Abstract
Influenza A viruses continue to cause widespread morbidity and mortality. There is an added concern that the highly pathogenic H5N1 influenza A viruses, currently found throughout many parts of the world, represent a serious public health threat and may result in a pandemic. Intervention strategies to halt an influenza epidemic or pandemic are a high priority, with an emphasis on vaccines and antiviral drugs. In these studies, we demonstrate that a 20-amino-acid peptide (EB, for entry blocker) derived from the signal sequence of fibroblast growth factor 4 exhibits broad-spectrum antiviral activity against influenza viruses including the H5N1 subtype in vitro. The EB peptide was protective in vivo, even when administered postinfection. Mechanistically, the EB peptide inhibits the attachment to the cellular receptor, preventing infection. Further studies demonstrated that the EB peptide specifically binds to the viral hemagglutinin protein. This novel peptide has potential value as a reagent to study virus attachment and as a future therapeutic.
Influenza A viruses are a major cause of morbidity and mortality in the United States. It has been estimated that influenza A viruses are associated with approximately 31,000 deaths in the United States annually (30). Although annual vaccination is the primary strategy for preventing infections, influenza antiviral drugs play an important role in a comprehensive approach to controlling illness and transmission (12).
Two classes of antiviral drugs, the adamantane derivatives (amantadine and rimantadine) and neuraminidase inhibitors (NAIs; zanamivir and oseltamivir), have been approved for treatment and prophylaxis of influenza (24). For maximum benefits, the drugs must be administered within 48 h of symptom onset. The adamantanes bind to and block the function of the influenza A virus M2 ion channel protein, preventing virus replication within the infected cell (34). Resistance to adamantanes can emerge during treatment by a single point mutation in an M2 residue at position 26, 27, 30, 31, or 34 (2, 11). These resistant viruses are typically fully pathogenic and transmissible (12). The rate of adamantane resistance in the United States has increased significantly from below 2% from 1995 to 2002 to an alarming 92.3% in the initial months of the 2005 to 2006 influenza seasons (4).
The NAIs block the enzymatic activity of the viral neuraminidase (NA), preventing viral exit from the infected cell. Influenza viruses with low susceptibility to the NAIs have been isolated in vitro and in vivo. Resistance involves either a mutation in the active site of the NA, altering its sensitivity to inhibition, or a mutation in the hemagglutinin (HA) (24). Mutations in HA that confer drug resistance decrease the affinity of the protein for the cellular receptor, thus enabling virus to escape from infected cells without the need for viral NA. To date, few viruses with altered susceptibility to NAIs have been recovered from patients (24, 35). This may be because NAI-resistant variants often show impaired growth in cell culture and reduced infectivity and transmissibility in animal infections. However, several recent influenza isolates from patients with H5N1 influenza infection have demonstrated resistance to both classes of antiviral drugs (8, 17, 26). The increasing appearance of resistant strains of influenza virus highlights our need to identify new antiviral drugs.
Novel peptides have been identified that can deliver covalently linked moieties into cells (1, 7, 10, 20). Surprisingly, several of these cell-penetrating peptides demonstrated antiviral activity against herpes simplex virus type 1 (HSV-1) by inhibiting entry, inactivating virions, and inducing cell resistance to infection depending on the peptide concentration (5, 6; C. Brandt et al., unpublished data). In an effort to identify new antiviral therapies effective against influenza viruses, we tested a panel of cell-penetrating peptides for anti-influenza activity. In these studies we show that a peptide denoted EB (entry blocker), derived from the fibroblast growth factor 4 (FGF-4) signal sequence, displayed significant broad-spectrum antiviral activity against influenza viruses both in vitro and in vivo. The peptide functions by preventing attachment to host cells, potentially through an interaction with the HA protein.
MATERIALS AND METHODS
Viruses, cells, and viral infections.
A/PuertoRico/8/34 ([PR/8] strain H1N1), A/Swine/Indiana/1726/88 (strain H1N1), A/Singapore/1/57 (strain H2N2), A/Aichi/2/68 (strain H3N2), A/Turkey/England/69 (strain H3N2), A/Turkey Ontario/6828/67 ([Tk/Ont] strain H5N9), and A/Turkey/Oregon/71 (strain H7N3) were propagated in the allantoic cavity of 10-day specific-pathogen-free embryonated chicken eggs (Sunnyside Farms, Beaver Dam, WI) at 37°C. The allantoic fluid was harvested, centrifuged for clarification, and stored at −70°C. A/New Caledonia/20/99 (strain H1N1), A/Hong Kong/483/97 ([HK/483] strain H5N1), A/Hong Kong/156/97 ([HK/156] strain H5N1), A/Vietnam/1203/04 (strain H5N1), A/Chicken/Vietnam/NCVD-8/03 (strain H5N1), and B/Lee/40 were propagated on Madin-Darby canine kidney (MDCK) cells, and culture supernatants were harvested 24 to 72 h postinfection (hpi) and stored at −70°C. Viral titers were determined by the 50% tissue culture infective dose (TCID50) analysis in MDCK cells and evaluated by the method of Reed and Muench (27). MDCK cells were cultured in modified Eagle's medium (MEM; CellGro, Herndon, VA) supplemented with 4.5 g of glucose per liter, 2 mM glutamine, and 10% fetal bovine serum (Harlan, Madison, WI) at 37°C in 5% CO2.
Virus concentration and fluorescent labeling.
