Abstract
Walleye dermal sarcoma virus (WDSV) is a complex retrovirus associated with dermal sarcomas in walleye fish. A WDSV accessory gene encodes a cyclin homolog or retroviral cyclin (rv-cyclin). WDSV rv-cyclin was found to be associated with transcription complexes and to affect transcription in a cell-type and promoter-dependent manner. It inhibited the WDSV promoter in walleye fibroblasts and activated transcription from GAL4 promoters when fused to the GAL4 DNA binding domain, and an activation domain (AD) has been localized to 30 amino acids in the carboxyl region. rv-cyclin can block the pulldown of transcription coactivators by the AD of VP16, and the isolated rv-cyclin AD interferes specifically with the interaction between the carboxyl halves of the VP16 AD, VP16C, and TATA-binding protein-associated factor 9 (TAF9). The carboxyl region and isolated AD can bind TAF9 directly in assays of protein-protein interaction in vitro. Furthermore, rv-cyclin and the isolated rv-cyclin AD interfere specifically with the function of VP16C in transcription assays. A previously identified motif within the VP16C sequence mediates TAF9 binding, and this motif is present in the activation domains of a variety of TAF9-binding transcriptional activators. A similar motif is present in the rv-cyclin AD, and point mutations within this motif affect rv-cyclin function and protein-protein interactions. The results support a model of transcription regulation by direct interaction with TAF9.
Walleye dermal sarcoma virus (WDSV), a member of the epsilonretroviruses in the family Retroviridae, is a complex retrovirus etiologically associated with dermal sarcomas in walleye fish (Sander vitreus, formerly Stizostedion vitreum) (13, 14, 26-28). The seasonal appearance of this disease is characterized by the expression of low levels of spliced accessory gene transcripts A1 and B during the period of tumor development and high levels of full-length and spliced transcripts during tumor regression (17, 20).
Transcript A1 encodes a retroviral cyclin (rv-cyclin) (OrfA protein) with a cyclin box motif but with limited homology to host cyclins (10). WDSV rv-cyclin was able to induce cell-cycle progression in G1-cyclin-deficient yeast (Saccharomyces cerevisiae) (10) and was also associated with the induction of hyperplastic skin lesions in transgenic mice when expressed from a keratin promoter (9). In mammalian or piscine cell culture, WDSV rv-cyclin localized in the nucleus and was concentrated in interchromatin granule clusters (20), nuclear domains enriched in proteins necessary for mRNA transcription and processing. The first 112 amino acids (aa) were necessary for localization in interchromatin granule clusters. The rv-cyclin was found, by coimmunoprecipitation and by glutathione S-transferase (GST) pulldown, to be specifically associated with active transcription complexes, with cofactors of transcription, and with cyclin-dependent kinase 8, cyclin C, and Sur-2, components of the Mediator complex (21, 22). Mediator serves as a general cofactor of transcription (reviewed in references 1, 3, and 19).
In piscine cells, exogenous rv-cyclin inhibited transcription from the WDSV promoter in a luciferase reporter system (22, 29), and this inhibition was independent of cis-acting DNA sequences within the virus long terminal repeat (21). However, other viral promoters, such as that of simian virus 40 (SV40), were found to be activated by exogenous rv-cyclin in the same cells (22). In select mammalian cells, rv-cyclin activated the WDSV promoter and inhibited the SV40 promoter, demonstrating its ability to either inhibit or activate transcription in a manner dependent on both the promoter and the cell type (22). rv-cyclin activated transcription from a GAL4 promoter when fused directly to the GAL4 DNA binding domain (DBD), and a minimal, 30-aa activation domain (AD) was identified in the carboxyl region of the protein (21). The AD, when fused to GST, pulled down CBP, p300, and Sur-2, and point mutations that altered its activation function also altered its capacity for these protein interactions (21). Furthermore, mutations that altered AD function and protein interactions reduced the capacity of the full-length protein to inhibit transcription from the WDSV promoter.
The rv-cyclin AD was first identified by sequence alignment with the acidic activation domain of VP16 (21). Subsequent comparisons of the transcription functions and protein-protein interactions of the rv-cyclin AD with those of the two subdomains of the VP16 AD, VP16N and VP16C, indicated similarities specifically with the VP16C subdomain (21). The VP16C subdomain shares a conserved motif with a number of transcriptional activators which are capable of binding to TATA binding protein-associated factor 9 (TAF9) (2), a component of the TFIID transcription factor required for transcription initiation by RNA polymerase II. Here, the comparisons of the rv-cyclin AD with VP16C are extended to TAF9 binding and its effects on the regulation of transcription.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis.
All rv-cyclin sequences, whether full length or partial, were derived from the pKH3OrfA construct (20). Full-length subclones were prepared directly from the BamHI and EcoRI insertion sites of rv-cyclin in pKH3 vector when possible. When necessary, and for the cloning of all subfragments, PCR primers were designed to obtain the desired fragment sequence and to bestow restriction sites and the appropriate reading frame for each vector system. GAL4 DNA binding domain fusions for assays of metazoan tissue culture cells were generated in pFA-CMV vector (Stratagene). GST fusion proteins were generated in pGEX-2T vector (Pharmacia). The VP16 AD (aa 413 to 490) and mutated VP16 AD (F442S and F475A) were expressed from the vectors pGEX-2T-VP16 and -VP16M, which were generous gifts from Andrew P. Rice, Department of Molecular Virology and Microbiology, Baylor College of Medicine. The VP16 AD subregions VP16N (aa 413 to 452; H1) and VP16C (aa 453 to 490; H2) and their corresponding mutations, VP16N F442S and VP16C F475A, were derived by PCR from pGEX2T-VP16 and -VP16M and cloned in pGEX-2T. Amino-terminal poly-His fusions were generated in pET-19 vector (Novagen).
