Abstract
Lytic replication of Kaposi's sarcoma-associated herpesvirus (KSHV) is essential for viral propagation and pathogenicity. In Kaposi's sarcoma lesions, constant lytic replication plays a role in sustaining the population of latently infected cells that otherwise are quickly lost by segregation of latent viral episomes as spindle cells divide. Lytic DNA replication initiates from an origin (ori-Lyt) and requires trans-acting elements. Two functional ori-Lyts have been identified in the KSHV genome. Some cis-acting and trans-acting elements for ori-Lyt-dependent DNA replication have been found. Among these, K8 binding sites, a cluster of C/EBP binding motifs, and a replication and transcription activator (RTA) responsive element (RRE) are crucial cis-acting elements. Binding of K8 and RTA proteins to these motifs in ori-Lyt DNA was demonstrated to be absolutely essential for DNA replication. In the present study, functional roles of RTA in ori-Lyt-dependent DNA replication have been investigated. Two distinct functions of RTA were revealed. First, RTA activates an ori-Lyt promoter and initiates transcription across GC-rich tandem repeats. This RTA-mediated transcription is indispensable for DNA replication. Second, RTA is a component of the replication compartment, where RTA interacts with prereplication complexes composed of at least six core machinery proteins and K8. The prereplication complexes are recruited to ori-Lyt DNA through RTA, which interacts with the RRE, as well as K8, which binds to a cluster of C/EBP binding motifs with the aid of C/EBP α. The revelation of these two functions of RTA, together with its role in initiation of a transcriptional cascade that leads to transcription of all viral lytic genes, shows that RTA is a critical initiator and regulator of KSHV lytic DNA replication and viral propagation.
Like all herpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV) has two distinct life cycles, namely, latent and lytic cycles. In different life cycles, the virus replicates its genome by different replication mechanisms. During latency, viral genomes are maintained as extrachromosomal episomes (plasmids) and replicated in synchrony with cell division (3). The terminal repeat (TR) sequence in the KSHV genome is necessary and sufficient for episome persistence and serves as the origin of latent plasmid replication (ori-P) (4). The virally encoded latency-associated nuclear antigen specifically binds to the cis-acting TR DNA and acts in trans on ori-P to mediate episome persistence (4, 12).
Lytic DNA replication of herpesviruses differs from latent DNA replication in at least three aspects. First, latent DNA replication initiates at ori-P and proceeds bidirectionally, while viral lytic replication initiates from a distinct origin (ori-Lyt) and proceeds via a rolling circle mechanism. The lytic rolling circle DNA replication results in the generation of concatemeric genomic DNA which is subsequently cleaved into monomeric units and packaged into virions. Second, latent DNA replication depends on host cellular DNA polymerase and accessory factors, while viral lytic replication utilizes its own DNA polymerase and other factors. Third, latent DNA replication occurs in synchrony with the host cell to maintain a stable number of viral DNA episomes in each cell, while viral DNA is amplified 100- to 1,000-fold in lytic replication.
Two duplicated copies of the lytic DNA replication origin [referred to as ori-Lyt (L) and ori-Lyt (R)] were recently identified in the KSHV genome (2, 20). They are located in the KSHV genome between open reading frames K4.2 and K5 and between K12 and ORF71, respectively. These two ori-Lyt regions share an almost identical 1.1-kb core component sequence and 600-bp GC-rich repeats that are represented as 20-bp and 30-bp tandem arrays (20). A systematic mutational analysis of the core component domain of a KSHV ori-Lyt defined several crucial elements in ori-Lyt (31). The first component contains eight C/EBP binding motifs that organize as four spaced C/EBP palindromes. Substitution mutagenesis of these C/EBP motifs showed that these C/EBP palindromes are required for binding of K8 protein in ori-Lyt DNA and for DNA replication. The second component is an 18-bp AT palindrome, which is believed to facilitate the localized unwinding of double-stranded DNA during the initiation of replication. The third component is a previously unidentified sequence and is required for DNA replication. The fourth component consists of an ORF50/replication and transcription activator (RTA) responsive element (RRE) and a TATA box. The presence of a functional RRE and a downstream TATA box suggests that this region serves as an ORF50/RTA-responsive promoter and that a transcription event may be necessary for ori-Lyt-dependent DNA replication. Using a luciferase reporter system, we demonstrated that the region of the RRE and TATA box indeed constitutes an RTA-dependent promoter. A polyadenylated RNA of 1.4 kb was identified downstream of the promoter (31).
Herpesviral ori-Lyt-dependent DNA replication relies on virally encoded proteins. The proteins essential for DNA replication have been identified in herpes simplex virus type 1 (HSV-1), cytomegalovirus (CMV), Epstein-Barr virus (EBV), and KSHV (9, 23, 32, 33). Six core replication proteins are conserved among all herpesviruses, and they are the DNA polymerase (POL), the polymerase processivity factor (PPF), the single-stranded DNA binding protein (SSB), and a trivalent helicase-primase complex which consists of a helicase (HEL), a primase (PRI), and a primase-associated factor (PAF). Additional proteins, such as origin binding proteins, regulatory proteins, and auxiliary factors, were identified and found to be unique to each individual virus (7, 8, 10, 28). In terms of KSHV, six core replication proteins and two regulatory proteins, namely, RTA and K8, were demonstrated to be necessary and sufficient for KSHV ori-Lyt-dependent DNA replication (1). RTA binds to a consensus RRE in ori-Lyt, and K8 associates with the ori-Lyt DNA through interaction with C/EBP α molecules bound on a cluster of C/EBP binding motifs (31).
The participation of RTA and K8 in ori-Lyt-dependent DNA replication appears to be the most salient feature in the event. Wu et al. (33) reported that six KSHV core replication proteins could replicate EBV ori-Lyt-containing plasmid in the presence of EBV-encoded protein ZTA. In the process, ZTA functioned as a DNA binding initiator protein responsible for loading core replication machinery complexes onto the EBV ori-Lyt DNA (10). KSHV K8 protein is a distant evolutionary homologue of ZTA and was found to be incorporated into viral replication compartments (VRC) in the nuclei of KSHV-infected primary effusion lymphoma cells (33). In view of this, we wanted to find out if K8 is a functional analog of ZTA of EBV and functions as a DNA binding initiator protein in KSHV ori-Lyt DNA replication. Furthermore, the role of RTA in ori-Lyt-dependent DNA replication was also very intriguing. It may contribute to ori-Lyt replication through its transcriptional activity. It may also contribute to the DNA replication through other mechanisms, such as serving as an origin binding protein (OBP) and DNA replication initiator protein because of its DNA binding nature. In an effort to elucidate the mechanisms underlying the regulation of KSHV ori-Lyt-dependent DNA replication, we investigated the roles of RTA and K8 in viral lytic DNA replication and studied the contribution of the ori-Lyt-associated transcription to viral DNA replication. The results demonstrated a tight coupling of the DNA replication and transcription events. ori-Lyt-dependent DNA replication is inhibited when the RNA transcription is prematurely terminated. The interplay between KSHV DNA replication and transcription has provided a basis for further investigation into the regulation of DNA replication in DNA viruses and eukaryotic cells. Furthermore, RTA and K8 were found to be components of the viral replication compartment, where they interact with core replication proteins and recruit the replication complexes to ori-Lyt DNA.
MATERIALS AND METHODS
Cell culture.
The primary effusion lymphoma cell line BCBL-1, which carries latently infected KSHV and was established by Renne and colleagues (26), was obtained from the NIH AIDS Research and Reference Reagent Program. The cells were grown in RPMI 1640 medium (Gibco-BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gibco-BRL). BC-1 cells, which carry KSHV and EBV and were established by Cesarman et al. (5), were purchased from the American Type Culture Collection (ATCC) and grown in RPMI 1640 medium supplemented with 15% fetal bovine serum. BJAB cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. All cultures contained penicillin-streptomycin (50 U/ml) and 1.25 μg/ml amphotericin B and 1.25 μg/ml sodium deoxycholate (Fungizone).
Plasmids and constructs.