Allantoic fluid was centrifuged at 3,000 × g for 20 min at 4°C, and the resulting supernatant was subjected to ultracentrifugation in a Beckman SW-28 rotor at 19,000 rpm for 2 h at 15°C. The viral pellet was resuspended in 1 ml of cold phosphate-buffered saline (PBS) and labeled with fluorescein isothiocyanate (FITC) using an EZ-Label FITC protein labeling kit (Pierce, Rockford, IL) according to the manufacturer's instructions. To confirm that the FITC-labeled virus remained infectious, TCID50 analysis was performed as described above.
Laboratory facility.
All experiments with H5N1 and H2N2 viruses were performed in a biosafety level 3+ containment laboratory approved for such use by the U.S. Department of Agriculture and the Centers for Disease Control and Prevention. Investigators were required to wear appropriate respirator equipment (RACAL; Health and Safety Inc., Frederick, MD). Mice were housed in HEPA-filtered negative pressure cages (M.I.C.E. racks; Animal Care Systems, Littleton, CO).
Peptides.
Peptide synthesis and analysis were performed at the University of Wisconsin-Madison Biotechnology Center as previously described (5, 6). The TAT series included TAT-C, derived from the protein transduction domain of the human immunodeficiency virus (HIV) Tat protein with an additional C-terminal cysteine and with a sequence-altered control peptide containing two norleucine substitutions at residues 50 and 51 (n50,51TAT-C) (Table 1) (5, 6, 18). The EB series included EB, composed of the FGF-4 signal sequence with a positively charged amino terminal sequence added to increase solubility, EB-d prepared with d-amino acids (to inhibit degradation by host proteases), and a sequence-scrambled control EBX (5, 6, 33). Unless otherwise stated, virus was pretreated with peptide (0 to 30 μM) for 1 h at 37°C prior to use in assays. EB-d and EB had similar 50% inhibitory concentration (IC50) values in all in vitro assays.
TABLE 1.
Peptides utilized in the present study
| Peptide | Sequencea | Source |
|---|---|---|
| TAT-C | NH2-GRKKRRQRRRC-COOH | HIV Tat protein |
| n50,51TAT-Cb | NH2-GRnnRRQRRRC-COOH | HIV Tat protein |
| EB | NH2-RRKKAAVALLPAVLLALLAP-COOH | FGF-4 signal sequence |
| EBXb | NH2-RRKKLAALPLVLAAPLAVLA-COOH | FGF-4 signal sequence |
| EB-d | NH2-RRKKAAVALLPAVLLALLAP-COOH | FGF-4 signal sequence |
Charged residues are shown in bold letters.
Underlining highlights structural differences among peptides
Inhibition of cell death and viral replication assays.
MDCK cells were inoculated with medium alone or peptide-treated (0 to 30 μM) HK/483 (multiplicity of infection [MOI] of 0.05) for 1 h at 37°C. Following adsorption, monolayers were washed and incubated in MEM containing 1% bovine serum albumin (BSA). Cytopathic effect was monitored by light microscopy and quantitated by CellTiter 96 Aqueous One Solution following the manufacturer's instructions (Promega, Madison, WI). To assess viral replication, MDCK cells were inoculated with peptide-treated or untreated HK/483 at an MOI of 0.05, supernatants were collected at 72 hpi, and viral titers were determined by TCID50 analysis on MDCK cells. The IC50 value was estimated by interpolation of the dose-response curve.
Immunofluorescence assay.
MDCK cells seeded at a density of 5 × 104 on glass coverslips were inoculated with medium alone, HK/483 (MOI of 0.5), or peptide-treated (0 to 30 μM) virus for 1 h on ice and then washed with cold PBS to remove unbound virus. For entry-based assays, untreated virus was allowed to attach on ice for 1 h, followed by washing and addition of medium containing peptide (0 to 30 μM). Cells were shifted to 37°C for 6 h, fixed with 1% paraformaldehyde, and stained for nucleoprotein (ATCC clone HB-65; Manassas, VA) and DNA (4′,6′-diamidino-2-phenylindole [DAPI], 1:1000; Sigma). Secondary antibody was anti-mouse Alexa 594 (Molecular Probes, Eugene, OR) diluted 1:200 in PBS containing 0.1% Tween-20 and 1% BSA for 1 h at room temperature. Coverslips were mounted in ProLong Gold (Molecular Probes), and fluorescence was examined on a Zeiss Axiovert 100TV microscope at a total magnification of ×100. Three randomly selected fields of 100 cells each per experimental group were counted on the DAPI setting, and the number of nucleoprotein (NP)-positive cells within this group was determined. Data are representative of at least three independent experiments.
Hemagglutination inhibition assays.
The hemagglutination inhibition assay was employed to evaluate the effects of the peptides on viral adsorption to target cells. Peptides (0 to 30 μM) in serial twofold dilutions in PBS were mixed with a standardized HA concentration of influenza virus and incubated for 1 h at 37°C, and 50 μl of the solution was mixed with an equal volume of a 0.5% chicken red blood cell suspension for 30 min at room temperature. Results are represented as percentages of the virus-alone sample (100% agglutination).
Flow cytometry.
MDCK cells in suspension (2 × 105) were incubated with MEM alone (mock) or FITC-labeled peptide-treated Tk/Ont (32 HA units; 3 × 104 TCID50) for 45 min on ice. Following extensive washing, cells were fixed with 4% paraformaldehyde, and binding was determined by flow cytometric analysis on an LSR-II benchtop flow cytometer (BD Biosciences, Franklin Labs, NJ). Single cell populations of mock-infected cells were gated based upon forward and side scatter characteristics, and 5,000 events from each experimental group were recorded. Experiments on all samples were performed in quadruplicate.
HA binding assay.