For nuclear localization of peptides expressed in cell culture, a pKH3 vector was prepared with an SV40 large T antigen nuclear localization signal (nls) (PKKKRKV) cloned in frame with a triple hemagglutinin tag for fusion at the amino terminus of the expressed protein.
All point mutations in rv-cyclin were made with a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The resulting mutations and the sequences of all constructs were confirmed by sequence analysis.
Protein expression.
GST and His fusion proteins were expressed in BL21(DE3)-RIL cells (Stratagene) by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. Bacteria were lysed by sonication, and soluble proteins were purified twice by affinity chromatography on glutathione Sepharose (Amersham Pharmacia) or nickel-charged His-bind resin (Novagen) according to the manufacturers' instructions. Insoluble His-tagged proteins were solubilized in 6 M guanidine and refolded by rapid dilution in 1 M nondetergent sulfobetaine 201 (Calbiochem) (5). Full-length His-rv-cyclin was further purified by gel filtration on Superose 6 Sepharose (Amersham Pharmacia).
Reporter-gene constructs and luciferase assays.
pFR-Luc reporter vector (Stratagene) contains five tandem repeats of the yeast GAL4 DNA binding site upstream of the luciferase gene. HeLa cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum at 37°C in 5% CO2. W12 cells (a WDSV-negative walleye fibroblast cell line established in the laboratory of Paul Bowser at Cornell University by Danielle Martineau) were maintained in Liebowitz 15 medium with 10% fetal calf serum (Gibco), 4 mM glutamine, 100 units penicillin ml−1, and 100 μg streptomycin ml−1 at 20°C. Cells were seeded into 24-well plates in a volume of 1 ml medium and transfected using 0.3 μg of pFR-Luc vector, 0.01 μg (W12) or 0.0001 μg (HeLa) of pRL-TK (Promega), 0.025 μg pFA-VP16N or pFA-VP16C, and 0.1 μg pKH3nls or pKH3nls240-270 with FuGENE 6 (Roche) according to the manufacturers' suggestions. For the pKH3OrfA titration experiment, cells were transfected with 0.3 μg pFR-luc, 0.01 μg pRL-TK, 0.025 μg pFA-VP16N or pFA-VP16C, and a total of 0.1 μg of pKH3 and/or pKH3OrfA. Experiments were performed using a dual-luciferase reporter assay system (Promega). Cell lysates were harvested 48 h (HeLa) or 72 h (W12) after transfection with passive lysis buffer and then centrifuged at 20,000 × g for 5 min at 4°C. Luciferase activities were obtained with a Beckman Coulter LD400 luminometer. Luciferase activity from the reporter vector was normalized for transfection efficiency with values from the cotransfected pRL-TK vector. All transfections were performed in quadruplicate. Student's t test and 95% confidence intervals based on a t distribution were used for statistical analyses. A P value of less than 0.01 was considered significant.
GST pulldown.
Nuclear extracts from HeLa cells were prepared by hypotonic lysis and KCl extraction of nuclei as described by Mayeda and Krainer (15). Extracts were diluted to 2 μg/μl in D100 buffer (10 mM HEPES [pH 8.0], 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5% NP-40) and preadsorbed with 10 μg purified GST protein and 50 μl glutathione Sepharose CL-4B per mg total protein extract overnight at 4°C. Aliquots (75 μg) of preadsorbed extracts were rotated overnight with 2.5 μg equivalents of glutathione Sepharose-bound GST fusion proteins (10 to 20 μl of a 10% suspension) in siliconized microcentrifuge tubes. Relative quantities of bound input fusion proteins were confirmed by Western analysis with anti-GST antibody (Pierce). Glutathione Sepharose-bound proteins were pelleted for 8 s in an Eppendorf microcentrifuge and washed five times with 250 μl D100 buffer. The final pellets were suspended in 8 μl sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading dye and heated, the soluble portion was separated on 4% to 12% denaturing polyacrylamide gradient gels, and Western blots were probed with the indicated antibodies. Multiple proteins were detected sequentially after treating blots with strip buffer (30 mM Tris-HCl [pH 6.8], 150 mM NaCl, 1 M β-mercaptoethanol, 0.2% sodium dodecyl sulfate) for 1 h at 45°C. The antibodies used for Western blots included a monoclonal antibody to Sur-2 (Med 23; PharMingen) and Santa Cruz antibodies reactive to CBP (C-1; monoclonal antibody), p300 (N-15; rabbit), and TAF II p32 (TAF9; N-16; goat). The CBP and p300 antibodies do not cross-react.
In order to block GST pulldowns, 20 μg or indicated quantities of purified poly-His-tagged proteins were incubated overnight at 4°C with 75 μg of nuclear extract in a final volume of 300 μl D100 buffer. The overnight His-protein-nuclear extract preparations were centrifuged at 20,000 × g for 15 min at 4°C, and the supernatant was subjected to GST pulldown and analysis as described above. Control samples (no block) were treated identically but with no His protein. One-fifth of the supernatant remaining after GST pulldown, corresponding to 4 μg input poly-His fusion protein, was subjected to trichloroacetic acid precipitation and Western blot analysis with anti-poly-His antibody (Sigma) in order to confirm relative quantities of blocking proteins.
For the in vitro assay of protein-protein interactions, His-tagged proteins were suspended at 1 μg/100 μl TAF9 binding buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, 10% glycerol, 1 μg/ml bovine serum albumin, 0.1 mM EDTA, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.1% NP-40) and preadsorbed with 1 μg equivalents of glutathione Sepharose-bound GST fusion protein overnight at 4°C. Aliquots of 1 μg were rotated overnight with 1 μg equivalents of glutathione Sepharose-bound GST fusion proteins (1 to 5 μl of a 10% suspension) in siliconized microcentrifuge tubes. Relative quantities of bound input fusion proteins were confirmed by Western analysis with anti-GST antibody (Pierce). Glutathione Sepharose-bound proteins were pelleted for 8 s and washed five times with 250 μl TAF9 binding buffer. Final pellets were suspended in 8 μl sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading dye and heated, the soluble portion was separated on 12% denaturing polyacrylamide gels, and Western blots were probed with mouse anti-TAF9 (N-16; Santa Cruz) or anti-poly-His (Sigma).
