Abstract
Dominant epitope-specific CD8+ T-lymphocyte responses play a central role in controlling viral spread. We explored the basis for the development of this focused immune response in simian immunodeficiency virus (SIV)- and simian-human immunodeficiency virus (SHIV)-infected rhesus monkeys through the use of two dominant (p11C and p199RY) and two subdominant (p68A and p56A) epitopes. Using real-time PCR to quantitate T-cell receptor (TCR) variable region beta (Vβ) family usage, we show that CD8+ T-lymphocyte populations specific for dominant epitopes are characterized by a diverse Vβ repertoire, whereas those specific for subdominant epitopes employ a dramatically more focused Vβ repertoire. We also demonstrate that dominant epitope-specific CD8+ T lymphocytes employ TCRs with multiple CDR3 lengths, whereas subdominant epitope-specific cells employ TCRs with a more restricted CDR3 length. Thus, the relative dominance of an epitope-specific CD8+ T-lymphocyte response reflects the clonal diversity of that response. These findings suggest that the limited clonal repertoire of subdominant epitope-specific CD8+ T-lymphocyte populations may limit the ability of these epitope-specific T-lymphocyte populations to expand and therefore limit the ability of these cell populations to contribute to the control of viral replication.
It is well established that dominant epitope-specific CD8+ T-lymphocyte responses play a central role in controlling viral replication, while subdominant epitope-specific responses contribute minimally to effective antiviral immunity (1, 15, 19, 24, 28, 32, 42, 46, 52). This is dramatically illustrated in human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) infections, where it has been demonstrated that a loss of a single dominant epitope-specific CD8+ T-lymphocyte response can abrogate effective control of virus replication, leading to uncontrolled viremia and death (4, 39). Moreover, although dominant epitope-specific CD8+ T lymphocytes exert sufficient immunologic pressure to select for HIV/SIV escape variants, subdominant epitope-specific CD8+ T lymphocytes do not impose enough immune pressure to select for such mutations. These findings have led to the suggestion that CD8+ T-lymphocyte-based vaccines for HIV would be most effective if the vaccines increased the breadth of epitope-specific CD8+ T-lymphocyte responses, effectively increasing the number of epitopes of the virus that are recognized as dominant. In order to shape the development of strategies that will accomplish this, we sought to elucidate the basis for establishment of epitope dominance hierarchies in CD8+ T-lymphocyte responses.
A number of mechanisms have been proposed to explain why CD8+ T-lymphocyte responses are so focused and hierarchical (11, 44, 53). Studies have reported associations between epitope dominance hierarchies and limitations in the infected cell's ability to process certain peptides, the relative binding affinities of epitope peptides for the major histocompatibility complex (MHC) class I molecule, and T-lymphocyte competition for as-yet-undefined molecules (18, 30, 35, 37, 45, 48, 53). However, these various proposed mechanisms do not explain the intense immune pressure mediated by dominant epitope-specific CD8+ T-lymphocyte responses and therefore may provide only a partial explanation for the phenomenon of epitopic immunodominance.
Dominance hierarchies of CD8+ T-lymphocyte epitopes have been defined in nonhuman primate models. Rhesus monkeys expressing the MHC class I molecule Mamu-A*01 that are chronically infected with SIV or the chimeric simian-human immunodeficiency virus (SHIV) develop CD8+ cytotoxic T-lymphocyte responses directed against a Gag epitope (p11C) and a Pol epitope (p68A) (2, 16, 25, 29). The magnitude of these epitope-specific CTL responses can be measured in peripheral blood using tetramer-binding and functional assays (16, 25). Consistently, these infected monkeys develop strong responses to p11C and very weak responses to p68A (6, 8, 47). Similarly, SIV- or SHIV-infected rhesus monkeys expressing the MHC class I molecule Mamu-A*02 generate strong responses against the Nef epitope p199RY and much weaker responses against another Nef epitope, p56 (34). Interestingly, both Nef epitopes share similar lengths and terminal amino acid sequences. Genetically selected rhesus monkeys infected with SIV/SHIV therefore provide a powerful model for exploring the basis for CD8+ T-lymphocyte epitope immunodominance hierarchies.
Since each T-lymphocyte clone expresses a unique T-cell receptor (TCR), the definition of the TCR repertoire employed by an epitope-specific T-lymphocyte population provides a useful approach for determining the clonality of these cells (14, 49). We have employed this strategy to explore the contribution of clonality to the relative dominance of CD8+ T-lymphocyte epitopes in SIV-infected monkeys. In these studies, we show that the distinction between epitope dominance and subdominance in these infected monkeys reflects the clonal diversity of the CD8+ T-lymphocyte responses to those epitopes.
MATERIALS AND METHODS
Animals.
The rhesus monkeys used in this study were maintained in accordance with the guidelines of the Institutional Animal Care and Use Committee for Harvard Medical School and the Guide for the Care and Use of Laboratory Animals (32a). Monkeys were screened for the presence of the Mamu-A*01 and Mamu-A*02 alleles using a PCR-based technique as previously described (22, 26). DNA sequence analysis was performed on all potential positive samples to confirm identity with the established Mamu-A*01 and Mamu-A*02 sequence (29). The Mamu-A*01+ monkeys were vaccinated with plasmid DNA followed by recombinant poxvirus immunogens and challenged with SHIV-89.6P (41). The Mamu-A*02+ monkeys were vaccinated with plasmid DNA followed by recombinant adenovirus immunogens and challenged with SHIV-89.6P (43) or were naïve monkeys infected with SIVmac251 (34).
Antibodies, tetramers, and peptides.
