Abstract
Antiviral innate immune responses can be triggered by accumulation of intracellular nucleic acids resulting from virus infections. Double-stranded RNA (dsRNA) can be detected by the cytoplasmic RNA helicase proteins RIG-I and MDA5, two proteins that share sequence similarities within a caspase recruitment domain (CARD) and a DExD/H box RNA helicase domain. These proteins are considered dsRNA sensors and are thought to transmit the signal to the mitochondrial adapter, IPS-1 (also known as MAVS, VISA, or CARDIF) via CARD interactions. IPS-1 coordinates the activity of protein kinases that activate transcription factors needed to induce beta interferon (IFN-β) gene transcription. Another helicase protein, LGP2, lacks the CARD region and does not activate IFN-β gene expression. LGP2 mRNA is induced by interferon, dsRNA treatments, or Sendai virus infection and acts as a feedback inhibitor for antiviral signaling. Results indicate that LGP2 can inhibit antiviral signaling independently of dsRNA or virus infection intermediates by engaging in a protein complex with IPS-1. Experiments suggest that LGP2 can compete with the kinase IKKi (also known as IKKɛ) for a common interaction site on IPS-1. These results provide the first demonstration of protein interaction as an element of negative-feedback regulation of intracellular antiviral signaling by LGP2.
The innate immune system is triggered by pathogen-associated molecular patterns and represents the first line of defense against a variety of infectious microorganisms (21). The Toll-like receptors (TLR) 3, 7, 8, and 9 are expressed in immune cells and function as transmembrane pattern recognition receptors to detect foreign nucleic acids (1, 10). TLR detection of viral RNA or DNA can trigger downstream signal transduction, resulting in production of type I interferons (IFN), including beta interferon (IFN-β) and alpha interferon (IFN-α) isoforms (13). These IFNs can initiate autocrine and paracrine signal amplification via the JAK/STAT pathway to produce a potent generalized antiviral state that protects the target cell from virus infection and also assists in subsequent activation of adaptive immune responses (7, 8, 17).
In addition to TLRs, intracellular pattern recognition receptors have been described as critical elements of pathogen detection in mammalian cells (9, 19, 24, 30, 31). Intracellular RNA helicase proteins that participate in innate immune responses are ubiquitously expressed and recognize double-stranded RNA (dsRNA) produced during virus replication or from synthetic sources (24, 30, 31). One cytoplasmic RNA helicase, called retinoic acid-inducible gene I (RIG-I), is an intracellular sensor of dsRNA that can induce IFN production independently of TLR signaling (31). RIG-I is a member of a protein family characterized by two recognized domains. The amino terminus contains two regions similar to the caspase activation and recruitment domain (CARD) and acts as a protein interaction motif to facilitate associations with downstream components. The carboxyl terminus contains homologies with the DExD/H box RNA helicase family and is implicated in dsRNA binding and ATP-dependent unwinding. The second member of this CARD-helicase family, melanoma differentiation-associated gene 5 (MDA5), is similar in domain architecture and also responds to virus infection or intracytoplasmic dsRNA to activate IFN antiviral responses (2). Recent studies of mice harboring targeted disruptions in RIG-I and MDA5 have demonstrated differential roles for these proteins in the recognition of RNA viruses (11, 12). Of the viruses tested, RIG-I was found to be essential for the production of IFNs in response to several RNA viruses, whereas MDA5 was critical only for detection of picornaviruses.
A third protein, LGP2, is similar to RIG-I and MDA5 in the DExD/H box RNA helicase domain but differs from the other two, as it lacks the homologous CARD region. The LGP2 amino acid sequence shares 30% identity to the helicase domain of RIG-I and 40% identity to that of MDA5. In vitro RNA binding analysis suggests that all of these helicase domain-containing proteins are capable of binding to dsRNAs but not single-stranded RNAs (24, 30).
A primary outcome of dsRNA sensing by CARD-helicase proteins is transcriptional activation of the IFN-β promoter through the action of immediate responding transcription factors, including interferon regulatory factor 3 (IRF-3) and nuclear factor kappa B (NF-κB) (28). Currently available evidence suggests a model wherein RIG-I and MDA5 are held inactive by an autoinhibitory mechanism, but virus infection or dsRNA transfection is sensed by direct RNA-protein interactions with the helicase domain (31). Binding to RNA may induce a structural or conformational change, freeing the CARD to interact with downstream signaling molecules. In support of this model, a CARD-containing signaling intermediate, IPS-1 (also known as MAVS, VISA, or Cardif) (14, 22, 25, 29) was recently demonstrated to associate with the activated CARD-helicase proteins.
The IPS-1 protein is a mitochondrial resident, localized by a C-terminal transmembrane domain (25). At its N terminus, IPS-1 also contains a CARD, which is recognized by the activated RIG-I and MDA5 proteins. The CARD interaction initiates the activity of cytoplasmic kinase complexes containing the TBK1, IKKi (also known as IKKɛ), and IKK α, β, and γ proteins, which function in the phosphorylation-mediated activation of IRF-3 and NF-κB, respectively (14, 22, 25, 29). These transcription factors translocate to the nucleus and participate in transcriptional activation of the IFN-β gene.
For RIG-I, mutations to the putative ATP binding site can render the protein defective in dsRNA signaling (31), implying that helicase enzymatic activity is required for dsRNA signal transduction. Truncation of RIG-I from the C terminus produces an isolated CARD fragment (called ΔRIG-I [31], N-RIG [27], or RIG-IN [30]) that can constitutively activate downstream signaling. Truncation of RIG-I from the N terminus to remove the CARD produces a dominant negative variant (called C-RIG [27] or RIG-IC [31]) that is thought to function by competing with RIG-I for dsRNA substrates. The absence of a CARD region in LGP2 coupled with its ability to bind well to dsRNA in vitro has led to the supposition that LGP2 acts like the RIG-IC reagent to sequester intracellular dsRNA and prevent activation of the CARD-helicase proteins (24, 30). Indeed, LGP2 expression is induced by antiviral stimuli and has the properties of a negative feedback regulator of intracellular dsRNA signaling (24, 30) (see Fig. 1).