HA binding assays were performed as previously described with slight modifications (15, 19). Briefly, microtiter wells were coated with increasing concentrations of peptide (0 to 75 μM) and washed with PBS containing 0.1% Tween-20, and nonspecific binding sites were blocked with 2% Fraction V BSA (Fisher Scientific) in PBS-0.1% Tween-20. Purified recombinant Hong Kong/1203 HA (rHA; Protein Sciences, Meriden, CT) was added to each well at a concentration of 0.01 μg, incubated for 2 h at room temperature, and washed extensively. Peptide-bound rHA was detected by incubation with anti-HAV5 goat serum (National Institute of Allergy and Infectious Diseases, Bethesda, MD) and donkey anti-goat immunoglobulin G (1:2,000; Southern Biotech, Birmingham, AL), followed by quantification using tetramethylbenzidine (R&D Systems). Absorbance was measured at 405 nm (A405 nm) with an A605 correction on a SpectraMax 250 Spectraphotometer (Molecular Devices). Specific binding was determined by calculating the relative change (n-fold) of rHA binding over wells precoated with BSA (2%). To test competition, 0.1 μg/ml rHA was combined with 10 μM peptide and added to 10 μM EB-coated wells (1:1 molar competition concentration). Data are presented as the mean value ± standard error of A405 nm/A605 nm (n = 3).
Animals.
Four- to six-week-old BALB/c mice (Charles River Laboratories, Stoneridge, NY) were lightly anesthetized by halothane inhalation and intranasally inoculated with 25 μl of PBS (control), untreated HK/156 (104 TCID50), or EB-treated (2 mM) HK/156 virus. To evaluate the therapeutic use of EB, additional groups of infected mice were anesthetized and administered 2 mM EB-d or rimantadine hydrochloride (40 mg/kg; Sigma) (32) intranasally 6 hpi, with readministration daily for 5 days. Animals were scored for clinical signs of infection (0, no visible signs of illness; 1, ruffled coat, hunched posture; 2, slowed movement, shivering; 3, labored breathing, anorexia, little to no movement; or 4, paralysis, moribund [23]), and individual body weights were recorded for each group (10 mice per group) on 0 to 11 days postinfection (dpi). At days 2 and 4 postinfection, two mice from the control group and three mice from each experimental group were euthanized, and the lungs were collected. The tissues were weighed and homogenized in 1 ml of cold PBS and stored at −70°C until use. Titers were evaluated by serial titration on MDCK cells. The peptides alone exhibited no toxicity in vivo as measured by clinical signs and weight loss (data not shown).
Statistical analysis.
All data were determined in triplicate and are representative of at least three separate experiments. The results represent the means ± standard deviations of triplicate determinations. Statistical significance of the data was determined by using analysis of variance or a Student's t test.
RESULTS
EB inhibits influenza virus-induced cell death and replication in vitro.
Cytotoxicity studies demonstrated that the EB and TAT peptide series did not induce significant cytotoxicity in MDCK cells until concentrations exceeded 50 μM in medium containing 1% BSA (data not shown). Thus, subsequent in vitro studies were performed with peptide concentrations ranging from 0 to 30 μM.
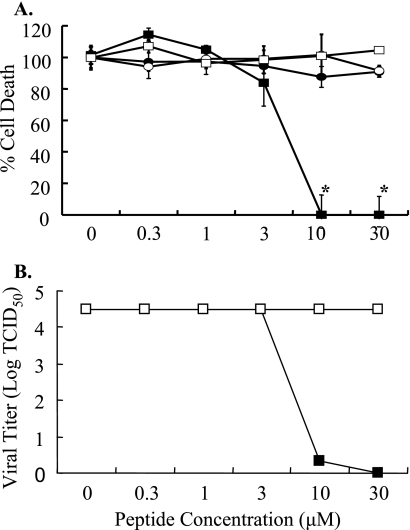
Peptides were evaluated for the ability to inhibit viral-induced cell death using a cytotoxicity assay. Briefly, MDCK cells were mock inoculated (medium alone) or inoculated with peptide-treated (0 to 30 μM) HK/483 (MOI of 0.05) virus, and cell death was evaluated at 48 hpi. In the absence of peptide, HK/483 induced significant death and was scored as 100%. All other samples were normalized to this value. Pretreatment with EB decreased HK/483-induced cell death in a dose-dependent manner (Fig. 1A). Complete inhibition occurred at concentrations greater than 10 μM. Based on dose-response curves, the IC50 for EB was approximately 4.5 μM. In contrast, EBX (scrambled control peptide) and the TAT series peptides had no effect on viral-induced cell death (Fig. 1A). To confirm that EB inhibited viral replication, MDCK cells were infected with peptide-treated (0 to 30 μM) HK/483 (MOI of 0.05), cellular supernatants were collected at 72 hpi, and viral titers were determined by TCID50 assay (Fig. 1B). The EB peptide inhibited HK/483 replication in a dose-dependent manner with an estimated IC50 of 4.5 μM. Again, EBX and the TAT series peptides had no effect on viral titers, indicating that the antiviral effect was specific to the EB peptide (Fig. 1B and data not shown). These results were not limited to H5N1 viruses. EB also inhibited H5N9 and H1N1 strains (data not shown), indicating that EB was a broad-spectrum inhibitor of influenza virus replication.
FIG. 1.
Antiviral activity of peptides. (A) MDCK cells were inoculated with untreated HK/483 virus or HK/483 treated with increasing concentrations of TAT-C (•), n50,51TAT-C (○), EB (▪), or EBX (□), and cell death was determined by Promega CellTiter assay at 48 hpi. Samples are compared and normalized to measurements of virus alone (100% lethal). The results represent the means ± standard deviations of triplicate determinations. *, statistical significance (P < 0.01). (B) MDCK cells were inoculated with untreated HK/483 virus (0 value) or EB (▪)- or EBX (□)-treated HK/483 virus, and viral titers were determined on supernatants 72 hpi. The results represent the means ± standard deviations of triplicate determinations. *, statistical significance (P < 0.01).
