Abstract
Chronic wasting disease (CWD), a prion disease of deer and elk, is highly prevalent in some regions of North America. The establishment of mouse-adapted CWD prions has proven difficult due to the strong species barrier between mice and deer. Here we report the efficient transmission of CWD to transgenic mice overexpressing murine PrP. All mice developed disease 500 ± 62 days after intracerebral CWD challenge. The incubation period decreased to 228 ± 103 days on secondary passage and to 162 ± 6 days on tertiary passage. Mice developed very large, radially structured cerebral amyloid plaques similar to those of CWD-infected deer and elk. PrPSc was detected in spleen, indicating that murine CWD was lymphotropic. PrPSc glycoform profiles maintained a predominantly diglycosylated PrP pattern, as seen with CWD in deer and elk, across all passages. Therefore, all pathological, biochemical, and histological strain characteristics of CWD appear to persist upon repetitive serial passage through mice. These findings indicate that the salient strain-specific properties of CWD are encoded by agent-intrinsic components rather than by host factors.
Mammalian prion diseases are believed to be caused by the misfolding of a host-encoded cellular protein, PrPC, into an aggregated, beta-sheet-rich, insoluble isoform, PrPSc, which self-propagates and leads to fatal neurodegeneration (25, 38). Insoluble PrPSc aggregates are detectable in the central nervous system and skeletal muscle and also throughout the lymphoid system in a subset of diseases, including variant (22, 47) and sporadic (19) Creutzfeldt-Jakob disease in humans, scrapie in sheep and goats (21), and chronic wasting disease (CWD) in cervids (34, 43).
CWD is a geographically widespread and locally prevalent prion disease of North American cervids. CWD occurs naturally in wild and captive mule deer, white-tailed deer, Rocky Mountain elk (51), and wild moose (M. W. Miller, unpublished data). The origins of CWD and its relationship to other prion diseases remain uncertain (33). Mouse-adapted prion strains of sheep scrapie and bovine spongiform encephalopathy (BSE) have proven very useful for studying and comparing prion strain properties (7, 28) and for understanding the peripheral pathogenesis of prion disease (11, 23, 29, 30, 37). However, the transmission of CWD to wild-type mice is inefficient (12) in contrast to transgenic mice expressing the deer or elk PrP sequences (4, 12, 24). This suggests that a species barrier exists between cervids and mice. Here we used a transgenic mouse model that overexpresses murine PrP to develop a murine-adapted CWD strain of prion. We report that this strain induces a unique histopathological phenotype and displays biochemical and biophysical properties similar to those of deer CWD but distinct from those of a Rocky Mountain Laboratory (RML) strain of mouse-adapted scrapie. These strain-specific properties were stable over three generations of serial transmission in mice. These newly generated murine CWD prions provide a tool for further studying the biophysical nature of prion strains, e.g., by identifying differential PrPSc-interacting proteins.
MATERIALS AND METHODS
Tga20 mice and prion infection.
Transgenic mice overexpressing murine PrP (17) were intracerebrally inoculated in the left parietal cortex with 30 μl of a 10% brain homogenate (5% for subsequent passages) from a terminal, CWD-infected mule deer or an uninfected control deer (mock). Mice were monitored every second day, and scrapie was diagnosed according to clinical criteria, including ataxia, kyphosis, and hind leg paresis. Mice were sacrificed at the onset of terminal disease. Mice were maintained under specific pathogen-free conditions, and all experiments were performed in accordance with the animal welfare guidelines of the Kanton of Zürich.
Histochemical and immunohistochemical stains.
Tissues were fixed in 4% buffered formalin, treated with 98% formic acid for 1 h, and postfixed for ≥24 h prior to paraffin embedding. Two-micrometer-thick sections were cut on positively charged glass slides and stained with hematoxylin and eosin or immunostained using antibodies for PrP (SAF84), astrocytes (glial fibrillary acidic protein [GFAP]), or microglia (Iba1). For PrP staining, sections were deparaffinized and incubated for 6 min in 98% formic acid and then washed in distilled water for 5 min. Sections were heated to 100°C in citrate buffer (pH 6.0), cooled for 3 min, and then washed in distilled water for 5 min. Immunohistochemical stains were performed on an automated Nexus staining apparatus (Ventana Medical Systems) using an IVIEW DAB detection kit (Ventana). After incubation with protease 1 (Ventana) for 16 min, sections were incubated with anti-PrP SAF-84 (SPI bio; 1:200) for 32 min. Sections were counterstained with hematoxylin. GFAP immunohistochemistry for astrocytes (1:1,000 for 24 min; DAKO) and Iba1 (1:2,500 for 32 min; Wako Chemicals) for microglia were similarly performed, however, with antigen retrieval by heating to 100°C in EDTA buffer (pH 8.0). Histoblot analyses were performed as previously described (45).
For thioflavin T and Congo red staining, frozen brain sections were fixed in 70% ethanol for 10 min. For thioflavin staining, sections were immersed in 1% thioflavin T for 3 min, followed by 1% acetic acid for 10 min and then distilled water for 5 min. For Congo red staining, slides were immersed in an alkaline solution and then stained with Congo red solution for 20 min.
Lesion profile.
Twelve anatomic brain regions were selected in accordance with previous strain typing analysis protocols (13, 18) by using three to five mice per passage. Spongiosis was evaluated on a scale of 0 to 5 (not detectable, mild, moderate, severe, and status spongiosis). Gliosis and PrPSc content were scored on a scale of 0 to 3 (not detectable, mild, moderate, and severe). A sum of the three scores resulted in the value that was employed in order to obtain the lesion profile for the individual animal. Histological analysis was performed by independent investigators blinded to animal identification.