Analysis of protein sequences.
Protein sequences used for comparative alignments were retrieved from the NCBI nr database (National Center for Biotechnology Information [GenBank]; http://www.ncbi.nlm.nih.gov) and included human herpesvirus 1 (herpes simplex virus type 1) alpha gene trans-inducing factor (alpha-TIF, VP16, Vmw65; accession no. NP_044650) and WDSV rv-cyclin (OrfA protein; AAA99528). TAF9 binding proteins identified by Choi et al. (2) included ALL-1 (acute lymphocytic leukemia protein; Q03164); HSF-1 (heat shock transcription factor 1; AAP36015); ESX (epithelial restricted with serine box, also known as E74-like factor 3 [ELF3] and epithelial-specific Ets protein; AAB58075); NFAT1 (nuclear factor of activated T cells; AAC50886); NF-κB p65 (nuclear factor kappa-B; AAA36408); NF-IL-6 (nuclear factor for interleukin-6 expression; S12788); and p53 (tumor protein 53; NP_000537). The accession numbers for TAF9 cDNAs include human (BT019652), spotted green pufferfish (Tetraodon nigroviridis; CR674522), and zebrafish (Danio rerio; BC076279). All protein sequences were aligned with MacVector multiple sequence alignment software (Accelrys) using the Clustal W algorithm (23) with the BLOSUM 30 matrix, an open gap penalty of 10.0, and an extend gap penalty of 0.1.
Cloning of walleye TAF9.
RNA was isolated from cultured walleye dermal sarcoma tissue with RNAzol (Tel-Test, Inc.), and cDNAs were prepared with random hexamer primers and Superscript III according to instructions of the manufacturer (Invitrogen). An internal TAF9 sequence was amplified from the cDNA by use of primers designed using highly conserved regions of zebrafish and pufferfish sequences (5′ primer, 5′-ATCAC[G/A]GAGTACGA[G/A]CCCAGAGT-3′; 3′ primer, 5′-GC[A/G]TC[A/G]TAGTC[A/G]TC[C/G]TC[A/C]TCTTCATGCTTC-3′). The remaining unknown walleye TAF9 sequence was then determined by RNA ligase-mediated and oligonucleotide-capping rapid amplification of cDNA ends (RACE) according to the instructions of the manufacturer (GeneRacer kit; Invitrogen). Internal, walleye-specific RACE primers were as follows: 5′ RACE primer, 5′-GGAAGCATGAAGATGAGGATGACTAT-3′; 3′ RACE primer, 5′-GGGGAGTCGGGGGCCGGTGTAGGGTTT-3′.
Nucleotide sequence accession number.
The final cDNA sequence is a consensus of 15 individual clones and has been assigned GenBank accession number DQ768754.
RESULTS
His-tagged rv-cyclin interferes with VP16 AD protein interactions.
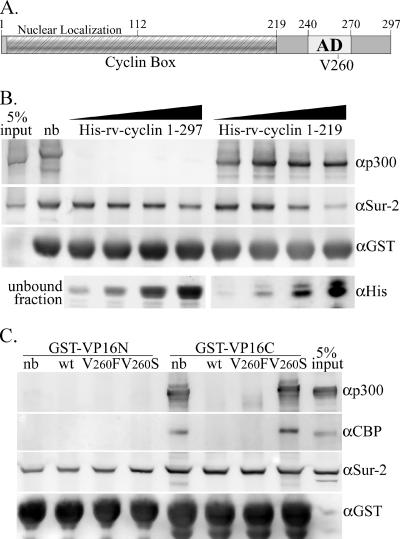
As shown previously, a GST-VP16 AD fusion protein has the capacity to pull down a number of proteins from HeLa nuclear extracts such as CBP/p300 and the entire Mediator complex (6, 7, 21). WDSV rv-cyclin protein when fused to GST has also been shown to have the capacity to pull down a variety of nuclear proteins, including CBP/p300 and a component of Mediator, Sur-2 (Med 23) (21). The possibility that rv-cyclin shares specific binding sites with the VP16 AD was tested by incubation of nuclear extracts with rv-cyclin prior to incubation and pulldown with GST-VP16 AD (aa 413 to 490). Figure 1B shows the results of preincubation of nuclear extracts with serial twofold dilutions of either full-length, His-tagged rv-cyclin (aa 1 to 297) or a carboxy-truncated form (aa 1 to 219), which excludes the rv-cyclin AD (aa 240 to 270). Both proteins had slight effects on the pulldown of Sur-2 at their highest concentrations, but only full-length rv-cyclin had the capacity to block VP16 pulldown of p300, and blocking was efficient at all concentrations of His-rv-cyclin. These data suggest a shared p300 binding site between the carboxyl domain of rv-cyclin and the VP16 AD.
FIG. 1.
WDSV rv-cyclin blocks pulldown of proteins by GST-VP16. A. Model of rv-cyclin illustrating regions characterized by sequence analysis (Cyclin Box) or by functional assays (nuclear localization and AD). Numbers represent amino acid positions. Valine at position 260 within the defined AD is indicated (V260). B. Western blot of pulldown by GST-VP16 AD from 75 μg HeLa nuclear extract (nb [no block]) and from matching extracts preincubated with increasing twofold concentrations (20 to 160 μg) of recombinant His-rv-cyclin (aa 1 to 297) or carboxy-truncated His-rv-cyclin (aa 1 to 219). Blots were probed consecutively with antibodies reactive to p300 (αp300), Sur-2 (αSur-2), and GST (αGST), which recognizes the input GST-VP16 AD fusion protein. A portion of the unbound fraction, representing 20% of total input, was subsequently precipitated and run separately to assess His-tagged proteins with anti-poly-His antibody (αHis; bottom panels). C. Pulldowns by GST-VP16N (aa 413 to 452) and GST-VP16C (aa 453 to 490) from 75 μg HeLa nuclear extract (nb) and from matching extracts preincubated with 20 μg wild-type (wt) or mutated (V260F and V260S) His-rv-cyclin carboxyl region (aa 219 to 297). Blots were probed consecutively with antibodies reactive to CBP (αCBP), p300 (αp300), Sur-2 (αSur-2), and GST (αGST), which recognizes the input GST-VP16N and GST-VP16C fusion proteins. The antibodies used for CBP and p300 are specific for these proteins and do not cross-react.