Antibodies used in this study were directly coupled to fluorescein isothiocyanate (FITC), phycoerythrin (PE)-labeled Texas red (ECD), or allophycocyanin. The following monoclonal antibodies (MAbs) were used: ECD-conjugated anti-CD8α (clone 7PT3F9; Beckman Coulter), ECD-conjugated anti-CD8αβ (Beckman Coulter), FITC-conjugated anti-CD3 (clone FN18; BioSource International, Camarillo, CA), or FITC-conjugated anti-CD3 (clone SP34; BD PharMingen, San Diego, CA). Mamu-A*01/p11C, C-M/β2m (SIVmac Gag), Mamu-A*01/p68A/β2m (SIVmac Pol), Mamu-A*02/p199RY/β2m (SIVmac Nef), and Mamu-A*02/p56/β2m (SIVmac Nef) tetramer complexes were prepared as previously described (2, 16, 25, 33). Phycoerythrin-labeled ExtrAvidin (Sigma) was mixed stepwise with biotinylated Mamu-A*01/peptide or Mamu-A*02/peptide complexes at a molar ratio of 1:4 to produce the tetrameric complexes. Gag p11C (CTPYDINQM), Pol p68A (STPPLVRLV), Nef p199RY (YTSGPGIRY), and Nef p56 (YTYEAYVRY) peptides were obtained from QCB/Biosource (Hopkinton, MA). Lyophilized peptides were dissolved in a minimum volume of dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO), diluted to a stock peptide concentration of 15 mg/ml in water containing 5 mM dithiothreitol (Sigma-Aldrich), and then frozen at −70°C in aliquots. Before use, peptides were diluted to a working concentration in RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with glutamine, 12% fetal calf serum, penicillin, streptomycin, and gentamicin.
Flow cytometry.
Peripheral blood mononuclear cells (PBMCs) cultured in the presence of peptide and interleukin-2 were harvested on days 10 to 14 and separated over a Ficoll layer (Ficoll-Paque Plus; Amersham-Pharmacia Biotech, Uppsala, Sweden). Cultured cells or whole-blood specimens were stained with Mamu-A*01/p11C, C-M/β2m, Mamu-A*01/p68A/β2m, Mamu-A*02/p199RY/β2m, or Mamu-A*02/p56/β2m tetramer for 30 min at room temperature. Cells were then stained with a mixture of anti-CD3 and anti-CD8 antibodies for 30 min. Cultured cells were fixed in 1% formaldehyde. Whole-blood specimens were lysed using a Coulter Immunoprep reagent system and a Q-prep workstation (Beckman-Coulter) before being fixed in 1.5% formaldehyde. Samples were analyzed on a Coulter EPICS XL-MCL located in a dedicated biosafety level 3 area (Beckman Coulter) or a FACSCalibur flow cytometer (BD Biosciences).
Peptide binding assay.
Peptide binding assays for Mamu-A*01-restricted peptides were performed by measuring inhibition of the iodinated p11C analog peptide (ATPYDINQM) or iodinated p68A native peptide (ATPPLVRLV) binding to 721.221 cells that express the MHC class I molecule Mamu-A*01. Similarly, peptide binding assays for Mamu-A*02-restricted peptides were performed by measuring inhibition of the iodinated p199RY native peptide (YTSGPGIRY) or iodinated p56 native peptide (YTYEAYVRY) binding to 721.221 cells that express the MHC class I molecule Mamu-A*02. Cells (2 × 106) were incubated overnight at 26°C with 3 μg of human β2 microglobulin per ml. Iodinated peptides (at 1.5 × 105 cpm) and serial half-log dilutions of unlabeled test peptides were then incubated with washed cells for 4 h at 20°C. Cells were then washed three times, and the radioactivity of the cell pellet was measured with a scintillation counter. The concentration of test peptide required for inhibition of binding of the index peptide by 50% was designated the 50% inhibitory concentration (IC50).
Generation of cDNA.
RNA was extracted from Mamu-A*01/p11C, C-M/β2m, Mamu-A*01/p68A/β2m, Mamu-A*02/p199RY/β2m, and Mamu-A*02/p56/β2m tetramer-binding CD8+ T-lymphocyte populations according to the instructions supplied with the RNeasy Mini extraction kit from QIAGEN (Valencia, CA). The integrity of the RNA was confirmed using an Agilent 2100 bioanalyzer. cDNA was then synthesized from the extracted RNA, as outlined in the Super SMART PCR cDNA synthesis kit from Clontech Laboratories (Palo Alto, CA). Briefly, the single-stranded cDNA reaction was catalyzed by using Moloney murine leukemia virus reverse transcriptase with the 3′ SMART CDS primer II A and Smart II A oligonucleotide primers provided in the Super SMART cDNA synthesis kit. Preamplified double-stranded cDNA libraries were made by a 10- to 20-cycle PCR, utilizing the 5′ PCR primer IIA primer and reagents also provided in the Clontech kit. The optimal number of cycles of preamplification was found by performing a test run in the presence of SYBR green to determine the maximum number of PCR cycles that could be performed in the log-linear amplification range.
Primers and sequencing.
Primers used for the real-time PCR assay and for spectratyping (see data at http://www.viralpath.org/Portals/0/Supplementary_Figure_1.jpg) were ordered through Biosource International, manufactured by Keystone Labs (Camarillo, CA), and purified by high-performance liquid chromatography. The real-time Taqman probes were synthesized at Biosearch Technologies, Inc. (Novato, CA) and purified by high-performance liquid chromatography. Primers specific for the variable and constant region of the TCR β chain were designed from rhesus monkey TCR sequences obtained from GenBank and our laboratory. The specificity of each of the primer pairs was confirmed by gel electrophoresis and sequencing of PCR products.
Quantitative PCR.
Briefly, cDNA derived from each sample was equally distributed into 34 individual PCR mixtures which contained a sense Vβ family-specific primer, an antisense Cβ-specific primer, and the TaqMan Cβ probe. PCRs were carried out using Sure-Start Taq (Stratagene). The real-time PCR was carried out for 55 cycles on an MX4000 QPCR machine (Stratagene) using the following cycling program: 95°C for 10 min followed by 55 cycles of 95°C for 10 s and 58°C for 30 s, followed by reading of fluorescence, and then 72°C for 30 s. To confirm the results of Vβ family expression, the identified Vβ families in each cDNA sample were assessed for complementarity-determining region 3 (CDR3) profiles through Genescan-based spectratyping.
Spectratyping.
CDR3 profiles were analyzed by Genescan-based spectratyping (38, 54). cDNA generated for use in the quantitative PCR assay was used as template for second round PCRs utilizing individual Vβ primers and a 5′-6-carboxyfluorescein-labeled Cβ primer (Biosource International, Inc., Camarillo, CA). The cDNA was amplified for 30 cycles in a Perkin-Elmer 9600 GeneAmp PCR system under the following conditions: 95°C for 10 s, 57°C for 30 s, and 68°C for 60 s. One microliter of each reaction product was mixed with deionized formamide and a TAMRA-500 size standard and then electrophoresed on a 5% acrylamide gel on a 377 DNA sequencer (Applied Biosystems). Data were analyzed for size and fluorescence intensity by using Genescan software version 3.1 (Applied Biosystems). Further cloning of PCR products into the pGEM-T Easy vector (Promega) for sequencing, in conjunction with the CDR3 length display, allowed for the prediction of CDR3 lengths. These lengths were expressed as predicted numbers of amino acids spanning the portion of the VDJ joining segments.