Efficacy in vivo.
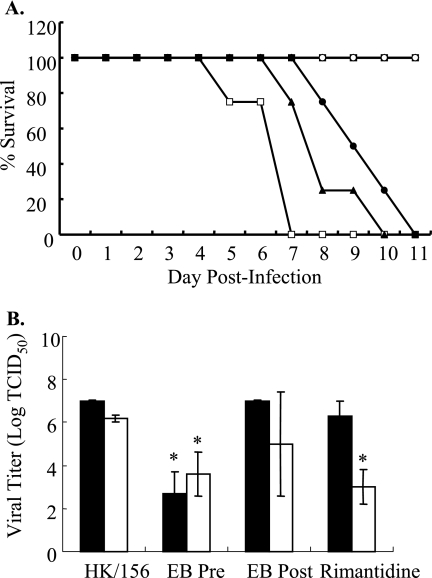
To evaluate the efficacy of EB in vivo, BALB/c mice were lightly anesthetized and inoculated intranasally with PBS (control), untreated HK/156 virus (104 TCID50 units), or EB (2 mM)-treated HK/156 virus and monitored daily for morbidity. Mice administered EB alone exhibited no toxicity as monitored by weight loss and clinical signs of illness (data not shown). All HK/156-infected mice exhibited a clinical score of 3 by 5 dpi, and all mice either succumbed to infection or were euthanized by 7 dpi (Fig. 2A). Pretreatment of HK/156 with 2 mM EB resulted in 100% survival with all mice retaining a clinical score of 0 throughout the experiment (Fig. 2A). These studies demonstrated that pretreatment with EB protected mice from lethal H5N1 infection.
FIG. 2.
Efficacy of EB in vivo. Mice (10 per group) were intranasally inoculated with PBS (▪; control), untreated HK/156 virus (□; 104 TCID50), or EB-d (○; 2 mM)-treated HK/156 virus and monitored for morbidity. Additionally, 10 mice per group were infected with HK/156 and then treated with 2 mM EB-d (•) or 40 mg/kg rimantadine (▴) at 6 hpi and then daily for 5 dpi. Results are representative of four separate experiments. (A) Mice were observed daily for morbidity and clinical signs of illness through an 11-day observation period. (B) On days 2 (solid bars) and 4 (open bars) postinfection, three mice from each group were sacrificed, lungs were harvested, and viral titers were determined from pooled homogenates by TCID50 analysis on MDCK cells. The results represent the means of triplicate determinations. *, statistical significance (P < 0.01).
To evaluate the therapeutic use of EB, BALB/c mice were intranasally administered 2 mM EB-d or rimantadine (40 mg/kg [31]) 6 hpi infection with HK/156 and then monitored daily for 5 dpi. There was a significant delay in mortality and clinical signs in EB-d- and rimantadine-treated groups (Fig. 2A). There was 0% survival by 7 dpi in HK/156-infected mice with a clinical score of 4. In contrast, the EB-d posttreatment group had 100% survival and a clinical score of 2 at 7 dpi. Survival and clinical signs decreased beginning at day 8 postinfection, with 0% survival observed by day 11 postinfection. A similar trend was observed with rimantadine treatment. This begs the question whether continued administration would result in complete protection, and such studies are under way.
On days 2 and 4 postinfection, lungs were isolated and homogenized, and titers were determined on MDCK cells by TCID50 analysis. No virus was isolated from the lungs of mock-inoculated mice (data not shown). Viral titers were significantly lower in the EB pretreated group by day 2 postinfection (Fig. 2B). However, the rimantadine- and EB-d-treated groups did not have a significant decrease in viral titers at 2 dpi and were only slightly decreased at 4 dpi compared to untreated virus (Fig. 2B). Although the EB-treated group did not have a statistically significant decrease, individual mice within the group did have decreased viral titers. EB also protected mice infected with HK/483 and PR/8, suggesting that EB provides broad-spectrum protection in vivo (data not shown).
EB blocks early protein expression.
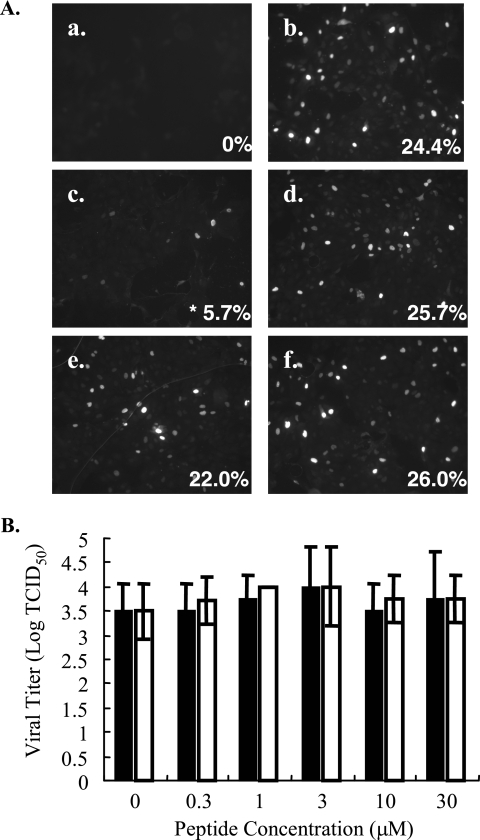
To determine if EB inhibited an early or late stage of viral replication, NP expression was monitored by immunofluorescence microscopy. Briefly, untreated or peptide-treated (0 to 30 μM) HK/483 virus (MOI of 0.5) was allowed to bind to MDCK cells for 1 h at 4°C to synchronize infection. Unbound virus was removed by washing, the temperature was shifted to 37°C to allow internalization, and the cells were stained for NP expression at 6 hpi. More than 24% of cells inoculated with untreated virus or EBX-treated virus were NP positive at 6 hpi (Fig. 3A, frame b) compared to mock-inoculated controls (Fig. 3A, frame a). Pretreatment of HK/483 with EB, but not EBX, resulted in a dose-dependent decrease in NP expression. At 10 μM, less than 6% of the cells were NP positive, with less than 2% of cells NP positive at 30 μM (Fig. 3A, frames c and d, and data not shown). Based on dose-response curves, an IC50 of ∼7 μM was determined. This is similar to the IC50 value obtained for inhibiting viral replication (Fig. 1A).