Plasmid pOri-A and its mutants (pOri-Δ15.7, pOri-Δ16.2, pOri-M12, pOri-M1256, etc.) were constructed and described previously (31). pOri-polyA1 and pOri-polyA3 were generated by inserting a simian virus 40 (SV40) late poly(A) signal sequence at nucleotides 24271 and 25067 (according to the numbering of Russo et al. [27]), respectively, in the pOri-A plasmid. pOri-polyA2 is a derivative of pOri-polyA1 in which the polyadenylation consensus sequence AAUAAA has been deleted by using a PCR-based mutagenesis system, namely, ExSite (Stratagene). In brief, a pair of phosphorylated oligonucleotides in opposite directions was used in a high-fidelity PCR with pOri-polyA1 plasmid as a template. After PCR, the template DNA was removed by complete digestion with DpnI, which does not degrade PCR-synthesized DNA. The PCR products were self-ligated and used to transform Escherichia coli competent cells. pOri-ΔTR and pOri-mTR were generated by deleting the entire fragment of GC-rich tandem repeats (the PmlI-MulI fragment between nucleotides 24252 and 25069) from pOri-A and by replacing this fragment with a PmlI-MulI fragment from pGL3 plasmid (between nucleotides 747 and 16) (Promega), respectively.
pOri-PmU6, pOri-PSV40, and pOri-PCMV were constructed by replacing the RRE-associated promoter region (between nucleotides 24022 and 24252) with mouse U6 (mU6) promoter (315-bp), SV40 early promoter (454-bp), or human CMV (HCMV) immediate-early (IE) promoter (596-bp) sequence, respectively.
pCR3.1-ORF50 was constructed by cloning the cDNA sequence of the ORF50 coding region into pCR3.1 vector (Invitrogen). The construct was described in detail by Lin et al. (20). The expression vectors for Gal4 fusion proteins (pSG-C50 and pSG-424) were described by Lukac et al. (21). The internal deletion mutants of pSG-C50 and pCR3.1 were generated using the ExSite system as described above.
Transient-transfection DNA replication assay.
To transfect BCBL-1 cells, 5 μg pOri-A or its mutant plasmids and 5 μg pCR3.1-ORF50 (or pCR3.1 vector) were mixed with 107 cells in OPTI-MEM medium (GIBCO-BRL) and electroporated (200 V, 960 μF) with a Genepulser II (Bio-Rad, Hercules, CA). Electroporated cells were then transferred to RPMI 1640 medium supplemented with 10% serum and grown for 72 h.
Extrachromosomal DNAs were prepared from cells by using the Hirt DNA extraction method as follows. Cells were lysed in 700 μl lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM EDTA, and 0.6% sodium dodecyl sulfate [SDS]). Chromosomal DNA was precipitated at 4°C overnight by adding 5 M NaCl to a final concentration of 0.85 M. Cell lysates were centrifuged at 4°C at 14,000 rpm for 30 min. The supernatant containing extrachromasomal DNA was subjected to phenol-chloroform extraction, followed by ethanol precipitation. The DNA was treated with RNase A at 25°C for 30 min and then with proteinase K at 50°C for 30 min. DNA (5 μg) was digested with KpnI/SacI or KpnI/SacI/DpnI (New England Biolabs). The DNAs were separated by electrophoresis on 1% agarose gels and transferred onto GeneScreen membranes (Perkin Elmer, Boston, MA). The Southern blots were hybridized with 32P-labeled pBluescript plasmid in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt's solution, 1% SDS, and 50 μg/ml denatured salmon sperm DNA at 68°C.
Northern blotting and hybridization.
Total RNA was isolated from transfected 293 cells with Trizol reagent (Gibco-BRL) and separated by electrophoresis in 1% agarose-6% formaldehyde gel in 20 mM morpholinepropanesulfonic acid (MOPS) buffer, pH 7.0. Each lane was loaded with mRNA from 2 × 107 cells. The RNA was transferred to a Nytran membrane (Schleicher & Schuell, Keene, N.H.) and hybridized with a single-stranded 32P-labeled probe. The probe was prepared by asymmetric PCR with PmlI-digested pOri-A plasmid as the template and the oligonucleotide OriT-1 (5′-TGTTTATTTCAAGAGCCTATGCTCG-3′, nucleotides 25428 to 25403) as the primer. The labeling reaction was performed in 15 μl of reaction solution (1× Taq polymerase buffer, which consisted of 16.67 μM [each] dATP, dGTP, and dTTP, 1.67 μM dCTP, 5 μl of [32P]dCTP [800 Ci/mmol, 10 μCi/μl; Amersham], 100 ng of DNA, 20 pmol of primer, and 2.5 U of Taq polymerase). The PCR was initiated with a denaturing step of 2 min at 94°C, followed by 15 cycles of sequential steps of 1 min at 94°C, 1 min at 50°C, and 3 min at 74°C. Finally, the reaction was extended for 10 min at 74°C. The RNA loading equivalence was controlled by probing with β-actin cDNA. A 0.24- to 9.5-kb RNA ladder (Gibco-BRL) was included in each agarose-formaldehyde gel and detected in Northern blots by hybridization with labeled DNA.
Reporter plasmids and luciferase assays.
Reporter plasmids (pK8-DE250 and its derivatives, 13F and its mutants) were constructed and described previously (30, 31). Ten micrograms luciferase reporter construct, 1 μg pRL-TK plasmid (Promega), and 3 μg pCR3.1-ORF50 (or pCR3.1 empty vector) were mixed with 107 BJAB or BCBL-1 cells in OPTI-MEM medium (GIBCO-BRL) and electroporated (250 V, 960 μF) with a Genepulser II (Bio-Rad, Hercules, CA). Electroporated cells were then transferred to RPMI 1640 medium supplemented with 10% serum and grown for 48 h. The pRL-TK plasmid was included as an internal control that constitutively expresses Renilla luciferase. A dual luciferase reporter assay system (Promega) was used to examine the responsiveness of the promoters to ORF50/Rta. Transfected cells were washed once with 1× PBS and suspended in 400 μl of 1× passive lysis buffer. Cells were broken by one freeze-thaw cycle and centrifuged in a mirocentrifuge for 1 min. Supernatants were assayed for firefly luciferase and Renilla luciferase activities by use of a TD-20/20 luminometer with a dual auto injector (Turner Designs). The luciferase assays were carried out according to the manufacturer's instruction (Promega).
DNA affinity purification and assays.
Various biotinylated DNA fragments were synthesized using PCR with pOri-A DNA or its mutants as templates and two oligonucleotides as primers. The two oligonucleotides for 3F DNA fragments were 3F (5′-CGGCAAAGCTAATTTGCATG-3′) and Biotin-7R (5′-biotin-ACTGGAATAGGGGCTGCGATGACTC-3′). The oligonucleotides for 9F DNA were 9F (5′-CAATTCTATAATTAAACAAGGTAGAA-3′) and Biotin-ID13R (5′-biotin-CGCCACCGAACAACCCCGTGGACAG-3′). The oligonucleotides for 11F DNAs were 11F (5′-TAGGGCCCGATGAGTCATGGGGTT-3′) and Biotin24280R (5′-biotin-ACGGGTAAATCCAAGAGATCCGTCCC-3′). The resultant biotinylated PCR fragments were coupled to streptavidin-conjugated magnetic beads (Dynal, Oslo, Norway) and then incubated with nuclear extracts prepared from 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced (and uninduced) BCBL-1 cells for 45 min at 25°C. The bound material was washed four times in D150 buffer (20 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM KCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.05% NP-40) and then progressively eluted with D300 buffer (same as D150 buffer, except with 300 mM KCl), D500 buffer (with 500 mM KCl), and D1000 buffer (with 1 M KCl). The affinity-purified materials were assayed by Western blotting with different antibodies.
Coimmunoprecipitation assay.
Approximately 5 × 107 cells were collected by centrifugation and washed with cold PBS. The cells were suspended in 0.5 ml of immunoprecipitation (IP) buffer (120 mM potassium acetate, 20 mM Tris-acetate [pH 7.9], 5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.1% Nonidet P-40, and one complete protease inhibitor cocktail tablet per 25 ml), placed on ice for 30 min, and sonicated for 5 s. The cell lysates were clarified by centrifugation at 4°C for 10 min. Immunoprecipitation was performed by the addition of 2 μl of monoclonal anti-ORF50 (a gift from Keiji Ueda at Osaka University, Japan) or monoclonal anti-K8 antibody to the cell lysates (0.5 ml) with gentle agitation at 4°C for 60 min. Then, 100 μl of protein G-coated paramagnetic beads (Dynal Biotech, Oslo, Norway) was added and the mixtures were incubated at 4°C overnight. A reaction with 2 μl of mouse immunoglobulin G (IgG) (Sigma) was performed as a control. The beads were then washed five times with cold IP buffer. The precipitates were resuspended in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled for 10 min, and loaded onto SDS-PAGE gels. To eliminate DNA-mediated protein interactions, ethidium bromide (EtBr) was added to aliquots of some cell lysates and maintained at a concentration of 50 or 100 μg/ml throughout the entire IP process, including wash steps.