RESULTS
Tetramer staining and SHIV peptide binding affinities for MHC class I.
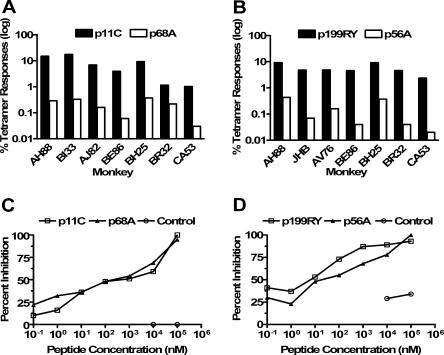
The frequency of SIV and SHIV epitope-specific CD8+ T lymphocytes was measured in PBMCs of seven Mamu-A*01+ and Mamu-A*02+ rhesus monkeys by staining with Mamu-A*01/peptide or Mamu-A*02/peptide tetramers (25) (Fig. 1A and B). Consistent with our prior observations, the p11C-specific CD8+ T-lymphocyte populations in fresh peripheral blood of Mamu-A*01+ monkeys varied between 1.03 and 17.53% of total CD8+ T lymphocytes, while the p68A-specific populations varied between 0.03 and 0.37%. In the Mamu-A*02+ monkeys, p199RY-specific CD8+ T-lymphocyte populations varied between 2.36 and 9.31%, while p56-specific populations varied between 0.02 and 0.43% of total CD8+ T lymphocytes. Thus, p11C and p199RY are dominant and p68A and p56 are subdominant epitopes.
FIG. 1.
Immunodominant and subdominant SIV/SHIV epitopes have similar peptide binding affinities for MHC class I. (A) Peak tetramer responses for the dominant p11C and subdominant p68A epitopes in SIV-infected Mamu-A*01-positive rhesus monkeys. (B) Peak tetramer responses for the dominant p199RY and subdominant p56 epitopes in SIV-infected Mamu-A*02-positive monkeys. (C) Binding affinities of p11C and p68A for Mamu-A*01. (D) Binding affinities of p199RY and p56 for Mamu-A*02.
To evaluate the mechanism underlying the immunodominance hierarchy of these epitopes, we assessed the relative binding affinities of these epitope peptides for the Mamu-A*01 or Mamu-A*02 molecules (3). Interestingly, the binding affinities of both the dominant and subdominant epitope peptides for the MHC class I molecules that present them to CD8+ T lymphocytes were comparable at the IC50 (Fig. 1C and D). We have previously shown that Mamu-A*01-restricted epitope peptides are processed and presented equivalently when the immune system is exposed to these peptides via a polyepitope DNA vaccine (47). Thus, differences in viral epitope peptide binding to MHC class I molecules do not explain the dominance hierarchy of the epitope-specific CD8+ T-lymphocyte populations.
Evaluation of Vβ TCR repertoires by quantitative PCR.
We then sought to examine the contribution of clonal diversity to determining the magnitude of these epitope-specific CD8+ T-lymphocyte responses. We did this by evaluating the TCR repertoire of each epitope-specific CD8+ T-lymphocyte population, reasoning that each distinct TCR present in a cell population represents a distinct CD8+ T-lymphocyte clone. Since only a small number of the available anti-human TCR β-chain family-specific monoclonal antibodies cross-react with rhesus monkey TCRs (www.bidmc.harvard.edu/display.asp?node_id = 3770), we were unable to use cell staining and flow cytometric analysis to determine TCR usage. Therefore, we employed a PCR-based approach to assess the TCR repertoire of these rhesus monkey cell populations. Based on data generated through extensive sequencing of rhesus monkey TCR β-chain cDNAs (7, 9, 12, 27), we developed a series of 34 forward primers specific for β-chain variable region (Vβ) gene families and subfamilies (see data at http://www.viralpath.org/Portals/0/Supplementary_Figure_1.jpg). These primers, when paired with a single reverse primer designed to hybridize to the proximal portion of the β constant chain region (Cβ), yielded PCR products of 200 to 300 bp. A hybridization site for a single fluorescent TaqMan probe was placed between the primer binding sites, within the conserved C region.
Two-step amplification of cellular RNA does not bias the TCR repertoire.
For analysis of Vβ gene usage in selected T-lymphocyte populations, poly(A)+ cDNA was prepared from total cellular RNA of those cells. This cDNA was preamplified for 10 to 15 cycles using a universal primer. The optimal number of cycles of preamplification was found by performing a test run in the presence of SYBR green to determine the maximum number of PCR cycles that could be performed in the log-linear amplification range. This information allowed us to select amplification conditions that would not distort quantitation of the relative abundances of individual cDNA species. Preamplified cDNAs were then used as templates for real-time PCR analysis using the Vβ-specific primers and probes.
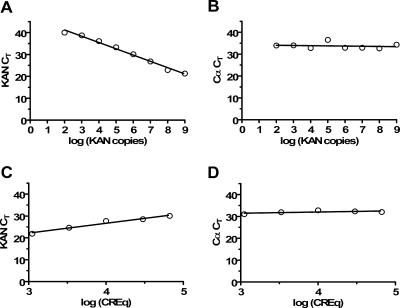
To verify that this two-step amplification did not significantly bias the apparent TCR repertoire of a lymphocyte population, we used this approach to measure the copy numbers of a cellular RNA (TCR α chain) and an exogenously added kanamycin control RNA (13). Total cellular RNA extracted from rhesus monkey CD8+ T lymphocytes was evenly divided among eight tubes, and 102 to 109 copies of kanamycin RNA were added to the tubes. Separate cDNA synthesis reactions were performed on each sample, and all samples were individually preamplified, with the number of cycles determined by the previously described SYBR green test run. These preamplified samples were then analyzed using the TCR α-chain and kanamycin probes (see data at http://www.viralpath.org/Portals/0/Supplementary_Figure_1.jpg). As shown in Fig. 2A and B, the detection assay was linear over the entire range of kanamycin RNA concentrations tested. Moreover, the threshold cycle number for detection of TCR α was the same in each sample.
