Abstract
Nipah virus (NiV) and Hendra virus (HeV) are paramyxoviruses capable of causing considerable morbidity and mortality in a number of mammalian species, including humans. Case reports from outbreaks and previous challenge experiments have suggested that cats were highly susceptible to NiV infection, responding with a severe respiratory disease and systemic infection. Here we have assessed the cat as a model of experimental NiV infection and use it in the evaluation of a subunit vaccine comprised of soluble G glycoprotein (sG). Two groups of two adult cats each were inoculated subcutaneously with either 500 or 5,000 50% tissue culture infective dose(s) (TCID50) of NiV. Animals were monitored closely for disease onset, and extensive analysis was conducted on samples and tissues taken during infection and at necropsy to determine viral load and tissue tropism. All animals developed clinical disease 6 to 9 days postinfection, a finding consistent with previous observations. In a subsequent experiment, two cats were immunized with HeV sG and two were immunized with NiV sG. Homologous serum neutralizing titers were greater than 1:20,000, and heterologous titers were greater than 1:20,000 to 16-fold lower. Immunized animals and two additional naive controls were then challenged subcutaneously with 500 TCID50 of NiV. Naive animals developed clinical disease 6 to 13 days postinfection, whereas none of the immunized animals showed any sign of disease. TaqMan PCR analysis of samples from naive animals revealed considerable levels of NiV genome in a wide range of tissues, whereas the genome was evident in only two immunized cats in only four samples and well below the limit of accurate detection. These results indicate that the cat provides a consistent model for acute NiV infection and associated pathogenesis and an effective subunit vaccine strategy appears achievable.
Nipah virus (NiV) is a recently emerged paramyxovirus and, together with Hendra virus (HeV), comprises the newly classified genus Henipavirus. In contrast to all other paramyxoviruses, the henipaviruses cause zoonotic infections exhibiting a broad species tropism and the ability to cause fatal disease in a variety of animal species, including humans (reviewed in reference 17). HeV was recognized in eastern Australia in 1994 and was transmitted to humans from infected horses (reviewed in reference 27). NiV emerged in 1998 and 1999 in peninsular Malaysia associated with a much larger outbreak and was primarily transmitted to humans from infected pigs (reviewed in reference 11). NiV has continued to reemerge, with outbreaks in Bangladesh in 2001 and 2003 (4, 25) and further outbreaks in early 2004 and 2005 that were associated with a higher incidence of acute respiratory distress syndrome in conjunction with encephalitis, person-to-person transmission, and higher case fatality rates (∼75%) (2, 3, 5, 25). NiV is classified as a select agent by the U.S. Centers for Disease Control and Prevention and, unlike most notable viral agents of biodefense concern such as smallpox or Ebola virus, NiV can be isolated from natural sources (14, 29, 32) and readily grown in cell culture to high titers (15). It may be transmitted via the respiratory tract and can be amplified and spread in livestock serving as a source for transmission to humans (18, 24).
There is currently a paucity of accepted animal models for henipavirus infection and the restriction of live NiV and HeV experimentation to biological safety level 4 (BSL4) containment has significantly hampered their rapid development. The implementation of typical laboratory animal models has also been problematic. NiV and HeV do not cause disease in mice after subcutaneous administration, although they are lethal if administered intracranially (G. Crameri and B. T. Eaton, unpublished data). Further, there was no serological evidence of NiV infection in rodents from Malaysia (11, 39) or of HeV infection in rodents from Australia (30). To our knowledge, rabbits have been examined only with respect to HeV, and no clinical disease was observed (36). Guinea pigs have proven considerably variable in respect to both NiV- and HeV-induced pathology (reviewed in reference 17). Symptoms of NiV infection in humans range from fever and headache to either severe acute febrile encephalitis (predominant) or acute respiratory distress syndrome (less frequent) (2, 13, 20), whereas the porcine disease is primarily a febrile respiratory illness (24). In both humans and pigs, NiV pathogenesis presents as a systemic vasculitis (26, 38). Both cats and dogs were observed to be susceptible to disease during the initial NiV outbreak (24). The clinical and pathological features of cats naturally (24) or experimentally infected resembled those seen in humans and pigs (26), particularly with respect to tropism for arterial endothelial cells and respiratory epithelium. Moreover, experimental infection of cats and hamsters has revealed a similar underlying pathology (26, 37). While the latter study indicated a role for hamsters in respect to a NiV disease model for the central nervous system (37), encephalitis has not been identified in cats from limited field or experimental data.
Currently, there are no approved active or passive therapeutic treatment modalities available for the henipaviruses (reviewed in reference 7). Although there is some evidence that ribavirin therapy may be of clinical benefit (10), recent experimental infection models have not supported this (19). Ribavirin therapy also has limited value due to the toxic side effects seen at therapeutic dose rates, mainly involving hemolytic anemia (16). In regard to vaccination approaches, it is the major viral envelope glycoproteins against which virtually all neutralizing antibodies are directed, and neutralizing antibodies are the key vaccine-induced protective mechanisms in the case of mumps and measles viruses (21, 28), with the attachment glycoproteins often serving as the predominant target. Previously, it has been shown that vaccinia virus expressed fusion (F) or attachment (G) glycoproteins from NiV can elicit virus-neutralizing antibodies (31), and studies using a hamster model have indicated that a vaccination approach using recombinant vaccinia virus encoding NiV F or G may be effective (22). However, such a vaccine is unlikely to receive regulatory approval. Most recently, a recombinant canarypox vaccine candidate for swine encoding NiV F and G glycoproteins has been shown to protect animals against challenge (34).
We sought to explore the possibility of a subunit vaccine approach for henipaviruses and have produced and extensively characterized recombinant expressed, soluble versions of the G glycoprotein (sG) from HeV and NiV (8). Purified sG has been shown to retain a number of important native structural, functional, and antigenic features, including the ability to bind virus receptor (ephrinB2) (6), block virus-mediated membrane fusion and virus infection, and elicit a robust polyclonal neutralizing antibody response in rabbits and mice (8). The sG glycoprotein can also capture and isolate virus-specific neutralizing human monoclonal antibodies (MAbs) from naive recombinant libraries (40). These attributes make sG potentially suitable as a vaccine candidate. We also wanted to evaluate a species capable of developing systemic disease as a consequence of natural exposure to NiV, analogous to an epidemic or outbreak scenario and, because cats were also susceptible to disease caused by HeV, both directly and via contact with affected animals (35), we chose to develop cats as an animal model of NiV infection. The U.S. Food and Drug Administration recently implemented the Animal Efficacy Rule in 2002, which applies to the development of therapeutic products when human efficacy studies are not possible (17a). In short, evidence derived from animal studies on product effectiveness in preventing or treating disease may be relied on when particular criteria are met, such as a well-understood mechanism for the pathogenicity of the agent in conjunction with the underlying mechanism of the product. Importantly, however, the therapeutic effect must be demonstrated in more than one animal species. By characterizing a highly susceptible animal model that provides consistent clinical outcomes, it may be exceedingly useful in future studies for the screening and evaluation of potential therapeutic intervention strategies for henipavirus infection.
MATERIALS AND METHODS
Animals, accommodation, handling, and biosafety.