IFA.
The primary antibodies used in the immunofluorescence assay (IFA) included anti-K8 monoclonal IgG (1:300 dilution; raised using purified glutathione S-transferase [GST]-K8 α fusion protein), anti-ORF50 monoclonal antibody (1:300 dilution; provided by Keiji Ueda at Osaka University, Japan), anti-ORF50 polyclonal antibody (1:300 dilution; provided by David Lukac at UMDNJ), anti-PPF polyclonal antibody (1:1,000 dilution; provided by Robert Ricciardi at the University of Pennsylvania), anti-SSB polyclonal antibody (1:300 dilution; provided by Gary Hayward at Johns Hopkins), and sheep antibromodeoxyuridine (anti-BrdU) antibody (1:100 dilution; purchased from Capralogics, Inc., Hardwick, MA).
BCBL-1 cells were induced with TPA (20 ng/ml) for 48 h. The cells were washed with PBS and fixed with 2% paraformaldehyde in PBS for 10 min at room temperature. The fixed cells were spun onto Shandon double cytoslides at 1,000 rpm for 5 min. The slides were immersed in PBS for 5 min and permeabilized in 0.2% Triton X-100 in PBS for 20 min on ice. For the double-label experiment, the primary mouse monoclonal and rabbit polyclonal antibodies were diluted together and incubated with cells on the slides at room temperature for 50 min. After wash, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG and Texas Red-conjugated anti-mouse antibodies (Vector Laboratories, Inc., Burlingame, CA) at room temperature for 50 min.
Triple-label IFA was performed to visualize replication-active DNA in the nucleus as well as viral DNA replication proteins. TPA-treated BCBL-1 cells were incubated in the media containing 10 μM BrdU (Sigma) for 60 min before fixation. The pulse-labeled cells were fixed with 2% paraformaldehyde in PBS for 20 min at room temperature and then incubated with 2 N HCl for 30 min at room temperature in order to expose incorporated BrdU residues. After four washes with PBS, the cells were incubated with primary mouse monoclonal anti-RTA or anti-K8, rabbit polyclonal anti-SSB, and sheep anti-BrdU antibodies at room temperature for 50 min, followed by incubation with a combination of secondary antibodies (FITC-conjugated anti-rabbit IgG, Cy5-conjugated anti-mouse IgG [Rockland Immunochemicals, Inc., Gilbertsville, PA], and Texas Red-conjugated anti-sheep IgG [Vector Laboratories, Inc.]). All secondary antibodies were diluted at 1:250.
The slides were examined with a Leica TCS SPII confocal laser scanning system. Two or three channels were recorded simultaneously or sequentially. Data acquisition was controlled for possible breakthrough between the green and red channels and between the blue and red channels.
RESULTS
KSHV ori-Lyt-associated transcription is absolutely required for DNA replication.
Virally encoded transcriptional activator ORF50/RTA binds to KSHV ori-Lyt, and the binding is absolutely required for DNA replication (31). What role(s) does RTA play in the replication? We have shown previously that the RRE in ori-Lyt constitutes an RTA-dependent promoter and directs transcription of a 1.4-kb RNA (31). Thus, to understand the role(s) of RTA in the ori-Lyt-dependent DNA replication, we first asked if the RTA-mediated transcription from ori-Lyt is required for the DNA replication. An SV40 polyadenylation signal sequence was inserted at different locations of the ori-Lyt-associated transcription units to prematurely terminate transcription (Fig. 1A). When the 155-bp polyadenylation signal sequence was placed between the transcription initiation site and the GC-rich tandem repeats at nucleotide 24265, transcription from ori-Lyt in the resulting plasmid (designated pOri-polyA1) was efficiently terminated and no transcript was detected in the GC-rich repeat region by Northern analysis (Fig. 1B). To confirm the role of the polyadenylation signal in termination of the transcription, the AATAAA consensus sequence was deliberately deleted in the pOri-polyA1 plasmid (Fig. 1A) and a transcript over the GC-rich repeats was detectable in the resulting plasmid (designated pOri-polyA2) (Fig. 1B).
FIG. 1.
Effect of premature termination of ori-Lyt-associated transcription on ori-Lyt-dependent DNA replication. (A) Schematic diagram of the constructs in which the SV40 late polyadenylation sequence was inserted at different locations in pOri-A plasmid. (B) Transcription from ori-Lyt in pOri-A and its derivatives, as indicated, was examined by Northern analysis. In addition to the polyadenylation signal insertion mutants, pOri-Δ15.7 and pOri-Δ16.2, in which the RRE and the TATA box in ori-Lyt have been deleted, respectively (31), are included in the experiment as negative controls. pOri-ΔTR and pOri-mTR are derivatives of pOri-A in which the GC-rich tandem repeats have been deleted or replaced with an irrelevant sequence, respectively. Total RNAs were isolated from transfected cells, separated on a 1.0% agarose-formaldehyde gel, and transferred onto a Nytran membrane. The membrane was probed with 32P-labeled GC-rich tandem repeat sequence. (C) The abilities of these constructs and their parental wild-type plasmid, pOri-A, to mediate DNA replication were tested in BCBL-1 cells by using a transient replication assay as described in Materials and Methods. KSHV lytic replication is induced by expression of RTA. Extrachromosomal DNAs were prepared using the Hirt extraction method and used for the assay. DpnI-resistant products of DNA replication were detected by Southern blotting with 32P-labeled pBluescript (pBlue) plasmid. Rep'd, replicated.
The abilities of these constructs to support ori-Lyt-dependent DNA replication were examined by a transient DNA replication assay. Briefly, wild-type ori-Lyt-containing plasmid (pOri-A) and its polyadenylation signal insertion derivatives were introduced into BCBL-1 cells along with the RTA expression vector (pCR3.1-ORF50). DNA was isolated from the transfected cells 72 h posttransfection and digested with KpnI/SacI and KpnI/SacI/DpnI. Replicated plasmid DNA was distinguished from input plasmid by DpnI restriction digest, which cleaves input DNAs that have been methylated indam+ E. coli but leaves intact the DNA that has been replicated at least one round in eukaryotic cells. In other words, newly replicated plasmid DNA in BCBL-1 cells is resistant to DpnI digestion and can be detected in Southern analysis. As shown in Fig. 1C, pOri-polyA1 was unable to be replicated in the lytic phase in BCBL-1 cells, while in pOri-polyA2, restoration of DNA replication was observed. These data suggest that termination of ori-Lyt-associated transcription between the transcription initiation site and the GC-rich tandem repeats abolished DNA replication. In addition, when the SV40 polyadenylation sequence was inserted after the GC-rich repeats but before a potential open reading frame (31), the resulting plasmid (designated pOri-polyA3) maintained the ability to replicate in the lytic phase of BCBL-1 cells (Fig. 1C).
The results presented above led to a hypothesis that a transcription event through a GC-rich tandem repeat sequence is required for DNA replication to occur. To prove the hypothesis, two more constructs, one in which the GC-rich tandem repeats were deleted (pOri-ΔTR) and one in which the GC-rich tandem repeats were replaced with an irrelevant DNA sequence of similar length (pOri-mTR), were made. The results showed that both plasmids were incompetent for DNA replication in lytic BCBL-1 cells (Fig. 1C). Taken together, our study strongly suggests that assembly of a transcription complex in ori-Lyt is necessary but not sufficient to support DNA replication; active transcription through a GC-rich tandem repeat sequence is needed for KSHV ori-Lyt-dependent DNA replication.
Can the RRE-dependent promoter in ori-Lyt be replaced by other Pol II or Pol III promoters?