Ultrastructural characterization.
Small samples (1 mm3) were retrieved from formalin-fixed, paraffin-embedded blocks and reversed to electron microscopy as previously described (3). Grids stained with lead citrate and uranyl acetate were observed and photographed with a JEM 100C transmission electron microscope at 80 kV.
Western blots.
Ten-percent brain or spleen homogenates were prepared in phosphate-buffered saline using a ribolyzer, and 80 to 90 μg protein was diluted with Tris lysis buffer (10 mM Tris, 10 mM EDTA, 100 mM NaCl, 0.5% NP-40, and 0.5% deoxycholate) and digested with 100 μg/ml proteinase K for 30 min at 37°C. Sodium dodecyl sulfate-based loading buffer was then added, and samples were heated at 95°C for 5 min prior to electrophoresis through a 12% Bis-Tris precast gel (Invitrogen), followed by transfer to a nitrocellulose membrane by wet blotting. Proteins were detected with the following panel of “POM” anti-PrP antibodies (36): POM1 (epitope amino acids [aa] 121 to 230), POM2 (epitope aa 58 to 64, 66 to 72, 74 to 80, and 82 to 88), POM3 (epitope aa 95 to 100), and POM11 (epitope aa 64 to 72 and 72 to 80). Signals were visualized with an ECL detection kit (Pierce).
RESULTS
Incubation period and clinical disease.
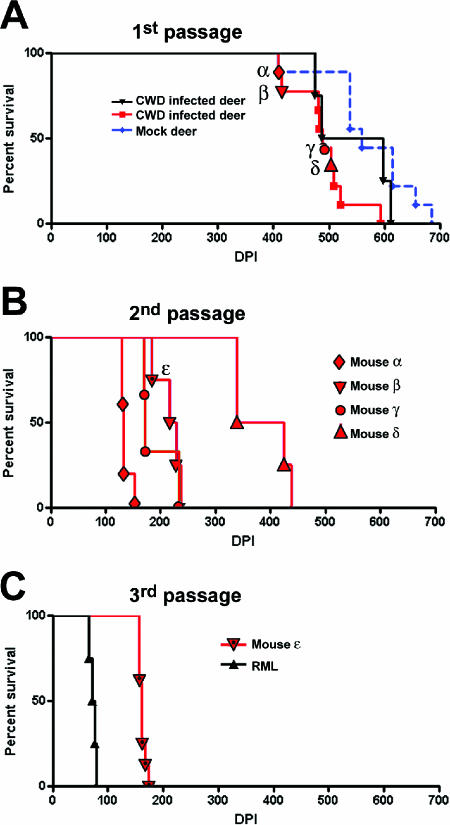
tga20 transgenic mice overexpress mouse PrP owing to a Prnp-encoding vector known as the “half-genomic construct,” which is based on a murine PrP minigene lacking intron 2 (17). tga20 mice were inoculated intracerebrally with CWD from a terminally infected mule deer. All inoculated mice developed neurologic disease with an incubation period (IP) of 500 ± 62 (range, 408 to 521) days postinoculation (Fig. 1A). Remarkably, the clinical attack rate was 100% (10/10). This contrasts with the poor infectibility of wild-type mice with CWD and may be attributed to the higher PrPC expression levels in tga20 mice.
FIG. 1.
Survival of tga20 PrP-overexpressing mice challenged with CWD prions. (A) First passage of mule deer CWD performed on two separate occasions shows similar survival curves. Four mice were used for subpassage into additional tga20 mice (indicated by α, β, γ, and δ). Mock-inoculated mice were periodically sacrificed at the time points indicated. (B) Second passage of CWD-infected brains from four mice. Mouse ɛ was used for the third subpassage. (C) Third passage of CWD. An RML scrapie survival curve is shown for comparison.
To determine whether the new murine CWD strain would adapt further in the mouse, we performed a second passage of CWD. This was accomplished by injecting brain homogenate from CWD-infected, terminally sick mice into another group of tga20 mice. The second passage led to an abbreviated IP: mice developed terminal disease with an IP of 228 ± 103 dpi (range, 139 to 438) (Fig. 1B). By the third passage, the incubation period dropped to 162 days, with little variability among the mice (±6 days) (Fig. 1C). In contrast, tga20 mice inoculated with 3 × 103 50% lethal dose units of RML scrapie developed terminal neurologic disease after 74 days (±5 days) (Fig. 1C).
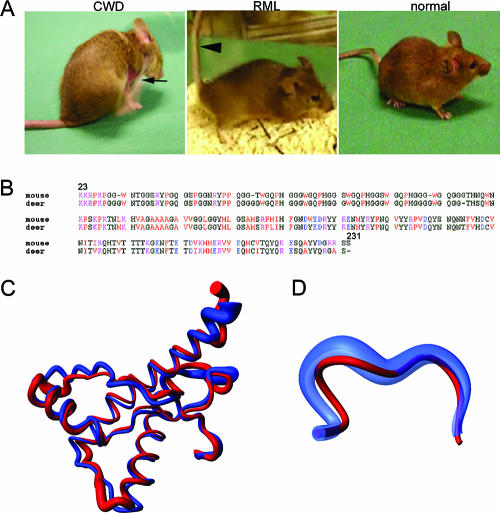
Clinical signs were similar among mice receiving primary deer CWD and murine-adapted CWD and included kyphosis, ataxia, tremors, and intense pruritus (Fig. 2 and see the supplemental material). CWD-infected mice often showed a clinical phenotype of obsessive pruritus. Mice continued scratching, even with palliative care of open abrasions, making euthanasia unavoidable. Exogenous causes of ulceration were not evident in the skin; no ectoparasites or other infectious agents were observed.
FIG. 2.