Four adult (12- to 24-month-old) domestic shorthair cats (three castrated males and one entire female) were used to assess susceptibility to NiV and six adult cats (five entire females and one castrated male) were used for the vaccine efficacy study. The animal husbandry methods and experimental design were endorsed by the CSIRO Australian Animal Health Laboratory's Animal Ethics Committee. Animals were housed in a single room at BSL4. Room temperature was maintained at 22°C with 15 air changes per hour. Humidity varied between 40 and 60%. The room was fitted with a surveillance camera that allowed constant monitoring of the animals. Cats were housed individually in cages 1.4 m wide, 0.9 m high, and 0.6 m deep, within visual and auditory contact of each other. The cats were fed twice daily with a complete premium dry food and provided with water ad libitum. Before inoculation or collection of specimens, animals were immobilized with a mixture of ketamine HCl (2 mg/kg; Ketamil; Ilium, Smithfield, Australia) and medetomidine (20 μg/kg; Domitor; Novartis, Pendle Hill, Australia) by intramuscular injection. For reversal, atimepazole (Antisedan; Novartis) was given intramuscularly at 50% of the dose used for medetomidine. Staff wore fully encapsulated suits with breathing apparatus while in the animal room. Serology, virus isolations, and the initial stages of RNA extraction were carried out at BSL4.
Nipah virus infections.
Two cats were inoculated subcutaneously with 1 ml of phosphate-buffered saline (PBS) containing 500 50% tissue culture infective dose(s) (TCID50) and two with 5,000 TCID50 of NiV (stock virus titer, 1.1 × 107 TCID50/ml) prepared as described previously (15). Cats were assessed daily and scored out of 10 for a range of clinical observations, including alertness, grooming behavior, curiosity, depression, food eaten, feces production, and respiration rate. Clinical scores for each day were combined to provide a cumulative daily score. Rectal temperatures and body weights were recorded during sampling periods, which occurred every second day. Once the cats were clinically assessed to be exhibiting signs of disease, animals were then immobilized as described above. Blood was collected via intracardiac puncture, and the animals were immediately euthanized by intravenous injection of sodium pentabarbitone.
Immunization and challenge.
Four adult cats were immunized three times with a 100-μg dose of recombinant, sG (8); two animals received HeV sG, and two received NiV sG. Each dose was prepared in CSIRO triple adjuvant (60% [vol/vol] Montanide, 40% [vol/vol] sG [combined with Quil A, 3 mg/ml, and DEAE-dextran, 30 mg/ml] in water) (8). Animals were inoculated subcutaneously, at 2-week intervals, and bled approximately 2 weeks and 2 months after the final immunization. Immunizations took place on days 0, 13, and 28. The first bleed was on day 48; the second bleed was on day 93. NiV challenge occurred on day 104. Serum was assessed for antibody production by using a serum neutralization test as described below. Prior to virus challenge, all animals for the vaccine study were implanted with radiotelemetry transmitters for continuous, remote monitoring of body temperature as described below. The four vaccinated animals and two unvaccinated controls were inoculated subcutaneously with 500 TCID50 of NiV and then monitored twice daily for clinical disease onset and sampled over a range of times as described above.
Radiotelemetry measurements of body temperature.
With the animals under anesthesia as described above, single-stage transmitters fitted with an internal loop antenna and coated with an inert two-pot epoxy resin (Sirtrack, Havelock North, New Zealand) were implanted subcutaneously between the shoulder blades (final dimensions, 25 by 15 by 6 mm). Briefly, transmitters use a thermistor to control the pulse rate and emit pulses at discrete frequencies (e.g., 150.100 MHz) in proportion to temperature. All transmitters were calibrated by submersion in a water bath, followed by chemical sterilization (chlorhexidine followed by 70% alcohol and, finally, sterile saline) prior to implantation. Pulses were detected with a PCR-1000 scanning receiver (Icom, Inc., Osaka, Japan) linked to a standard PC via an RS232 connection. An in-house Visual Basic code was written to control the receiver and log the telemetry data. Transmitters were consecutively scanned for 1 min each (6-min cycle time), providing near real-time monitoring of the body temperature for all animals.
Sample collection.
Specimens for virus isolation (heparinized blood and oral swabs) and RNA isolation (EDTA blood and oral swabs) were collected every second day postinfection (p.i.) and just prior to euthanasia. Urine was only collected from female cats by urethral catheterization at each sampling time and from all cats at necropsy. Each swab for virus isolation was immediately placed into 1 ml of viral transport medium (PBS containing 100 U of penicillin, 100 μg of streptomycin/ml, and 500 μg of amphotericin B [Fungizone]/ml), while all samples for RNA isolation were mixed with 1 ml of RNAlater (Ambion, Inc., Austin, TX). All specimens were held at −80°C until isolation attempts were made. After euthanasia, the samples were collected aseptically from the lung (apical and diaphragmatic lobes); brain (olfactory and occipital lobes); heart; prescapular, retropharyngeal, or thoracic lymph nodes; spleen; liver; kidney; bladder; and adrenal gland. In addition, in female cats, the uterine horn and ovaries were collected. Tissues were either fixed in 10% neutral buffered formalin for 48 h prior to histological processing or submerged in RNAlater or viral transport medium and then stored at −80°C until processing for RNA or viral isolation, respectively.
Virus isolation.
NiV was isolated by using Vero cells grown in Eagle's minimal essential medium (EMEM) supplemented with 1% fetal calf serum (FCS) and antibiotics as described above. A 10% homogenate of each tissue sample was prepared in PBS. Blood was diluted 1:10 and 1:100 in media (EMEM plus 1% FCS). Fluids from swabs were tested neat. Each sample (200 μl) was inoculated onto one well of a 24-well plate containing 80% confluent Vero cells, followed by incubation for 60 min. The monolayers were washed with PBS and incubated with 2 ml of EMEM containing 1% FCS and antibiotics. Samples were observed for the NiV cytopathic effect (CPE) over a period of 4 days. All samples were passaged twice before being considered negative. The identity of NiV was confirmed by immunofluorescence staining as described previously (9).
RNA isolation.
RNA was isolated from blood cells by using the RiboPure-Blood kit (Ambion, Inc.); from swabs, serum, and urine by using the QIAamp viral RNA kit (QIAGEN Pty, Ltd., Clifton Hill, Australia); and from tissues by using the RNeasy Minikit (QIAGEN Pty, Ltd.). Briefly, under BSL4 conditions, 300 μl of blood was added to 800 μl of RiboPure lysis buffer and 50 μl of sodium acetate. Then, 140 μl of swab solution or urine was added to 560 μl of AVL viral lysis buffer (QIAamp). Tissues were homogenized by removing the tissue from RNAlater and adding it to tubes containing 600 μl of RLT lysis medium (RNeasy) containing 6 μl of β-mercaptoethanol and 250 μl of 1-mm zirconia/silica beads (BioSpec Products, Inc., Bartlesville, OK). Samples were then homogenized for two cycles of 30 s by using a Mini-Bead beater (Biospec Products, Inc.) and centrifuged to pellet the cell debris, and the supernatant was transferred to a new tube. All samples were then removed from the BSL4 lab and processed according to the manufacturer's instructions supplied with each kit.
NiV genome TaqMan PCR assay.