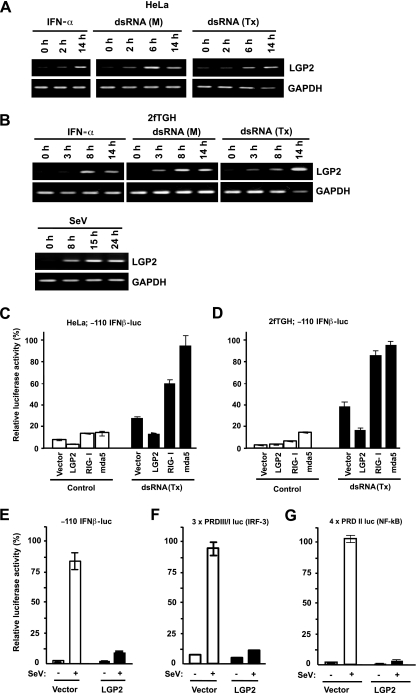
FIG. 1.
LGP2 is a feedback inhibitor of the antiviral system. (A and B) HeLa (A) or 2fTGH (B) cells were treated with 1,000 U/ml IFN-α or poly(I:C) {both extracellular [dsRNA (M)] and intracellular [dsRNA (Tx)] } or infected with Sendai virus (MOI = 1) for the indicated time course. Total RNA was isolated from cells and subjected to reverse transcriptase (RT)-PCR with the gene-specific primer sets indicated. (C and D) HeLa (C) or 2fTGH (D) cells were transiently transfected with the −110 IFNβ-luc reporter gene and the indicated RNA helicase vector. The cells were mock treated (control) or transfected with poly(I:C) for 6 h [dsRNA (Tx)]. (E) 2fTGH cells transfected with the LGP2 expression vector or empty vector (vector) along with the −110 IFNβ-luc reporter gene were mock treated or infected with Sendai virus (MOI = 1) for 12 h. (F) Similar analysis as for panel E was carried out with the 3× PRD III/I luc reporter gene. (G) Similar analysis as for panel E was carried out with the 4× PRDII luc reporter gene. In all cases, the data were normalized to results for the cotransfected Renilla luciferase reporter and the bars indicate the average (n = 3) ± the standard deviation, plotted as the percent control for each reporter gene. SeV, Sendai virus.
To investigate negative regulation in the intracellular dsRNA-sensing innate immune systems, the function of the LGP2 protein was explored. In support of a feedback inhibitory role in IPS-1-mediated dsRNA signaling, LGP2 mRNA synthesis is rapidly induced in human cells by IFN-α, dsRNA, or virus infection. Results indicate that LGP2 can inhibit antiviral signaling independently of dsRNA or virus infection intermediates, suggesting that LGP2 is capable of targeting downstream molecule(s) for inhibition. Mechanistic studies reveal that LGP2 coprecipitates with IPS-1 in a virus-independent manner that requires both mitochondrial localization and C-terminal residues of IPS-1, but the CARD region is not involved. LGP2 can coprecipitate CARD-helicase proteins MDA5 and RIG-I only following Sendai virus infection, suggesting that LGP2 can associate with a multimeric complex. Competition analysis demonstrates that LGP2 can interfere with IKKi recruitment to IPS-1, indicating that protein interaction is an alternative to dsRNA sequestration as a mechanistic basis for attenuation of dsRNA signaling.
MATERIALS AND METHODS
Plasmids and antibodies.
LGP2 cDNA was obtained by PCR amplification of Human Universal Quick Clone II (Clontech) and cloned into p3xFLAG-CMV10 (Sigma), pcDNA3 (Invitrogen) with an N-terminal hemagglutinin (HA)-epitope sequence, and pcDNA4/HisMAX (Invitrogen). For subcloning of IPS-1, cDNA was amplified by PCR, using the IMAGE clone with GenBank accession number BC044952, and cloned into p3xFLAG-CMV10 (Sigma) and pcDNA3 (Invitrogen) with a HA sequence. Various regions (1 to 118, 1 to 200, 118 to 500, 118 to 540, 118 to 535, 118 to 540, 300 to 540, 444 to 540, 500 to 540, and 509 to 540) of IPS-1 were amplified by PCR and subcloned into the glutathione S-transferase mammalian expression vector, pEBG (a gift from B. Mayer, University of Connecticut), or p3xFLAG-CMV10 (Sigma). pEFBos-FLAG-RIG-I and pEFBos-FLAG-MDA5 expression plasmids were provided by M. Gale, Jr. (UT Southwestern). pcDNA3.0 Zero-FLAG-IKKi was provided by J. Hiscott (McGill University). For the C-terminal Myc-tagged RIG-I and MDA5 vector, the entire coding region was amplified by PCR and cloned into pcDNA4/myc-His (Invitrogen). The IFN-β-luciferase (luc) reporters, −110 IFNβ-luc, PRD III/I (3×) luc, and PRD II (4×) luc, were provided by D. Thanos. To create pcDNA4-6xHis-RIG-IC, the region that encodes amino acids 218 to 925 was amplified by PCR and cloned into pcDNA4/HisMAX (Invitrogen). A Renilla luciferase vector was purchased from Promega. The FLAG and His antibodies are from Sigma. HA and Myc antibodies were purchased from Roche. Anti-Cardif (IPS-1) antibodies were obtained from Axxora (San Diego, CA). Rabbit polyclonal antibody against LGP2 was raised using bacterially expressed GST-LGP2 fusion protein (Covance, PA). Anti-GST antibody was removed with a GST affinity column, and the immunoglobulin G (IgG) fraction was purified by a protein G column. For immunoprecipitation of endogenous proteins, HEK 293T cells were treated with Sendai virus (MOI = 10) for 20 h. The cells were lysed with RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM sodium chloride, 1% Triton X-100, 1 mM dithiothreitol, 0.l % sodium dodecyl sulfate, 0.5% sodium deoxycholate, 10% glycerol, 1 mM sodium orthovanadate, 1 mM sodium fluoride), and after being precleared with control IgG and protein G beads (Roche Diagnostics), the lysate was incubated with LGP2 antibody or control rabbit IgG overnight. The precipitated proteins were analyzed by Western blotting with IPS-1 antibody and LGP2 antibody.