FIG. 3.
EB is neither virucidal nor blocks influenza virus entry. (A) To monitor the effects of EB on early gene expression, MDCK cells (5 ×104) on glass coverslips were inoculated with medium alone (mock) (a), untreated HK/483 (MOI of 0.5) (b), EB-treated HK/483 (10 μM) (c), or EBX-treated HK/483 virus (10 μM) (d) for 1 h on ice. To examine the effect on entry, MDCK cells were infected with HK/483 virus (MOI of 0.5) for 1 h on ice, followed by washing and the addition of EB (e) or EBX (f) peptide (10 μM) for 15 min on ice. All cells were shifted to 37°C, and NP expression was determined 6 h after the temperature shift. To quantitate NP expression, five fields of 100 cells each were counted, and percentages of NP-positive cells were determined. *, statistical significance (P < 0.01). (B) HK/483 was incubated with medium alone (0 value) or increasing concentrations of EB (solid bars) or EBX (open bars) for 1 h at 37°C. After extensive dialysis, resulting viral titers were determined by TCID50 analysis on MDCK cells. The results represent the means of triplicate determinations. *, statistical significance (P < 0.01).
To further define the antiviral activity, we asked if the EB peptide simply inactivated the virus. To test this, H/483 virus (105.8 TCID50 units) was incubated with increasing concentrations of EB or EBX (0 to 30 μM) for 1 h, followed by dialysis to remove the peptide. The titer of the resulting virus was determined on MDCK cells. Similar titers were obtained with untreated and peptide-treated virus, suggesting that EB did not inactivate virus in solution (Fig. 3B). Further, pretreatment of cells with EB failed to inhibit influenza infection, suggesting that the antiviral effects were independent of any effects on the cell (data not shown).
To investigate the effects on viral entry, MDCK cells were incubated with medium alone or inoculated with HK/483 virus (MOI of 0.5) at 4°C to allow virus attachment to cells but not entry. After 1 h, cells were washed extensively and then incubated with peptide (0 to 30 μM) for 15 min at 4°C, followed by incubation at 37°C for 6 h. The cells were stained for NP expression and examined by immunofluorescence microscopy. Both EB and EBX had no effect on viral entry when added postattachment at concentrations below 10 μM (Fig. 3A, frames e and f, respectively). These assays suggest that EB inhibits early viral replication independent of whether virus is inactivated or virus entry is prevented.
EB inhibits viral replication by preventing attachment to cells.
Two independent assays, agglutination of blood cells and flow cytometry, were used to test the hypothesis that EB inhibited viral attachment. Initially, the inhibition of viral-induced agglutination of chicken red blood cells (cRBCs) by the EB peptide was monitored. Twofold dilutions of untreated or peptide-treated (0 to 30 μM) HK/483 virus were incubated with cRBCs, and agglutination was measured. The EB peptide inhibited HK/483 cRBC agglutination in a dose-dependent manner at concentrations greater than 3 μM, with an estimated IC50 of 8 μM (Fig. 4A). In contrast, EBX had no effect on agglutination. These results were not restricted to HK/483. As shown in Table 2, the incubation of representative human, swine, and avian influenza A H1N1, H2N2, H3N2, H5N1, H5N9, H7N3 strains and influenza B viruses with EB inhibited cRBC agglutination, with IC50 values ranging from 3 to 20 μM (Table 2). These studies confirmed the broad-spectrum activity of EB.
FIG. 4.
The EB peptide inhibits binding. (A) Twofold dilutions of untreated HK/483 (0 value) and EB (closed bars)- or EBX (open bars)-treated HK/483 were incubated with 0.5% cRBC for 45 min at room temperature to allow hemagglutination. All measurements were performed in triplicate, and results are normalized to measurements of virus alone (100% agglutination). The results represent the means of triplicate determinations. *, statistical significance (P < 0.01). (B) MDCK cells in suspension (2 × 105 cells) were incubated with PBS (heavy line), 32 HA units of untreated FITC-labeled Tk/Ont (thin line), or EB- or EBX-treated Tk/Ont (shaded area) for 45 min at 4°C. Following extensive washing and fixation, viral binding to cells was determined by flow cytometry. Gating was determined using forward and side scatter determinants and 5,000 single cell events recorded from each gate. The results are the means of quadruplicate measurements with the average percent FITC-positive cells indicated in each histogram and are representative of three separate experiments. *, statistical significance (P < 0.001).
TABLE 2.