The primers and TaqMan probe to detect NiV genome sequences were designed from the N gene sequence of NiV AF212302 (12) using specified criteria in the Primer Express Software (version 1.5; Applied Biosystems, Foster City, CA). The specific NiV oligonucleotide primers were Nipah-N1198F (5′-TCAGCAGGAAGGCAAGAGAGTAA-3′) and Nipah-N1297R (5′-CCCCTTCATCGATATCTTGATCA-3′). The fluorogenic 5′ nuclease (TaqMan) probe was Nipah 1247comp (5′-FAM [6-carboxy-fluorescein]-TGCTGGCACCAGACTTGCCCTC-3′-TAMRA [6-carboxy-tetramethyl rhodamine]). 18S rRNA was used to normalize for variations in RNA extraction. The 18S oligonucleotide primers were 18SrRNAF (5′-CGGCTACCACATCCAAGGAA-3′) and 18SrRNAR (5′-GCTGGAATTACCGCGGCT-3′). The fluorogenic 5′ nuclease (TaqMan) probe for 18SrRNA was 5′-VIC-CCTCCAATGAGCACACCTCCTGCAG-3′-TAMRA.
Assays were performed in triplicate using a one-step protocol consisting of an initial reverse transcription reaction followed immediately by cDNA amplification. All TaqMan reagents were purchased from Applied Biosystems except the primers, which were obtained from Geneworks (Hindmarsh, Australia). RNA (2 μl) was added to 23 μl of PCR mix in each well of a MicroAmp optical reaction plate containing 12.5 μl of TaqMan One-Step PCR Mastermix, 0.625 μl of 40× Multiscribe/RNase inhibitor, 5.75 μl of distilled water, 1.25 μl each of 18 μM Nipah N1198F and N1297R primers, 1.25 μl of 5 μM Nipah-1247-comp-FAM-labeled probe, 0.125 μl each of 10 μM 18SrRNAF and 18SrRNAR, and 0.125 μl of 40 μM 18SrRNA-VIC-labeled probe. The samples were amplified in a GeneAmp 7700 sequence detection system (Applied Biosystems) using the following program: 48°C for 30 min, 1 cycle; 95°C for 10 min, 1 cycle; and 95°C for 15 s and 60°C for 60 s, 50 cycles. Primer and probe optimization experiments were performed as recommended in the AB7700 sequence detection system protocol. A dilution series was made of each NiV primer (900 to 50 nM) and probe (250 to 25 nM) to determine the optimum concentrations (data not shown).
Standard curves were obtained for NiV-specific cDNA in triplicate for each assay. The threshold cycle (CT) values obtained from the standard dilutions were plotted against the input cDNA concentration to create a standard curve (data not shown). Linear regression analysis was used to quantify the NiV RNA. To correct for sample variation, CT values for NiV genome in samples were normalized against 18S rRNA expression, and NiV genome was expressed relative to a standard NiV cDNA dilution (3.6 pg of NiV cDNA).
Serum neutralization test.
Virus neutralization antibody titers were determined as previously described (8). Briefly, sera were diluted in EMEM containing 10% FCS (EMEM-10) and subjected to doubling dilution starting at 1:20. Then, 50 μl of serum was added to each well in quadruplicate in a 96-well plate, followed by 50 μl of virus containing 200 TCID50 of either HeV or NiV; the plates were then incubated at 37°C for 30 min. A total of 2 × 104 Vero cells was added to all wells in 150 μl of EMEM-10, followed by incubation at 37°C for 4 days in a humidified 5% CO2 atmosphere. Neutralization titers were determined by the presence of CPE and recorded as the serum dilution where at least one of the replicate wells showed no virus-induced CPE.
Histopathology and immunohistochemistry.
Tissues were routinely dehydrated through graded alcohols, cleared and embedded in paraffin wax, sectioned at 3 to 4 μm, and stained with hematoxylin and eosin. Further sections were prepared for immunohistochemistry by using a rabbit polyclonal anti-NiV antiserum (26).
RESULTS
Nipah virus infection of naive cats.
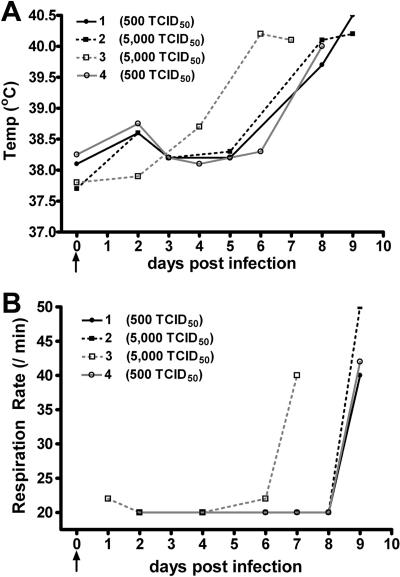
The evaluation of any potential therapeutic approach for treating or preventing henipavirus infection will likely require the establishment of at least two accepted animal models. Here, we sought to examine the outcomes of lower challenge doses of NiV in order to determine the minimal dose of virus that consistently gave rise to disease, thus more closely mimicking a natural infection. We used two lower doses of NiV (500 and 5,000 TCID50), which were administered subcutaneously to four cats. All of the cats developed an acute febrile disease comparable to that seen previously in experimental infection of cats given 50,000 TCID50 of NiV orally and intranasally (26). Cat 3, which received 5,000 TCID50 of NiV, was noted to be febrile (>39°C) on day 6 p.i., while the other animal infected with 5,000 TCID50 and the two that received 500 TCID50 of NiV became febrile on day 8 p.i. (Fig. 1A). The rise in temperature observed in all animals preceded an increase in respiratory rate by approximately 24 h (Fig. 1B). A rise in cumulative daily clinical scores was observed in all cats in parallel to the increase in respiratory rate (data not shown). As companion animals, cats were not allowed to progress to serious clinical disease and, based on the development of an increased respiratory rate, the animals were euthanized on either day 7 p.i. (cat 3, 5,000 TCID50 of NiV) or day 9 p.i. Upon necropsy, three of the four animals exhibited scattered 1- to 3-mm hemorrhagic nodular lesions on the visceral pleura, while one animal (cat 1, 500 TCID50 of NiV) had no gross pathological changes. Cat 2, a female, had marked edema of the bladder serosa with dilation of the serosal lymphatic vessels.
FIG. 1.
NiV susceptibility study body temperatures and respiration rates. (A) Rectal temperatures obtained at various days postinfection in cats inoculated with either 500 TCID50 of NiV (circles) or 5,000 TCID50 of NiV (squares). All cats became febrile 6 to 8 days postinfection. (B) Respiration rates determined for cats infected with either 500 TCID50 NiV (circles) or 5,000 TCID50 NiV (squares). Immediately prior to euthanasia, all cats exhibited a marked increase in respiration rate.