Cells, DNA, and RNA transfection and luciferase assays.
2fTGH, HEK 293T, and HeLa cells were cultured in Dulbecco's modified eagle medium supplemented with calf cosmic serum (10%; HyClone), penicillin (100 U/ml), and streptomycin (100 μg/ml). Poly(I:C) was purchased from Amersham-Pharmacia. Sendai virus (Cantell strain) was obtained from Tom Moran (Mt. Sinai, NY). SMART pool siRNAs were purchased from Dharmacon (Lafeyette, CO). 2fTGH, HeLa, and HEK 293T cells were transiently transfected with SuperFect (QIAGEN) or Effectene (QIAGEN) as described by the manufacturer. Poly(I:C) was transfected with lipofectamine or Lipofectamine2000 (Invitrogen). The Dual-Luciferase Assay System (Promega) was used for luciferase assays. As an internal control, the Renilla luciferase construct was used to normalize luciferase activity.
RNA interference.
Small interfering RNA (siRNA) Smart Pools were obtained from Dharmacon, and transfection of siRNAs targeting LGP2, RIG-I, or MDA5 and control scrambled siRNA was carried out using siLentFect reagent (Bio-Rad) according to the manufacturer's instructions.
mRNA analysis.
Total RNA was prepared by using TRIzol (Invitrogen) treated with DNase I (amplification grade; Invitrogen) and subjected to reverse transcriptase with SuperScript III RNase H-reverse transcriptase (Invitrogen). The PCR primers used for semiquantitative PCR were as follows. For LGP2, the forward primer was 5′-CAACTGAAGGTAGCCGGGA-3′, and the reverse primer was 5′-CCACAGCCACCATGCAGTTGAT-3′; for IFN-β, the forward primer was 5′-GATTCATCTAGCACTGGCTGG-3′, and the reverse primer was 5′-CTTCAGGTAATGCAGAATCC-3′; for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), the forward primer was 5′-ACCACAGTCCATGCCATCAC-3′, and the reverse primer was 5′-TCCACCACCCTGTTGCTGTA-3′; for RIG-I, the forward primer was 5′-CAGTATATTCAGGCTGAG-3′, and the reverse primer was 5′-GGCCAGTTTTCCTTGTC-3′; and for MDA5, the forward primer was 5′-AGTTTGGCAGAAGGAAGTGTC-3′, and the reverse primer was 5′-GGAGTTTTCAAGGATTTGAGC-3′.
Quantitative real-time PCR was carried out by using the 7900 HT Fast Real-Time PCR system with SYBR GREEN PCR Master Mix (Applied Biosystems). The relative abundances of mRNA was obtained by calculating the difference in threshold cycles of the target and control samples, commonly known as the ΔΔCT method, using GAPDH for normalization. Primers used for quantitative real time PCR were as follows. For LGP2, the forward primer was 5′-CTTTGACTTCCTGCAGCATT-3′, and the reverse primer was 5′-CAATGAGGTGGTCAGTCCAG-3′; for IFN-β, the forward primer was 5′-CATTACCTGAAGGCCAAGGA-3′, and the reverse primer was 5′-CAATTGTCCAGTCCCAGAGG-3′; for GAPDH, the forward primer was 5′-ACAGTCAGCCGCATCTTCTT-3′, and the reverse primer was 5′-ACGACCAAATCCGTTGACTC-3′; for RIG-I, the forward primer was 5′-CTCTGCAGAAAGTGCAAAGC-3′, and the reverse primer was 5′-GGCTTGGGATGTGGTCTACT-3′; and for MDA5, the forward primer was 5′-TGGTCTCGTCACCAATGAAA-3′, and the reverse primer was 5′-CTCCTGAACCACTGTGAGCA-3′.
Transfection of siRNA and mRNA analysis were performed in at least two independent experiments for each variable, and results of the representative experiments are shown in the figures.
Protein interaction analysis.
HEK 293T cells transfected with expression plasmids were lysed with WCE buffer (50 mM Tris-HCl [pH 8.0]-280 mM NaCl-0.5% NP-40-0.2 mM EGTA-2 mM EDTA-10% glycerol-1 mM dithiothreitol-1 mM NaF-1 mM sodium orthovanadate) after being mock infected or infected with Sendai virus. The clarified cell lysate was incubated with M2 agarose beads (Sigma) or HA agarose beads (Roche) to purify FLAG or HA-tagged proteins by immunoaffinity. Copurified proteins were analyzed by immunoblot analysis.
RESULTS
LGP2 is an inhibitor of RIG-I and MDA5 signaling.