The VP16 AD contains two functional subdomains, VP16N (aa 413 to 452; H1 subdomain) and VP16C (aa 453 to 490; H2 subdomain) (7, 18), with different capacities for protein interactions. VP16N binds Mediator but not CBP/p300, and VP16C binds Mediator and CBP/p300 (6, 7, 21). The differential blocking of CBP/p300 pulldown by His-tagged rv-cyclin suggested specific interference with the VP16C subdomain, and the dependence of this block on the presence of the rv-cyclin carboxyl region (aa 219 to 297) suggested a specific interaction by this region. A His-tagged carboxyl region protein (His-219-297) was tested for the capacity to block the pulldown of proteins by GST fusion constructs containing either VP16 subdomain (Fig. 1C). As observed previously, GST-VP16N was unable to pull down either CBP or p300 (first lane marked nb, where nb indicates “no block”), and His-219-297 did not interfere with GST-VP16N pulldown of Sur-2. Preincubation with His-219-297 did block GST-VP16C pulldown of CBP and p300 (Fig. 1C). Additionally, mutations of valine at position 260 in the region comprising positions 219 to 297 were tested. These mutations within the defined AD (aa 240 to 270) had been previously shown to affect AD function and protein binding (21). V260F is a conservative mutation that enhanced AD function and protein interactions, and V260S reduced function and interactions. Here, the V260F mutation of His-219-297 blocked GST-VP16C pulldown of CBP and p300 and the V260S mutation did not. His-219-297 did not interfere with the pulldown of Sur-2 by GST-VP16C. These results show that the rv-cyclin carboxyl region can interfere with the interactions of VP16C with CBP and p300 in nuclear extracts and suggest a specific role for the rv-cyclin AD in this interference.
GST-rv-cyclin AD pulls down TAF9.
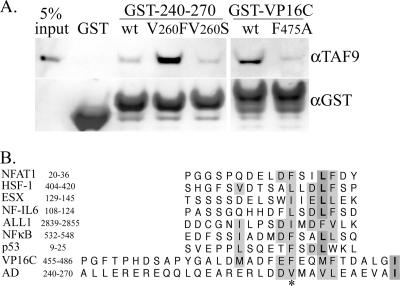
Previously, the rv-cyclin AD, fused to GST, was able to pull down a number of cellular proteins from HeLa nuclear extracts, including p300, CBP, Sur-2, and cdk8, but its pulldown capacity was reduced from that of the entire carboxyl region (aa 219 to 297) (21). Mutation V260F enhanced the GST-AD pulldowns and allowed pulldown of TATA-binding protein (TBP), and mutation V260S greatly reduced interactions with CBP and p300 (21). The capacity of the GST-rv-cyclin AD to pull down CBP and p300 was comparable to that of the GST-VP16C fusion protein (6, 7, 21), and VP16C had been shown to bind directly to human TAF9 (2). The result of a test of GST-rv-cyclin AD pulldown of TAF9 from HeLa nuclear extracts was positive (Fig. 2A). GST-VP16C also pulled down TAF9, as expected. The rv-cyclin AD-TAF9 interaction was enhanced by the V260F mutation but only slightly reduced by the V260S mutation. The V260S mutation of the rv-cyclin AD yielded results similar to the F475A mutation of VP16C. These results indicate that the rv-cyclin AD interacts with TAF9 or with TAF9-containing complexes in HeLa nuclear extracts.
FIG. 2.
GST-rv-cyclin AD pulls down TAF9. A. Pulldowns from 75 μg HeLa nuclear extract by GST protein, by wild-type GST-rv-cyclin AD (GST-240-270) (wt) and mutated forms V260F and V260S, and by GST-VP16C and corresponding mutated form F475A. The Western blot was probed with antibody reactive to the amino terminus of human TAF9 (aTAF9) and reprobed with anti-GST (αGST; bottom panel), which recognizes the input GST and GST fusion proteins. The lane labeled “input” represents 5% of the total extract used for each pulldown. B. Alignment of TAF9-binding motifs identified by Choi et al. (2) with the VP16C AD (aa 455 to 486) and rv-cyclin AD (aa 240 to 270). The relative amino acid position of each motif within its respective protein is indicated after each name. Similar and identical residues are shaded. An asterisk indicates the relative positions of VP16C F475 and rv-cyclin V260. ALL-1, acute lymphocytic leukemia protein; HSF-1, heat shock transcription factor 1; ESX, epithelial restricted with serine box, also known as E74-like factor 3 [ELF3] and epithelial-specific Ets protein; NFAT1, nuclear factor of activated T cells; NFκB p65, nuclear factor kappa-B; NF-IL-6, nuclear factor for interleukin-6 expression; and p53, tumor protein 53.
The VP16C AD shares a conserved TAF9 binding domain with other transcriptional activators (2). Figure 2B shows several TAF9 binding motifs, predicted and confirmed by Choi et al. (2), aligned with the VP16C and WDSV rv-cyclin ADs. Valine 260 lies within the motif and aligns with phenylalanine 475 of the VP16C AD. F475 was originally identified as an effective target for mutation to reduce the ability of the VP16 AD to activate transcription (18).
His-tagged rv-cyclin AD interferes with VP16C protein interactions.
