion that consisted of the stop codon immediately followed by a single FRT site. In the case of truncations made after codons 3022, 3054, and 3078, the UL36 coding sequence downstream of the insertion was replaced by the 37-nt insertion. In-frame deletions within the UL36 gene were also made using the RED-GAM/Flp method. However, stop codons were not included in the primer sequences, resulting in a single FRT site residing at the site of deletion, with two additional nucleotides present to maintain the reading frame. As a result, the 12 amino acids GSSYSLESIGTS are encoded at the site of each in-frame deletion. Primer sequences used for RED-GAM mutagenesis are available upon request. All RED-GAM/Flp recombinations were introduced into the pGS443 (GFP-capsid) full-length infectious clone.
A virus carrying the in-frame deletion lacking codons 6 to 225 of the UL36 gene was remade using a two-step RED-GAM recombination protocol (29). Using this protocol, the deletion was made without leaving behind an FRT site in the viral genome (codons 5 and 226 were abutted directly). The primers used for this protocol were 5′-CCCACGCGCGTGTGTTATTTCAGCCATGACGGCCGACGCGCCCTCCGTGCACCCGATGGCAGGATGACGACGATAAGTAGGG and 5′-CCGCCGGGTCCGGCGGCAGGGCCATCGGGTGCACGGAGGGCGCGTCGGCCGTCATGGCTGCAACCAATTAACCAATTCTGA TTAG. The primers contain homology to the UL36 gene both upstream and downstream of the sequence to be deleted and homology to the pEPkan-S template plasmid (underlined) (29). The recombination was used to modify the pGS847 infectious clone, which encodes the previously described mRFP1-VP26 (red capsid) allele (27). The resulting recombinant infectious clone, pGS1651, was confirmed by sequencing. Transfection of pGS1651 into PK15 cells resulted in production of virus PRV-GS1651, which typically grew to a titer of 1 × 105 PFU/ml. To make a revertant of the PRV-GS1651 virus, a segment of the UL36 gene was subcloned with 616 nt of flanking sequence upstream and 1,822 nt downstream of the deletion (pGS765). This construct was transiently transfected into Vero cells, and the cells were infected with PRV-GS1651 the next day. Virus was harvested at 3 days postinfection and passaged two additional times, and a single isolate was plaque purified. The resulting virus stock, PRV-GS1651R, was confirmed to carry the full-length UL36 gene by restriction analysis and DNA sequencing.
Subcloning.
The 3′-end 189 nt of the UL36 gene (encoding the carboxy-terminal 62 amino acids and stop codon of VP1/2) was amplified with primers 5′-GCAGATCTCGCGTGGTGGAGTCG and 5′-GCAAGCTTAACCGAGAATCAGGCG. The resulting PCR product was subcloned into pEGFP-C1 (Clontech) by using BglII and HindIII restriction sites (underlined), resulting in pGS1163. The UL36 insert was confirmed by DNA sequencing. The full-length UL36 gene was isolated from the viral genome by using a variation of the RED-GAM recombination procedure, which allowed us to avoid the possibility of introducing PCR errors into the ∼9.3-kbp open reading frame. Briefly, a plasmid encoding an R6K origin of replication and beta-lactamase (ampicillin resistance) was PCR amplified with primers 5′-CCAAATAAAAAGATTTTTCCCCCACGCGCGTGTGTTATTTCAGCCGATTTTTATCGAATTCGTCATCCATATCACCACG and 5′-ACTGATTACGATAGCCGACGACCACCGCGTCGGCCGTCAAAGCTTCCACATGTGGAATTCCCAT (underlined sequences encode BsaBI, EcoRI and HindIII restriction sites). The resulting PCR product was recombined immediately upstream of the UL36 start methionine codon by using RED-GAM as described above (the 5′ end of each primer encodes the homology to the UL36 gene). A resulting ampicillin-resistant recombinant was digested with BsaBI to liberate a viral genomic fragment containing the PCR product adjacent to the UL36 gene, as well as downstream sequences that include the UL35 gene and a small segment of the UL34 gene (which encodes a second BsaBI restriction site). Self-ligation of the fragment resulted in a plasmid that was cloned into S17λpir Escherichia coli, allowing for replication based off the R6K origin. The UL36 gene was then subcloned in frame downstream of the enhanced GFP open reading frame within the pEGFP-C1 mammalian expression vector (Clontech) by using the primer-encoded EcoRI and HindIII sites. The resulting plasmid, pGS1521, expresses GFP fused to aa 2 to 3084 of VP1/2 (the entire protein minus the start methionine).