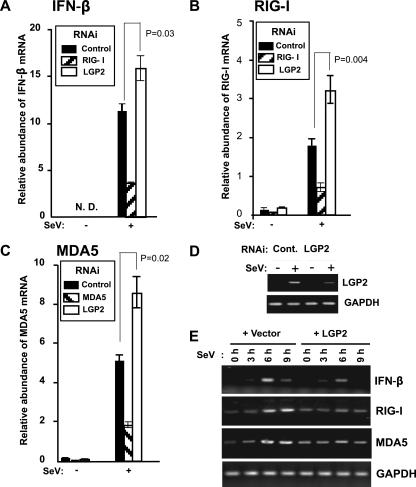
Expression of the RIG-I helicase fragment (RIG-IC) produces a protein that can act as a dominant negative inhibitor for dsRNA responses (31). The similar domain arrangement of LGP2 and RIG-IC suggested it was a negative regulator of intracellular dsRNA antiviral signaling (24, 30). In accordance with these reports, we observe that LGP2 mRNA is induced in a time-dependent manner by IFN-α-treatment, addition of dsRNA [poly(I:C)] to the culture medium, dsRNA transfection, or Sendai virus infection (Fig. 1A and B). LGP2 expression inhibits Sendai virus- or dsRNA transfection-mediated activation of the human IFN-β gene promoter through attenuation of dsRNA and virus signal transduction to IRF-3 and NF-κB (Fig. 1C to G), and interference with LGP2 expression increased not only IFN-β mRNA but also RIG-I and MDA5 mRNA induction by Sendai virus (Fig. 2A -D). Correspondingly, expression of LGP2 inhibited the Sendai virus-mediated induction of endogenous IFNβ, RIG-I, and MDA5 mRNA (Fig. 2E). These data support the characterization of LGP2 as a feedback inhibitor of antiviral signaling and are in general agreement with earlier reports (24, 30).
FIG. 2.
LGP2 negatively regulates endogenous antiviral gene expression. (A to C) Effect of LGP2 interference on antiviral response gene induction. 2fTGH cells were transfected with siRNA for LGP2, RIG-I, or MDA5 (as indicated in inset boxes), and total RNA was processed for real time RT-PCR. The relative abundances of IFN-β mRNA (A), RIG-I mRNA (B), and MDA5 mRNA (C) were quantified by real time RT-PCR. The Student's t test was used for statistical analysis, and P values are indicated. These results were representative of at least two independent transfection experiments. Error bars indicate the standard deviations for the results of duplicate PCRs. N.D., not detected. (D) Demonstration of the efficacy of siRNA-mediated LGP2 knockdown. 2fTGH cells were transfected with a control scrambled siRNA duplex or LGP2-specific siRNA and then infected with Sendai virus or mock infected. Total RNA was prepared and subjected to RT-PCR amplification with LGP2- or GAPDH-specific primers. (E) 2fTGH cells transfected with empty vector (+ Vector) or LGP2 expression vector (+ LGP2) were infected with Sendai virus for the indicated time course. Total RNA was isolated from cells and subjected to RT-PCR with the gene-specific primer sets indicated. SeV, Sendai virus.
LGP2 inhibits the RIG-I pathway independently of virus or dsRNA.
Truncation of the CARD from RIG-I produces a dominant negative variant, RIG-IC, that is thought to function by competing with RIG-I for dsRNA substrates (31). The apparent primary structural similarity between RIG-IC and LGP2 has suggested that LGP2 may also compete with RIG-I for dsRNA substrates (24, 30). Expression of either LGP2 or RIG-IC inhibits Sendai virus-induced IFN-β promoter activity (Fig. 3A). To compare the inhibitory activities of LGP2 and RIG-IC in the absence of either virus infection or dsRNA stimulation, expression of the RIG-I CARD fragment, RIG-IN, was used to constitutively induce IFN-β-luciferase reporter gene activity (31). Expression of RIG-IN efficiently activated the IFN-β-luciferase reporter gene (Fig. 3B), but expression of RIG-IC had no effect. In contrast, LGP2 expression efficiently blocked the signaling activity of RIG-IN, preventing IFN-β-luciferase activity. This result demonstrates that LGP2 can negatively regulate RIG-I signaling independently of virus infection, dsRNA treatment, or the presence of the RIG-I helicase domain.
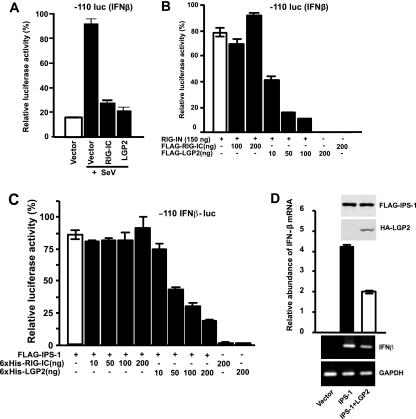
FIG. 3.
LGP2 inhibits RIG-I signaling independently of dsRNA or virus infection. (A) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 100 ng of the expression vector for RIG-IC or LGP2. Twenty-four hours after being transfected, the cells were infected with Sendai virus (SeV) for 8 h. Lysates were prepared and subjected to luciferase assays, plotted as the percent positive control. (B) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 150 ng of the expression vector for RIG-IN alone or with increasing amounts of the expression plasmid for LGP2 or RIG-IC, as indicated. The lysates were prepared and subjected to luciferase assays. (C) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 100 ng of the expression vector for IPS-1 alone or with increasing amounts of the expression plasmid for LGP2 or RIG-IC, as indicated. (D) HEK 293T cells were transfected with the expression vector for FLAG-tagged IPS-1 alone or along with the expression vector for HA-tagged LGP2. After 24 h, total RNA was isolated for quantitative real time RT-PCR (top graph) or nonquantitative RT-PCR followed by agarose gel electrophoresis and ethidium bromide staining (bottom). The relative abundance of IFN-β mRNA was normalized to the abundance of GAPDH mRNA. The expression of proteins was confirmed by immunoblotting (inset). The data are representative of at least two independent transfection experiments. The error bars indicate the standard deviation for duplicate PCRs.