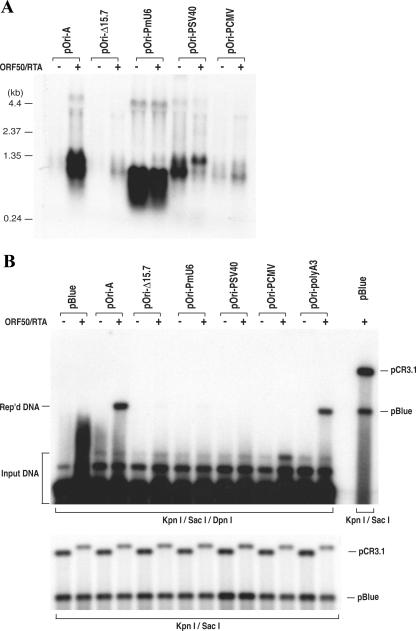
Since we discovered that transcription through a GC-rich tandem repeat sequence is essential for DNA replication, we were curious if replacement of the RRE-containing promoter with other promoters allows ori-Lyt-dependent DNA replication to take place. The RRE-containing promoter region from nucleotides 24025 to 24252 was replaced with an HCMV immediate-early promoter, an SV40 early promoter (both are polymerase II [Pol II] promoters), and a mouse U6 snRNA promoter (a Pol III promoter) in pOri-A. These mutants, as well as the wild-type parental plasmid pOri-A, were introduced into BCBL-1 and 293 cells along with the RTA expression vector. Transcription of these mutant plasmids was analyzed in 293 cells by Northern analyses. As illustrated in Fig. 2A, the U6 promoter was able to strongly and constitutively initiate transcription. The size of the U6-transcribed RNA was shorter than the wild-type transcript because the U6-driven transcript terminated at nucleotide 25301 (150 nucleotides earlier than the Pol II transcripts) and was not polyadenylated. The mutants with SV40 and HCMV promoters were also found to be transcribed, even though the levels of HCMV promoter-driven transcripts were lower than those driven by other promoters (Fig. 2A). All of the mutants were also analyzed for their ori-Lyt function in a transient replication assay in BCBL-1 cells. To our surprise, all of the mutants, including U6, SV40, and HCMV promoter replacement derivatives, failed to replicate in the lytic stage of BCBL-1 cells (Fig. 2B). This finding raised the possibility that in addition to supporting transcription, RTA bound on ori-Lyt may have another function(s) that is also critical for ori-Lyt-dependent DNA replication.
FIG. 2.
Replacement of the RRE-involved promoter in KSHV ori-Lyt with other promoters abolished ori-Lyt-dependent DNA replication. The ori-Lyt promoter region (nucleotides 24022 to 24252) was replaced by a mouse U6 snRNA promoter, an SV40 early promoter, or an HCMV IE promoter in pOri-A to generate pOri-PmU6, pOri-PSV40, and pOri-PCMV, respectively. (A) The transcription initiated by these promoters was examined by Northern analysis. Total RNAs were isolated from 293 cells that had been transfected with pOri-A and its derivatives, as indicated above each lane. These RNAs were separated on a 1.0% agarose-formaldehyde gel and transferred onto a Nytran membrane. The membrane was probed with a 32P-labeled GC-rich tandem repeat sequence. (B) The abilities of these constructs to mediate DNA replication were tested in BCBL-1 cells by using a transient replication assay as described in Materials and Methods. pBlue, pBluescript; Rep'd, replicated.
Mapping the RTA molecule for transcription and DNA replication function domains.
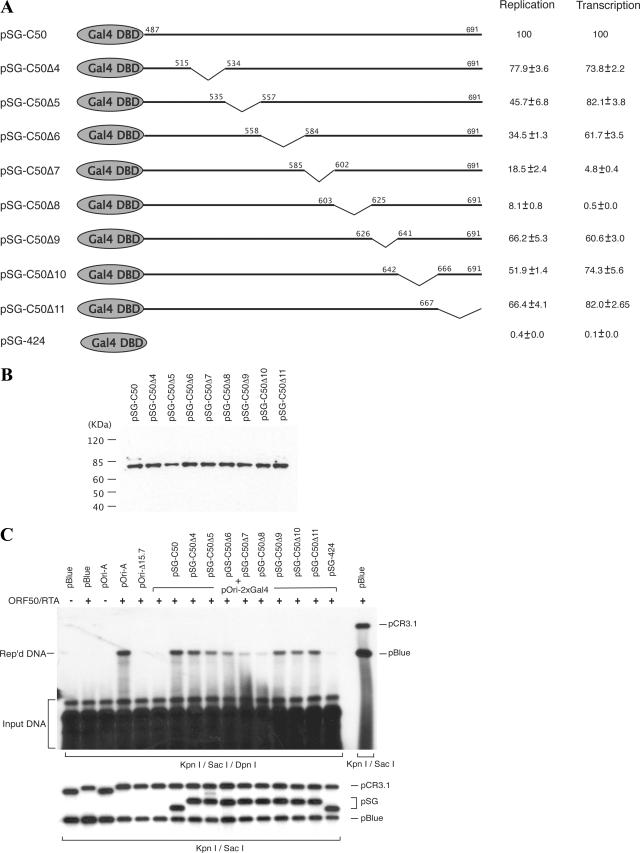
In order to further understand the role of RTA-mediated transcription from ori-Lyt in lytic DNA replication and to explore the other possible functions of RTA in DNA replication, we attempted to map the transcription function as well as the DNA replication function in the RTA molecule. If the transcription function structurally overlaps with the DNA replication function in the RTA molecule, a crucial contribution of the RNA transcription to the DNA replication would be suggested. On the other hand, identification of an additional domain in RTA, which is important for DNA replication but not essential for RNA transcription, would imply a new function of RTA in lytic DNA replication. Our previous study showed that when the RRE in ori-Lyt was replaced with two copies of yeast Gal4 binding motif (the construct was designated pOri-2xGal4), DNA replication was abolished. However, DNA replication could be fully restored if a Gal4-RTA fusion protein (SG-C50), in which the activation domain of RTA (205 carboxyl-terminal amino acids of RTA) is fused with the DNA binding domain of Gal4, was introduced to the cells (31). This result suggests that the replication function of RTA is associated with the activation domain that resides within the carboxyl-terminal 205 amino acids. Thus, we focused our study on the carboxyl-terminal region of 205 amino acids and again employed the Gal4-RTA fusion protein in the study. To map the replication function in greater detail, a series of internal deletions was introduced into the construct of Gal4-RTA fusion protein across the activation domain of RTA (Fig. 3A). These mutants were introduced into BCBL-1 cells, and expression of these mutants of the Gal4-RTA fusion protein was monitored by Western blotting with an antibody specific to Gal4 protein (Fig. 3B). Then, these deletion mutants were used to cotransfect BCBL-1 cells along with pOri-2xGal4 and the wild-type RTA expression vector for a transient DNA replication assay. Wild-type RTA induced the lytic cycle in BCBL-1 cells and activated viral lytic genes necessary for the lytic DNA replication. The transfected cells were collected 72 h posttransfection, and the effects of these deletions on ori-Lyt-dependent DNA replication were examined by using a transient DNA replication assay. This system allows an assessment of the contribution of each region that is missing from the Gal4-RTA fusion protein to ori-Lyt-dependent DNA replication. As illustrated in Fig. 3C, the Δ5 to Δ8 deletion mutants displayed obviously reduced replication function in comparison to that of the wild-type counterpart (SG-C50). Among them, the Δ8 mutant had the greatest effect on DNA replication. This deletion mutant lacks amino acids 603 to 625.
FIG. 3.
Identification of region of RTA required for DNA replication. (A) Schematic diagram of a set of deletion mutants of pSG-C50. Deleted amino acids are shown by the downward sloping lines. DBD, DNA binding domain. (B) Expression levels of SG-C50 and its derivatives were monitored by Western blotting with an antibody specific to Gal4 protein. (C) pOri-2xGal4 was introduced into BCBL-1 cells along with SG-C50 and a series of its deletion mutants. The RTA expression vectors were included to initiate lytic DNA replication. (C) Replicated (Rep'd) DNA was distinguished from input DNA by DpnI digest and detected by Southern blotting with 32P-labeled pBluescript (pBlue) plasmid. The replication rate of each mutant relative to that of wild-type pOri-A was calculated by comparing the normalized intensity of the replicated DNA band with that of wild-type pOri-A. Each number is the average of results from two independent experiments. The transcription ability of each mutant was determined by a luciferase assay with the 13F-2xGal4 reporter construct (31).
The deletion mutants of the Gal4-RTA fusion protein were also tested for their transactivation ability by use of the 13F-2xGal4-luciferase reporter construct in a transient expression assay. The reporter construct was cotransfected into BCBL-1 cells with wild-type pSG-C50 and its deletion mutants, and promoter activities mediated by the Gal4-RTA fusion protein and its derivatives were examined by a luciferase assay. The results showed that transcription activity was almost completely lost with the Δ8 mutant and severely impaired with the Δ7 mutant in transactivation (Fig. 3A). In addition, the internal deletions were also introduced into intact RTA expression vector pCR3.1-ORF50. The effects of these deletions on RTA-mediated transcription were analyzed using the ori-Lyt-associated promoter as well as the K8 delayed-early promoter in BJAB or 293 cells, and similar results were observed (Fig. 4). A previous report showed that the region of RTA between amino acids 607 to 627 is essential for RTA-mediated transcription (13), and our data were consistent with their result.