Cells, transfection, and infection.
Pig kidney epithelial cells (PK15) were used for viral propagation, including assays for single-step growth as previously described (28). Infectious clones of PRV were isolated following transfection of infectious-clone plasmids into PK15 cells as previously described (17). A high-titer stock of the virus lacking amino acids 6 to 225 (designated Δ6-225aa) (PRV-GS1651; see above) for use in single-step growth analysis was prepared by infecting 9.5 × 107 PK15 cells in roller bottles at a multiplicity of infection (MOI) of 0.01. At 3 days postinfection, the culture medium was harvested and cleared of cells by centrifugation at 3,950 × g for 30 min, and virions were concentrated by pelleting through a 10% Nycodenz cushion at 13,000 rpm in an SW28 rotor (Beckman). Pellets were resuspended in approximately 150 μl Dulbecco's modified Eagle's medium supplemented with 2% bovine growth serum (HyClone), and titers were determined using African green monkey kidney epithelial cells (Vero) by plaque assay. This typically resulted in a PRV-GS1651 titer of 1 × 107 PFU/ml.
Vero cells were used for subcellular localization studies and were transfected with polyethylenimine (Polysciences, catalog no. 23966) as follows: to 1 ml Dulbecco's modified Eagle's medium, 45 μl polyethylenimine solution (1 μg/ml) and 45 μl plasmid DNA (300 μg/ml) were added, mixed, and incubated at room temperature for 10 min. The mixture was added to a 10-cm dish of subconfluent cells and incubated for 8 to 18 h. The cells were then passaged onto coverslips for imaging. In a subset of experiments, the cells were infected with an RFP-capsid virus (PRV-GS847; see above) at an MOI of 10 prior to imaging at between 10 and 12 h postinfection (hpi).
Chicken embryonic dorsal root sensory neurons were isolated and cultured for studies of axonal transport, as previously described (23, 26). Neurons were infected after 2 to 3 days in culture. Time-lapse imaging of capsid transport following entry was done at between 0 and 1 hpi. Capsid transport velocities were measured as the instantaneous distance traveled between successive captured frames divided by the frame rate of 50 ms/frame. Capsid egress in axons following replication was imaged at 20 hpi, and axons were scored for the presence or absolute absence of capsids.
Fluorescence microscopy.
Wide-field fluorescence imaging was performed with living cells, using an inverted wide-field Nikon Eclipse TE2000-U microscope. The microscope was housed in a 37°C environmental box (Life Imaging Services) and was fitted with a Cascade:650 charge-coupled device (Roper Scientific). The Metamorph software package was used for image acquisition. Fluorescence and phase-contrast images used to examine cell-to-cell viral transmission were captured with a 10×/0.3 numerical aperture (NA) objective. A 60X/1.4NA objective was used to acquire GFP-VP1/2 or RFP-capsid fluorescence from cells in sealed chambers as previously described (26). Statistical analysis of capsid transport velocities was carried out using the Prism software package (GraphPad Software).
Confocal image Z-series were acquired on a LSM510 Meta microscope using a 63X/1.4NA objective with 488-nm argon, 543-nm HeNe, and 405-nm diode laser lines. Cells were incubated in 5 μg/ml Hoechst 33342 (Molecular Probes) for 30 min prior to imaging.
Western blot analysis, antibodies, and densitometry.