To verify this result using a downstream activator in the RIG-I pathway, expression of the adaptor IPS-1 was used to activate the IFN-β promoter in the absence of a signal. This IFN-β activation was also efficiently inhibited by LGP2 expression in a dose-dependent manner (Fig. 3C). Again, the IPS-1-dependent IFN-β luciferase activity was not prevented by RIG-IC (Fig. 3C). In confirmation, IPS-1-mediated induction of the endogenous IFN-β mRNA was also inhibited by LGP2 expression (Fig. 3D). We conclude that the natural cellular inhibitor, LGP2, differs mechanistically from the artificial RIG-IC construct in its ability to block IFN-β gene activation independently of either virus infection or dsRNA and at a step downstream of RNA sensing by RIG-I.
LGP2 coprecipitates with IPS-1.
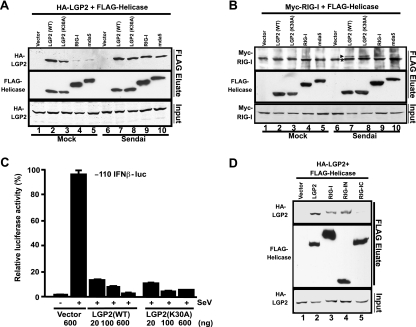
To investigate the mechanism of RNA- and virus-independent signaling inhibition by LGP2, protein associations were tested in coimmunoprecipitation assays. The ability of LGP2 to inhibit IPS-1-mediated signaling prompted us to test whether LGP2 might associate with IPS-1 or interfere with the ability to form a RIG-I/IPS-1 complex mediated by CARD-CARD interactions (14, 22, 25, 29). A coimmunoprecipitation assay was carried out using Myc-tagged RIG-I and FLAG-tagged IPS-1 expressed in the presence or absence of LGP2. IPS-1 coprecipitated with RIG-I (Fig. 4A, lane 3), and this complex was detected irrespective of the presence of LGP2 (Fig. 4A, lane 5) or a control protein, Yap (Fig. 4A, lane 4) (16). This experiment revealed that LGP2 was able to coprecipitate with IPS-1 (Fig. 4A, lane 5), and this interaction does not exclude RIG-I binding to IPS-1. To further confirm that LGP2 does not compete with RIG-I for IPS-1 binding, a titration experiment was performed (Fig. 4B). Even at saturation levels of LGP2 coprecipitation, robust RIG-I interactions were detected. These results suggest that a target for LGP2 inhibition may be IPS-1 itself.
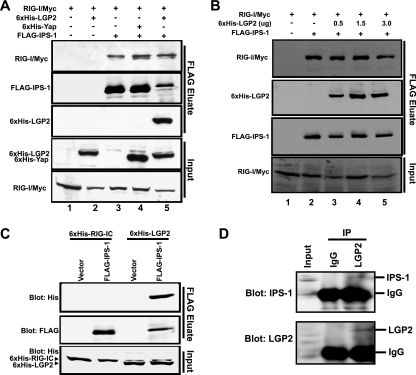
FIG. 4.
LGP2 coprecipitates with IPS-1 and RIG-I. (A) HEK 293T cells were transfected with the expression vector for Myc-tagged RIG-I and the expression vector for FLAG-tagged IPS-1 along with the expression vector for His-tagged LGP2 or His-tagged Yap. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads and immunoblotting for Myc or His tags. (B) HEK 293T cells were transfected with the expression vector for Myc-tagged RIG-I and the expression vector for FLAG-tagged IPS-1 along with increasing amounts of the expression vector for His-tagged LGP2. Lysates were subjected to FLAG immunoprecipitation as for panel A. (C) HEK 293T cells were transfected with the expression vector for His-tagged RIG-IC or LGP2 along with the expression vector for FLAG-tagged IPS-1 or empty vector. Lysates were subjected to FLAG immunoprecipitation followed by immunoblotting, using His-tag antibody. (D) Lysates from Sendai virus-infected HEK 293T cells were subjected to immunoprecipitation with LGP2 antibody or control rabbit IgG. The precipitated proteins were analyzed by anti-Cardif antibody, and the same blot was reprobed with LGP2 antibody.
To verify this result more directly and to test the specificity of LGP2 and IPS-1 coprecipitation, another precipitation assay was performed without RIG-I expression. LGP2 coprecipitation with IPS-1 was easily detected, but RIG-IC was not detected in association with IPS-1 (Fig. 4C). This selective coprecipitation of LGP2 and not RIG-IC reinforces the differential properties of these proteins observed in the antiviral signaling assays, wherein LGP2 but not RIG-IC inhibits RIG-IN- or IPS-1-mediated IFN-β promoter activity (Fig. 3A and B).
To confirm these conclusions from protein expression, endogenous LGP2-IPS-1 interactions were tested. A polyclonal antiserum directed against a bacterially expressed GST-LGP2 fusion protein or a control rabbit IgG were used for immunoprecipitation of whole-cell extracts from Sendai virus-infected cells. After gel separation, a commercial antiserum (Axxora, San Diego, CA) that recognizes IPS-1 was used to detect coprecipitation (Fig. 4D). Although these antisera only poorly detected the endogenous proteins prior to immunoprecipitation, the appearance of IPS-1- and LGP2-specific signals was detected only in the coprecipitation sample. Together, these results provide a body of evidence supporting LGP2 interaction with IPS-1.
ATP hydrolysis is dispensable for LGP2 inhibition.
If LGP2 is interfering with CARD-helicase signaling at the level of IPS-1, it might be predicted that RIG-I or MDA5 could be found in a multimeric complex with LGP2 following virus infection. To test this idea, physical association between LGP2 and RIG-I or MDA5 was assessed by coimmunoprecipitation (Fig. 5A). HA-tagged LGP2 was expressed along with FLAG-tagged LGP2, RIG-I, or MDA5, and the lysates from uninfected or Sendai virus-infected cells were subjected to immunopurification with anti-FLAG (M2) beads. Immunoblotting with HA antibody to detect the copurified HA-LGP2 revealed that LGP2 was able to self-associate, resulting in coprecipitation. However, only trace levels of the association between LGP2 and RIG-I or MDA5 were detected in the uninfected cells. In contrast, LGP2 efficiently coprecipitated both RIG-I and MDA5 from the cells infected with Sendai virus (Fig. 5A). This result indicates that Sendai virus infection induces the formation of a complex between LGP2 and CARD-helicase proteins.