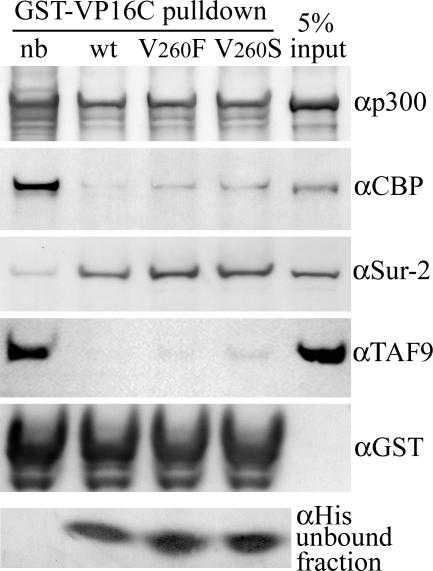
When a His-tagged rv-cyclin AD (aa 240 to 270) and corresponding V260 mutations were tested for their ability to block pulldowns by the GST-VP16C fusion protein, there was very little interference with the pulldown of p300 but significant interference with CBP pulldown (Fig. 3). This is in contrast to the interference by the larger carboxyl region, which blocked pulldown of both p300 and CBP (Fig. 1C). As expected, there was no interference with Sur-2 pulldown. The isolated rv-cyclin AD did demonstrate a complete block of TAF9 pulldown. Mutations at V260 had little effect on blocking of CBP and TAF9 pulldown by GST-VP16C; the V260S mutant was able to block most but not all. These results suggest that the rv-cyclin AD competes directly with VP16C for TAF9 and CBP binding. Regions outside of the isolated AD may be responsible for more-extensive protein interactions, as observed previously (21) and reflected here in a greater capacity to interfere with VP16 interactions. The effects of mutations at V260 were also greater in the context of the entire carboxyl region.
FIG. 3.
Pulldowns by GST-VP16C from 75 μg HeLa nuclear extract (nb [no block]) and from matching extracts preincubated with 20 μg wild-type (wt) and mutant (V260F and V260S) His-rv-cyclin AD (aa 240 to 270). Blots were probed consecutively with antibodies reactive to CBP (αCBP), p300 (αp300), Sur-2 (αSur-2), and GST (αGST), which recognizes the input GST-VP16 fusion protein. The antibodies used for CBP and p300 are specific for these proteins and do not cross-react. A portion of the unbound fraction, representing 20% of total input, was subsequently precipitated and run separately to assess His-tagged proteins with anti-poly-His antibody (αHis; bottom panel). The lane labeled “input” represents 5% of total untreated extract used for each pulldown.
rv-cyclin carboxyl region and isolated AD bind TAF9 in vitro.
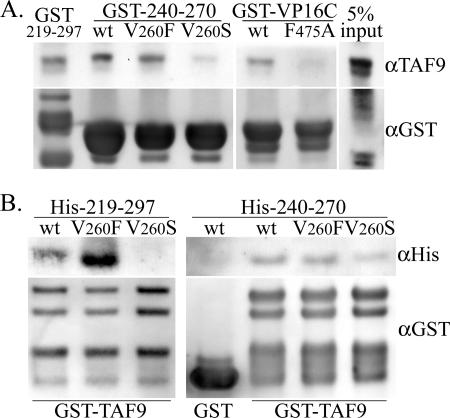
A cDNA construct was prepared in order to express and purify a His-tagged protein containing the first 140 aa of human TAF9. This portion of TAF9 had been shown previously to include the VP16C binding region and to bind to GST-VP16C in protein-protein interaction assays (2). Figure 4A demonstrates the pulldown of recombinant His-TAF9 (aa 1 to 140) with GST fusions of the rv-cyclin carboxyl region (aa 219 to 297), with wild-type and mutated forms of the rv-cyclin AD (aa 240 to 270), and with wild-type and mutated VP16C. The carboxyl domain and isolated AD both pulled down His-TAF9. The V260S mutation reduced the pulldown compared to the results seen with the wild type and the V260F mutation. The GST-VP16C construct also bound His-TAF9 compared to the results seen with signal from the mutated form, F475A. The reduction in signal with the V260S mutation was comparable to the effect of the F475A mutation of VP16C.
FIG. 4.
Interaction in vitro of recombinant TAF9 (aa 1 to 140) and rv-cyclin carboxyl regions. A. Pulldown of recombinant, His-tagged TAF9 by GST fusion proteins rv-cyclin aa 219 to 297 (GST-219-297), by wild-type GST-rv-cyclin AD (GST-240-270) (wt) and mutated forms V260F and V260S, and by wild-type GST-VP16C (wt) and corresponding mutated form, F475A. Western blots were probed with antibody reactive to the amino terminus of TAF9 (αTAF9) and reprobed with anti-GST (αGST; bottom panel), which recognizes the input GST fusion proteins. B. Reciprocal pulldown in vitro of wild-type His-tagged rv-cyclin aa 219 to 297 (His-219-297) (wt) and aa 240 to 270 (His-240-270) (wt) and mutated forms of each protein, V260S and V260F, with GST-TAF9 fusion protein (aa 1 to 140). The results of a control pulldown of wild-type His-240-270 (wt) with GST protein alone are shown in the first lane of the right panel. Western blots were probed with antibody reactive to poly-His (αHis) and reprobed with anti-GST (αGST; bottom panel), which recognizes the input GST and GST-TAF9 fusion protein.
In reciprocal experiments, the His-tagged rv-cyclin carboxyl region (aa 219 to 297) and AD (aa 240 to 270) proteins were subject to pulldown by a GST-TAF9 fusion protein (aa 1 to 140) in separate experiments (Fig. 4B). Only the wild-type and V260F-mutated forms of His-219-297 were pulled down by GST-TAF9, and the V260F mutation enhanced pulldown of His-219-297. The effects of the V260 mutations on the pulldown of His-240-270 were less apparent, with only a small decrease in pulldown from equivalent quantities of the His-240-270 V260S protein and no apparent enhancement of pulldown by the V260F mutation compared to wild-type results (Fig. 4B). As observed in blocking experiments, the effect of the V260S mutation was greater in the context of the larger His-219-297 protein than in that of the isolated AD, suggesting variations in the extent and total number of contacts these proteins have with TAF9.