EB inhibits cRBC agglutination by multiple influenza strains
| Virusa | Strain | Estimated EB IC50 (μM) |
|---|---|---|
| PR/8 | H1N1 | 20 |
| Sw/Indiana | H1N1 | 10 |
| New Caledonia | H1N1 | 5 |
| Singapore | H2N2 | 20 |
| Aichi | H3N2 | 20 |
| TK/Eng | H3N2 | 20 |
| HK/483 | H5N1 | 8 |
| VN/1203 | H5N1 | 3 |
| CK/VN/03 | H5N1 | 10 |
| Tk/Ont | H5N9 | 3 |
| TK/Oregon | H7N3 | 5 |
| B/Lee | Influenza B | 5 |
Sw/Indiana, A/Swine/Indiana/1726/88; New Caledonia, A/New Caledonia/ 20/99; Singapore, A/Singapore/1/57; Aichi, A/Aichi/2/68; Tk/Eng, A/Turkey/England/69; VN/1203, A/Vietnam/1203/04; CK/VN/03, A/Chicken/Vietnam/NCVD-8/03; Tk/Oregon, A/Turkey/Oregon/71; B/Lee, B/Lee/40.
To directly demonstrate that EB inhibited viral attachment to cells, MDCK cells were incubated with 32 HA units of untreated FITC-labeled Tk/Ont or peptide-treated (0 to 30 μM) FITC-labeled virus for 45 min on ice. Following extensive washing and fixation, single-cell populations were gated by forward and side scatter, and 5,000 events from each experimental group were recorded by flow cytometry. At the viral concentration used, more than 99% of cells inoculated in the absence of peptide displayed surface attachment of fluorescently labeled virus, displaying a binding peak compared to the fluorescence intensity from noninfected cells (Fig. 4B). Treatment with EBX had no effect on viral attachment to cells or the binding peak compared to virus alone (Fig. 4B). Similar results were obtained with virus treated with 1 and 3 μM EB. In contrast, there was a statistically significant (P < 0.001) shift in virus attachment from 99% to 64.5% in the presence of 10 μM EB. Attachment was further reduced to 46.5% when virus was treated with 30 μM EB (Fig. 4B). Viral attachment was completely inhibited by 30 μM EB when fewer viral particles were used (data not shown). As controls, MDCK cells were incubated with 30 μM EB or EBX and demonstrated minimal background fluorescence (data not shown). Based on the agglutination and flow cytometry data, we propose that the antiviral activity of EB is due to inhibition of viral attachment to cells.
EB interacts with HA.
Based on the ability of EB to inhibit viral attachment, we hypothesized that the peptide interacted with either NA or HA since changes to either surface glycoprotein can alter fitness of the virus (22). To determine if EB inhibited NA enzymatic activity, untreated or peptide-treated Tk/Ont was tested for enzymatic activity by MUNANA [2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid]. Untreated and peptide-treated Tk/Ont had similar enzymatic activity, suggesting that EB peptide had no effect on NA activity (data not shown).
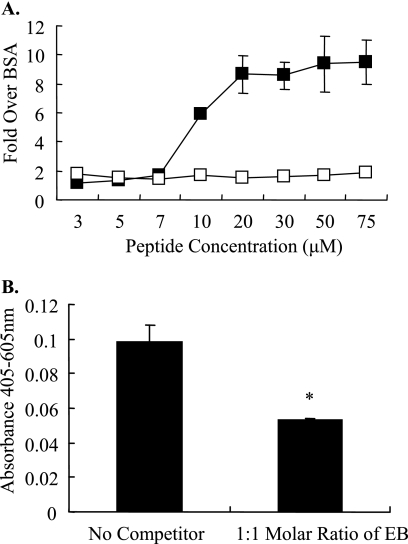
To determine if EB interacted directly with HA, binding assays were performed with baculovirus-derived rHA (Vietnam/1203). Briefly, wells were coated with EB, EBX, TAT (0 to 75 μM), or BSA; they were washed, incubated with rHA, and detected with an anti-H5 antibody. Binding was determined by calculating the relative change (n-fold) of rHA binding over wells coated with BSA. rHA bound to EB peptide in a dose-dependent manner with saturation achieved at the 20 μM EB concentration (about ninefold over background) (Fig. 5A). Limited binding to HA was detected with the EBX peptide. In addition, competition with 1:1 molar soluble EB decreased HA binding by 50% (Fig. 5B). These data suggest that influenza virus HA binds to EB in a specific and saturable manner.
FIG. 5.
EB binds to HA. (A) Increasing concentrations of rHA (Vietnam/1203) were incubated with microtiter plates coated with EB (▪) or EBX (□) and washed, and binding was determined by enzyme-linked immunosorbent assay. Relative binding (n-fold) over nonspecific binding to BSA was determined by recombinant HA in wells that had been precoated with blocking buffer alone. All samples were tested in triplicate, and results are representative of at least three separate experiments. *, statistical significance (P < 0.01). (B) Competition of HA binding to EB was accomplished by adding a 1:1 molar ratio of free HA immediately prior to addition to a well containing immobilized EB or BSA. Data are expressed as relative absorbances and are presented as the mean values ± standard errors of A405 nm/A605 (n = 3). *, statistical significance (P < 0.01).
DISCUSSION
We have identified a novel peptide inhibitor with broad-spectrum antiviral activity against influenza viruses at low micromolar concentrations. The EB peptide demonstrated low cytotoxicity, especially in the presence of serum (data not shown), with a therapeutic index of at least 22. Pretreatment with EB provided 100% protection against numerous subtypes including the highly pathogenic H5N1 viruses in vivo and in vitro, demonstrating its broad-spectrum activity. Protection was supported by a decrease in viral titers in the lungs of infected animals. Although postinfection treatment with peptide was not as effective as pretreatment, it should be noted that the EB peptide was given only once per day in these studies and was as effective as rimantadine in protecting mice from H5N1 infection. Additional studies are under way to further define the dose and treatment schedules required for optimum protection and to determine if peptide treatment inhibits transmission to naïve animals. The use of EB in combination with rimantadine and NAIs is also being examined. These results suggest that EB or modified versions of EB may have potential as a broad-spectrum influenza virus therapy.