FIG. 5.
LGP2 coprecipitates RIG-I and MDA5 following virus infection. (A) HEK 293T cells were transfected with expression vectors for HA-tagged LGP2 and FLAG-tagged LGP2, RIG-I, or MDA5. The cells were infected with Sendai virus or mock infected for 6 h. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads. The HA-LGP2 proteins copurified with FLAG-tagged RNA helicases were analyzed by immunoblotting, using HA antibody (Roche). (B) Similar to procedure for panel A but using Myc-tagged RIG-I along with FLAG-tagged LGP2, LGP2 mutant (K30A), RIG-I, or MDA5. FLAG eluates were analyzed by immunoblotting using Myc antibody (Roche). An asterisk indicates a nonspecific band, and an arrowhead indicates Myc-RIG-I. (C) The LGP2 Walker A motif mutation K30A retains inhibitory activity. A luciferase assay was carried out as for Fig. 3A, comparing the activities of wild-type (WT) and mutant (K30A) LGP2 resulting from transfection of increasing amounts of expression vectors from 20 to 600 ng, as indicated. (D) HEK 293T cells were transfected with expression vectors for HA-tagged LGP2 and FLAG-tagged LGP2, RIG-I, RIG-IN, or RIG-IC and infected with Sendai virus for 6 h. Lysates were subjected to FLAG immunoprecipitation as in for panel A. Coprecipitated LGP2 protein was detected by immunoblotting, using HA antibody (Roche).
To verify this result in a complementary experiment and to test the possibility of constitutive CARD-helicase oligomerization, a similar assay using FLAG-tagged LGP2, RIG-I, or MDA5 along with Myc-tagged RIG-I was carried out (Fig. 5B). No evidence was obtained suggesting constitutive homo-oligomerization of RIG-I or hetero-oligomerization with MDA5 in this experiment. However, FLAG-LGP2 was able to coprecipitate Myc-RIG-I only from virus-infected cells (Fig. 5B).
In addition to wild-type LGP2, a variant protein with a mutation in the predicted Walker A motif of the ATP binding site (K30A) was tested in these experiments. Mutation of this residue prevents LGP2 ATP hydrolysis activity (D. Bamming, A. Komura, and C. M. Horvath, unpublished observations) but did not disrupt LGP2 homo-oligomerization or virus-inducible hetero-oligomerization with RIG-I (Fig. 5A and B). Luciferase assays demonstrate that the K30A mutant did not disrupt the inhibitory activity of LGP2 during Sendai virus infections (Fig. 5C). This finding indicates that LGP2 ATP hydrolysis activity is not strictly required for its ability to engage in protein interactions leading to antiviral inhibition.
To test the role of the RIG-I catalytic domain, a coimmunoprecipitation assay was carried out using the two RIG-I fragments. LGP2 was found to coprecipitate both RIG-I and RIG-IN from Sendai virus-infected cells, but the RIG-IC helicase fragment did not interact with LGP2 (Fig. 5D). Therefore, coprecipitation of RIG-I with LGP2 is mediated by the CARD of RIG-I and independently of the helicase domains.
LGP2 interaction with IPS-1 fragments.
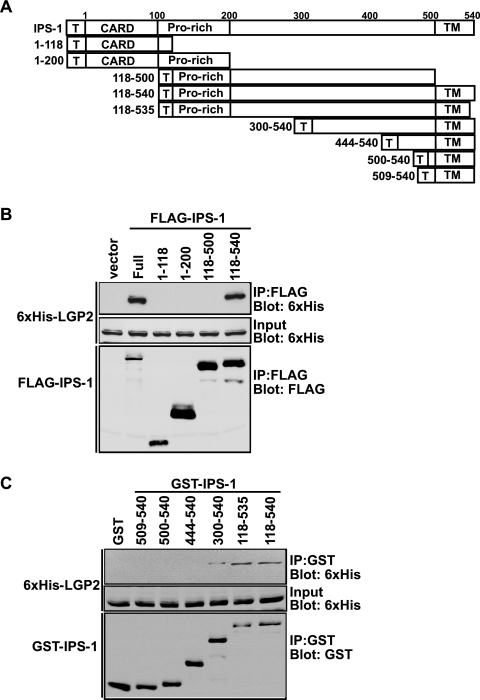
To delineate the region of IPS-1 required for coprecipitation with LGP2, IPS-1 fragments fused to the FLAG epitope tag (Fig. 6A) were coexpressed with His-LGP2, and FLAG proteins were affinity purified (Fig. 6B). Full-length IPS-1 copurified with LGP2, as did the CARD-deficient fragment containing amino acids 118 to 540. No association was observed with IPS-1 fragments representing the CARD alone (1 to 118), the CARD plus proline-rich region (1 to 200), or a CARD-deficient fragment lacking the 40-amino-acid C-terminal mitochondrial targeting transmembrane domain (118 to 500). These results indicate that neither the CARD nor the proline-rich region is needed for LGP2 interactions. To test if the transmembrane domain segment alone was sufficient for LGP2 interactions, C-terminal IPS-1 fragments were expressed in cells as GST fusion proteins. None of the C-terminal regions tested, encompassing residues 444 to 540, were capable of copurifying LGP2 (Fig. 6C). In contrast, increasing the fragment size to include amino acids 300 to 540 recovered the ability to copurify LGP2. Together, these results demonstrate that LGP2 requires both the mitochondrial targeting transmembrane domain and the residues between 300 and 444 for coprecipitation with IPS-1.