FIG. 4.
Identification of the region in RTA that is required for transcription activation. The RTA deletions illustrated in Fig. 3A were introduced into pCR3.1-ORF50. The RTA mutants (designated RtaΔ4, RtaΔ5, etc.) were examined for the ability to activate the ori-Lyt RRE-containing promoter as well as the K8 delayed-early promoter by use of luciferase reporters. The reporter plasmids containing a firefly luciferase gene under control of the ori-Lyt promoter or the K8 delayed-early promoter were introduced into BJAB cells by electroporation or into 293 cells by calcium phosphate precipitation. Renilla luciferase plasmid was included in each transfection as an internal control. At 48 h posttransfection, dual luciferase assays were performed with the lysates of transfected cells. Relative luciferase activities were calculated by dividing the normalized firefly luciferase activity of each reporter by that of pGL3 plasmid in pCR3.1-transfected cells.
Significant reductions in DNA replication were also found with the Δ5 and Δ6 mutants (Fig. 3C). However, the effects of these mutations on RNA transcription were trivial (Fig. 3A and 4). The separation of the transcription function and the DNA replication function in these mutants suggests that RTA may also contribute to ori-Lyt-dependent DNA replication by other mechanisms in addition to being a transcription activator.
RTA binds to ori-Lyt DNA and interacts with core replication protein complex.
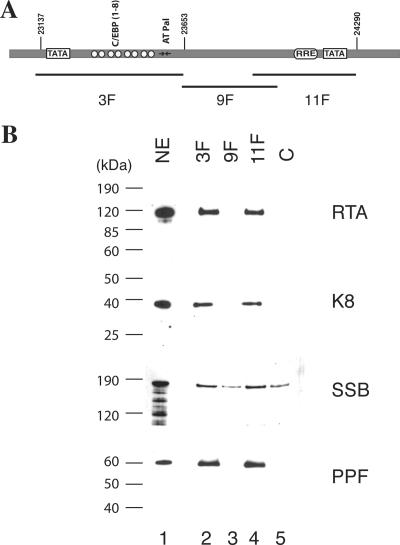
The results described above led to the speculation that RTA has multiple functions in ori-Lyt-dependent DNA replication. To explore the new function of RTA in DNA replication, we studied the interaction of RTA with ori-Lyt DNA as well as with viral core replication proteins by using a DNA affinity purification approach. Three overlapping DNA fragments representing the core domain of KSHV ori-Lyt were synthesized and labeled with biotin at their 5′ ends (Fig. 5A). The DNA was coupled to streptavidin-conjugated magnetic beads and then incubated with nuclear extract derived from TPA-induced BCBL-1 cells. Bound material was washed with 150 mM KCl-containing buffer and sequentially eluted with 300 mM, 500 mM, and 1 M KCl. Affinity-purified materials were resolved on 4 to 12% SDS-PAGE gels and probed with antibodies against RTA and other viral DNA replication proteins, such as K8, SSB, and PPF. As expected, K8 and RTA proteins were eluted with 500 mM KCl from the 3F and 11F DNA fragments, respectively. However, K8 was also eluted from the 11F DNA and RTA eluted from the 3F DNA. In addition, SSB and PPF bound to 3F and 11F DNAs and eluted with 500 mM KCl (Fig. 5B).
FIG. 5.
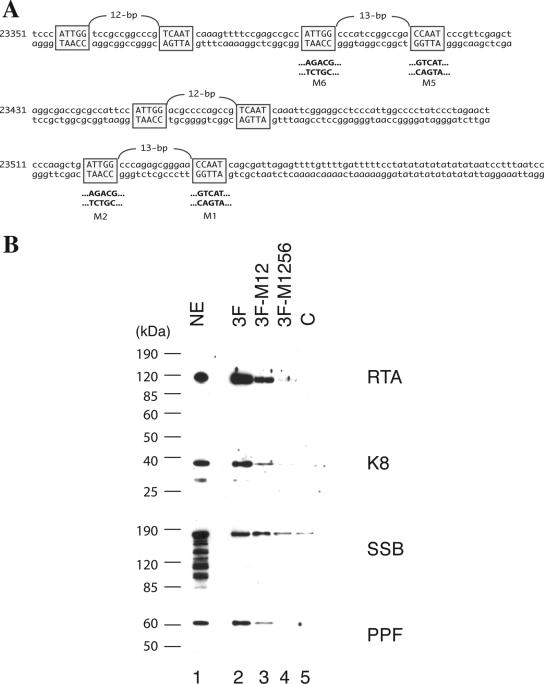
Binding of K8, RTA, and core replication proteins to KSHV ori-Lyt DNA. (A) Schematic of KSHV ori-Lyt core domain and the three DNA fragments that were used in the DNA affinity assay. The C/EBP cluster (K8 binding sites), AT palindrome (AT Pal), RRE, and TATA boxes are illustrated. (B) Nuclear extract (NE) from TPA-induced BCBL-1 cells was subjected to DNA affinity purification with each of the three DNA probes, and the D500 eluates were assayed by Western blotting with antibodies as indicated. An irrelevant DNA (ORF45 coding region) was included as a control, C.
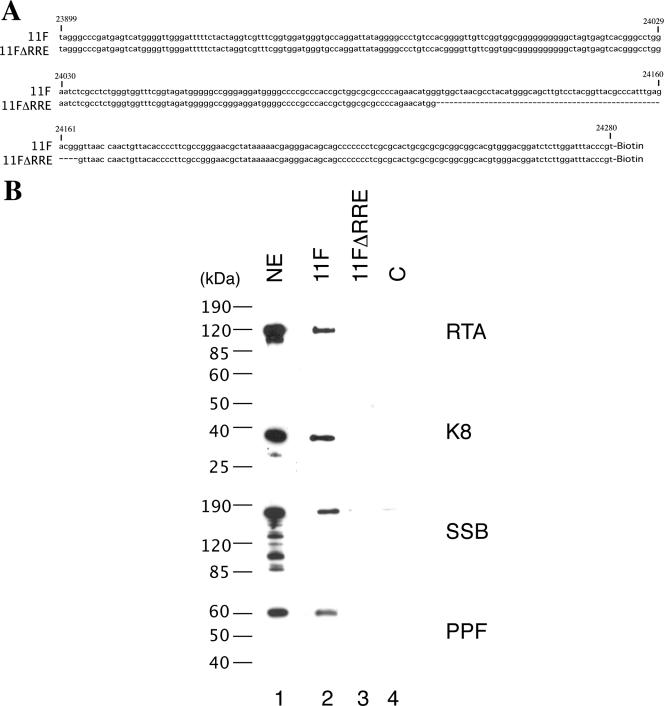
It has been demonstrated previously that RTA binds to ori-Lyt DNA through an RRE motif located within the 11F region, while K8 associates with ori-Lyt through a 240-bp C/EBP cluster region within the 3F sequence (31). However, it was not clear how K8, SSB, and PPF bind to 11F DNA and how RTA, SSB, and PPF interact with 3F DNA. Do K8, SSB, and PPF recognize certain DNA sequence motifs within the 11F fragment, or do they bind to the DNA through interaction with RTA? Similarly, does RTA bind to 3F DNA directly or through interaction with K8? We designed two lines of experiment to address these questions. First, we examined if the association of K8, SSB, and PPF with 11F DNA is dependent on RTA and if the binding of RTA, SSB, and PPF to 3F DNA is dependent on K8. If these replication machinery proteins bind to ori-Lyt DNA through interaction with RTA and K8, one can predict that deletion or mutation of the RRE motif or K8 binding region in ori-Lyt will not only result in failure of RTA or K8 to bind their respective sites in ori-Lyt but also prevent all of the components of the replication complexes from associating with ori-Lyt DNA. Therefore, we tested a mutant 11F DNA, in which the RRE motif was deliberately deleted (Fig. 6A), for its ability to associate with K8 and other replication proteins. The wild type (11F) and the RRE deletion mutant (11FΔRRE) were used for DNA affinity purification with nuclear extract from TPA-induced BCBL-1 cells. As shown in Fig. 6B, RTA, K8, SSB, and PPF bound to wild-type 11F but not to the RRE deletion mutant. Similarly, wild-type 3F and its mutants, in which the C/EBP cluster was mutated, were used in a DNA affinity assay. Previous studies demonstrated that three of eight C/EBP binding motifs in the cluster, namely, C/EBP-1, -2, and -6, contribute more to K8 binding than the other C/EBP motifs (34) and DNA replication (31). Thus, we designed a double mutation in C/EBP-1 and -2 (the M12 mutant) and a quadruple mutation in C/EBP-1, -2, -5, and -6 (the M1256 mutant) and examined the effects of these mutations on the binding of ori-Lyt DNA by K8, RTA, and other replication proteins. The results showed that not only did K8 fail to bind to the M1256 mutant but RTA and other replication proteins also did not bind at all or bound with reduced affinity. All of the proteins bind to the M12 mutant with compromised affinity (Fig. 7).