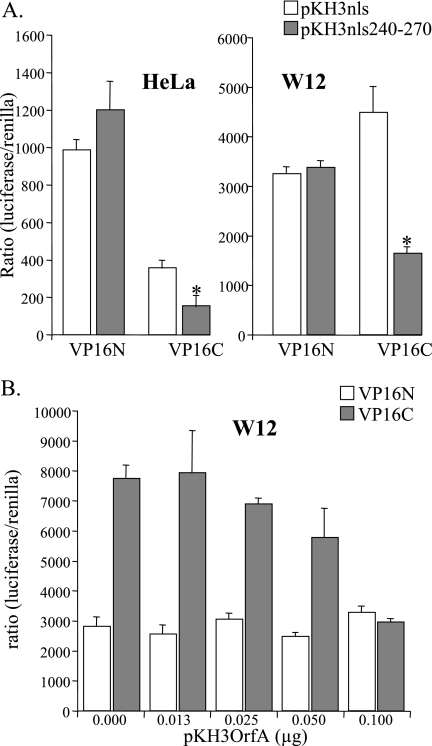
Expression of rv-cyclin interferes with VP16C activation of transcription.
The VP16 subdomains when fused to the GAL4 DBD activate transcription from pFR-luc, a construct containing five GAL4 DNA binding sites, to drive luciferase expression in cells. In order to localize expressed rv-cyclin AD (aa 240 to 270) to the nucleus, a cytomegalovirus-driven nls fusion construct, pKH3nls, was prepared and the coding sequence for the rv-cyclin AD cloned in frame (pKH3nls240-270). Coexpression of the nls-rv-cyclin AD in cells with either the GAL4DBD-VP16N or -VP16C construct and the luciferase reporter demonstrated significant inhibition only of VP16C activity in HeLa cells (Fig. 5A). In the W12 walleye cell line, the GAL4DBD-VP16 subdomains activate transcription from pFR-luc and the VP16C subdomain has greater activity than the VP16N subdomain (21). Coexpression of the nls-rv-cyclin AD with either GAL4DBD-VP16N or -VP16C demonstrated significant inhibition only of VP16C activity in walleye cells (Fig. 5A). Efforts to test the reversal of inhibition of GAL4DBD-VP16C activity by the overexpression of TAF9 were precluded by the inhibition of GAL4DBD-VP16C by exogenous TAF9 alone (data not shown).
FIG. 5.
Interference with the activation of transcription by GAL4-VP16C in human and walleye cells. A. HeLa cells or walleye W12 cells were cotransfected with a GAL4-luciferase reporter (pFR-luc), vectors expressing GAL4-DNA binding domain fusion of either VP16N or VP16C, control nls expression vector pKH3nls or an rv-cyclin aa 240 to 270-nls fusion protein (pKH3nls240-270), and Tk promoter-Renilla vector pRL-Tk. All transfections were performed and assayed in quadruplicate. *, statistically significant differences (P < 0.01) in the results of assays of pKH3nls versus pKH3nls240-270 expression. B. Walleye W12 cells were cotransfected with a GAL4-luciferase reporter (pFR-luc), vectors expressing GAL4-DNA binding domain fusion of either VP16N or VP16C, indicated quantities of pKH3OrfA vector, which expresses an hemagglutinin-tagged, full-length rv-cyclin protein, and pRL-Tk. All transfections were performed and assayed in quadruplicate. Results are reported as ratios of luciferase activity versus Renilla activity to correct for transfection efficiency.
Full-length rv-cyclin contains a nuclear localization domain in the amino end of the protein (20). WDSV rv-cyclin expressed from pKH3OrfA vector (20) was tested for its effect on the activation of luciferase expression by the GAL4DBD-VP16N and -VP16C constructs (Fig. 5B). Titration of the pKH3OrfA construct demonstrated specific inhibition of the activity of GAL4DBD-VP16C in walleye cells. These results demonstrate direct and specific interference with VP16C function in both human and walleye cells and suggest a common mechanism for the effects of the rv-cyclin across species: specific interaction with TAF9 in transcription complexes.
rv-cyclin carboxyl region and isolated AD bind walleye TAF9 in vitro.
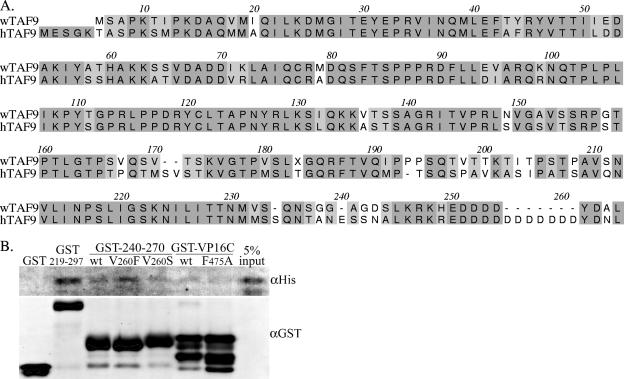
TAF9 is highly conserved among metazoan organisms. Walleye TAF9, cloned and sequenced from WDS tumor cell cDNA, was found to be 71% identical and 80% similar to human TAF9 (Fig. 6A). Variation was greater at the amino and carboxyl termini and at three areas in the carboxyl half of the sequence: aa 167 to 172, 193 to 211, and 238 to 243. Within the region of TAF9 used for in vitro assays of protein-protein interaction, aa 1 to 140, and the corresponding walleye TAF9 sequence, aa 1 to 136, the walleye and human sequences are 78% identical and 94% similar.
FIG. 6.