The antiviral activity was specific to EB and not a general property of cell-penetrating peptides. Peptides derived from the HIV Tat protein had no effect on viral replication. We also observed a lack of antiviral activity with the scrambled control peptide EBX compared to EB, suggesting that EB's antiviral activity may be sequence specific and not simply an interaction based upon charge or hydrophobic interactions. Studies are currently under way to determine if a minimal peptide sequence exists and to identify the critical residues required for the anti-influenza activity by preparing an EB library with site-directed residue changes as well as a deletion series. In this screen, we may identify a peptide of shorter length that is as effective or more effective, further shedding light on a structure-based interaction with HA.
The antiviral activity of the EB peptide is not unique to influenza virus. What is intriguing is that the EB peptide appears to act by distinct mechanisms depending on the virus. With HSV-1, EB irreversibly inactivates virions, prevents entry at a postattachment site (6), and induces a resistance to infection in cells treated with peptide (H. Bultmann and C. Brandt, unpublished data). The EB peptide did not interfere with HSV-1 attachment to cells, in contrast to our data with influenza virus, and in fact enhanced binding at concentrations between 10 and 50 μM due to aggregation (5, 6). Further support for a specific interaction between EB and influenza comes from the observation that the EB peptide showed no antiviral activity against HIV, human astrovirus (data not shown), or adenovirus type VI (unpublished data). Our studies demonstrated that EB inhibits influenza virus attachment to cells, leading to decreased viral replication. Although entry was affected, this occurred only at peptide concentrations three times higher than the in vitro IC50 values; thus, the primary antiviral mechanism is through inhibiting viral attachment.
One question under ongoing investigation is the mechanism whereby the EB peptide inhibits viral attachment. Our studies demonstrated that the EB peptide specifically bound to the viral HA protein. The sharp increase in antiviral activity (range of approximately 5 to 30 μM) suggests that there may be cooperative binding of the EB peptide to HA similar to the binding events described for the O2 binding hemoglobin proteins (28). This initial binding between EB and HA could be of weak affinity, with increasing affinity as more EB binds. The binding of EB to HA could lead to a conformation change in HA, decreasing the affinity for sialic acids on the cell surface. Another possibility is that EB interacts at or near the receptor binding pocket of the HA1 subunit, blocking the pocket or causing steric hindrance during receptor docking. Glycans present on HA1 could serve as sites of EB binding, and the variability of glycosylation patterns among HA subtypes could account for the differing IC50 values noted among the viruses tested in this study. The effect of glycosylation on the antiviral activity of EB is currently being examined. As a final possibility, the EB peptide could induce aggregation of the influenza virion, resulting in decreased binding to cells. Exploring these possibilities, in addition to the isolation and characterization of a peptide-resistant mutant, will further our understanding of the mechanism of EB anti-influenza activity.
The use of peptides such as defensins (14, 16) to inhibit bacterial infection have been widely described (reviewed in references 7, 9, and 29). More recent work has demonstrated the usefulness of peptides against numerous viral infections including respiratory syncytial virus (25), HIV (21), influenza virus (3), and general broad-spectrum antiviral activity (13). The demonstration of clinical antiviral efficacy by the HIV fusion inhibitor enfuvirtide (T-20) shows that synthetic peptides can be developed into effective antiviral drugs (21). The EB peptide exhibits a low therapeutic index and in vivo efficacy, suggesting its candidacy for structure-based drug design. In the near future, the EB peptide could be developed as a successful inhibitor of virus binding for use in the study of replication of influenza viruses.
In summary, we have identified a novel peptide with broad-spectrum inhibitory activity against influenza virus. Of great interest, the peptide inhibits the infection of the H5N1 viruses associated with increasing numbers of human deaths throughout Southeast Asia. EB inhibits viral attachment to cells, most likely by binding to HA. As we continue to cope with yearly influenza epidemics and the H5N1 viruses continue to spread, understanding how the viruses interact with cells and developing tools to block this earliest step in viral infection are of enormous public health importance. The EB peptide has potential as a valuable tool for the study of influenza viruses and their receptor-binding interactions, as well as therapeutic possibilities.
Acknowledgments
We thank Gary Case for production of peptides; Jon Woods, Laura Knoll, and Jenny Gumperz for advice and use of their equipment; and Kurt Kapcho and Barbara Israel for providing H5 HA. We gratefully acknowledge Martha McGregor, Chris Olsen, Kimberly Luke, Lindsey Moser, John Lindner, Christina Carlson, and Kevin O'Brien for technical advice, experimental assistance, and/or critical reviews of the manuscript.
E.A.T. was supported by a postdoctoral training fellowship in tumor virology through the McArdle Cancer Center at the University of Wisconsin (CA09075-28) and NIH NRSA grant F32 AI060292. This work was supported by funds from the University of Wisconsin—Madison Medical Education and Research Committee (S.S.C.), NIH grant P01-AI52049 (C.R.B.), DARPA grant MDA972-97-1-005 (C.R.B.), and Sigma Xi Grants-In-Aid of Research (J.C.J.).