VP5 and UL37 antigens were detected in PK15 cells infected with either PRV-GS847 or PRV-GS1651 at an MOI of <0.1. Infections were carried out in 10-cm dishes of confluent cells, which were harvested once all cells displayed a cytopathic effect. Cells were lysed in 1 ml 2× final sample buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100) containing 10% β-mercaptoethanol, and the samples were boiled for 5 min prior to electrophoresis of 10 μl of each sample through an 8% sodium dodecyl sulfate-polyacrylamide gel. Proteins were subsequently transferred onto a Hybond-P membrane (Amersham Biosciences), and Western blot analysis was performed as previously described (25). 3C10 is a mouse monoclonal antibody raised against the PRV VP5 protein and used at 1:1,000 dilution and was a kind gift of Lynn Enquist. D1789 is a rabbit antiserum raised against a peptide derived from the PRV UL37 sequence (REAADRVLGDYHE) and was used at a 1:2,500 dilution. The secondary goat anti-mouse and anti-rabbit horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch) were used at a 1:10,000 dilution. Horseradish peroxidase was detected with a luminol-coumeric acid-H2O2 chemiluminescence solution and exposed to film. For detection of VP5 and UL37 antigens in extracellular virions, equal volumes of purified virions (see above) and 2× final sample buffer were mixed and boiled for 5 min, and 15 μl of each sample was separated by electrophoresis on a 4 to 20% sodium dodecyl sulfate-polyacrylamide gel. Western blot analysis was carried out as described above.
Relative expression levels of UL37 protein were determined by digitizing exposed films with an EDAS 290 documentation system (Kodak). The intensities of UL37 and VP5 protein bands were quantitated using the ImageJ software package (1). UL37 protein expression was normalized using VP5 expression as a baseline, and the relative expression of UL37 was calculated as a percentage relative to virus encoding wild-type VP1/2.
RESULTS
Analysis of viruses encoding VP1/2 truncation mutations.
As a first step toward identifying regions of the VP1/2 protein that are important for the alphaherpesvirus infectious cycle, we aligned the predicted amino acid sequences of 10 different herpesvirus VP1/2 homologues. Although the sequences were largely conserved, two regions were notably divergent between viruses. These two regions, aa 226 to 299 and aa 2026 to 2970 in the PRV VP1/2 sequence, had high proline contents (33.78% and 28.15%, respectively) and are referred to here as proline-rich regions 1 and 2 (PRR1 and PRR2). Within the remainder of VP1/2, we noted an alanine-rich region (ARR) from aa 397 to 1293 that was, conversely, low in proline content (25.64% alanine and 3.68% proline). Because VP1/2 has no significant homologies to other proteins, the proline- and alanine-rich regions were used as the basis for mutational design.
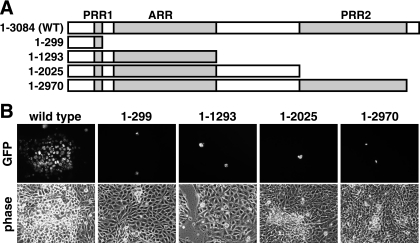
A series of four stop codons were independently introduced into the UL36 gene of the PRV infectious clone, positioned at either the beginning or the end of a PRR or ARR coding sequence (Fig. 1A). The infectious clone used in these studies encodes a GFP-VP26 fusion protein, which produces the GFP-capsid virus described previously (26). To examine the impact of each mutation on viral propagation, each clone was transfected into PK15 cells and GFP fluorescence and cytopathic effects were monitored. Although transfected cells became evident based on GFP-capsid emissions, viral spread to neighboring cells was absent with each of the mutant viruses. This is in contrast to the parent GFP-capsid virus, which spread to all cells within 3 days posttransfection (Fig. 1B). These findings indicated that the carboxy-terminal 114 amino acids of VP1/2 were essential for viral propagation.
FIG. 1.
Propagation of PRV encoding VP1/2 truncations. (A) Illustrations of predicted VP1/2 truncations expressed from recombinant PRV carrying stop codons in the UL36 gene. WT, wild type. (B) Detection of infected cells by GFP-capsid fluorescence at 2 days posttransfection with the corresponding infectious clone. Isolated fluorescent cells in the mutants indicate a lack of virus spread from the initially transfected cells.
Analysis of viruses carrying VP1/2 in-frame deletion mutations.
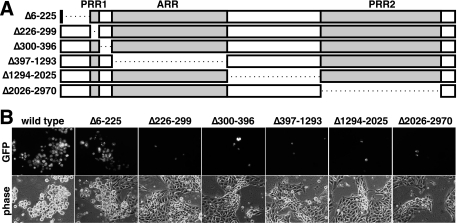
To determine if other regions of the VP1/2 tegument protein are required for viral propagation, a series of in-frame deletions were introduced into the UL36 gene (Fig. 2A). Viruses carrying deletions resulting in expression of VP1/2 proteins lacking either PRR1, PRR2, ARR, or the region between PRR1 and ARR or between ARR and PRR2 all failed to propagate in PK15 cells (Fig. 2B). In contrast, a virus encoding VP1/2 lacking the amino terminus (Δ6-225aa) was found to be viable.