Comparisons of walleye and human TAF9. A. Alignment of walleye TAF9 (wTAF9) and human TAF9 (hTAF9) amino acid sequences. Similar and identical residues are shaded. B. Pulldown of recombinant, His-tagged walleye TAF9 by GST or GST fusion proteins: rv-cyclin aa 219 to 297 (GST-219-297), by wild-type GST-rv-cyclin AD (GST-240-270) (wt) and mutated forms V260F and V260S, and by wild-type GST-VP16C (wt) and the corresponding mutated form, F475A. Western blots were probed with antibody reactive to poly-His (αHis) and reprobed with anti-GST (αGST; bottom panel), which recognizes the input GST fusion proteins.
The first 136 aa of the walleye TAF9 protein sequence were cloned for expression with a His tag. The purified, recombinant protein was then subjected to an in vitro protein-protein interaction assay with GST fusions of the rv-cyclin carboxyl region, isolated AD or mutated AD, and VP16C (Fig. 6B). As observed with His-tagged human TAF9 (aa 1 to 140), walleye TAF9 (aa 1 to 136) was pulled down by the GST-219-297 and GST-240-270 fusion proteins. The signal was greatest with the larger carboxyl region, and pulldown by the isolated AD was enhanced by the V260F mutation. GST-VP16C also pulled down walleye TAF9 but to a lesser degree, perhaps due to degradation of the GST fusion protein (Fig. 6B; αGST). The results demonstrate the direct interaction of the rv-cyclin AD with walleye TAF9 in vitro.
DISCUSSION
The outcome of these studies points to a common TAF9 binding motif for the rv-cyclin AD and the VP16C subdomain as well as a common association with CBP and p300. While the TAF9 binding can be attributed to the presence of a specific sequence motif, interactions with CBP and p300 may be the result of less-specific mechanisms arising from TAF9 interactions. This general mechanism is indicated by the inability of the isolated AD to block p300 pulldown and by significant blocking of CBP pulldown by GST-VP16C. CBP and p300 binding sites have not been identified in VP16 and VP16 binding sites have not been identified in CBP and p300, although GAL4-VP16 activation can be inhibited by deletions of the bromodomain or the CH3/E1A-binding domain of p300 (8). The differential pulldown results for p300 versus CBP may also represent specific interactions between TAF9 and CBP.
The importance of a specific contact between the rv-cyclin AD and TAF9 does not lie in its comparison to VP16 but in the functions of the other transcription activators listed in Fig. 2A that have known TAF9 binding motifs. Of particular interest is p53, which requires TAF9 interaction for transcription activation (11, 12, 24, 25). Interference with p53-TAF9 binding serves in the attenuation of p53 activity by MDM2 protein (25), and interference with p53 function is a hallmark of tumor induction by DNA tumor viruses. The possible alteration of p53 function by the presence of rv-cyclin awaits future investigation.
An alternative to a possible “p53-interference” mechanism in the promotion of oncogenesis would be a direct rv-cyclin function in the specific activation or inhibition of transcription of a subset of host genes. The first 219 aa of rv-cyclin contain a predicted cyclin box motif (10). This motif consists of two repeats of five alpha helical regions. Each of these regions defines a predicted cyclin box fold, a structure characterized by low sequence but high structural homology (16). The rv-cyclin box has no specific association with cis-acting sequences in the WDSV promoter or with the DNA binding proteins that contact these sequences (21). Although cooperative effects in collaboration with DNA binding proteins may yet be identified, as with VP16 and Oct-1, current data suggest that the regulation of gene expression by rv-cyclin is mediated by control of the composition of the transcription complex. Cyclin boxes are shared by cyclins and the transcriptional regulatory proteins retinoblastoma protein (pRb) and transcription factor IIB (TFIIB). The protein interactions of pRb and TFIIB are quite distinct, so there is no clear indication whether divergent rv-cyclin should mimic one of these two proteins or interfere with their function, but the combination of cyclin box with TAF9 binding provides the level of complexity necessary for function as a novel regulator of transcription. rv-cyclin has demonstrable effects on gene expression that are dependent upon both promoter sequence and cell type (21, 22).
The different results obtained with full-length rv-cyclin, the carboxyl region, and the isolated AD demonstrate the variety of physical interactions that these proteins must be involved in. In particular, the efficient blocking of VP16C pulldown of CBP/p300 by the full-length and carboxyl region proteins suggests that aa 219 to 297 make contacts in addition to the AD-TAF9 interaction. Such contacts may increase the affinity of the interaction with TAF9 or extend to other proteins within CBP- or p300-containing complexes. The GST-219-297 construct has been shown to pull down TBP when the isolated GST-AD could not, but the V260F mutation of the isolated AD conferred TBP pulldown (21). The current results indicate that theTAF9 interaction is key to the expanded protein-protein interactions of the carboxyl region and full-length proteins. The carboxyl region blocking of VP16C pulldown of p300/CBP was lost with the V260S mutation of the AD/TAF9 binding motif, and the His-tagged carboxyl region protein with the V260S mutation was unable to bind GST-TAF9 (aa 1 to 140). The isolated AD appeared less sensitive to mutation at position 260 except for in the pulldown of endogenous TAF9 from nuclear extracts, where the V260F mutation enhanced it. In blocking experiments, its inability to affect p300/CBP pulldown by VP16C precluded evaluation of the mutations and there was little difference in the results of blocking of TAF9 pulldown. Previous work with mutations in VP16C has shown only very modest changes in TAF9 binding in vitro (2), and the results with the F475A mutation, tested here with human and walleye TAF9, were modest as well. The degree of TAF9 binding by the isolated rv-cyclin AD appears in general to be smaller. In the case of blocking of VP16C pulldowns, the results may be due to greater steric hindrance by the larger protein, enhancing the outcome of slight variations in TAF9 affinity. Sequences outside of or overlapping aa 240 to 270 may also be responsible for greater TAF9 interaction.