FIG. 2.
Two-step amplification of cellular RNA does not bias the detected TCR repertoire. (A and B) Total cellular RNA extracted from CD8+ T lymphocytes was evenly divided among eight tubes, and 102 to 109 copies of kanamycin (KAN) RNA were added to the tubes. Separate cDNA synthesis reactions were performed on each sample, and all samples were individually preamplified, with the number of cycles determined by the previously described SYBR green test run. These preamplified samples were then analyzed using the TCR α-chain and kanamycin probes. CT, cycle threshold. (C and D) Various amounts of cellular RNA were used in five separate preamplification reactions, while the amount of exogenously added kanamycin was kept constant in each reaction. The number of cycles was again determined by a SYBR green test run. R2 values calculated from the linear regression of panels A and C are 0.9889 and 0.9637, respectively. CREq, cell RNA equivalent.
Next, we varied the amount of cellular RNA in each sample while keeping the amount of exogenously added kanamycin RNA constant. Because these samples contained different amounts of cellular RNA, the optimal number of preamplification cycles for each sample, determined by the SYBR green analysis, differed as well. As expected, the detection threshold cycle for kanamycin in samples that initially contained smaller starting amounts of RNA (and therefore required more cycles of preamplification) was lower than the detection threshold cycle for samples containing higher starting quantities of RNA (Fig. 2C). Importantly, however, the threshold cycle for the TCR α chain was approximately the same in each sample (Fig. 2D), indicating that differential preamplification had, in effect, corrected for differences in the total starting RNA in each reaction. In subsequent studies, we used cell numbers that would yield quantities of cellular RNA in the same range as those tested in the studies shown in Fig. 2C and D. In addition, whenever possible, we prepared cDNA from equivalent numbers of cells rather than relying upon differential preamplification to normalize input RNA.
Peptide-stimulated tetramer-sorted PBMCs are a subset of the total CD8+ T-lymphocyte population.
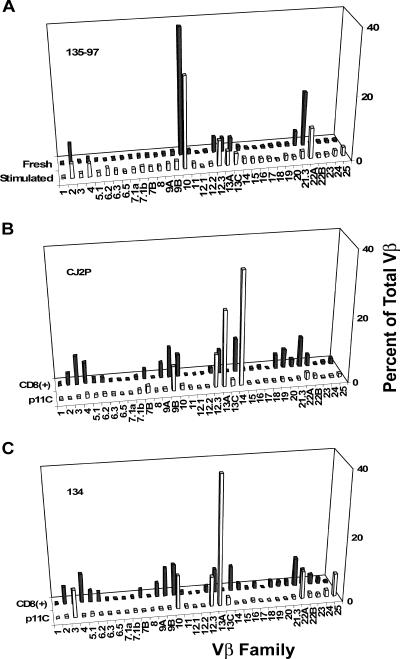
We next sought to analyze the TCR repertoires of populations of dominant and subdominant epitope-specific CD8+ T lymphocytes isolated by flow cytometric sorting from tetramer-stained PBMCs of SIV- or SHIV-infected Mamu-A*01+ and Mamu-A*02+ rhesus monkeys. We reasoned, however, that determination of the repertoires of subdominant epitope-specific cell populations that are found at very low frequencies in peripheral blood might present a major technical challenge. To obtain sufficient CD8+ T-cell lymphocytes specific for a subdominant epitope to analyze their TCR repertoire reliably, the CD8+ T-lymphocyte subpopulation in a PBMC sample might be expanded by in vitro stimulation with epitope peptide prior to staining with tetramer and cell sorting. However, this approach could possibly bias the detected TCR repertoire. Peptide-stimulated cells might also yield cDNA of greater complexity than unstimulated lymphocytes and thus obscure the contribution of rare cell populations to the epitope-specific T-cell response. To explore these possibilities, we compared the TCR Vβ repertoire of fresh p11C tetramer-binding CD8+ T lymphocytes, which could be obtained in sufficient numbers in PBMCs without resorting to in vitro peptide stimulation, to that of p11C-stimulated CD8+ p11C tetramer-binding CD8+ T lymphocytes from the same PBMC sample. As shown in Fig. 3A, only very minor differences in the Vβ repertoires were detected. The cDNA copy percentages of the most frequently employed Vβ subfamilies were as much as 1 or more orders of magnitude greater than those of the other subfamilies, and the patterns of these peaks did not vary between cDNA generated from fresh and peptide-stimulated cells. A similar study was not done to compare TCR repertoires of freshly sorted and in vitro-expanded subdominant epitope-specific CD8+ T-cell populations because insufficient RNA was available from the freshly sorted cells to carry out the analysis. To determine whether the pattern of Vβ usage in p11C-specific CD8+ T lymphocytes was merely a reflection of the pattern of Vβ usage in the aggregate CD8+ lymphocyte population, we compared the repertoires of p11C-stimulated CD8+ T lymphocytes with those of unstimulated total CD8+ T cells. In studies of PBMCs from two monkeys (Fig. 3B and C), the Vβ repertoires of these p11C-specific CD8+ T lymphocytes represented a distinct subset of the entire CD8+ T-lymphocyte Vβ repertoire.
FIG. 3.
p11C-stimulated tetramer-sorted PBMCs are a distinct subset of the total CD8+ T-lymphocyte population. (A) Real-time PCR Vβ quantitation of total peripheral blood cDNA generated from fresh p11C tetramer-sorted PBMCs and from 12-day p11C-stimulated, p11C tetramer-sorted PBMCs. (B and C), Vβ repertoire analysis from two monkeys (CJ2P and 134, respectively) of fresh sorted CD8+ T lymphocytes and 12-day p11C-stimulated, p11C tetramer-sorted PBMCs.
Subdominant epitope-specific populations have a restricted Vβ repertoire.