FIG. 6.
Effect of deletion of RRE from the ori-Lyt fragment (11F) on the recruiting of replication proteins to ori-Lyt DNA. (A) Sequences of 11F and 11FΔRRE DNA fragments used in the DNA affinity assay. (B) Biotinylated DNA fragments were prepared by PCR, with pOri-A (wild-type) DNA and pOri-Δ15.7 (an RRE deletion derivative, which was described by Wang et al. [31]) as templates. TPA-induced BCBL-1 nuclear extract (NE) was incubated with the DNA fragments conjugated on magnetic beads, washed, and eluted with D500 elution buffer. Samples were assayed by Western blotting with antibodies as indicated. An irrelevant DNA was included as a control, C.
FIG. 7.
Effects of mutations of the C/EBP motifs in the ori-Lyt fragment (3F) on recruiting of replication proteins to ori-Lyt DNA. (A) Schematic diagram of the sequences and locations of eight C/EBP binding sites that are organized as four spaced palindromes, as well as site-specific mutations on these C/EBP motifs. (B) Biotinylated DNA fragments were prepared by PCR with a pOri-A (wild-type) DNA template as well as pOri-M12 and pOri-M1256, mutations on certain C/EBP motifs, which were described by Wang et al. (31). TPA-induced BCBL-1 nuclear extract (NE) was incubated with the DNA fragments conjugated on magnetic beads, washed, and eluted with D500 elution buffer. Samples were assayed by Western blotting with antibodies as indicated. An irrelevant DNA was included as a control, C.
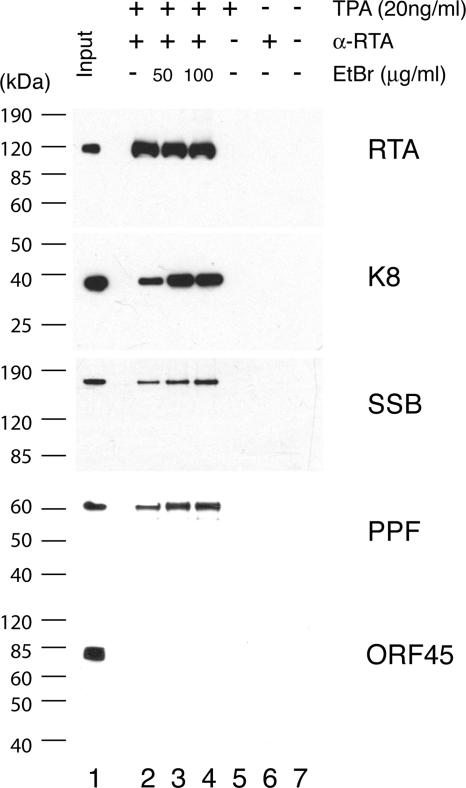
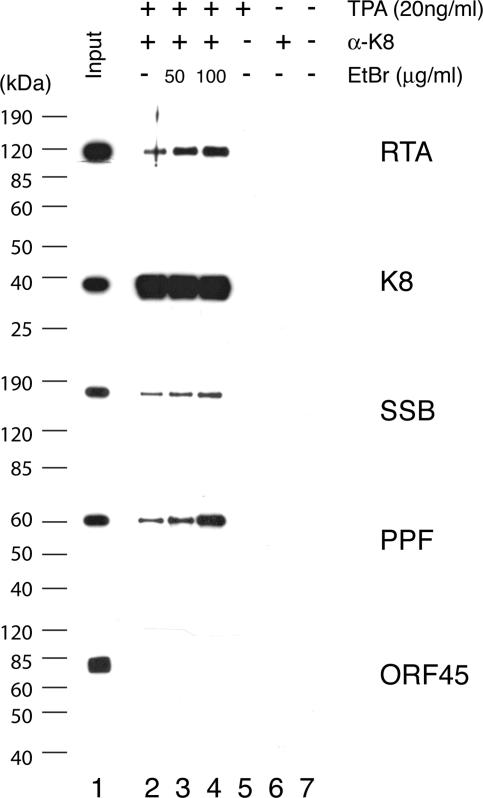
Second, to test if K8, SSB, and PPF bind to 11F DNA through protein-protein interaction with RTA and if RTA, SSB, and PPF bind to 3F DNA through K8 protein, coimmunoprecipitations were performed to confirm the interaction among RTA, K8, SSB, and PPF. Briefly, BCBL-1 cells, induced with TPA for 48 h, were lysed in IP buffer containing 0.1% Nonidet P-40, briefly sonicated, and clarified by centrifugation. The lysates were reacted with a mouse monoclonal anti-RTA antibody. Immunoprecipitated material was resolved by SDS-PAGE, followed by Western blot analysis with polyclonal antibodies specific to RTA, K8, SSB, and PPF. Appropriate controls, such as immunoprecipitation with unimmunized mouse IgG and use of lysates of uninduced BCBL-1 cells, were included in the assay (Fig. 8). The Western blot was also probed with an antibody against ORF45, a viral protein known to be not directly associated with viral DNA replication (35), as a control. Results showed that RTA, K8, SSB, and PPF were detected in the RTA-immunoprecipitated material (Fig. 8). Although the interaction between RTA and K8 has been reported previously (15), our data on coimmunoprecipitation of the four proteins suggest that RTA antibody may have brought down the entire replication complex, which consists of at least six viral core replication proteins (SSB, POL, PAL, HEL, PRI, and PPF) (33) as well as K8 and RTA. No ORF45 was detected in the precipitates (Fig. 8). Similarly, coimmunoprecipitation was also performed with anti-K8 antibody and the results showed that anti-K8 antibody coprecipitated SSB, PPF, RTA, and K8 proteins but not ORF45 (Fig. 9).
FIG. 8.
Coimmunoprecipitation of viral core replication proteins with RTA by using an anti-RTA monoclonal antibody. Immunoprecipitations were performed with TPA-induced or uninduced BCBL-1 lysates in the presence and absence of EtBr. Each sample was separated on SDS-PAGE gels and followed by Western analyses using polyclonal antibodies against proteins as indicated.
FIG. 9.
Coimmunoprecipitation of viral core replication proteins with K8 by using an anti-K8 monoclonal antibody. Immunoprecipitations were performed with TPA-induced or uninduced BCBL-1 lysates in the presence and absence of EtBr. Each sample was separated on SDS-PAGE gels and followed by Western analyses using polyclonal antibodies against proteins as indicated.
Some of the proteins found in anti-RTA and anti-K8 precipitates are intrinsic DNA binding proteins, so we asked whether the associations of core replication proteins to RTA and K8 were mediated by DNA-protein or protein-protein interactions. To address this question, we performed the immunoprecipitation in the presence of EtBr at concentrations of 50 and 100 μg/ml (18). Both concentrations of EtBr were tested and shown to be able to completely disrupt DNA-protein interaction (data not shown). The presence of EtBr did not disrupt the coprecipitation of SSB and PPF with either anti-RTA or anti-K8 antibody, confirming that the associations of the core replication proteins with RTA and K8 were not DNA mediated (Fig. 8 and 9). Furthermore, the addition of EtBr to the immunoprecipitation reaction mixtures increased the coprecipitation of the core replication proteins, suggesting that the association of core replication protein complexes (or prereplication complexes) with RTA and K8 is tighter without DNA. This could suggest that prereplication complexes, which consist of six core machinery proteins as well as RTA and K8, are assembled independently of ori-Lyt DNA. Once the complexes are recruited and loaded on ori-Lyt DNA, the prereplication complexes become less stable in order to convert to replication fork complexes.
Taken together, our results from the coimmunoprecipitation experiments and the DNA affinity purification study led to two implications: (i) RTA and K8 are components of KSHV prereplication complexes, and (ii) viral prereplication complexes bind to ori-Lyt DNA through RTA and K8, interacting with their respective DNA binding motifs.
RTA and K8 are components of replication compartments, and prereplication complexes are recruited to ori-Lyt DNA through RTA and K8 binding motifs.