FIG. 2.
Propagation of PRV encoding VP1/2 deletions. (A) Illustrations of predicted VP1/2 deletions expressed from recombinant PRV carrying in-frame deletions in the UL36 gene. (B) Detection of infected cells by GFP-capsid fluorescence 3 at days posttransfection with the corresponding infectious clone.
Characterization of a virus lacking the amino terminus of VP1/2.
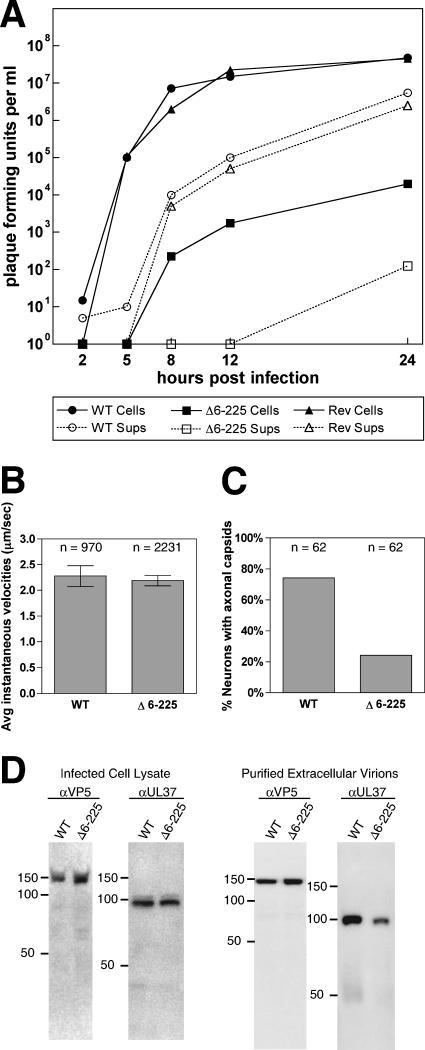
We noted that the Δ6-225aa virus propagated poorly relative to the parent virus. To confirm that this defect was not due to the FRT sequence present at the deletion site, a new virus, PRV-GS1651, that carried a seamless in-frame deletion of codons 6 to 225 was made using a recently described two-step recombination protocol (29). PRV-GS1651 grew to a typical titer of 1 × 105 PFU/ml (reduced approximately 3 logs from that of the parent virus). Single-step growth analysis showed a substantial decrease in both cell-associated and extracellular virus production. A revertant of PRV-GS1651 propagated with wild-type kinetics (Fig. 3A). The decrease in viral propagation did not affect the ability of viral particles to move intracellularly in axons of sensory neurons upon entering a cell: retrograde transport kinetics were indistinguishable from those of the VP1/2 wild-type virus (Fig. 3B). However, we noted a defect in the localization of progeny viral particles in axons following replication. In the majority of infected neurons, no capsids were detected egressing in axons of sensory neurons (Fig. 3C) (26). Overall, we observed a 50% reduction in egressing axonal capsids at a late time point postinfection (20 hpi).
FIG. 3.
Characterization of PRV lacking the VP1/2 amino terminus. (A) Single-step propagation kinetics. The following viruses were harvested from the medium (Sups) (dashed lines and open symbols) and PK15 cells (solid lines and filled symbols) at the indicated times: PRV-GS847 (wild type [WT] VP1/2; circles), PRV-GS1651 (Δ6-225 VP1/2; squares), and PRV-GS1651R (revertant [Rev] VP1/2; triangles). (B) Rates of retrograde capsid transport in axons following infection of primary sensory neurons in culture. Error bars are standard errors of the means. (C) Frequency of de novo assembled capsids observed in axons of cultured sensory neurons follow replication (20 hpi). (D) Western blot analysis of the level of expression of the UL37 tegument protein in infected cells and released extracellular virions.