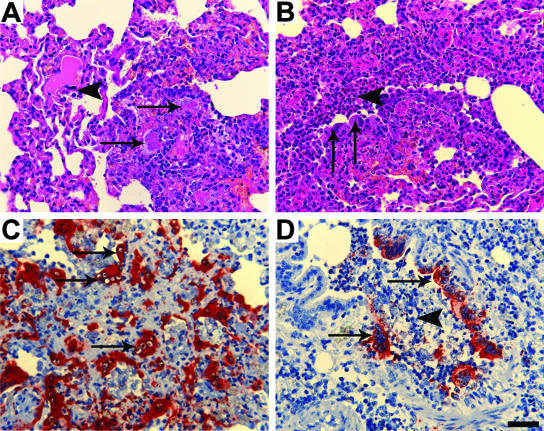
Each cat exhibited mild to moderately severe histological lesions consistent with that previously reported in NiV infection of cats. These comprised, variously, acute bronchiolitis and focal necrotizing alveolitis with syncytium formation in respiratory epithelial cells (Fig. 2A and B), pulmonary arteritis with fibrinoid necrosis of the vessel walls, endothelial syncytial cell formation, and intracytoplasmic eosinophilic inclusion bodies. All cats had focal hemorrhagic and necrotic lesions of the splenic red pulp associated with syncytia. Cat 2 had edema of the serosal, muscular, and submucosal layers of the bladder wall; submucosal hemorrhages with acute inflammatory cell infiltration; and endothelial syncytia. Necrotizing lymphadenitis was identified in cat 4. Positive immunostaining for NiV antigen was detected in bronchiolar and alveolar epithelial cells (Fig. 2C and D), in the pulmonary vascular endothelium and tunica media of pulmonary arteries, and within syncytial cells within the lungs and spleens of all four cats. Antigen was also identified in syncytial cells, connective tissue cells, and the endothelium of inflamed bladder submucosa and lymphoid tissue.
FIG. 2.
Histopathology and immunohistopathology associated with NiV infection in cats. (A) Necrotizing alveolitis (arrows) in NiV-infected cat and endothelial syncytial cell (arrowhead) (HE). (B) Broncho-alveolitis in NiV-infected cat, with bronchiolar epithelial syncytial cells (arrows) and intraluminal debris (arrowhead) (HE). (C) Positive staining for NiV antigen in regions of alveolitis, including endothelial syncytial cells (arrows) (anti-NiV polyclonal antibody). (D) Positive staining for NiV antigen in bronchiolar epithelial cells (arrows) and intraluminal debris (arrowhead) (anti-NiV polyclonal antibody). Scale bar for all images = 50 μm.
Detection of Nipah virus genome.
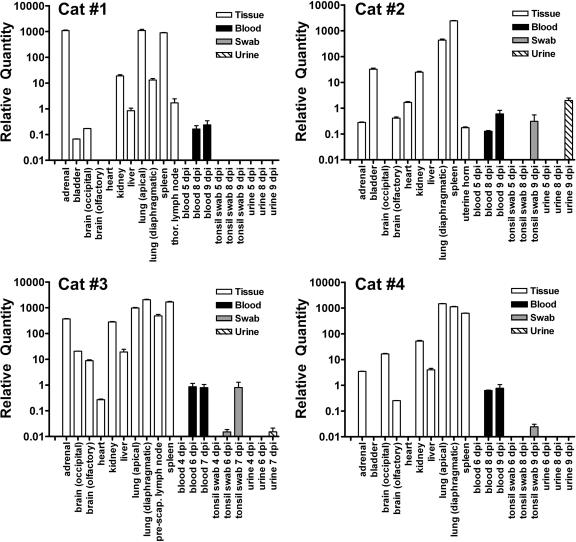
To examine the extent of virus replication, as well as viral tissue tropism, NiV genome was measured in blood, urine, and tonsil swabs sampled over the course of infection, as well as in tissues removed upon necropsy. Because NiV isolation can only be conducted under high biological containment, a TaqMan PCR assay was developed based on the detection of RNA corresponding to the N gene of NiV and used for initial screening for the presence of virus. In all animals, NiV genome was detected in the blood 1 to 2 days prior to and on the day of euthanasia and in oral swabs from three cats at similar times postchallenge (Fig. 3). A broad range of tissues revealed significant quantities of NiV genome (Fig. 3 and Table 1). NiV genome was only detected in urine from cats inoculated with 5,000 TCID50 of NiV (Table 1). The lungs appeared to be a site of extensive virus replication and highly vascularized lymphoid tissues (lymph nodes and spleen) revealed high levels of genome. In contrast, brain and heart tissue had relatively low amounts of NiV genome (50- to 100-fold less than the lung) similar to the levels observed circulating in blood. All cats did reveal NiV genome in brain tissue but, notably, no animals displayed any apparent neurological signs at the point of euthanasia. Consistent with the pathological observations, cat 2 revealed moderate genome levels in bladder (Table 1). Bladder tissue was not collected from the second cat inoculated with 5,000 TCID50 of NiV, and the remaining two animals also revealed low levels of NiV genome present in the bladder. In addition, NiV genome was readily demonstrated in kidneys from all cats, as well as from the urine from the two cats inoculated with 5,000 TCID50 of NiV.
FIG. 3.
NiV genome in cats detected by TaqMan PCR. Normalized, relative NiV genome levels in samples collecting during NiV infection in cats and at necropsy. TaqMan PCR CT values were determined in triplicate for the NiV genome and normalized by dividing this value by the 18S rRNA CT values for each sample. The relative NiV genome was determined by linear regression of NiV cDNA standard curves for each assay. Values are expressed as the average of all replicates. The adrenal gland, liver, lung, spleen, and lymph nodes consistently displayed the highest relative NiV genome levels, while the brain and heart frequently revealed the lowest. The genome was detectable in the blood in all cats 1 day prior to euthanasia but only detectable in the urine from cats infected with 5,000 TCID50 of NiV.
TABLE 1.
Normalized relative NiV genome levels detected by TaqMan PCR in NiV-infected naive catsa
| Sample | Location | dpi | Relative NiV genome level in cat (inoculum size)b:
|
|||
|---|---|---|---|---|---|---|
| 1 (500) | 2 (5,000) | 3 (5,000) | 4 (500) | |||
| Adrenal gland | PM | 1,083.866 | 0.282 | 369.042 | 3.461 | |
| Bladder | PM | 0.067 | 32.387 | NS | 0.009 | |
| Blood | 0 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 2 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| 3 | 0.000 | 0.000 | NS | NS | ||
| 4 | NS | NS | 0.000 | 0.000 | ||
| 5 | 0.000 | 0.000 | NS | NS | ||
| 6 | NS | NS | 0.854 | 0.000 | ||
| 7 | NS | NS | 0.786 | NS | ||
| 8 | 0.165 | 0.125 | NS | 0.628 | ||
| 9 | 0.237 | 0.599 | NS | 0.768 | ||
| Brain | Occipital lobe | PM | 0.172 | 0.000 | 0.622 | 16.624 |
| Olfactory lobe | PM | 0.000 | 0.411 | 0.578 | 0.251 | |
| Heart | PM | 0.000 | 1.669 | 7.512 | 0.000 | |
| Kidney | PM | 18.974 | 24.599 | 22.421 | 0.000 | |
| Liver | PM | 0.842 | 0.001 | 0.334 | 18.679 | |
| Lung | Apical lobe | PM | 1,094.862 | NS | 973.303 | 1,480.818 |
| Diaphragmatic lobe | PM | 13.143 | 432.096 | 2,027.956 | 1,124.226 | |
| Lymph node | Thoracic | PM | 1.695 | NS | NS | NS |
| Prescapular | PM | NS | NS | 486.245 | NS | |
| Skeletal muscle | PM | NS | NS | 0.001 | NS | |
| Spleen | PM | 890.187 | 2,438.948 | 1,667.516 | 628.465 | |
| Swab | 0 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 2 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| 4 | 0.000 | NS | 0.000 | 0.000 | ||
| 5 | NS | 0.000 | NS | NS | ||
| 6 | NS | NS | 0.010 | NS | ||
| 7 | NS | NS | 0.798 | NS | ||
| 8 | 0.000 | 0.000 | NS | NS | ||
| 9 | 0.000 | 0.311 | NS | 0.017 | ||
| Urine | 0 | NS | 0.000 | NS | NS | |
| 3 | NS | 0.000 | NS | NS | ||
| 7 | NS | NS | 0.018 | NS | ||
| 8 | NS | 0.000 | NS | NS | ||
| 9 | 0.000 | 1.977 | NS | 0.000 | ||
| Uterine horn | PM | NS | 0.176 | NS | NS | |
Relative values were determined by linear regression analysis using NiV cDNA standards and normalized using the ratio of NiV:18S rRNA CT values. PM, postmortem tissue; dpi, days postinfection; NS, no sample. Values are expressed as the fold difference from 3.6 pg of NiV cDNA standard and are the average of all replicates. Values in boldface indicate samples from which virus was isolated.