FIG. 6.
LGP2 interaction requires the C-terminal and the transmembrane regions of IPS-1. (A) Schematic representation of IPS-1 fragments used for mapping. Positions of FLAG or GST tags (T), CARD, the proline-rich region (Pro-rich), and the transmembrane domain (TM) are indicated. (B) HEK 293T cells were transfected with expression vectors for six-His-tagged LGP2 and the FLAG-tagged regions of IPS-1, as indicated. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads. His-LGP2 protein copurification with FLAG-tagged IPS-1 was analyzed by immunoblotting, using six-His antibody. (C) HEK 293T cells were transfected with expression vectors for His-tagged LGP2 and GST-tagged regions of IPS-1, as indicated. Cell lysates were subjected to glutathione bead affinity purification. Six-His-LGP2 protein copurification with GST-IPS-1 was analyzed by immunoblotting, using six-His antibody.
LGP2 and IKKi compete for IPS-1 interaction.
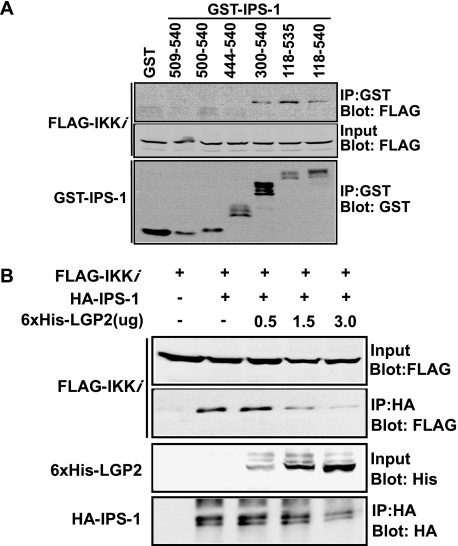
The C-terminal region of IPS-1 has been shown to interact with the kinase IKKi, which is essential for IRF-3 activation (22). Using the same set of GST-IPS-1 fragments, we mapped the IPS-1 region that recruits IKKi. Kinase interaction was observed with amino acids 300 to 540, but not 444 to 540, defining the IPS-1 site for kinase coprecipitation as identical with the LGP2 binding region (Fig. 7A).
FIG. 7.
LGP2 competes with IKKi for IPS-1binding. (A) HEK 293T cells were transfected with expression vectors for FLAG-tagged IKKi and GST-IPS-1 fusions, as indicated. Cell lysates were subjected to glutathione bead affinity purification. FLAG-tagged IKKi copurification with GST-IPS-1 was analyzed by immunoblotting, using FLAG antibody. (B) HEK 293T cells were transfected with expression vectors for FLAG-tagged IKKi and HA-tagged IPS-1 along with increasing amounts of the expression vector for six-His-tagged LGP2. Cell lysates were subjected to HA purification. FLAG-tagged IKKi purified with HA-IPS-1 was analyzed by immunoblotting, using FLAG antibody.
The overlapping protein interaction sites suggest that competitive displacement of IKKi might provide a plausible mechanistic basis for LGP2-dependent inhibition of antiviral signaling. To test this idea, a coprecipitation competition assay was carried out. IKKi coprecipitated with IPS-1 in the absence of LGP2, but increasing the amount of LGP2 coprecipitation correlated with decreased amounts of IKKi detected in the immune complexes (Fig. 7B). These data are consistent with a model in which LGP2 regulates IPS-1 signaling by preventing IKKi interaction (Fig. 8).
FIG. 8.
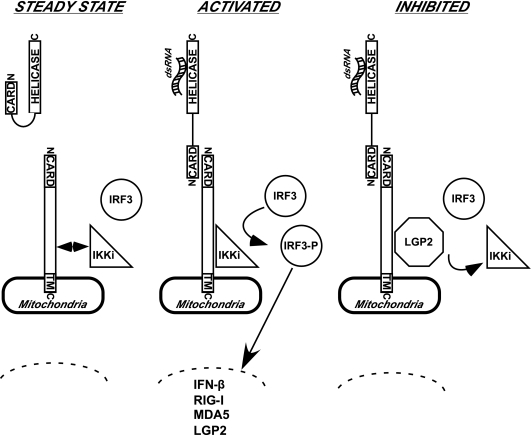
Model of LGP2 action in dsRNA- and virus-independent inhibition of antiviral signal transduction. Schematic diagrams are drawn to illustrate key features of the results and interpretation. For simplicity, a single CARD-helicase protein is illustrated. Many components of the antiviral response and additional unknown components that might bridge indirect protein interactions are not depicted. (A) In the steady state, the absence of virus infection or cytosolic dsRNA maintains the CARD-helicase RNA sensor protein in an autoinhibited quiescent state. IPS-1 is anchored in the mitochondria, where it may associate with IKKi. IRF3 is unphosphorylated and in its latent cytoplasmic location. (B) Once activated by virus infection or cytosolic dsRNA, the CARD of the RNA sensor is activated to make heterotypic contact with the CARD of IPS-1. This activates IKKi to phosphorylate IRF3, inducing its nuclear translocation and transcriptional activation of target genes, including IFN-β, RIG-I, MDA5, and LGP2. (C) LGP2 accumulation in the cell leads to signal inhibition. LGP2 interaction displaces IKKi from the IPS-1 C terminus to disengage signal propagation. In this case, the CARD-helicase can remain in association with the IPS-1 plus LGP2 complexes.