Based on our previous observations, the deletion in PRV-GS1651 may decrease upstream UL37 expression, which in turn may contribute to the defects seen with this virus (18). Western blot analysis of UL37 expression confirmed that UL37 expression was reduced: UL37 expression levels in cells infected with Δ6-225aa were 54% of those in the wild type. This decreased expression resulted in a corresponding drop in the amount of UL37 incorporated into released viral particles (43% relative to the wild type) (Fig. 3D). Nevertheless, these results indicate that, similar to the case for betaherpesviruses, the alphaherpesvirus cysteine protease present in the amino terminus of VP1/2 is dispensable for viral propagation in cell culture (13, 22, 32).
A conserved carboxy terminus of VP1/2 is targeted to nuclear capsid assemblies.
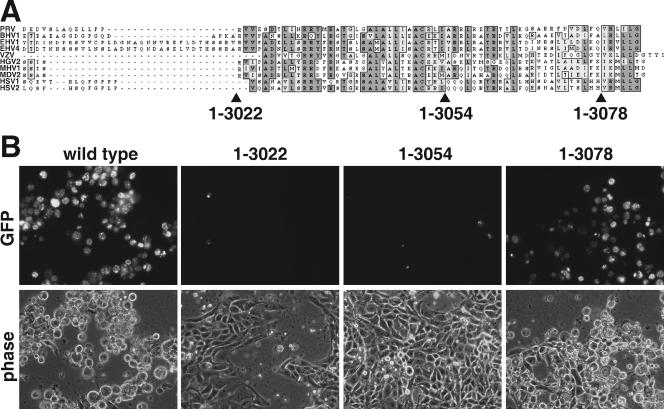
To further map the carboxy-terminal essential domain, three additional stop codons were inserted into the 3′ end of the UL36 gene (Fig. 4A). The last 62 amino acids of VP1/2 are reasonably conserved; therefore, the stop codons were positioned at the beginning, at the middle, and near the end of this region. Following transfection of the infectious clones into PK15 cells, only the clone encoding aa 1 to 3078 resulted in a productive infection (Fig. 4B). These results confirm that the carboxy terminus performs an essential function, as removal of only the last six amino acids allowed for a productive infection (which produced a viral stock with a titer of 1 × 107 PFU/ml). The essential domain was further confirmed by introducing two small in-frame deletions in the UL36 gene to produce viruses that express VP1/2 lacking either aa 3024 to 3052 or aa 3053 to 3077. Upon transfection into PK15 cells, both mutants failed to spread from cell to cell (data not shown).
FIG. 4.
Propagation of PRV encoding carboxy-terminal truncations in VP1/2. (A) Alignment of the predicted amino acid sequences of the VP1/2 carboxy termini from 10 alphaherpesviruses. Sites of three PRV truncation mutations are indicated. (B) Detection of infected cells by GFP-capsid fluorescence at 3 days posttransfection with the corresponding infectious clone.
Attempts to propagate viruses with truncations or deletions in the UL36 gene by using a previously described VP1/2-complementing cell line were unsuccessful due to a high frequency of recombination leading to repair of the viral genomes (data not shown) (18). As an alternative approach to examining the function of the carboxy-terminal domain, the 3′ end of the UL36 gene was subcloned in frame into a GFP expression vector. The resulting construct encoded GFP fused to the amino terminus of the VP1/2 fragment from aa 3023 to 3084 [designated GFP-VP1/2(aa3023-3084)].