Clinical signs, genetics, and structural biology of murine CWD. (A) Mice with clinical signs of CWD are characterized by kyphosis, pruritus, ataxia, and tremors in contrast to mice with RML scrapie which are primarily ataxic with stiff tails (arrowhead). The CWD-infected mouse is scratching (arrow indicates skin abrasion). (B) A PrP gene sequence alignment for the mouse and mule deer shows 90.2% homology. Amino acids are color coded as follows: red, small and hydrophobic; blue, acidic; magenta, basic; green, hydroxyl and amine and basic. (C and D) Superposition of the mean nuclear magnetic resonance (NMR) structures of the polypeptide segment of aa 125 to 229 (C) and 165 to 172 (D) in ePrP(121-230) (red) and mPrP(121-230) (blue). A spline function was drawn through the Cα positions. The radius of the cylindrical rods representing the polypeptide chains is proportional to the mean backbone displacement per residue (XX), as evaluated after superposition for best fit of the atoms N, Cα, and C′ in the 20 energy-minimized conformers used to represent the NMR structures (9). The PrP structure from the CWD source deer used herein may vary slightly from the elk.
Genetic and structural considerations.
The prion sequence of the mule deer was compared to that from mouse. When comparing the sequences from residues 23 to 231, there was a 90.2% sequence homology between the deer and the mouse (Fig. 2B). A comparison of the three-dimensional structures of the elk and mouse prion proteins shows that both proteins have the same globular fold, consisting of a flexibly disordered domain of ∼100 residues and a C-terminal domain of similar size, with three α-helices and a short antiparallel β-sheet (20). However, the loop region that connects the second β strand with the second α-helix is very precisely defined in the elk prion protein but disordered in mouse PrPC (Fig. 2C and D). Elk and mule deer amino acid sequences are identical at this loop region.
Disease phenotype and lesion profiles.
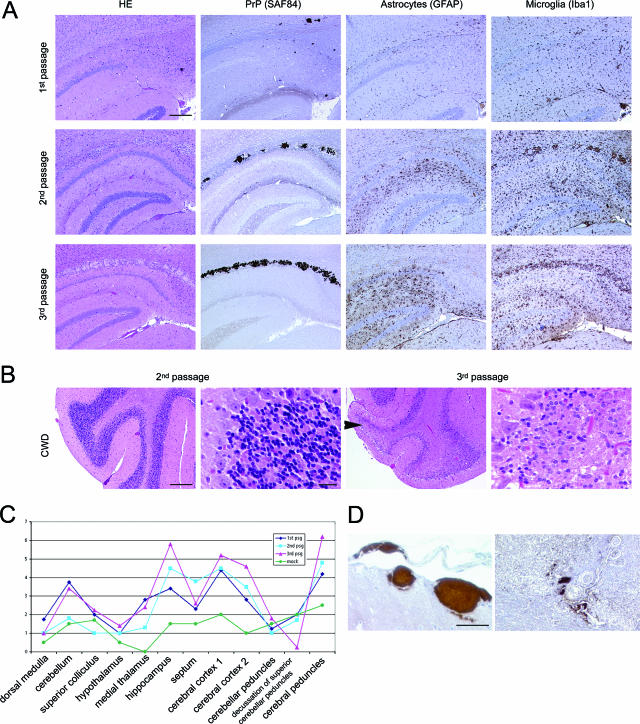
We performed a detailed investigation of the histopathologic lesions in each of the serial passages of CWD (Fig. 3A). Spongiform lesions were most prominent in the neuopil of thalamus, hippocampus, and basal ganglia. Some cases also displayed extensive damage to the cerebral cortex and the white matter tracts. Lesions appeared to acquire greater severity with each serial passage (Fig. 3A). By the third passage, mice developed a subtotal elective loss of cerebellar granular cells, a finding absent in the first and second passages (Fig. 3B). Astrogliosis and microglial activation were often severe and regionally extensive in CWD-infected mice (Fig. 3A). The findings described above were quantified using a standardized lesion-profiling methodology (35) and indicated that the severity of spongiform degeneration, astrogliosis, and PrPSc deposition increased with each serial passage (Fig. 3C). Some mock-inoculated animals had mild-to-moderate vacuolization of white matter tracts, but no neuronal loss or extensive astrogliosis, microgliosis, or plaques.
FIG. 3.
Histopathology and PrPSc plaques in CWD-infected mice. (A) Brain at hippocampus. PrP plaques are visible in hematoxylin and eosin (HE) stains and by immunohistochemical labeling for PrP, particularly in the corpus callosum. Extensive astrocytosis and microglial activation is evident. Note the increased severity of gliosis, microglial activation, and plaque deposition with each passage. Bar = 200 μm. (B) Severe degeneration with massive loss of the cerebellar granular cells occurred by the third passage. Bar = 200 μm or 20 μm. (C) Lesion profile of CWD infected mice, comparing first through third passage. Profile scores are determined by spongiform change, gliosis, and PrPSc deposits in the specific brain regions indicated. (D) Vascular plaques in meninges and brain parenchyma of CWD-infected mice. Bar = 50 μm.