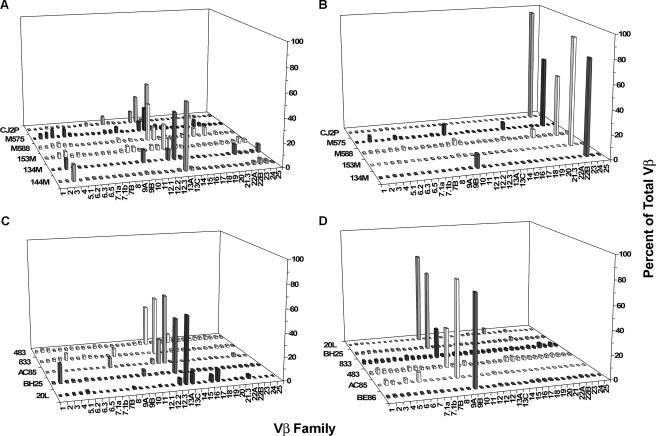
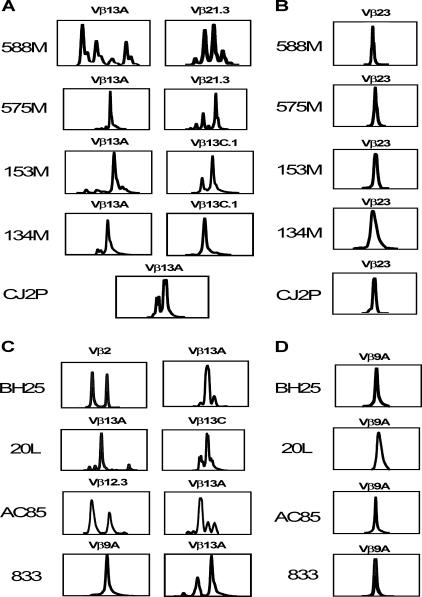
We then analyzed the TCR Vβ repertoires of dominant and subdominant epitope-specific CD8+ T lymphocytes of a cohort of chronically infected Mamu-A*01+ and Mamu-A*02+ rhesus monkeys. As shown in Fig. 4A and C, the repertoires of the dominant p11C and p199RY epitope-specific CD8+ T lymphocytes from each monkey included a number of different Vβ gene families. An assessment of the Vβ gene usage by the dominant CD8+ T-lymphocyte populations of six different Mamu-A*01+ and five different Mamu-A*02+ monkeys showed a predominance of Vβ13 family usage, a feature of the p11C-specific CD8+ T-lymphocyte repertoire that we have previously described (10). In addition, several less frequently used Vβ gene families were represented in the repertoires of these dominant epitope-specific cells. Although the patterns varied between the different monkeys, a preference for the use of Vβ3, -10, -12.3, and -21 was evident. In contrast, the subdominant p68A- and p56-specific CD8+ T-lymphocyte repertoires from all of the monkeys tested were strikingly limited. In fact, the repertoire was almost identical for every monkey with the same MHC class I haplotype that was studied, consisting almost entirely of Vβ23 in the p68A-specific populations and Vβ9 in the p56-specific populations (Fig. 4B and D). Therefore, the dominant epitope-specific CD8+ T lymphocytes were comprised of clones employing a much greater diversity of Vβ genes than the subdominant epitope-specific CD8+ T lymphocytes.
FIG. 4.
CD8+ T-lymphocyte populations recognizing dominant Mamu-A*01- or Mamu-A*02-restricted epitopes consist of a diverse Vβ repertoire, while those recognizing subdominant epitopes have a more restricted Vβ repertoire. (A) Mamu-A*01-restricted p11C-specific T-lymphocyte Vβ repertoires. (B) Mamu-A*01-restricted p68A-specific T-lymphocyte Vβ repertoires. (C) Mamu-A*02-restricted p199RY-specific T-lymphocyte Vβ repertoires. (D) Mamu-A*02-restricted p56-specific T-lymphocyte Vβ repertoires.
Vβ families of CD8+ T lymphocytes specific for subdominant epitopes have a restricted CDR3 length.
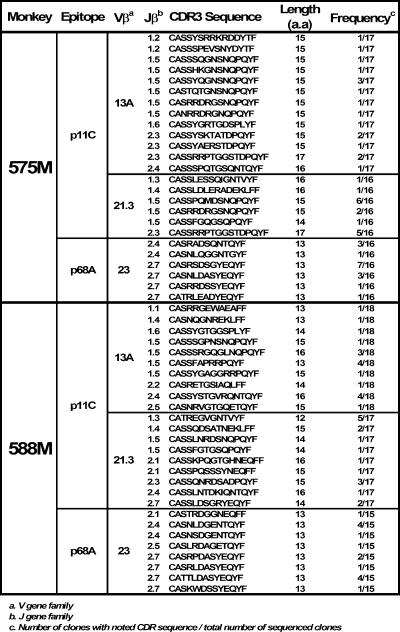
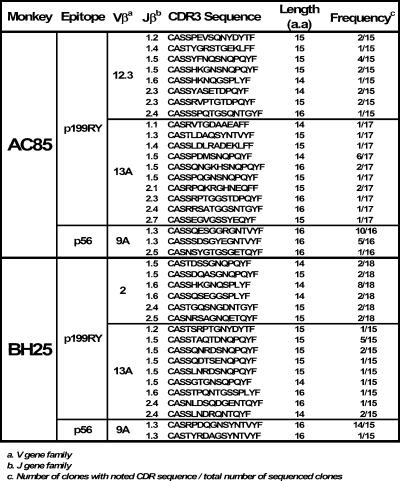
We then further examined the diversity of the TCR repertoires of these dominant and subdominant epitope-specific CD8+ T-lymphocyte populations. Simply measuring the contribution of a particular Vβ family or subfamily to the epitope-specific repertoire may underestimate the true diversity of the repertoire, since CDR3 diversity is not detected by this approach. We therefore evaluated the diversity of the TCR repertoires of these CD8+ T-lymphocyte populations by examining the CDR3 lengths of the Vβ genes using spectratyping (38) (Fig. 5) and sequencing (Fig. 6 and 7). Analysis of the CDR3 length polymorphisms of Vβ23 and Vβ9 in the cDNA derived from p68A-specific and p56-specific CD8+ T lymphocytes, respectively, revealed a striking uniformity of CDR3 lengths (Fig. 5B and D). In contrast, almost all Vβ spectratypes for the dominant p11C and p199RY epitope-specific CD8+ T-lymphocyte populations showed substantial diversity of CDR3 lengths (Fig. 5A and C). In addition to this spectratyping, clones were generated and sequenced from these same cDNAs from two of the Mamu-A*01+ and two of the Mamu-A*02+ monkeys (Fig. 6 and 7). As seen in the spectratyping analyses, sequencing of the CDR3 regions of the TCR β genes of the CD8+ T lymphocytes specific for subdominant epitopes had a strikingly restricted length: in the case of p68 Vβ23, 13 amino acids, and for p56 Vβ9, 16 amino acids. For Vβ genes employed by the dominant epitope-specific CD8+ T lymphocytes, CDR3 sequences were of variable lengths.