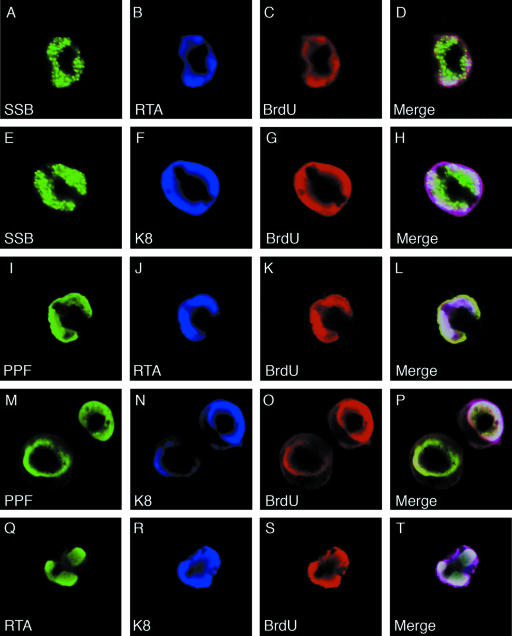
Viral replication compartments (VRC) are intranuclear domains, which are formed when viral replication successively annexes a large portion of the nucleus, and are the locations where viral DNA replication takes place in the host cell nucleus (24). In herpesviruses, VRC are visible in immunofluorescence assays as large, irregularly shaped nuclear domains. We examined whether RTA is an integral component of the replication compartment. BCBL-1 cells were treated with TPA for 48 h to induce lytic DNA replication. The cells were fixed and examined by double-label IFA. Replication compartment-like large nuclear domains, detected with rabbit polyclonal antibodies against SSB and PPF, were seen in about 47% of the positive cells (17% of cells with smaller granular bodies/domains and 36% of cells with diffuse nuclear staining). In these cells, K8 and RTA, detected with mouse monoclonal antibodies against them, were found to be completely colocalized with SSB and PPF in the nuclear domains (data not shown because they are similar to data shown in Fig. 10). In addition, double label with polyclonal RTA and monoclonal K8 antibodies showed complete colocalization of RTA and K8 in the VRC-like nuclear domains.
FIG. 10.
Visualization and characterization of VRC in TPA-induced BCBL-1 cells and localization of RTA and K8 in VRC. BCBL-1 cells were treated with TPA for 48 h and pulse-labeled with BrdU for 60 min. The cells were subjected to triple-label IFA using mouse monoclonal anti-RTA or anti-K8 antibody (Cy5), rabbit polyclonal anti-SSB or anti-PPF antibody (FITC), and sheep anti-BrdU antibody (Texas Red). The triple-label IFA shows that RTA and K8 are colocalized with core replication machinery proteins (SSB and PPF) as well as newly synthesized DNA (BrdU incorporated) in VRC in TPA-induced BCBL-1 cells.
To further investigate whether the VRC-like nuclear structures are mature VRC and viral DNA synthesis indeed takes place in the structure, we pulse-labeled TPA-induced BCBL-1 cells with BrdU for 60 min and performed triple-label IFA on these cells with rabbit polyclonal anti-SSB or anti-PPF, mouse monoclonal anti-RTA or anti-K8, and goat polyclonal anti-BrdU antibodies. The results showed that the nuclear domains in which SSB, PPF, K8, and RTA colocalized are the sites of BrdU incorporation, as indicated by staining with anti-BrdU antibody (Fig. 10). It has been shown that in virally infected cells the formation of VRC leads to marginalization of host cell chromatin, which forms a dense layer surrounding the VRC, and that the incorporation of BrdU in VRC denotes the initiation of viral DNA replication (22). Therefore, nuclear domains in which RTA colocalized with SSB and BrdU are functional viral replication compartments and RTA is an integral component of VRC.
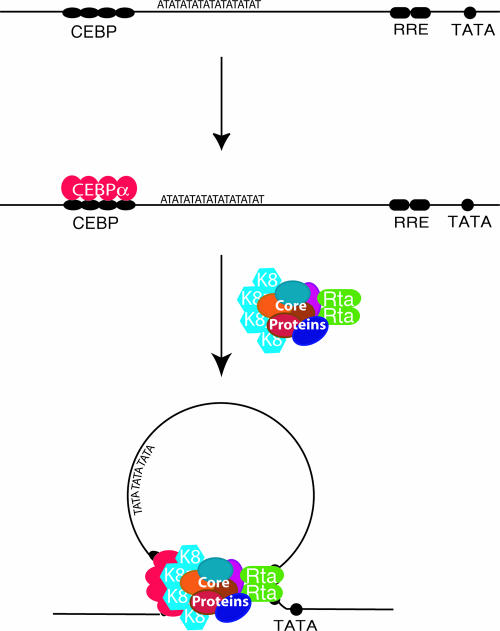
In summary, several lines of evidence, including the binding of K8 and RTA to ori-Lyt DNAs, interactions of K8 and RTA with other replication complex proteins, and colocalization of K8 and RTA and other core replication machinery proteins in viral replication compartments, all led to the conclusion that both K8 and RTA are components of replication complexes in VRC. This notion, together with the result that the association of core replication machinery proteins with ori-Lyt DNA requires the K8 and RTA binding motifs in ori-Lyt, strongly suggests that one of the functions of RTA, as well as that of K8, is interacting with prereplication complexes and recruiting the complexes to ori-Lyt DNA through interactions with their cis-acting motifs, i.e., the C/EBP binding site cluster and the RRE. Figure 11 presents a model for the formation of the replication complex in ori-Lyt, in which a prereplication complex interacts with ori-Lyt DNA through two contact points mediated by K8 and RTA.
FIG. 11.
Model for recruitment of core replication proteins to KSHV ori-Lyt DNA through RTA and K8. Six core replication machinery proteins (core proteins), RTA, and K8 form a prereplication complex regardless of the presence of ori-Lyt DNA. The prereplication complex is loaded at a KSHV ori-Lyt by a two-point contact through RTA and K8 that interacts with their respective sites on ori-Lyt.
DISCUSSION
Herpesviral lytic DNA replication requires cis-acting elements, i.e.,replication origins, and trans-acting elements, including six core replication machinery proteins (POL, SSB, HEL, PRI, PAF, and PPF), origin binding protein(s), and auxiliary factors. In KSHV, six core replication machinery proteins plus RTA and K8 are necessary and sufficient for ori-Lyt-dependent DNA replication in a transient cotransfection assay (1). RTA and K8 have been demonstrated to bind to ori-Lyt DNA in a direct DNA binding manner and through interaction with C/EBP α, respectively. The associations of both proteins to ori-Lyt DNA were shown to be essential for DNA replication (31). Thus, it is conceivable that RTA and K8 play key roles in initiation of KSHV ori-Lyt-dependent DNA replication. However, the details regarding their involvement in the DNA replication were not known. In this report, we attempted to reveal the roles of RTA and K8 in DNA replication as well as the mechanisms of their actions. The salient features of this report are as follows.
(i) We previously showed that one function of RTA in ori-Lyt is to initiate ori-Lyt-associated transcription in DNA replication (31). In this report, we assessed the contribution of RTA-mediated ori-Lyt transcription to DNA replication. The results showed that premature termination of the transcription before the GC-rich tandem repeats abolished DNA replication. However, if the transcription was terminated after the GC-rich repeats (before a putative open reading frame), little effect on DNA replication was observed. These data indicate that assembly of a transcription complex in ori-Lyt is necessary but not sufficient to support DNA replication; active transcription through GC-rich tandem repeats is required for DNA replication. The sequences of the tandem repeats in two ori-Lyt regions [ori-Lyt (L) and ori-Lyt (R)] are quite different, but both are GC-rich repeats of approximately 600 bp and organized as 20- to 30-nucleotide tandem arrays. It suggests that transcription over a particular DNA pattern, rather than a specific sequence, is required for initiation of DNA replication. The importance of the transcription event in the DNA replication was also confirmed by the finding that the RTA transactivation domain is critical for the DNA replication function. The participation of transcription factors and involvement of RNA transcription in the regulation of DNA replication have been observed for viruses as well as eukaryotic cells (6, 14, 29). For example, a powerful transcription enhancer region resides in an Epstein-Barr virus ori-Lyt, which contains DNA binding sites for EBV transcription factors RTA and ZTA. This enhancer region can be functionally replaced with an HCMV major immediate-early enhancer (14). Recently, Jenke and colleagues demonstrated that an active transcription unit and a downstream nuclear scaffold/matrix-attached region (S/MAR) are sufficient for DNA replication and maintenance of a mammalian episome (16). Although the roles of transcriptional elements in DNA replication have not been fully understood, the findings regarding the interplay between transcription and DNA replication have provided a basis for further investigation into the functional roles of RNA transcription in regulation of DNA replication in DNA viruses and eukaryotic cells.