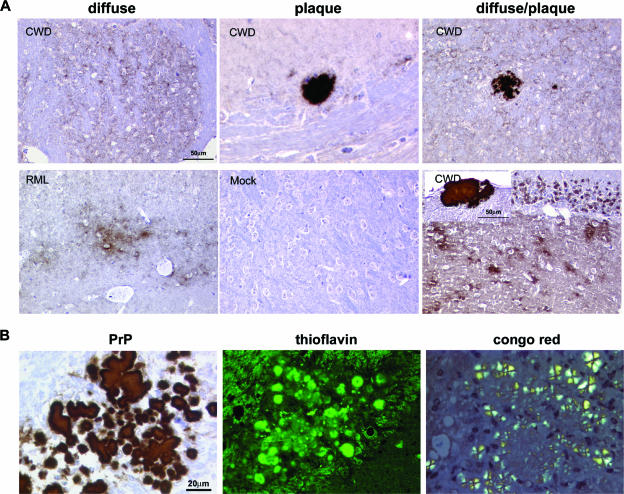
Four of nine (44%) first-passage, CWD-inoculated mice had multiple large (50- to 300-μm) plaques visible in the hematoxylin-and-eosin-stained sections, which immunolabeled for PrP. Plaques appeared in the basal ganglia, thalamus, and hippocampus and separated white matter fiber tracts in the corpus callosum and cerebellum. Extensive plaques are not a feature of RML mouse scrapie (A. Aguzzi and C. J. Sigurdson, unpublished data) but are common with cervid CWD (5, 44). Remarkably, PrP plaques and deposits were often perivascular (Fig. 3D) as in CWD in deer and elk but were also within vessel walls. However, a subset of mice developed only small, diffuse PrP deposits typical of RML mouse scrapie or developed both plaques and diffuse deposits (Fig. 4A). Third passage of the “plaque” strain led to plaque formation in all infected mice. Plaques were composed of dense deposits similar to those seen with the first passage.
FIG. 4.
Plaque characterization in CWD-infected mice. (A) PrPSc deposits vary from small, diffuse aggregates to dense plaques, often occurring as a mixture of both deposit types. Bar = 50 μm. (B) Plaques were thioflavin positive and showed birefringence after Congo red staining indicative of amyloid formation. Bar = 20 μm.
Plaque structure.
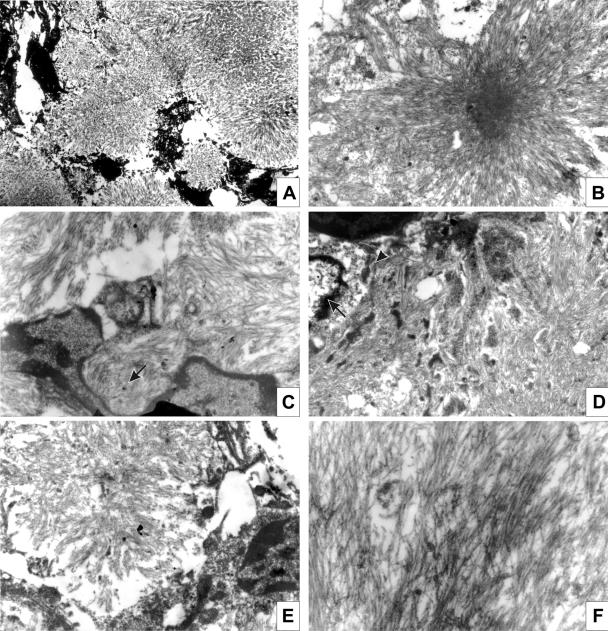
By light microscopy, murine CWD plaques were composed of fiber bundles emanating from a dense core in stellate arrangement and were unicentric or multicentric. Thioflavin T positivity and Congo red birefringence indicated that the plaques had a highly organized amyloid fibril cross-β fold structure (Fig. 4B). By electron microscopy, plaques were composed of compact amyloid bundles and appeared as subpial (Fig. 5A), unicentric (Fig. 5B and E), or perivascular deposits (Fig. 5D). Overall, the morphologies of plaques recapitulated that observed in mule deer with CWD (27) and were similar to the human prion disease Gerstmann-Straussler-Scheinker disease (GSS). As with GSS, microglial cells were in close contact with the amyloid bundles (Fig. 5C) (6, 26).
FIG. 5.
Ultrastructural analysis of murine CWD plaques. (A) Subpial deposits were composed of numerous unicentric plaques. Magnification, ×2,000. (B) Unicentric kuru-like plaque. Magnification, ×6,000. (C) A plaque margin showing microglial cells. Amyloid bundles are in close contact with the cell (arrow). Magnification, ×13,000. (D) Perivascular plaque. The nucleus (arrow) and cytoplasm (arrowhead) of the pericyte is visible. Magnification, ×8,300. (E) Unicentric plaque. Magnification, ×8,300. (F) High-power view of separate amyloid bundles. Magnification, ×20,000.
Histoblot analysis.
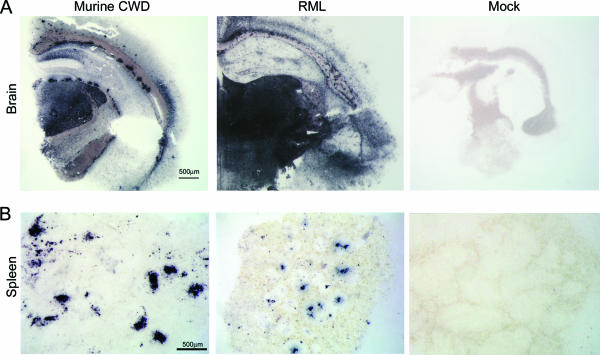
To visualize proteinase K (PK)-resistant PrPSc in brain and spleen, we performed histoblot analyses of frozen tissue sections pressed onto nitrocellulose, proteinase K treated, and labeled with anti-PrP antibodies. Here again we found abundant PrPSc plaques and diffuse deposits in the brain in a pattern that varied from those of RML mice (Fig. 6A). Mice inoculated with second and third passages of the “plaque” strain developed PrPSc deposits in the spleen in follicular patterns similar to those of RML-infected mice (Fig. 6B).
FIG. 6.