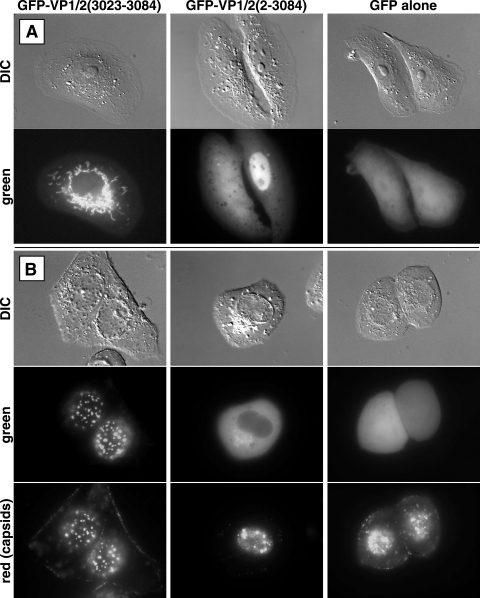
In Vero cells transiently transfected with the GFP-VP1/2(aa3023-3084) construct, fluorescence was exclusively cytoplasmic and was localized to long tubular structures (Fig. 5A). These structures were determined to be mitochondria, based on a MitoTracker fluorescence counterstain (Molecular Probes) (data not shown). The relevance of this localization was not clear. GFP fused to the full-length VP1/2 protein, [GFP-VP1/2(aa2-3084)], never displayed mitochondrial localization and was instead diffuse throughout cells, often showing a enriched nuclear localization (Fig. 5A). However, infection of the GFP-VP1/2(aa3023-3084)-transfected cells with a virus that encodes red fluorescent capsids but is otherwise unaltered (PRV-GS847) resulted in a dramatic redistribution of GFP fluorescence to capsid assemblies within the nucleus, previously termed “assemblons” (Fig. 5B and 6) (9, 27, 33). All infected cells containing capsid assemblons displayed this redistribution of GFP-VP1/2(aa3023-3084), whereas the fusion was never observed in the nuclei of uninfected cells. All mRFP1-capsid assemblies emitted detectable GFP-VP1/2(aa3023-3084) fluorescence, although the amount of GFP fluorescence associated with capsid assemblies varied. When examined by confocal microscopy, the nuclear GFP and mRFP1 emissions were confirmed to colocalize and were noted to be predominantly clustered near the nuclear rim, consistent with previous observations of HSV capsid assemblons (see movie S1 in the supplemental material) (33). Unlike cells transfected with the GFP-VP1/2(aa3023-3084) construct, we noted that the majority of GFP-V1/2(aa2-3084)-transfected cells were not permissive for infection. In those that were infected, the full-length GFP-VP1/2(aa2-3084) did not localize to assemblons upon infection and was in fact excluded from the nucleus altogether (Fig. 5). GFP protein alone (not fused to VP1/2 sequences) was diffuse throughout the cytoplasm and nuclei of transfected cells and did not relocalize upon infection with PRV.
FIG. 5.
Expression of VP1/2 fused to GFP in Vero cells. Cells transfected with either GFP-VP1/2(aa3023-3084), GFP-VP1/2(aa2-3084), or unfused GFP are shown. (A) Uninfected cells; (B) cells subsequently infected with PRV expressing red-fluorescent capsids. Images were captured from a basal focal plane of the cells, thereby revealing capsid assemblies near the bottom of the nucleus. DIC, differential interference contrast.
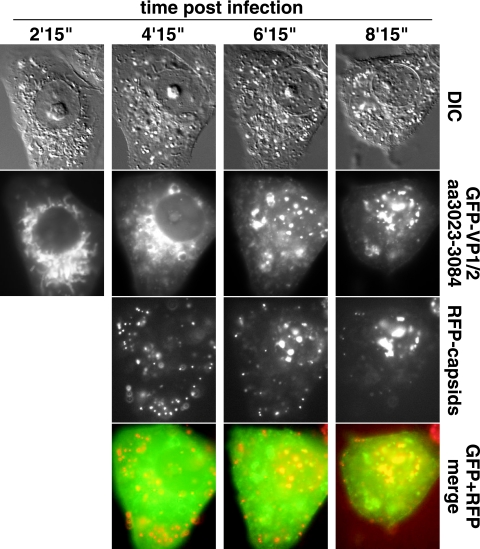
FIG. 6.
Time course of GFP-VP1/2(aa3023-3084) redistribution. A Vero cell transiently transfected with the GFP-VP1/2(aa3023-3084) construct was infected with PRV-GS847 (expresses mRFP1-capsids) and imaged for GFP emissions at 2-h intervals beginning at 2 h 15 min postinfection. RFP-capsid images were captured beginning at 4.25 hpi. DIC, differential interference contrast.