FIG. 5.
Spectratyping analysis reveals CDR3 diversity within represented Vβ families of CD8+ T-lymphocyte populations specific for dominant Mamu-A*01- and Mamu-A*02-restricted CD8+ T-lymphocyte epitopes and a single CDR3 length for Vβ families of CD8+ T lymphocytes specific for subdominant epitopes. Shown are spectratyping results for representative Vβ families found in (A) p11C-specific, (B) p68A-specific, (C) p199RY-specific, and (D) p56-specific CD8+ lymphocyte populations.
FIG. 6.
TCR Vβ CDR3 sequences of CD8+ T lymphocytes specific for the Mamu-A*01-restricted p11C and p68A epitopes. CDR3 sequences are arranged in order of corresponding Jβ families. a.a, amino acids.
FIG. 7.
TCR Vβ CDR3 sequences of CD8+ T lymphocytes specific for the Mamu-A*02-restricted p199RY and p56 epitopes. CDR3 sequences are arranged in order of corresponding Jβ families. a.a, amino acids.
These findings therefore indicate that the subdominant epitope-specific CD8+ T lymphocytes were comprised of a very limited number of clones with highly constrained CDR3 regions, while the dominant epitope-specific CD8+ T lymphocytes were comprised of a diversity of clones with considerable variability in their CDR3 regions. This striking difference between the subdominant and dominant epitope-specific CD8+ T-lymphocyte populations suggests that the relative dominance of an epitope may be determined, at least in part, by the diversity of CD8+ T-lymphocyte clones that can recognize that epitope peptide/MHC class I molecular complex.
DISCUSSION
In the present study, we show that CD8+ T-lymphocyte populations specific for dominant epitopes are characterized by a diverse Vβ repertoire, whereas those specific for subdominant epitopes employ a more focused Vβ repertoire. We also demonstrate that dominant epitope-specific CD8+ lymphocytes utilize TCRs with multiple CDR3 lengths, whereas subdominant epitope-specific cells utilize TCRs with a more restricted CDR3 length. Thus, the relative dominance of a viral epitope reflects the clonal diversity of the CD8+ lymphocyte response to that epitope.
This analysis of CD8+ T-lymphocyte TCRs has focused entirely on the diversity of the β chain. Although the α chain of the TCR certainly contributes to the diversity of the overall CD8+ T-lymphocyte response, this contribution is difficult to evaluate. There are a large number of human and rhesus monkey Vα gene families. In fact, more Vα-chain sequences than Vβ-chain sequences have been defined. Thus, an exhaustive evaluation of both Vβ and Vα gene usage by TCRs that recognize a single epitope would be a very large undertaking (31). Of greater importance, CD8+ T lymphocytes can express more than one α chain. Whereas a single T lymphocyte expresses one unique TCR β chain, up to 20% of human T lymphocytes express two distinct α chains (17, 21, 36). Because of this, a substantial fraction of the expressed α chains in an epitope-specific T-lymphocyte population may not be used in epitope recognition. Therefore, in order to analyze the contribution of the Vα repertoire, it would be necessary to define the α chains that are consistently coexpressed with the β chains, a challenging technical problem. We have, therefore, assumed in the present study that a T-lymphocyte population with a single Vβ sequence represents a distinct clone of cells.
Epitope-specific T lymphocytes undergo clonal expansion following exposure to their cognate epitope peptides. The mechanisms that regulate the magnitude of this expansion in vivo are not fully understood, but there is evidence to suggest that, once triggered to divide, each clone is programmed to undergo a fixed number of cell divisions (40, 50). The relative dominance of a particular epitope-specific T-lymphocyte population may therefore reflect the number of CD8+ T-lymphocyte clones that can recognize that epitope in the immunologically naïve individual. Epitopes such as p68 and p56 may be subdominant because the germ line-encoded TCR β-chain sequences of rhesus monkeys capable of generating TCRs able to recognize such epitopes are limited in number. It is also possible that T lymphocytes bearing β chains that might recognize these epitopes are negatively selected during thymic maturation. By whichever mechanism, the present data suggest that only limited numbers of clones of naïve epitope-specific CD8+ T lymphocytes can expand to form cell populations that recognize a subdominant epitope.
The striking CDR3 diversity in the CD8+ T lymphocytes that recognize the dominant epitopes and the lack of CDR3 diversity in those cells that recognize the subdominant epitopes suggest another possible factor that differentiates dominant from subdominant epitopes. Since TCRs with CDR3 regions that differ in both length and sequence can recognize a dominant epitope, it is possible that the peptide/MHC class I complexes that comprise dominant epitopes have a three-dimensional conformation and charge distribution that is particularly conducive to the engagement of a diversity of TCRs (51). On the other hand, because of the extreme limitations in length and sequence in the CDR3 regions of TCRs that recognize subdominant epitopes, the conformation and charge distribution of the peptide/MHC class I complex of these subdominant epitopes may be particularly idiosyncratic and therefore can bind only using a very limited number of TCRs. Structural assessment of these peptide/MHC class I complexes will be needed to evaluate these possibilities.
A diversity of clonal cytotoxic T-lymphocyte (CTL) populations with subtly different recognition specificities for the same viral epitope peptide/MHC class I complex may confer optimal immune protection against HIV-1 or SIV replication. We have previously shown that while two different CTL clones can recognize the same dominant Gag epitope of SIV, each of those clones has a different capacity to recognize closely related mutant sequences of that dominant epitope (6). The likelihood of HIV/SIV immune escape at a particular epitope through mutation should therefore be diminished if the T-lymphocyte response to that epitope is highly polyclonal. Thus, the greater the diversity of the TCRs recognizing a particular epitope sequence, the smaller the chance that an epitope sequence variant will go unrecognized.