Histoblots of brain and spleen reveal differences in neural PrPSc distribution in murine CWD (overall less PrPSc) compared to RML mouse scrapie. Abundant PrPSc deposits were present in spleen of some CWD-infected mice. No PrPSc stain was seen in the mock-inoculated mouse. Bar = 500 μm.
Biochemical profile of murine CWD.
We analyzed proteinase K-resistant PrPSc in brain infected with murine CWD by Western blotting and compared it with that of a standard murine RML scrapie isolate and with that of CWD in deer. We found that CWD in deer was characterized by a predominant diglycosylated isoform of PrPSc as described previously (39). In contrast, RML murine scrapie presents with a dominant monoglycosylated PrPSc isoform. In most CWD-infected mice with the “plaque” strain, the diglycosylate-rich glycoform pattern was faithfully propagated. However, two of seven mice developed nearly equal levels of di- and monoglycosylated isoforms on the third passage (Fig. 7). Murine first-passage strains of sheep scrapie and BSE (also in tga20) showed glycoform ratios similar to that found with the third passage of murine CWD (Fig. 7). The PK cutting site in murine CWD was then defined by differential antibody immunoreactivity of PrP27−30 to be between amino acids 73 to 82, as POM11 (epitope aa 64 to 72 and 72 to 80) did not recognize PrP27−30 (data not shown).
FIG. 7.
Biochemical characterization of PrPSc in murine CWD (A and B). Western blots and triplot of murine CWD-infected brain showing a predominant diglycosylated band in most mice on first passage. By the third passage, the glycoform pattern shifted slightly with an increase in the monoglycosylated PrP and was similar to deer CWD, murine BSE, and murine sheep scrapie, but unlike the mouse-adapted RML. −, absence of; +, presence of.
DISCUSSION
Prion transmission between species is often inefficient and goes along with prolonged incubation periods. However, serial passages within the new species often lead to a shortening of the incubation period (48), a phenomenon that has been termed adaptation. Neither the molecular underpinnings of the species barrier nor those of prion adaptation are fully understood. The primary amino acid sequence of host PrPC clearly contributes to the species barrier effect (40, 50), yet additional factors intrinsic to the infectious agent, be they PrPSc conformation, PrPSc aggregational state, and/or ancillary molecules (49), also affect susceptibility and disease phenotype (15, 16, 46). Because of such agent-autonomous factors, species barriers are specified by the pathogenic prions in addition to the genotype of the hosts: bovine spongiform encephalopathy is, e.g., readily transmissible to species that are hardly susceptible to sheep scrapie (32).
Although Prnp is very well conserved among most species, the mouse and deer PrP amino acid sequences are relatively divergent. Amino acid identity is only 90.2% between residues 23 to 231, and the respective PrPC structures strikingly diverge between amino acids 165 to 172 (20). This finding may underlie the inefficient CWD prion conversion in wild-type mice. However, even a high degree of homology between PrP molecules is not predictive for efficient conversion, as single-amino-acid substitutions can create a strong species barrier (31).
Despite the structural constraints listed above, we found that overexpression of murine PrP of four- to sixfold sufficed to enable the efficient transmission of deer CWD to mice. This was unexpected since the expression levels of Prnp were not found to be rate limiting for ecotropic prion replication in other systems (14). In contrast, these findings suggest that host PrPC expression levels may significantly affect the robustness of species barriers.
CWD manifested itself in tga20 mice with features that are remarkably similar to those of CWD in deer and strikingly different from those of the commonly used RML strain of mouse-adapted sheep prions. Some CWD-infected mice developed very large multicentric extracellular amyloid plaques that were intensely stained by thioflavin and Congo red, often positioned within white matter tracts or around vessels, similar to those described for deer with CWD (44) and for humans with GSS (26). In contrast, plaques were never seen in RML-infected tga20 mice or RML-infected wild-type C57BL/6 inbred, 129Sv inbred, or C57BL/6 × 129Sv crossbred mice (A. Aguzzi, unpublished data).
The plaques were reminiscent of those shown by Scott et al. in mice expressing bovine PrP and challenged with BSE or sheep scrapie prions (41). In mice with plaques, the plaque phenotype was stable and persisted even after the third serial passage of CWD in mice.
The second passage of CWD in the tga20 mice revealed broadly divergent disease phenotypes. First, the plaque phenotype was not present in all second-passage mice, as a subset of four mice (inoculated with isolates from a mouse that did not have plaques) developed only diffuse granular PrP stain deposits as seen by immunohistochemistry, and PrPSc signals in the brain by Western blotting was low. Second, one group of second-passage mice developed disease after a remarkably long incubation period. Third, there was also an inconsistent detection of splenic PrPSc in the second-passage mice. These findings suggest that there may be multiple CWD strains present in the deer, with each maintaining a presence in the tga20 mice. Alternatively, it is also possible that the variability is intrinsically linked to the mechanism of strain adaptation, with the strain phenotype diversifying as a process of the adaptation.
We considered the possibility that the above variability may have resulted from the contamination of the inoculum with our RML laboratory strain. This, however, is extremely unlikely, as the incubation periods for RML are much shorter than those reported here. When exposed to limiting infectious doses of RML, tga20 mice consistently develop disease in <180 days (10, 42) or resist infection completely. In addition, the clinical signs for RML differ strikingly from those detected with murine CWD. Finally, the time lag between the first clinical signs and terminal disease is only a few days in RML-infected tga20 mice, whereas it extended over weeks in CWD-infected mice.