DISCUSSION
VP1/2 is the largest herpesvirus-encoded protein, is conserved throughout the Herpesviridae, and is critical for the propagation of all herpesviruses examined (16, 24, 35). Little is known regarding the specific functions performed by this protein. Recently, the amino terminus of VP1/2 was found to have a conserved proteolytic activity that functions as a deubiquitinase (13, 22). This activity is not required for betaherpesvirus propagation in cell culture, which is consistent with our findings obtained using an alphaherpesvirus (32). In fact, our data demonstrate that the VP1/2 amino-terminal region (aa 6 to 225) is dispensable as a whole, but the kinetics of propagation are decreased and viral titers are typically reduced 1,000-fold relative to those of the wild type. The decrease in propagation is not associated with a decrease in the kinetics of retrograde transport of incoming viral particles to the nucleus, as assayed in cultured neurons. The decrease in propagation may in part be attributed to a polar effect resulting in decreased expression of the upstream UL37 gene, which encodes a tegument protein that is in turn incorporated at reduced levels into extracellular virions. As a side note, overexpression of UL37 was previously shown not to affect the copy number of the UL37 tegument protein in extracellular virions, suggesting that the abundance of the protein in virions is controlled during assembly (19). Our finding that reduced UL37 expression leads to reduced UL37 protein incorporation in virions suggests that protein abundance is restricted only by an upward limit of UL37 incorporation.
Unlike removal of the amino terminus, removal of other regions of the VP1/2 protein failed to result in productive viral infections. All viruses propagated and spread cell to cell to various degrees on a previously described UL36-complementing cell line, indicating that loss of VP1/2 functionality was the cause of lethality in each case (data not shown) (18). We cannot currently discriminate whether loss of viability corresponds to losses of essential functions throughout the remainder of VP1/2 or to a general loss of protein folding and stability. However, mutants with truncations and in-frame deletions which removed all or part of the last 62 aa of the carboxy terminus are nonviable, consistent with this conserved region of the protein performing an essential function during infection. In support of this conclusion, a recent study also identified the carboxy 62 aa of VP1/2 as being essential for propagation of PRV (3). We find that fusion of the carboxy-terminal 62 aa of VP1/2 to GFP is sufficient to redirect GFP from the cytoplasm to the nucleus and for it to associate with dynamic sites of capsid assembly, referred to as assemblons, during infection (9, 33). Interestingly, full-length VP1/2 did not display this property, suggesting either that the carboxy terminus is sequestered within the context of the intact VP1/2 protein or that the full-length protein is simply excluded from the nucleus during infection. These findings reveal a function for the carboxy terminus of VP1/2 that likely explains the essential property of this domain. We note that the carboxy 62-aa domain fused to GFP was excluded from the nucleus in uninfected cells, but time-lapse imaging demonstrated that this domain was actively recruited into the nucleus upon infection. This active relocalization upon infection indicates that a viral product, such as a capsid or capsid-associated protein, may associate with the VP1/2 carboxy terminus. Identifying the protein interactions that link the capsid and tegument and identifying where in the cell tegument assembly is initiated will be fundamental to furthering our understanding of herpesvirus morphogenesis.
Supplementary Material
Acknowledgments
We are grateful to Mindy Leelawong for help with imaging neuron infections, Jennifer Klabis for help with transfections and infections, Sarah Haverlock-Moyns and Kelly Coller for help with subcloning, Lynn Enquist for the gift of VP5 antibody, Teng-Leong Chew and the Cell Imaging Facility at Northwestern University Feinberg School of Medicine for help with confocal imaging, and Sarah Rice for helpful discussions of the data.
This work was supported by NIH grant 1RO1AI056346 (to G.A.S.). J.I.L. was supported by viral replication training grant NIHT32AI060523.
Footnotes
Published ahead of print on 27 September 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, J., and B. Roizman. 1989. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J. Virol. 63:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, C., A. J. Davison, A. R. MacLean, N. S. Taus, and J. D. Baines. 2000. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 74:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, A. J. 1993. Herpesvirus genes. Rev. Med. Virol. 3:237-244. [Google Scholar]
- 7.del Rio, T., C. J. DeCoste, and L. W. Enquist. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429-431. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 14.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLauchlan, J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J Gen. Virol. 78:189-194. [DOI] [PubMed] [Google Scholar]
- 20.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 66:7581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, C. L. 1998. Cultures from chick peripheral ganglia, p. 261-287. In G. Banker and K. Goslin (ed.), Culturing nerve cells. MIT Press, Cambridge, Mass.
- 24.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 72:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 30.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, M. L., and C. H. Chen. 1998. Evidence for the internal location of actin in the pseudorabies virion. Virus Res. 56:191-197. [DOI] [PubMed] [Google Scholar]
- 35.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.