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157-183. [DOI] [PubMed] [Google Scholar]
- 2.Belshe, R. B., M. H. Smith, C. B. Hall, R. Betts, and A. J. Hay. 1988. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J. Virol. 62:1508-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bovin, N. V., A. B. Tuzikov, A. A. Chinarev, and A. S. Gambaryan. 2004. Multimeric glycotherapeutics: new paradigm. Glycoconj. J. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 4.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295:891-894. [DOI] [PubMed] [Google Scholar]
- 5.Bultmann, H., and C. R. Brandt. 2002. Peptides containing membrane-transiting motifs inhibit virus entry. J. Biol. Chem. 277:36018-36023. [DOI] [PubMed] [Google Scholar]
- 6.Bultmann, H., J. S. Busse, and C. R. Brandt. 2001. Modified FGF4 signal peptide inhibits entry of herpes simplex virus type 1. J. Virol. 75:2634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dathe, M., and T. Wieprecht. 1999. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1462:71-87. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, M. D., T. T. Thanh, T. H. Khanh, V. M. Hien, G. J. D. Smith, N. V. Chau, B. V. Cam, P. T. Qui, D. Q. Ha, Y. Guan, J. S. M. Peiris, T. T. Hien, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T., and R. I. Lehrer. 1998. Antimicrobial peptides of vertebrates. Curr. Opin. Immunol. 10:41-44. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, B., T. S. Levchenko, and V. P. Torchilin. 2005. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv. Drug Deliv. Rev. 57:637-651. [DOI] [PubMed] [Google Scholar]
- 11.Hay, A. J., M. C. Zambon, A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1986. Molecular basis of resistance of influenza A viruses to amantadine. J. Antimicrob. Chemother. 18(Suppl. B):19-29. [DOI] [PubMed] [Google Scholar]
- 12.Hayden, F. G. 2006. Respiratory viral threats. Curr. Opin. Infect. Dis. 19:169-178. [DOI] [PubMed] [Google Scholar]
- 13.Horne, W. S., C. M. Wiethoff, C. Cui, K. M. Wilcoxen, M. Amorin, M. R. Ghadiri, and G. R. Nemerow. 2005. Antiviral cyclic d,l-α-peptides: targeting a general biochemical pathway in virus infections. Bioorg. Med. Chem. 13:5145-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izadpanah, A., and R. L. Gallo. 2005. Antimicrobial peptides. J. Am. Acad. Dermatol. 52:381-390. [DOI] [PubMed] [Google Scholar]
- 15.Katz, J. M., X. Lu, S. A. Young, and J. C. Galphin. 1997. Adjuvant activity of the heat-labile enterotoxin from enterotoxigenic Escherichia coli for oral administration of inactivated influenza virus vaccine. J. Infect. Dis. 175:352-363. [DOI] [PubMed] [Google Scholar]
- 16.Koczulla, A. R., and R. Bals. 2003. Antimicrobial peptides: current status and therapeutic potential. Drugs 63:389-406. [DOI] [PubMed] [Google Scholar]
- 17.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. L. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Science 437:1108. [DOI] [PubMed] [Google Scholar]
- 18.Lin, Y. Z., S. Y. Yao, R. A. Veach, T. R. Torgerson, and J. Hawiger. 1995. Inhibition of nuclear translocation of transcription factor NF-κB by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem. 270:14255-14258. [DOI] [PubMed] [Google Scholar]
- 19.Lu, X., M. Renshaw, T. M. Tumpey, G. D. Kelly, J. Hu-Primmer, and J. M. Katz. 2001. Immunity to influenza A H9N2 viruses induced by infection and vaccination. J. Virol. 75:4896-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magzoub, M., and A. Graslund. 2004. Cell-penetrating peptides: small from inception to application. Q. Rev. Biophys. 37:147-195. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215-225. [DOI] [PubMed] [Google Scholar]
- 22.Mishin, V. P., D. Novikov, F. G. Hayden, and L. V. Gubareva. 2005. Effect of hemagglutinin glycosylation on influenza virus susceptibility to neuraminidase inhibitors. J. Virol. 79:12416-12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton, D. B. 2000. A systematic approach for establishing humane endpoints. ILAR J. 41:80-86. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson, K. G., J. M. Wood, and M. Zambon. 2003. Influenza. Lancet 362:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastey, M. K., T. L. Gower, P. W. Spearman, J. E. Crowe, Jr., and B. S. Graham. 2000. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat. Med. 6:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puthavathana, P., P. Auewarakul, P. C. Charoenying, K. Sangsiriwut, P. Pooruk, K. Boonnak, R. Khanyok, P. Thawachsupa, R. Kijphati, and P. Sawanpanyalert. 2005. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 86:423-433. [DOI] [PubMed] [Google Scholar]
- 27.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 28.Riggs, A. F. 1988. The Bohr effect. Annu. Rev. Physiol. 50:181-204. [DOI] [PubMed] [Google Scholar]
- 29.Simmaco, M., G. Mignogna, and D. Barra. 1998. Antimicrobial peptides from amphibian skin: what do they tell us? Biopolymers 47:435-450. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179-186. [DOI] [PubMed] [Google Scholar]
- 31.Tumpey, T. M., A. Garcia-Sastre, A. Mikulasova, J. K. Taubenberger, D. E. Swayne, P. Palese, and C. F. Basler. 2002. Existing antivirals are effective against influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 99:13849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tumpey, T. M., A. Garcia-Sastre, J. K. Taubenberger, P. Palese, D. E. Swayne, and C. F. Basler. 2004. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 101:3166-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vives, E., P. Brodin, and B. Lebleu. 1997. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272:16010-16017. [DOI] [PubMed] [Google Scholar]
- 34.Wang, C., K. Takeuchi, L. H. Pinto, and R. A. Lamb. 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67:5585-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen, H.-L., L. M. Herlocher, E. Hoffmann, M. N. Matrosovich, A. S. Monto, R. G. Webster, and E. A. Govorkova. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49:4075-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]