Clues to prion strain differences lie in the conformational state of the misfolded prion protein and in the ratio of the PrP glycoforms (7, 8). Several techniques were employed to explore these features, including the quantitation of the relative proportion of the three glycoforms by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and determination of the proteinase K cleavage site (8) with a panel of anti-PrP antibodies recognizing various epitopes amino- and carboxy-proximal to the putative cleavage site (36, 53). We found that the murine CWD glycoform profile shifted slightly with serial passage from a highly abundant diglycosylated PrPSc on first passage to an almost equal ratio of di- and monoglycosylated PrPSc by the third passage, a ratio which was close to that of CWD in deer, murine BSE, and murine sheep scrapie and varied from that of RML. The PK cleavage site was assessed in the third passage of murine CWD and localized to amino acids 73 to 82. This is consistent with the PK-cutting sites for elk CWD at Gly78 and Gly82 (52) and suggests that features of the CWD PrPSc structure are maintained in the mouse.
CWD prions were serially passaged three times. With each passage, the incubation period decreased and the brain lesions and PrP plaques developed to a greater severity. This is consistent with other instances of strain adaptation, in which prion virulence tended to increase with time. However, the biochemical, the histopathological, and even the clinical characteristics of the disease remained remarkably constant, suggesting that the phenotype-specifying information is enciphered more within the agent rather than in the host. The stability of the phenotypic traits over three passages suggests that the phenotype-specifying information undergoes replication along with the infectious principle.
Within the framework of the protein-only hypothesis, the above phenomenon might be explained by postulating that CWD prions possess a particular steric conformation that is unique, distinct from that of RML scrapie prions, and capable of imparting the same conformation onto mouse PrPC, which will then propagate this unique conformation indefinitely upon serial passage (1).
Alternatively, PrPSc aggregates derived from CWD-infected deer may be packed in a specific quaternary assembly state that grows appositionally in a growth pattern similar to that of a crystal. Such quasicrystals might be shaped differently from RML aggregates and might incorporate murine PrPSc upon passaging to mice (2). Eventually, repeated serial passages would lead to the propagation of an aggregated moiety that consists exclusively of murine PrPSc but retains the unique quaternary structure of CWD prions.
A further model predicates that conventional replicative molecules, such as DNA, RNA, or micro-RNA, might be incorporated into PrPSc. While prion infectivity is solely specified by PrPSc (the “apoprion”), the strain-specific characteristics would be propagated by such “coprions” (49).
Finally, CWD prions may incorporate specific subsets of host molecules, be they proteins, lipids, nucleic acids, or other constituents, according to a unique stoichiometry. Prion replication upon cross-species passage might reproduce this stoichiometry, thereby preserving the strain-specific traits. The propagation of a well-characterized strain of CWD prions within genetically homogeneous hosts yields a powerful tool for testing each of the above hypotheses. A particularly promising avenue of future work consists of utilizing contemporary proteomic tools, such as differential mass spectrometry with isotope-coded affinity tags, to enumerate non-PrPSc components that are associated with mouse-passaged CWD prions and may contribute to their unique phenotypic traits.
Supplementary Material
Acknowledgments
We thank M. Heikenwälder and P. Nilsson for critical reviews of the manuscript and F. Heppner for optimization of the microglial immunohistochemical stain. We gratefully acknowledge the animal care staff and the histopathology technical support at the University of Zurich.
This study was supported by grants from the European Union (Apopis and TSEUR), the Swiss National Foundation, the National Competence Center for Research on Neural Plasticity and Repair (to A.A.), the National Institutes of Health (K08-AI01802 to C.J.S.), the Foundation for Research at the Medical Faculty, University of Zurich (to C.J.S.), and the United States National Prion Research Program (to C.J.S. and A.A.).
Footnotes
Published ahead of print on 4 October 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aguzzi, A. 1998. Protein conformation dictates prion strain. Nat. Med. 4:1125-1126. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi, A. 2004. Understanding the diversity of prions. Nat. Cell Biol. 6:290-292. [DOI] [PubMed] [Google Scholar]
- 3.Alwasiak, J., B. Mirecka, L. Wozniak, and P. P. Liberski. 1991. Neuroblastic differentiation of metastases of medulloblastoma to extracranial lymph node: an ultrastructural study. Ultrastuct. Pathol. 15:647-654. [DOI] [PubMed] [Google Scholar]
- 4.Angers, R. C., S. R. Browning, T. S. Seward, C. J. Sigurdson, M. W. Miller, E. A. Hoover, and G. C. Telling. 2006. Prions in skeletal muscles of deer with chronic wasting disease. Science 311:1117. [DOI] [PubMed] [Google Scholar]