Immunologic pressure exerted by CD8+ T-lymphocyte populations on a dominant epitope of HIV/SIV will, however, ultimately induce virus mutational escape from immune recognition at that epitope (5, 20). Despite the clonal diversity of the p11C and p199RY epitope-specific CD8+ T-lymphocyte populations, escape from CD8+ T-lymphocyte recognition eventually occurs, resulting first in a change in the virus quasispecies in the infected individual and then in the loss of these dominant epitope-specific responses. Such escape events have been associated with rapid clinical deterioration and death in infected monkeys and humans (4, 23).
This observation has led to the suggestion that vaccination to increase the magnitude of CD8+ T-lymphocyte responses against subdominant epitopes may transform them into dominant epitope-specific responses, enhancing control of viral replication during acute infection and facilitating subsequent immune control in the event that there is a mutational escape from CTL recognition at another epitope. However, our present findings suggest that the limited clonal repertoire of subdominant epitope-specific CD8+ T-lymphocyte populations may limit the ability of these epitope-specific T-lymphocyte populations to expand and therefore limit the ability of these cell populations to contribute to the control of viral replication.
Acknowledgments
We thank Ediane Dutra for her help with the spectratyping analysis.
This work was supported by grants AI20729, AI058882, and AI048400 from the National Institutes of Health.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. R. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 3.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Powers, D. M. Truitt, M. G. Kishko, J. C. Arthur, F. W. Peyerl, M. J. Kuroda, D. A. Gorgone, M. A. Lifton, C. I. Lord, V. M. Hirsch, D. C. Montefiori, A. Carville, K. G. Mansfield, K. J. Kunstman, S. M. Wolinsky, and N. L. Letvin. 2005. Dynamic immune responses maintain cytotoxic T lymphocyte epitope mutations in transmitted simian immunodeficiency virus variants. Nat. Immunol. 6:247-252. [DOI] [PubMed] [Google Scholar]
- 6.Charini, W. A., M. J. Kuroda, J. E. Schmitz, K. R. Beaudry, W. Lin, M. A. Lifton, G. R. Krivulka, A. Necker, and N. L. Letvin. 2001. Clonally diverse CTL response to a dominant viral epitope recognizes potential epitope variants. J. Immunol. 167:4996-5003. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z. W., Z. C. Kou, L. Shen, K. A. Reimann, and N. L. Letvin. 1993. Conserved T-cell receptor repertoire in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 151:2177-2187. [PubMed] [Google Scholar]
- 8.Chen, Z. W., Y. Li, X. Zeng, M. J. Kuroda, J. E. Schmitz, Y. Shen, X. Lai, L. Shen, and N. L. Letvin. 2001. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J. Immunol. 166:4525-4533. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z. W., L. Shen, M. D. Miller, S. H. Ghim, A. L. Hughes, and N. L. Letvin. 1992. Cytotoxic T lymphocytes do not appear to select for mutations in an immunodominant epitope of simian immunodeficiency virus gag. J. Immunol. 149:4060-4066. [PubMed] [Google Scholar]
- 10.Chen, Z. W., L. Shen, J. D. Regan, Z. Kou, S. H. Ghim, and N. L. Letvin. 1996. The T cell receptor gene usage by simian immunodeficiency virus gag-specific cytotoxic T lymphocytes in rhesus monkeys. J. Immunol. 156:1469-1475. [PubMed] [Google Scholar]
- 11.Choi, E. Y., G. J. Christianson, Y. Yoshimura, T. J. Sproule, N. Jung, S. Joyce, and D. C. Roopenian. 2002. Immunodominance of H60 is caused by an abnormally high precursor T cell pool directed against its unique minor histocompatibility antigen peptide. Immunity 17:593-603. [DOI] [PubMed] [Google Scholar]
- 12.Currier, J. R., K. S. Stevenson, and M. A. Robinson. 2000. Molecular cloning and sequencing of cDNA encoding eight new rhesus macaque TCRBV gene families. Immunogenetics 51:387-391. [DOI] [PubMed] [Google Scholar]
- 13.de Rozieres, S., C. H. Swan, D. A. Sheeter, K. J. Clingerman, Y. C. Lin, S. Huitron-Resendiz, S. Henriksen, B. E. Torbett, and J. H. Elder. 2004. Assessment of FIV-C infection of cats as a function of treatment with the protease inhibitor, TL-3. Retrovirology 1:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty, P. J., C. M. Roifman, S. H. Pan, U. Cymerman, S. W. Ho, E. Thompson, S. Kamel-Reid, and A. Cohen. 1991. Expression of the human T cell receptor V beta repertoire. Mol. Immunol. 28:607-612. [DOI] [PubMed] [Google Scholar]
- 15.Draenert, R., C. L. Verrill, Y. Tang, T. M. Allen, A. G. Wurcel, M. Boczanowski, A. Lechner, A. Y. Kim, T. Suscovich, N. V. Brown, M. M. Addo, and B. D. Walker. 2004. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J. Virol. 78:630-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egan, M. A., M. J. Kuroda, G. Voss, J. E. Schmitz, W. A. Charini, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Use of major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lymphocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J. Virol. 73:5466-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, J. I., and D. M. Altmann. 1995. Dual T cell receptor alpha chain T cells in autoimmunity. J. Exp. Med. 182:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, T. 1997. Transporter associated with antigen processing. Adv. Immunol. 65:47-109. [PubMed] [Google Scholar]
- 19.Gotch, F., A. McMichael, G. Smith, and B. Moss. 1987. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J. Exp. Med. 165:408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 21.Heath, W. R., F. R. Carbone, P. Bertolino, J. Kelly, S. Cose, and J. F. Miller. 1995. Expression of two T cell receptor alpha chains on the surface of normal murine T cells. Eur. J. Immunol. 25:1617-1623. [DOI] [PubMed] [Google Scholar]
- 22.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 23.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 24.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, M. J., J. E. Schmitz, D. H. Barouch, A. Craiu, T. M. Allen, A. Sette, D. I. Watkins, M. A. Forman, and N. L. Letvin. 1998. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J. Exp. Med. 187:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 27.Levinson, G., A. L. Hughes, and N. L. Letvin. 1992. Sequence and diversity of rhesus monkey T-cell receptor beta chain genes. Immunogenetics 35:75-88. [DOI] [PubMed] [Google Scholar]
- 28.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, M. D., H. Yamamoto, A. L. Hughes, D. I. Watkins, and N. L. Letvin. 1991. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J. Immunol. 147:320-329. [PubMed] [Google Scholar]
- 30.Momburg, F., and G. J. Hammerling. 1998. Generation and TAP-mediated transport of peptides for major histocompatibility complex class I molecules. Adv. Immunol. 68:191-256. [DOI] [PubMed] [Google Scholar]
- 31.Moss, P. A., W. M. Rosenberg, E. Zintzaras, and J. I. Bell. 1993. Characterization of the human T cell receptor alpha-chain repertoire and demonstration of a genetic influence on V alpha usage. Eur. J. Immunol. 23:1153-1159. [DOI] [PubMed] [Google Scholar]
- 32.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 32a.National Academy of Sciences. 1996. Guide for the care and use of animals. National Academy Press, Washington, D.C.