(ii) Further investigation of the requirement of RTA in ori-Lyt-dependent DNA replication showed a multiple-function property of RTA in DNA replication. This notion was first suggested by the promoter replacement experiment, which showed that when the RRE-containing promoter in ori-Lyt was replaced with heterogenic promoters, such as an SV40 early promoter, an HCMV immediate-early promoter, and a mouse U6 snRNA promoter, ori-Lyt DNA replication was abolished even though RNA transcription activities remained. Furthermore, mutagenesis of RTA demonstrated that in addition to the domain in RTA that is known to be essential for transactivation activity, the other region in the activation domain of the molecule was found to contribute to ori-Lyt DNA replication (Fig. 3). Therefore, RTA may play multiple roles in KSHV ori-Lyt-dependent DNA replication.
A new function of RTA, other than RNA transactivation, was revealed in this report. RTA was found to be a component of the viral replication compartment in the nuclei of infected cells and shown to interact with SSB, PPF, and K8 by a coimmunoprecipitation assay. It is likely that RTA interacts with the core replication machinery complex (or prereplication complex), which is composed of at least six core replication proteins. This assumption is made based on the finding by Wu et al. (33) that assembly of a complex of KSHV core replication machinery proteins can occur independently of its attachment to origin DNA. Furthermore, the loading of the core replication machinery complexes on ori-Lyt DNA appears to be mediated by RTA and be dependent upon the RRE in ori-Lyt, as the deletion of the RRE abolished the association of core replication proteins with ori-Lyt DNA. These data strongly suggest that RTA plays a role in recruiting the core replication machinery complexes to ori-Lyt DNA in order to initiate DNA replication.
(iii) Our study also showed that K8 binds to the other end of ori-Lyt (with the aid of C/EBP α) and has a function similar to that of RTA in interaction with core replication proteins and loading prereplication complexes to ori-Lyt DNA. It was shown previously that EBV-encoded ZTA, a distant evolutionary homologue of K8 protein, serves an essential origin binding and DNA replication initiation function (10). ZTA can mediate EBV ori-Lyt DNA replication with the core replication proteins of EBV, HSV-1, and KSHV (10, 33). The replication function of ZTA is mediated through interactions with the core replication proteins (11, 19). Therefore, it was not surprising to find that K8 exerts a function similar to that of its EBV counterpart in interaction with the core replication proteins and recruitment of prereplication complexes to ori-Lyt DNA. In a preliminary study, we found that K8 was able to interact with HEL (ORF41) and PAP (ORF40/41) in a GST pulldown assay. No other core replication protein was found to be pulled down by GST-K8 fusion protein in that assay (our unpublished data). In the coimmunoprecipitation assay presented in this report, SSB and PPF were detected in the K8 precipitates. Thus, K8 is likely to interact with the entire core replication complexes (through direct contacts with helicase primer proteins), thereby recruiting the complexes to ori-Lyt DNA.
(iv) Both RTA and K8 were found to interact with the prereplication complex, and both are responsible for bringing the complexes to ori-Lyt DNA. Extrapolating from the observation that both RTA and K8 are essential for ori-Lyt-dependent DNA replication (1, 31), we speculate that the formation of a functional DNA replication initiation complex with an ori-Lyt DNA requires the efforts of both RTA and K8 in loading prereplication complexes on ori-Lyt DNA. Thus, the prereplication complexes apparently bind to ori-Lyt DNA through two contact points, one at the K8 binding region (the C/EBP cluster) near one end of ori-Lyt and the other at the RRE near the opposite end (Fig. 11). The two-contact-point binding may bring two ends of an ori-Lyt element together, looping the DNA between the K8 and RTA binding sites, as illustrated in Fig. 11. An 18-bp AT palindrome sequence is located in the looped region.
(v) A similar loop structure, in which two UL9 protein dimers (OBP) bind to two binding sites (boxes I and II) in ori-S and protein-protein interactions between the UL9 molecules loop the origin DNA centered by an AT palindrome sequence, has been seen in the HSV-1 DNA replication system (17). Examination of binding of UL9 protein to ori-S DNA by electron microscopy revealed the presence of nucleoprotein complexes that were frequently involved in intermolecular and intramolecular interactions in which two DNA molecules or two regions of a DNA molecule were jointed by protein complexes(25). The looping leads to bending and distortion of ori-Lyt DNA, facilitating unwinding of the DNA sequence. The looping and DNA distortion/unwinding are separate activities, and the latter activities can be enhanced by SSB (17). Overall, UL9 protein functions as a replication initiator protein whose role is to make the DNA at the origin accessible to the DNA replication machinery. RTA and K8 may function in a manner similar to UL9 protein, serving as DNA replication initiators in KSHV ori-Lyt-dependent DNA replication.
However, the classic replicon model for herpesviruses, postulated based primarily on studies of HSV-1, is that the core origin region is first bound by a virus-specific OBP, such as HSV-1 UL9, that recruits the core replication machinery. The core replication machinery proteins are recruited to the origin in an ordered and sequential fashion. In contrast, our results clearly suggest that assembly of the KSHV core replication complex occurs independently of its attachment to ori-Lyt DNA and that the entire complex is loaded on ori-Lyt through two components, K8 and RTA, that have integrated in the complex. This may reflect the mechanistic difference between HSV-1 and KSHV in DNA replication or suggest that more studies are needed to truly understand the processes and mechanisms of DNA replication for herpesviruses.
Acknowledgments
We thank Keiji Ueda (Osaka University) for the anti-RTA monoclonal antibody, David Lukac (UMDNJ) for the anti-RTA polyclonal antibody, Jae Jung (Harvard Medical School) for the anti-K8 polyclonal antibody, Gary Hayward (Johns Hopkins University) for the anti-SSB polyclonal antibody, and Robert Ricciardi (University of Pennsylvania) for the anti-PPF antibody. We thank all members of the Yuan lab for constructive discussion, suggestions, and critical reading of the manuscript.
This work was supported by research grants from the National Institutes of Health (R01AI52789, R01CA86839, and R01AI41136).
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 6.DePamphilis, M. L. 1993. Origins of DNA replication that function in eukaryotic cells. Curr. Opin. Cell Biol. 5:434-441. [DOI] [PubMed] [Google Scholar]
- 7.Elias, P., C. M. Gustafsson, and O. Hammarsten. 1990. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J. Biol. Chem. 265:17167-17173. [PubMed] [Google Scholar]
- 8.Elias, P., and I. R. Lehman. 1988. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 85:2959-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, Z., A. Krithivas, J. E. Finan, O. J. Semmes, S. Zhou, Y. Wang, and S. D. Hayward. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 13.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 15.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenke, A. C., I. M. Stehle, F. Herrmann, T. Eisenberger, A. Baiker, J. Bode, F. O. Fackelmayer, and H. J. Lipps. 2004. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc. Natl. Acad. Sci. USA 101:11322-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koff, A., J. F. Schwedes, and P. Tegtmeyer. 1991. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J. Virol. 65:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, J., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZIP protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of the open reading frame 50 of Kaposi's-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monier, K., J. C. Gonzalez Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 23.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 25.Rabkin, S. D., and B. Hanlon. 1991. Nucleoprotein complex formed between herpes simplex virus UL9 protein and the origin of DNA replication: inter- and intramolecular interactions. Proc. Natl. Acad. Sci. USA 88:10946-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 27.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 91:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehle, I. M., M. F. Scinteie, A. Baiker, A. C. Jenke, and H. J. Lipps. 2003. Exploiting a minimal system to study the epigenetic control of DNA replication: the interplay between transcription and replication. Chromosome Res. 11:413-421. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., O. T. Chong, and Y. Yuan. 2004. Differential regulation of K8 gene expression in immediate-early and delayed-early stages of Kaposi's sarcoma-associated herpesvirus. Virology 325:149-163. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 78:8615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, C. A., N. J. Nelson, D. J. McGeoch, and M. D. Challberg. 1988. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Gen. Virol. 62:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, F. Y., S. E. Wang, Q.-Q. Tang, M. Fujimuro, C.-J. Chiou, Q. Zheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein α and p21CIP-1. J. Virol. 77:8893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, F. X., X. Li, F. Zhou, S.-J. Gao, and Y. Yuan. 2006. Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J. Virol. 80:12187-12196. [DOI] [PMC free article] [PubMed] [Google Scholar]