- 5.Bahmanyar, S., E. S. Williams, F. B. Johnson, S. Young, and D. C. Gajdusek. 1985. Amyloid plaques in spongiform encephalopathy of mule deer. J. Comp. Pathol. 95:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Barcikowska, M., P. P. Liberski, J. W. Boellaard, P. Brown, D. C. Gajdusek, and H. Budka. 1993. Microglia is a component of the prion protein amyloid plaque in the Gerstmann-Straussler-Scheinker syndrome. Acta Neuropathol. 85:623-627. [DOI] [PubMed] [Google Scholar]
- 7.Baron, T., C. Crozet, A. G. Biacabe, S. Philippe, J. Verchere, A. Bencsik, J. Y. Madec, D. Calavas, and J. Samarut. 2004. Molecular analysis of the protease-resistant prion protein in scrapie and bovine spongiform encephalopathy transmitted to ovine transgenic and wild-type mice. J. Virol. 78:6243-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen, R. A., and R. F. Marsh. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68:7859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billeter, M., A. D. Kline, W. Braun, R. Huber, and K. Wuthrich. 1989. Comparison of the high-resolution structures of the alpha-amylase inhibitor tendamistat determined by nuclear magnetic resonance in solution and by X-ray diffraction in single crystals. J. Mol. Biol. 206:677-687. [DOI] [PubMed] [Google Scholar]
- 10.Brandner, S., S. Isenmann, A. Raeber, M. Fischer, A. Sailer, Y. Kobayashi, S. Marino, C. Weissmann, and A. Aguzzi. 1996. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature 379:339-343. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K. L., K. Stewart, D. L. Ritchie, N. A. Mabbott, A. Williams, H. Fraser, W. I. Morrison, and M. E. Bruce. 1999. Scrapie replication in lymphoid tissues depends on prion protein-expressing follicular dendritic cells. Nat. Med. 5:1308-1312. [DOI] [PubMed] [Google Scholar]
- 12.Browning, S. R., G. L. Mason, T. Seward, M. Green, G. A. Eliason, C. Mathiason, M. W. Miller, E. S. Williams, E. Hoover, and G. C. Telling. 2004. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J. Virol. 78:13345-13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruce, M. E., I. McConnell, H. Fraser, and A. G. Dickinson. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72:595-603. [DOI] [PubMed] [Google Scholar]
- 14.Büeler, H., A. Raeber, A. Sailer, M. Fischer, A. Aguzzi, and C. Weissmann. 1994. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol. Med. 1:19-30. [PMC free article] [PubMed] [Google Scholar]
- 15.Caughey, B. 2003. Prion protein conversions: insight into mechanisms, TSE transmission barriers and strains. Br. Med. Bull. 66:109-120. [DOI] [PubMed] [Google Scholar]
- 16.Chien, P., J. S. Weissman, and A. H. DePace. 2004. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 73:617-656. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, M., T. Rülicke, A. Raeber, A. Sailer, M. Moser, B. Oesch, S. Brandner, A. Aguzzi, and C. Weissmann. 1996. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15:1255-1264. [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, H., and A. G. Dickinson. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol 78:301-311. [DOI] [PubMed] [Google Scholar]
- 19.Glatzel, M., E. Abela, M. Maissen, and A. Aguzzi. 2003. Extraneural pathologic prion protein in sporadic Creutzfeldt-Jakob disease. N. Engl. J. Med. 349:1812-1820. [DOI] [PubMed] [Google Scholar]
- 20.Gossert, A. D., S. Bonjour, D. A. Lysek, F. Fiorito, and K. Wuthrich. 2005. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc. Natl. Acad. Sci. USA 102:646-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heggebo, R., C. M. Press, G. Gunnes, L. Gonzalez, and M. Jeffrey. 2002. Distribution and accumulation of PrP in gut-associated and peripheral lymphoid tissue of scrapie-affected Suffolk sheep. J. Gen. Virol. 83:479-489. [DOI] [PubMed] [Google Scholar]
- 22.Hill, A. F., M. Zeidler, J. Ironside, and J. Collinge. 1997. Diagnosis of new variant Creutzfeldt-Jakob disease by tonsil biopsy. Lancet 349:99. [DOI] [PubMed] [Google Scholar]
- 23.Klein, M. A., P. S. Kaeser, P. Schwarz, H. Weyd, I. Xenarios, R. M. Zinkernagel, M. C. Carroll, J. S. Verbeek, M. Botto, M. J. Walport, H. Molina, U. Kalinke, H. Acha-Orbea, and A. Aguzzi. 2001. Complement facilitates early prion pathogenesis. Nat. Med. 7:488-492. [DOI] [PubMed] [Google Scholar]
- 24.Kong, Q., S. Huang, W. Zou, D. Vanegas, M. Wang, D. Wu, J. Yuan, M. Zheng, H. Bai, H. Deng, K. Chen, A. L. Jenny, K. O'Rourke, E. D. Belay, L. B. Schonberger, R. B. Petersen, M. S. Sy, S. G. Chen, and P. Gambetti. 2005. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J. Neurosci. 25:7944-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legname, G., I. V. Baskakov, H. O. Nguyen, D. Riesner, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2004. Synthetic mammalian prions. Science 305:673-676. [DOI] [PubMed] [Google Scholar]
- 26.Liberski, P. P., and H. Budka. 1995. Ultrastructural pathology of Gerstmann-Straussler-Scheinker disease. Ultrastuct. Pathol. 19:23-36. [DOI] [PubMed] [Google Scholar]
- 27.Liberski, P. P., D. C. Guiroy, E. S. Williams, A. Walis, and H. Budka. 2001. Deposition patterns of disease-associated prion protein in captive mule deer brains with chronic wasting disease. Acta Neuropathol. 102:496-500. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd, S. E., J. M. Linehan, M. Desbruslais, S. Joiner, J. Buckell, S. Brandner, J. D. Wadsworth, and J. Collinge. 2004. Characterization of two distinct prion strains derived from bovine spongiform encephalopathy transmissions to inbred mice. J. Gen. Virol. 85:2471-2478. [DOI] [PubMed] [Google Scholar]