- 33.Newberg, M. H., M. J. Kuroda, W. A. Charini, A. Miura, C. I. Lord, J. E. Schmitz, D. A. Gorgone, M. A. Lifton, K. Kuus-Reichel, and N. L. Letvin. 2002. A simian immunodeficiency virus nef peptide is a dominant cytotoxic T lymphocyte epitope in Indian-origin rhesus monkeys expressing the common MHC class I allele mamu-A*02. Virology 301:365-373. [DOI] [PubMed] [Google Scholar]
- 34.Newberg, M. H., K. J. McEvers, D. A. Gorgone, M. A. Lifton, S. H. Baumeister, R. S. Veazey, J. E. Schmitz, and N. L. Letvin. 2006. Immunodomination in the evolution of dominant epitope-specific CD8+ T lymphocyte responses in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 176:319-328. [DOI] [PubMed] [Google Scholar]
- 35.Niedermann, G., S. Butz, H. G. Ihlenfeldt, R. Grimm, M. Lucchiari, H. Hoschutzky, G. Jung, B. Maier, and K. Eichmann. 1995. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity 2:289-299. [DOI] [PubMed] [Google Scholar]
- 36.Padovan, E., G. Casorati, P. Dellabona, S. Meyer, M. Brockhaus, and A. Lanzavecchia. 1993. Expression of two T cell receptor alpha chains: dual receptor T cells. Science 262:422-424. [DOI] [PubMed] [Google Scholar]
- 37.Pamer, E., and P. Cresswell. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 16:323-358. [DOI] [PubMed] [Google Scholar]
- 38.Pannetier, C., J. Even, and P. Kourilsky. 1995. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today 16:176-181. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 40.Rocha, B., N. Dautigny, and P. Pereira. 1989. Peripheral T lymphocytes: expansion potential and homeostatic regulation of pool sizes and CD4/CD8 ratios in vivo. Eur. J. Immunol. 19:905-911. [DOI] [PubMed] [Google Scholar]
- 41.Santra, S., D. H. Barouch, B. Korioth-Schmitz, C. I. Lord, G. R. Krivulka, F. Yu, M. H. Beddall, D. A. Gorgone, M. A. Lifton, A. Miura, V. Philippon, K. Manson, P. D. Markham, J. Parrish, M. J. Kuroda, J. E. Schmitz, R. S. Gelman, J. W. Shiver, D. C. Montefiori, D. Panicali, and N. L. Letvin. 2004. Recombinant poxvirus boosting of DNA-primed rhesus monkeys augments peak but not memory T lymphocyte responses. Proc. Natl. Acad. Sci. USA 101:11088-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 43.Seaman, M. S., F. W. Peyerl, S. S. Jackson, M. A. Lifton, D. A. Gorgone, J. E. Schmitz, and N. L. Letvin. 2004. Subsets of memory cytotoxic T lymphocytes elicited by vaccination influence the efficiency of secondary expansion in vivo. J. Virol. 78:206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sercarz, E. E., P. V. Lehmann, A. Ametani, G. Benichou, A. Miller, and K. Moudgil. 1993. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 11:729-766. [DOI] [PubMed] [Google Scholar]
- 45.Sette, A., A. Vitiello, B. Reherman, P. Fowler, R. Nayersina, W. M. Kast, C. J. Melief, C. Oseroff, L. Yuan, J. Ruppert, et al. 1994. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153:5586-5592. [PubMed] [Google Scholar]
- 46.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 197:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subbramanian, R. A., M. J. Kuroda, W. A. Charini, D. H. Barouch, C. Costantino, S. Santra, J. E. Schmitz, K. L. Martin, M. A. Lifton, D. A. Gorgone, J. W. Shiver, and N. L. Letvin. 2003. Magnitude and diversity of cytotoxic-T-lymphocyte responses elicited by multiepitope DNA vaccination in rhesus monkeys. J. Virol. 77:10113-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka, K., N. Tanahashi, C. Tsurumi, K. Y. Yokota, and N. Shimbara. 1997. Proteasomes and antigen processing. Adv. Immunol. 64:1-38. [DOI] [PubMed] [Google Scholar]
- 49.Turner, S. J., K. Kedzierska, N. L. La Gruta, R. Webby, and P. C. Doherty. 2004. Characterization of CD8+ T cell repertoire diversity and persistence in the influenza A virus model of localized, transient infection. Semin. Immunol. 16:179-184. [DOI] [PubMed] [Google Scholar]
- 50.von Boehmer, H., and K. Hafen. 1993. The life span of naive alpha/beta T cells in secondary lymphoid organs. J. Exp. Med. 177:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wucherpfennig, K. W. 2005. The structural interactions between T cell receptors and MHC-peptide complexes place physical limits on self-nonself discrimination. Curr. Top. Microbiol. Immunol. 296:19-37. [DOI] [PubMed] [Google Scholar]
- 52.Yewdell, J. W., and J. R. Bennink. 1992. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule-restricted T lymphocytes. Adv. Immunol. 52:1-123. [DOI] [PubMed] [Google Scholar]
- 53.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, D., Y. Shen, L. Chalifoux, D. Lee-Parritz, M. Simon, P. K. Sehgal, L. Zheng, M. Halloran, and Z. W. Chen. 1999. Mycobacterium bovis bacille Calmette-Guerin enhances pathogenicity of simian immunodeficiency virus infection and accelerates progression to AIDS in macaques: a role of persistent T cell activation in AIDS pathogenesis. J. Immunol. 162:2204-2216. [PubMed] [Google Scholar]