The NiV inoculum (TCID50) for each cat is indicated in parentheses.
Isolation of Nipah virus from infected cats.
Virus isolation was only attempted on samples that were determined to be positive for the NiV genome by TaqMan PCR (raw CT < 40). For whole blood, all 1:10 dilutions were toxic to Vero cell culture, whereas all 1:100 dilutions were negative for virus isolation. In all other nontissue samples, virus could only be isolated from a single oral swab obtained from cat 3 immediately prior to euthanasia (Table 1). This may represent the limit of detection for this assay since this sample contained the highest level of NiV genome for any nontissue sample (approximately three relative units). However, in tissue samples, virus could be isolated consistently from the lung and spleen and, less frequently, from the kidney and lymph nodes. In addition, virus was also isolated from the bladder of cat 2, a finding consistent with the significant pathological observations, as well as TaqMan results indicating a high viral load.
With the exception of one lymph node (cat 1) and one oral swab (cat 2), all samples from which virus could be recovered had relative NiV genome levels at least 25-fold greater than the reference NiV cDNA. Clearly, these data underscore the poor sensitivity of direct virus isolation compared to TaqMan PCR as techniques for virus detection. This anomaly between TaqMan PCR and virus isolation data was most notable in the adrenal samples, which revealed very high relative genome levels but did not enable virus isolation.
In summary, the results of this susceptibility study indicated that NiV infection in cats induced a reproducible pathology with either a 500 or a 5,000 TCID50 of virus. Consistent with previous observations of natural and experimental infection in a range of species, NiV pathogenesis was mediated primarily by a vasculitis, followed by progressive pulmonary epithelial involvement. Using TaqMan PCR, detection of viral genome was possible from a broad range of tissues and longitudinal samples over a predictable disease induction period. Importantly, we have defined 500 TCID50 of NiV as a sufficient inoculum to establish infection and produce clinical disease in cats. With these parameters in place, it was decided that for subsequent studies infection would be determined by the detection of NiV genome in blood (<40 CT), and clinical disease is defined as a temperature of >39°C sustained for more than 12 h, with or without an increase in cumulative daily clinical score.
sG immunization and Nipah virus challenge.
Previously, we had demonstrated that the administration of purified sG as an immunogen in rabbits could elicit a potent neutralizing antibody response (8). For use as a vaccine in cats, we chose a standard immunization protocol using purified sG and CSIRO triple adjuvant (see Materials and Methods). Four animals (cats 5, 6, 7, and 8) were immunized with three doses of sG at 2-week intervals, and the animals were challenged with 500 TCID50 of NiV 2 months after the last immunization. Two unvaccinated naive cats (animals 9 and 10) were infected with 500 TCID50 of NiV concurrently. Immediately after the immunization protocol, vaccinated animals exhibited very high homologous serum neutralizing titers greater than 1:20,000, and heterologous titers ranged from >1:20,000 to 16-fold lower (Table 2). Retesting serum 2 months after the final immunization revealed no decrease in homologous titers, whereas there was a two- to eightfold drop in heterologous titers (Table 2).
TABLE 2.
Serum neutralization titers in naive and sG-immunized cats
| Cat no. (immunogen) | SNTa
|
|||
|---|---|---|---|---|
| HeV at:
|
NiV at:
|
|||
| 2 wk | 2 mo | 2 wk | 2 mo | |
| 5 (NiV sG) | 1:1,280 | 1:640 | 1:20,480 | 1:20,480 |
| 6 (HeV sG) | 1:20,480 | 1:20,480 | 1:20,480 | 1:2,560 |
| 7 (HeV sG) | 1:20,480 | 1:20,480 | 1:20,480 | 1:10.240 |
| 8 (NiV sG) | 1:2,560 | 1:1,280 | 1:20,480 | 1:20,480 |
| 9 (control) | ND | <1:20 | ND | <1:20 |
| 10 (control) | ND | <1:20 | ND | <1:20 |
Serum neutralization titers (SNT) were determined from serum samples at 2 weeks and 2 months after the final immunization with sG. SNTs were determined for both HeV and NiV. ND, not determined.
Upon challenge, both naive animals (cats 9 and 10) developed the early stages of an acute respiratory syndrome comparable to that seen in the susceptibility study described above. The body temperature of cat 10 gradually increased from 4 days p.i., and the animal became febrile on day 7 p.i. and was euthanized on day 8 p.i. (Fig. 4A). Cat 9 appeared normal until day 11, became febrile on day 12, and was euthanized on day 13 p.i. Clinically, both animals presented with depression and loss of appetite 1 to 2 days prior to euthanasia. Increases in cumulative clinical scores were observed in cat 9 over days 11 to 12 and on day 7 in cat 10. Gross respiratory pathology in both animals was confined to focal 1- to 3-mm hemorrhage nodules scattered over the visceral pleura of the lungs. Incidentally and unexpectedly, cat 9 was noted to be approximately 4 weeks pregnant at postmortem examination. Similar histological lesions, including pulmonary arterial vasculitis and acute necrotizing alveolitis, were identified in control cats 9 and 10 as was noted in the animals of the infection susceptibility study described above. Interestingly, pulmonary vasculitis was also observed in the fetus of cat 9, and degenerative placentitis was also present. These observations confirmed the pathogenicity of the viral inoculum administered to the immunized cats. None of the sG-vaccinated animals exhibited any clinical signs of NiV infection up to 24 days p.i. (Fig. 4B), and subsequent necropsy examination revealed no gross pathology. No significant histological lesions were detected in any vaccinated cats, apart from chronic cystitis without features of NiV infection noted in cat 5. In addition, similar patterns of immunostaining were observed in affected tissues from control cats 9 and 10 as found in the four animals of the susceptibility study, with the additional finding of antigen within degenerating lesions of the placenta, the fetal placenta, and inflamed fetal pulmonary blood vessels (data not shown). NiV antigen was not detected in any tissue from any of the sG-vaccinated animals (cats 5, 6, 7, and 8), including the lungs and spleen.
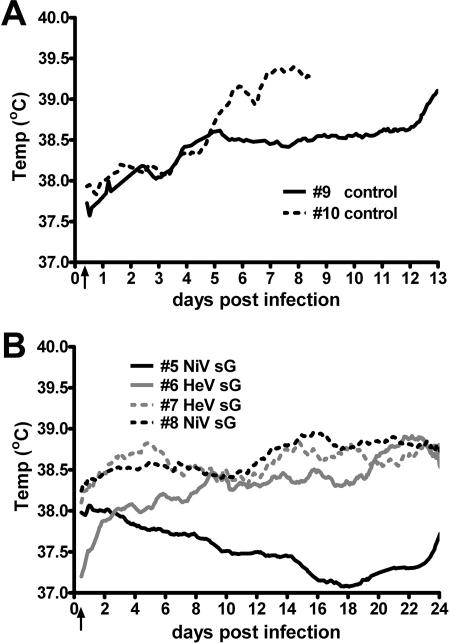
FIG. 4.