- 29.Mabbott, N. A., J. Young, I. McConnell, and M. E. Bruce. 2003. Follicular dendritic cell dedifferentiation by treatment with an inhibitor of the lymphotoxin pathway dramatically reduces scrapie susceptibility. J. Virol. 77:6845-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maignien, T., C. I. Lasme Zas, V. Beringue, D. Dormont, and J. P. Deslys. 1999. Pathogenesis of the oral route of infection of mice with scrapie and bovine spongiform encephalopathy agents. J. Gen. Virol. 80:3035-3042. [DOI] [PubMed] [Google Scholar]
- 31.Manson, J. C., E. Jamieson, H. Baybutt, N. L. Tuzi, R. Barron, I. McConnell, R. Somerville, J. Ironside, R. Will, M. S. Sy, D. W. Melton, J. Hope, and C. Bostock. 1999. A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J. 18:6855-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKintosh, E., S. J. Tabrizi, and J. Collinge. 2003. Prion diseases. J. Neurovirol. 9:183-193. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. W., and E. S. Williams. 2004. Chronic wasting disease of cervids. Curr. Top. Microbiol. Immunol. 284:193-214. [DOI] [PubMed] [Google Scholar]
- 34.O'Rourke, K. I., D. Zhuang, A. Lyda, G. Gomez, E. S. Williams, W. Tuo, and M. W. Miller. 2003. Abundant PrP(CWD) in tonsil from mule deer with preclinical chronic wasting disease. J. Vet. Diagn. Investig. 15:320-323. [DOI] [PubMed] [Google Scholar]
- 35.Outram, G. W., H. Fraser, and D. T. Wilson. 1973. Scrapie in mice. Some effects on the brain lesion profile of ME7 agent due to genotype of donor, route of injection and genotype of recipient. J. Comp. Pathol. 83:19-28. [DOI] [PubMed] [Google Scholar]
- 36.Polymenidou, M., K. Stoeck, M. Glatzel, M. Vey, A. Bellon, and A. Aguzzi. 2005. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 4:805-814. [DOI] [PubMed] [Google Scholar]
- 37.Prinz, M., M. Heikenwalder, T. Junt, P. Schwarz, M. Glatzel, F. L. Heppner, Y. X. Fu, M. Lipp, and A. Aguzzi. 2003. Positioning of follicular dendritic cells within the spleen controls prion neuroinvasion. Nature 425:957-962. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136-144. [DOI] [PubMed] [Google Scholar]
- 39.Race, R. E., A. Raines, T. G. Baron, M. W. Miller, A. Jenny, and E. S. Williams. 2002. Comparison of abnormal prion protein glycoform patterns from transmissible spongiform encephalopathy agent-infected deer, elk, sheep, and cattle. J. Virol. 76:12365-12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott, M., D. Foster, C. Mirenda, D. Serban, F. Coufal, M. Waelchli, M. Torchia, D. Groth, G. Carlson, S. J. DeArmond, D. Westaway, and S. B. Prusiner. 1989. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59:847-857. [DOI] [PubMed] [Google Scholar]
- 41.Scott, M. R., D. Peretz, H. O. Nguyen, S. J. Dearmond, and S. B. Prusiner. 2005. Transmission barriers for bovine, ovine, and human prions in transgenic mice. J. Virol. 79:5259-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seeger, H., M. Heikenwalder, N. Zeller, J. Kranich, P. Schwarz, A. Gaspert, B. Seifert, G. Miele, and A. Aguzzi. 2005. Coincident scrapie infection and nephritis lead to urinary prion excretion. Science 310:324-326. [DOI] [PubMed] [Google Scholar]
- 43.Sigurdson, C. J., E. S. Williams, M. W. Miller, T. R. Spraker, K. I. O'Rourke, and E. A. Hoover. 1999. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J. Gen. Virol. 80:2757-2764. [DOI] [PubMed] [Google Scholar]
- 44.Spraker, T. R., R. R. Zink, B. A. Cummings, M. A. Wild, M. W. Miller, and K. I. O'Rourke. 2002. Comparison of histological lesions and immunohistochemical staining of proteinase-resistant prion protein in a naturally occurring spongiform encephalopathy of free-ranging mule deer (Odocoileus hemionus) with those of chronic wasting disease of captive mule deer. Vet. Pathol. 39:110-119. [DOI] [PubMed] [Google Scholar]
- 45.Taraboulos, A., K. Jendroska, D. Serban, S. L. Yang, S. J. DeArmond, and S. B. Prusiner. 1992. Regional mapping of prion proteins in brain. Proc. Natl. Acad. Sci. USA 89:7620-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorberg, I., M. H. Groschup, E. Pfaff, and S. A. Priola. 2003. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J. Virol. 77:2003-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadsworth, J. D. F., S. Joiner, A. F. Hill, T. A. Campbell, M. Desbruslais, P. J. Luthert, and J. Collinge. 2001. Tissue distribution of protease resistant prion protein in variant CJD using a highly sensitive immuno-blotting assay. Lancet 358:171-180. [DOI] [PubMed] [Google Scholar]
- 48.Weissmann, C. 2004. The state of the prion. Nat. Rev. Microbiol. 2:861-871. [DOI] [PubMed] [Google Scholar]
- 49.Weissmann, C. 1991. A ‘unified theory’ of prion propagation. Nature 352:679-683. [DOI] [PubMed] [Google Scholar]
- 50.Westaway, D., G. A. Carlson, and S. B. Prusiner. 1989. Unraveling prion diseases through molecular genetics. Trends Neurosci. 12:221-227. [DOI] [PubMed] [Google Scholar]
- 51.Williams, E. S. 2005. Chronic wasting disease. Vet. Pathol. 42:530-549. [DOI] [PubMed] [Google Scholar]
- 52.Xie, Z., K. I. O'Rourke, Z. Dong, A. L. Jenny, J. A. Langenberg, E. D. Belay, L. B. Schonberger, R. B. Petersen, W. Zou, Q. Kong, P. Gambetti, and S. G. Chen. 2006. Chronic wasting disease of elk and deer and Creutzfeldt-Jakob disease: comparative analysis of the scrapie prion protein. J. Biol. Chem. 281:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanusso, G., A. Farinazzo, F. Prelli, M. Fiorini, M. Gelati, S. Ferrari, P. G. Righetti, N. Rizzuto, B. Frangione, and S. Monaco. 2004. Identification of distinct N-terminal truncated forms of prion protein in different Creutzfeldt-Jakob disease subtypes. J. Biol. Chem. 279:38936-38942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.