Previous analyses of the effects of WDSV rv-cyclin on transcription from various promoters identified paradoxical variations with the same promoter in the cells of different vertebrate species. Although TAF9 and other key transcriptional regulatory proteins are highly conserved across these species, they are not identical. Indeed, a second, TAF9-like protein, TAF9b, has recently been identified exclusively in humans, and it imparts variations in the regulation of gene expression (4). While rv-cyclin may be able to contact either walleye or human TAF9 proteins functionally, variations in the extent and affinity of such contacts and in the content of the surrounding protein complexes can determine the difference between the activation and inhibition of transcription from the same promoter.
In walleye dermal sarcomas, the lack of full virus expression may be critical to immune evasion by the virus. The outcome of this function is the production of a mass of cells harboring WDSV provirus. During spawning, the virus expression pattern shifts to virus production and transmission is maximized by the regression of the tumor mass and the release of the viral load to the environment. rv-cyclin appears to function specifically in the suppression of virus expression during tumorigenesis, and yet its ability to enhance expression from different promoters in the same cell type may contribute to host cell proliferation.
Acknowledgments
This work was supported by National Institutes of Health grant RO1 CA095056 from the National Cancer Institute.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Blazek, E., G. Mittler, and M. Meisterernst. 2005. The mediator of RNA polymerase II. Chromosoma 113:399-408. [DOI] [PubMed] [Google Scholar]
- 2.Choi, Y., S. Asada, and M. Uesugi. 2000. Divergent hTAFII31-binding motifs hidden in activation domains. J. Biol. Chem. 275:15912-15916. [DOI] [PubMed] [Google Scholar]
- 3.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30:250-255. [DOI] [PubMed] [Google Scholar]
- 4.Frontini, M., E. Soutoglou, M. Argentini, C. Bole-Feysot, B. Jost, E. Scheer, and L. Tora. 2005. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol. 25:4638-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg, M. E., N. Expert-Bezancon, L. Vuillard, and T. Rabilloud. 1996. Non-detergent sulphobetaines: a new class of molecules that facilitate in vitro protein renaturation. Fold. Des. 1:21-27. [DOI] [PubMed] [Google Scholar]
- 6.Hardy, S., M. Brand, G. Mittler, J. Yanagisawa, S. Kato, M. Meisterernst, and L. Tora. 2002. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem. 277:32875-32882. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda, K., T. Stuehler, and M. Meisterernst. 2002. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 7:49-58. [DOI] [PubMed] [Google Scholar]
- 8.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lairmore, M. D., J. R. Stanley, S. A. Weber, and D. L. Holzschu. 2000. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proc. Natl. Acad. Sci. USA 97:6114-6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaPierre, L. A., J. W. Casey, and D. L. Holzschu. 1998. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J. Virol. 72:8765-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 92:5154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, H., J. Lin, J. Chen, and A. J. Levine. 1994. The regulation of p53-mediated transcription and the roles of hTAFII31 and mdm-2. Harvey Lect. 90:81-93. [PubMed] [Google Scholar]
- 13.Martineau, D., P. R. Bowser, R. R. Renshaw, and J. W. Casey. 1992. Molecular characterization of a unique retrovirus associated with a fish tumor. J. Virol. 66:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martineau, D., R. Renshaw, J. R. Williams, J. W. Casey, and P. R. Bowser. 1991. A large unintegrated retrovirus DNA species present in a dermal tumor of walleye Stizostedion vitreum. Dis. Aquat. Org. 10:153-158. [Google Scholar]
- 15.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 16.Noble, M. E. M., J. A. Endicott, N. R. Brown, and L. N. Johnson. 1997. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biol. Sci. 22:482-487. [DOI] [PubMed] [Google Scholar]
- 17.Quackenbush, S. L., D. L. Holzschu, P. R. Bowser, and J. W. Casey. 1997. Transcriptional analysis of walleye dermal sarcoma virus (WDSV). Virology 237:107-112. [DOI] [PubMed] [Google Scholar]
- 18.Regier, J. L., F. Shen, and S. J. Triezenberg. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579:909-915. [DOI] [PubMed] [Google Scholar]
- 20.Rovnak, J., J. W. Casey, and S. L. Quackenbush. 2001. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin). Virology 280:31-40. [DOI] [PubMed] [Google Scholar]
- 21.Rovnak, J., B. W. Hronek, S. O. Ryan, S. Cai, and S. L. Quackenbush. 2005. An activation domain within the walleye dermal sarcoma virus retroviral cyclin protein is essential for inhibition of the viral promoter. Virology 342:240-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rovnak, J., and S. L. Quackenbush. 2002. Walleye dermal sarcoma virus cyclin interacts with components of the mediator complex and the RNA polymerase II holoenzyme. J. Virol. 76:8031-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uesugi, M., O. Nyanguile, H. Lu, A. J. Levine, and G. L. Verdine. 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277:1310-1313. [DOI] [PubMed] [Google Scholar]
- 25.Uesugi, M., and G. L. Verdine. 1999. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA 96:14801-14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, R. 1969. Virus associated with epidermal hyperplasia in fish. Natl. Cancer Inst. Monogr. 31:195-207. [PubMed] [Google Scholar]
- 27.Yamamoto, T., R. K. Kelly, and O. Nielsen. 1985. Morphological differentiation of virus-associated skin tumors of walleye (Stizostedion vitreum vitreum). Fish Pathol. 20:361-372. [Google Scholar]
- 28.Yamamoto, T., R. D. MacDonald, D. C. Gillespie, and R. K. Kelly. 1976. Viruses associated with lymphocystis and dermal sarcoma of walleye (Stizostedion vitreum vitreum). J. Fish Res. Board Can. 33:2408-2419. [Google Scholar]
- 29.Zhang, Z., and D. Martineau. 1999. Walleye dermal sarcoma virus: OrfA N-terminal end inhibits the activity of a reporter gene directed by eukaryotic promoters and has a negative effect on the growth of fish and mammalian cells. J. Virol. 73:8884-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]