Temperature recordings of naive and sG-immunized cats infected with NiV. (A) Two cats in the vaccine efficacy study were implanted with radiotelemetry temperature transmitters prior to NiV inoculation (arrow). Cat 10 became febrile (>39°C) 7 days postinfection, while cat 9 remained clinically normal until day 11, becoming febrile on day 12, and was euthanized on day 13. Rectal temperatures were recorded during sampling periods and are indicated for each cat (circles). (B) Three cats showed gradual but slight increases in temperature after NiV infection (arrow), while the fourth showed a gradual decrease over the course of the study. All immunized cats remained within normal physiological parameters up to 24 days p.i.
Finally, NiV genome detection and virus isolation were also assessed. In both unvaccinated naive control animals (cats 9 and 10), as observed in the animals in the susceptibility study, NiV genome was detected in blood 1 to 2 days prior to and on the day of euthanasia, as well as in oral swabs, serum, and urine immediately prior to euthanasia (Table 3). NiV genome was detected at comparable levels and in a similar range of tissues in both animals as described above, confirming the highly reproducible nature of NiV disease pathology in cats. Placental and fetal tissues, in addition to placental fluid, were collected from the pregnant animal and revealed high levels of NiV genome by TaqMan PCR (data not show). In contrast to the naive animals, NiV genome was only detected in extremely low levels in two of the vaccinated cats (Table 3). In cat 6, NiV genome was observed in the adrenal gland, brain (occipital lobe), heart, and diaphragmatic lung, while in cat 5 NiV genome was detected in the brain (olfactory lobe). However, these values were well beyond the acceptable limit of detection of the TaqMan assay (>40 cycles) for all samples. Cats 7 and 8 had no detectable NiV genome present. As in the initial infection susceptibility studies, virus isolation was only attempted on samples positive for NiV genome and, consequently, no attempt was made to isolate virus from samples obtained from any vaccinated animals. In both unvaccinated naive controls, virus could be isolated consistently from the kidneys, lungs, spleen, and lymph nodes but not from blood, as was observed in the initial susceptibility studies (Table 3).
TABLE 3.
Normalized relative NiV genome levels in naive and sG-immunized catsa
| Sample | Location | dpi | Relative NiV genome level in cat (immunogen):
|
|||||
|---|---|---|---|---|---|---|---|---|
| 5 (NiV sG) | 6 (HeV sG) | 7 (HeV sG) | 8 (NiV sG) | 9 (control) | 10 (control) | |||
| Adrenal gland | 0.000 | 0.003 | 0.000 | 0.000 | 10.084 | 709.758 | ||
| Bladder | 0.000 | 0.000 | 0.000 | 0.000 | 18.265 | 1.218 | ||
| Blood | 0 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 6 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.146 | ||
| 8 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.706 | ||
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.108 | NS | ||
| 13 | NS | NS | NS | NS | 0.561 | NS | ||
| 24 | 0.000 | 0.000 | 0.000 | 0.000 | NS | NS | ||
| Brain | Occipital lobe | PM | 0.000 | 0.001 | 0.000 | 0.000 | 0.087 | 1.286 |
| Olfactory lobe | PM | 0.005 | 0.000 | 0.000 | 0.000 | 0.018 | 1.095 | |
| Heart | PM | 0.000 | 0.003 | 0.000 | 0.000 | 0.061 | 0.033 | |
| Kidney | PM | 0.000 | 0.000 | 0.000 | 0.000 | 12.957 | 43.692 | |
| Liver | PM | 0.000 | 0.000 | 0.000 | 0.000 | 41.321 | 2.419 | |
| Lung | Apical lobe | PM | 0.000 | 0.000 | 0.000 | 0.000 | 851.193 | 2,158.535 |
| Diaphragmatic lobe | PM | 0.000 | 0.075 | 0.000 | 0.000 | 868.615 | 1,874.764 | |
| Lung exudate | 13 | NS | NS | NS | NS | 1.465 | NS | |
| Lymph node | Thoracic | PM | 0.000 | 0.000 | 0.000 | 0.000 | 2,030.773 | NS |
| Retropharyngeal | PM | NS | NS | NS | NS | NS | 31.537 | |
| Ovary | PM | 0.000 | 0.000 | 0.000 | 0.000 | 51.300 | NS | |
| Serum | 6 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 8 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.068 | ||
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NS | ||
| 13 | NS | NS | NS | NS | 0.254 | NS | ||
| 24 | 0.000 | 0.000 | 0.000 | 0.000 | NS | NS | ||
| Spleen | PM | 0.000 | 0.000 | 0.000 | 0.000 | 1,630.840 | 890.055 | |
| Swab | 6 | NS | NS | NS | NS | 0.000 | 0.000 | |
| 8 | NS | NS | NS | NS | 0.000 | 0.311 | ||
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | NS | ||
| 13 | NS | NS | NS | NS | 0.055 | NS | ||
| 24 | 0.000 | 0.000 | 0.000 | 0.000 | NS | NS | ||
| Urine | 6 | NS | NS | NS | NS | 0.000 | NS | |
| 8 | NS | NS | NS | NS | 0.000 | 4.708 | ||
| 10 | 0.000 | 0.000 | 0.000 | 0.000 | 2.897 | NS | ||
| 13 | NS | NS | NS | NS | 13.914 | NS | ||
| 24 | 0.000 | 0.000 | 0.000 | 0.000 | NS | NS | ||
| Uterine horn | PM | 0.000 | 0.000 | 0.000 | 0.000 | 33.063 | NS | |
Relative values were determined by linear regression analysis using NiV cDNA standards and normalized using the ratio of NiV:18S rRNA CT values. PM, postmortem tissue; dpi, days postinfection; NS, no sample. Values are expressed as the fold difference from 3.6 pg of NiV cDNA standard and are the average of all replicates. Values in boldface indicate samples from which virus was isolated.
DISCUSSION
All naive cats infected with NiV in the present study developed an acute respiratory syndrome comparable to previous observations in experimental (26) and natural infection of cats (24). NiV disease in cats is an acute febrile reaction accompanied by subtle changes in behavior, followed by a severe respiratory disease. Gross respiratory pathology was comprised of numerous small hemorrhagic nodules in the lungs indicating the early stages of a developing respiratory disease. Although cumulative daily clinical scores provided some measure of disease progression, these changes were very subtle and were often preceded by a rise in body temperature. The implementation of continuous, remote monitoring of body temperature via radiotelemetry for the subsequent vaccine efficacy study enabled earlier intervention prior to respiratory distress and euthanasia based primarily on the observation of a sustained febrile reaction.
The underlying systemic vasculitis observed in the present study was consistent with NiV infection in all susceptible animals, including humans (24, 37, 38). NiV genome was detected in the blood and serum 6 to 9 days postinoculation and in oral swabs and urine immediately prior to euthanasia, but virus was isolated infrequently from oral swabs and not from blood. Previous studies have indicated that considerable quantities of virus may be shed via the nasopharynx and in the urinary tract in cats (26). Here, the low frequency of virus isolation from nontissue samples may reflect the required earlier intervention during the course of disease undertaken in these studies, perhaps indicating that peak virus shedding occurs when clinical disease is well established. In the present study, cat 9, which was euthanized 13 days p.i., did show a large increase in NiV genome in urine prior to euthanasia. However, the early interventions combined with the 2-log decrease in virus inoculum used here may have significantly reduced the incidence and quantity of viral shedding.
The development of a TaqMan assay for the detection and quantitation of NiV genome represents a considerable advance in the assessment of virus dissemination. Given that virus isolation was only successful in about one half of tissue samples, previous studies reliant on virus isolation data as the primary indicator of tissue replication may have underestimated the extent of NiV dissemination. As expected, the lungs appear to be a primary site of viral replication, and this was confirmed by extensive pulmonary pathology. In addition, lymphoid tissues (lymph nodes and spleen) and highly vascularized tissues displayed high relative quantities of NiV genome, confirming the description of NiV pathogenesis as a systemic vasculitis (26). Surprisingly, adrenal tissue revealed very high genome levels but appeared to be refractory to virus isolation attempts. While this observation requires a more detailed investigation, it may indicate a role for hormonal inhibition of NiV infection, both in vitro and in vivo. It was also noteworthy that no distinct differences were observed between the two dosage groups with respect to incubation period, lesion severity, or routes of viral shedding.
Consistent with previous observations of NiV infection in cats and in contrast to NiV infection in pigs and humans was the apparent lack of neurological signs, despite the demonstration of NiV genome in brain tissue. Middleton et al. (26) described a moderately severe, diffuse meningitis in cats infected oronasally with 50,000 TCID50 of NiV, an observation not seen in the present study, perhaps in part due to the earlier interventions used here. There are a number of other possible reasons for this discrepancy, such as differences in virus passage history, route of inoculation, stage of disease, and dose of inoculum. Previous experiments in cats utilized a low-passage human isolate, whereas here the same isolate after two rounds of plaque purification was used. Further, although Weingartl et al. (33) observed NiV infection of olfactory cells and cranial nerves in pigs, the oronasal route of administration and the high titers (50,000 TCID50) used differ considerably from the parenteral, low doses described here.
The present results indicate that cats are highly susceptible to NiV infection with doses as low as 500 TCID50 and that infection consistently results in severe clinical disease that retains a number of parallels with NiV infection in other species. NiV infection in cats reliably manifests as a systemic disease with arterial endothelial tropism and a tropism for respiratory epithelium, showing little individual animal variability. The routes of viral shedding and/or transmission evident in cats are comparable to those postulated for pigs (26), while recent NiV outbreaks in Bangladesh suggest the possibility of human-to-human transmission via exposure to infectious human secretions (5). The ability to compare clinical samples between several species will be important in evaluating the relevance of vaccine and therapy models.
Utilizing this animal model, we evaluated a subunit vaccine candidate based on the G glycoprotein. It is almost without exception that all neutralizing antibodies to enveloped viruses are directed against the virus' envelope glycoproteins. Anti-G MAbs have been shown to be exceptionally potent in neutralizing NiV- and HeV-mediated fusion, as well as live virus infection (40), and a greater potency of NiV anti-G over anti-F MAbs in conferring passive protection of hamsters from NiV disease has recently been demonstrated (23).
Here we have shown complete protection from NiV infection in cats by immunization with sG derived from either NiV (homologous) or HeV (heterologous). The present findings indicate that both NiV and HeV sG are highly immunogenic in cats, similar to findings obtained in mice and rabbits, yielding homologous serum neutralizing titers of greater than 1:20,000 in cats. These levels of anti-G antibody titer were generated by a moderate immunization protocol of only three 100-μg doses of protein in adjuvant. Despite the somewhat lower heterologous neutralizing titers observed here, in particular for animals immunized with HeV sG, both immunogens completely protected against subsequent NiV challenge, suggesting that a single vaccine may be effective against both viruses. It will be of interest to confirm this notion in an immunization and challenge study with HeV. The NiV genome was demonstrated at very low levels in one cat immunized with HeV G. While this animal had considerably lower neutralizing titers than its homologous counterparts, potentially explaining these observations, caution must be exercised in interpretation of such high CT values (>40). Certainly, the absence of detectable viral genome in blood samples obtained during the course of the experiment and the absence of viral antigen immunohistologically indicates that negligible infection had occurred in vaccinated animals. While the optimum vaccine preparation and immunization regime remains to be elucidated, the results shown here clearly demonstrate that a subunit vaccination strategy appears to be possible for the prevention of infection by these highly lethal zoonotic agents. At present, in the absence of any available antiviral therapeutic modalities, vaccination of high-risk populations against NiV may prevent associated epidemic disease, whether of natural or malicious origin.
Acknowledgments
This study was supported by NIH grant AI056423 and the Middle Atlantic Regional Center of Excellence (MARCE) for Biodefense and Emerging Infectious Disease Research (NIH AI057168) to C.C.B.
We thank S. Crameri, R. Van Driel, and D. Magoffin for help caring for the cats during the 8-week vaccination protocol; Mark Rechenberg for writing the Visual Basic code for radio telemetry implementation; and Dayna Johnson for implanting the temperature transmitters in cats for the vaccine efficacy study.
The views expressed here are solely those of the authors and do not represent official views or opinions of the U.S. Department of Defense or The Uniformed Services University of the Health Sciences.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Reference deleted.
- 2.Anonymous. 2004. Nipah encephalitis outbreak over wide area of western Bangladesh, 2004. Health Sci. Bull. 2:7-11. [Google Scholar]
- 3.Anonymous. 2005. Nipah virus outbreak from date palm juice. Health Sci. Bull. 3:1-5. [Google Scholar]
- 4.Anonymous. 2003. Outbreaks of viral encephalitis due to Nipah/Hendra-like viruses, Western Bangladesh. Health Sci. Bull. 1:1-6. [Google Scholar]
- 5.Anonymous. 2004. Person-to-person transmission of Nipah virus during outbreak in Faridpur District, 2004. Health Sci. Bull. 2:5-9. [Google Scholar]
- 6.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 102:10652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossart, K. N., and C. C. Broder. 2006. Developments toward effective treatments for Nipah and Hendra virus infection. Expert. Rev. Anti. Infect. Ther. 4:43-55. [DOI] [PubMed] [Google Scholar]
- 8.Bossart, K. N., G. Crameri, A. S. Dimitrov, B. A. Mungall, Y. R. Feng, J. R. Patch, A. Choudhary, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble g glycoprotein of Hendra virus. J. Virol. 79:6690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossart, K. N., B. A. Mungall, G. Crameri, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol. J. 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong, H. T., A. Kamarulzaman, C. T. Tan, K. J. Goh, T. Thayaparan, S. R. Kunjapan, N. K. Chew, K. B. Chua, and S. K. Lam. 2001. Treatment of acute Nipah encephalitis with ribavirin. Ann. Neurol. 49:810-813. [DOI] [PubMed] [Google Scholar]
- 11.Chua, K. B. 2003. Nipah virus outbreak in Malaysia. J. Clin. Virol. 26:265-275. [DOI] [PubMed] [Google Scholar]
- 12.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 13.Chua, K. B., K. J. Goh, K. T. Wong, A. Kamarulzaman, P. S. Tan, T. G. Ksiazek, S. R. Zaki, G. Paul, S. K. Lam, and C. T. Tan. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257-1259. [DOI] [PubMed] [Google Scholar]
- 14.Chua, K. B., C. Lek Koh, P. S. Hooi, K. F. Wee, J. H. Khong, B. H. Chua, Y. P. Chan, M. E. Lim, and S. K. Lam. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 15.Crameri, G., L. F. Wang, C. Morrissy, J. White, and B. T. Eaton. 2002. A rapid immune plaque assay for the detection of Hendra and Nipah viruses and anti-virus antibodies. J. Virol. Methods 99:41-51. [DOI] [PubMed] [Google Scholar]
- 16.De Franceschi, L., G. Fattovich, F. Turrini, K. Ayi, C. Brugnara, F. Manzato, F. Noventa, A. M. Stanzial, P. Solero, and R. Corrocher. 2000. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology 31:997-1004. [DOI] [PubMed] [Google Scholar]
- 17.Eaton, B. T., C. C. Broder, D. Middleton, and L. F. Wang. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 4:23-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Federal Register. 2002. New drug and biological drug products: evidence needed to demonstrate effectiveness of new drugs when human efficacy studies are not ethical or feasible. Fed. Regist. 67:37988-37998. [Online.] www.fda.gov/cber/rules/humeffic.htm. [PubMed] [Google Scholar]
- 18.Field, H., P. Young, J. M. Yob, J. Mills, L. Hall, and J. Mackenzie. 2001. The natural history of Hendra and Nipah viruses. Microbes Infect. 3:307-314. [DOI] [PubMed] [Google Scholar]
- 19.Georges-Courbot, M. C., H. Contamin, C. Faure, P. Loth, S. Baize, P. Leyssen, J. Neyts, and V. Deubel. 2006. Poly(I)-poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob. Agents Chemother. 50:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh, K. J., C. T. Tan, N. K. Chew, P. S. Tan, A. Kamarulzaman, S. A. Sarji, K. T. Wong, B. J. Abdullah, K. B. Chua, and S. K. Lam. 2000. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N. Engl. J. Med. 342:1229-1235. [DOI] [PubMed] [Google Scholar]
- 21.Griffin, D. E. 1995. Immune responses during measles virus infection. Curr. Top. Microbiol. Immunol. 191:117-134. [DOI] [PubMed] [Google Scholar]
- 22.Guillaume, V., H. Contamin, P. Loth, M. C. Georges-Courbot, A. Lefeuvre, P. Marianneau, K. B. Chua, S. K. Lam, R. Buckland, V. Deubel, and T. F. Wild. 2004. Nipah virus: vaccination and passive protection studies in a hamster model. J. Virol. 78:834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillaume, V., H. Contamin, P. Loth, I. Grosjean, M. C. Courbot, V. Deubel, R. Buckland, and T. F. Wild. 2006. Antibody prophylaxis and therapy against Nipah virus infection in hamsters. J. Virol. 80:1972-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper, P., S. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Middleton, D. J., H. A. Westbury, C. J. Morrissy, B. M. van der Heide, G. M. Russell, M. A. Braun, and A. D. Hyatt. 2002. Experimental Nipah virus infection in pigs and cats. J. Comp. Pathol. 126:124-136. [DOI] [PubMed] [Google Scholar]
- 27.Murray, K., B. Eaton, P. Hooper, L. Wang, M. Williamson, and P. Young. 1998. Flying foxes, horses, and humans: a zoonosis caused be a new member of the Paramyxoviridae, p. 43-58. In W. M. Scheld, D. Armstrong, and J. M. Hughes (ed.), Emerging infections. ASM Press, Washington, D.C.
- 28.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10:806-810. [DOI] [PubMed] [Google Scholar]
- 29.Reynes, J. M., D. Counor, S. Ong, C. Faure, V. Seng, S. Molia, J. Walston, M. C. Georges-Courbot, V. Deubel, and J. L. Sarthou. 2005. Nipah virus in Lyle's flying foxes, Cambodia. Emerg. Infect. Dis. 11:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers, R. J., I. C. Douglas, F. C. Baldock, R. J. Glanville, K. T. Seppanen, L. J. Gleeson, P. N. Selleck, and K. J. Dunn. 1996. Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Aust. Vet. J. 74:243-244. [DOI] [PubMed] [Google Scholar]
- 31.Tamin, A., B. H. Harcourt, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota. 2002. Functional properties of the fusion and attachment glycoproteins of Nipah virus. Virology 296:190-200. [DOI] [PubMed] [Google Scholar]
- 32.Wacharapluesadee, S., B. Lumlertdacha, K. Boongird, S. Wanghongsa, L. Chanhome, P. Rollin, P. Stockton, C. E. Rupprecht, T. G. Ksiazek, and T. Hemachudha. 2005. Bat Nipah virus, Thailand. Emerg. Infect. Dis. 11:1949-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weingartl, H., S. Czub, J. Copps, Y. Berhane, D. Middleton, P. Marszal, J. Gren, G. Smith, S. Ganske, L. Manning, and M. Czub. 2005. Invasion of the central nervous system in a porcine host by Nipah virus. J. Virol. 79:7528-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weingartl, H. M., Y. Berhane, J. L. Caswell, S. Loosmore, J.-C. Audonnet, J. A. Roth, and M. Czub. 2006. Recombinant Nipah virus vaccines protect pigs against challenge. J. Virol. 80:7929-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westbury, H. A., P. T. Hooper, S. L. Brouwer, and P. W. Selleck. 1996. Susceptibility of cats to equine morbillivirus. Aust. Vet. J. 74:132-134. [DOI] [PubMed] [Google Scholar]
- 36.Westbury, H. A., P. T. Hooper, P. W. Selleck, and P. K. Murray. 1995. Equine morbillivirus pneumonia: susceptibility of laboratory animals to the virus. Aust. Vet. J. 72:278-279. [DOI] [PubMed] [Google Scholar]
- 37.Wong, K. T., I. Grosjean, C. Brisson, B. Blanquier, M. Fevre-Montange, A. Bernard, P. Loth, M. C. Georges-Courbot, M. Chevallier, H. Akaoka, P. Marianneau, S. K. Lam, T. F. Wild, and V. Deubel. 2003. A golden hamster model for human acute Nipah virus infection. Am. J. Pathol. 163:2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong, K. T., W. J. Shieh, S. Kumar, K. Norain, W. Abdullah, J. Guarner, C. S. Goldsmith, K. B. Chua, S. K. Lam, C. T. Tan, K. J. Goh, H. T. Chong, R. Jusoh, P. E. Rollin, T. G. Ksiazek, and S. R. Zaki. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 161:2153-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yob, J. M., H. Field, A. M. Rashdi, C. Morrissy, B. van der Heide, P. Rota, A. bin Adzhar, J. White, P. Daniels, A. Jamaluddin, and T. Ksiazek. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Z., A. S. Dimitrov, K. N. Bossart, G. Crameri, K. A. Bishop, V. Choudhry, B. A. Mungall, Y. R. Feng, A. Choudhary, M. Y. Zhang, Y. Feng, L. F. Wang, X. Xiao, B. T. Eaton, C. C. Broder, and D. S. Dimitrov. 2006. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 80:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]