DISCUSSION
The findings presented here support a role for the DExD/H box RNA helicase protein, LGP2, as an inhibitor of intracellular signaling that is initiated by cytosolic dsRNA or by Sendai virus infection. Like most cellular stress responses, innate antiviral immunity must be tightly controlled to prevent unwanted toxicity due to hyperactive signaling (15). Accumulation of IFNs and their downstream effectors can have strong cytotoxic effects (4, 5), in many cases requiring their expression to be rapid and transient. LGP2 mRNA accumulates in cells in response to IFN stimulation, virus infection, or intracellular dsRNA, and results indicate that increased expression of LGP2 negatively regulates IFN-β, RIG-I, and MDA5 biosynthesis. These findings demonstrate that LGP2 interferes with a step upstream of transcription factor activation but downstream of the dsRNA-sensing apparatus.
Protein interaction studies indicate that LGP2 coprecipitates with the signaling adaptor IPS-1 and forms a multicomponent complex requiring both the mitochondrial transmembrane domain and the amino acids between residues 300 and 440 of IPS-1 but not its CARD region. The CARD of IPS-1 is still free to engage upstream dsRNA sensors RIG-I and MDA5, and these regulated associations allow coprecipitation of CARD-helicase proteins from virus-infected cells. Data indicate that coprecipitation of RIG-I can be achieved independently of its RNA helicase domain and in the absence of ATP hydrolysis by LGP2. We interpret these observations to indicate an RNA- and virus -independent inhibitory activity mediated by LGP2. While it is possible that at least some of the observed protein interactions are indirect and rely on intervening proteins or nucleic acids, the data clearly indicate that protein complexes can provide a foundation for at least some of the observed LGP2 inhibition of antiviral signaling (Fig. 8).
In support of this model, coprecipitation of the protein kinase IKKi with IPS-1 was also observed, and this interaction was found to rely on the same polypeptide regions that bind LGP2. Results from competition experiments demonstrate that increased LGP2 expression can interfere with kinase binding to IPS-1, providing a plausible mechanism for dsRNA- and virus-independent signaling inhibition by LGP2. Note that in addition to IKKi, TBK1 is involved in phosphorylation of IRF-3 and IRF-7 to execute antiviral responses (3, 20, 26). While studies of TBK1- and IKKi-knockout mice have suggested that TBK1 plays a major role while IKKi has an accessory activity (6, 23), TBK1-deficient mouse embryonic fibroblasts respond normally to Newcastle disease virus for IRF-3 and IFN-β promoter activation (31). In addition, IKKi but not TBK1 can colocalize with IPS-1 in the mitochondria. Mitochondrial IPS-1/IKKi colocalization is dissociated by hepatitis C virus NS3-4A protease cleavage of IPS-1, suggesting that the IPS-1/IKKi signaling complex is a key element of intracellular antiviral responses for some infections (18). It is noteworthy that LGP2 also inhibits activation of NF-κB activity downstream of dsRNAs (Fig. 1G). While this is apparent from reporter gene assays, no association of IPS-1 with the canonical IKKs (IKKα, β, γ) required for NF-κB activation was observed in our assays (data not shown). Possibly relevant is that the Fas-associated death domain (FADD) protein and receptor-interacting protein 1 (RIP1) have been implicated in intracellular dsRNA signaling to NF-κB. While these proteins have also been demonstrated to associate with the C terminus of IPS-1, these interactions were not observed in our experiments (data not shown). Therefore, while it is clear that LGP2 interferes with NF-κB activation, the precise mechanism of action remains to be clarified. However, if the FADD and RIP1 or the canonical IKKs binding sites on IPS-1 overlap with the LGP2 binding site, a similar competitive inhibition mechanism is plausible. The links between IPS-1 and IRF-3 kinases and IκB kinases and the mechanism of NF-κB inhibition by LGP2 remain to be elucidated.
As LGP2 does not contain the CARD region but does bind to dsRNA, it has been proposed that the mechanism by which LGP2 inhibits RIG-I or MDA5 involves sequestration of dsRNA, thereby preventing activation of CARD signaling (24, 30). While the present study is in agreement about the negative regulatory role of LGP2, differences were found in the proposed mechanism of inhibition. While differences in methodology, reagents, and cell lines may account for some of the disparities, it is most likely that LGP2 functions by a combination of activities directed at either RNA binding or protein interactions.
In this study, we demonstrate that LGP2 inhibits signaling induced by expression of RIG-IN or by expression of IPS-1, both of which induce transcriptional responses independently of dsRNA treatments or virus infections. This supports the concept that LGP2 does not merely sequester dsRNA but also blocks IPS-1 signaling at a step downstream of the CARD-helicase receptor and IPS-1 adaptor. It remains formally possible that RNA sequestration functions in parallel with protein interaction-based inhibition, but it is important to note that the RIG-IC-terminal fragment (RIG-IC) fails to antagonize the constitutive signaling induced by RIG-IN or IPS-1. This demonstrates that the similarities between the primary sequences of LGP2 and the artificial RIG-IC fragment are somewhat superficial and that data obtained with RIG-IC might not accurately reflect the function of the natural regulator, LGP2. In other words, RIG-IC might well function as an inhibitor by RNA sequestration, but this remains to be experimentally proven. Two findings may be relevant to the differential mechanisms of LGP2 and RIG-IC. First, LGP2 was found to be constitutively oligomeric, and second, the LGP2 K30A mutant remains active as a negative regulator despite the inability to hydrolyze ATP. For RIG-I, an analogous mutation fails to function in antiviral signaling and acts as a dominant negative inhibitor. Irrespective of the mechanistic differences between the present results and prior studies (24, 30), it is clearly recognized that only LGP2 is a native component of dsRNA signal transduction attenuation.
Acknowledgments
We are grateful to Michael Gale for providing reagents and advice, Dimitris Thanos and John Hiscott for plasmids, Tom Moran for Sendai virus Cantell, Joe Bass and Akira Kosaka for support and advice on real-time PCR, and Maylene Corpuz and Charita Wallace-Smith for technical assistance. We thank members of the Horvath lab for discussions and comments on the manuscript.
This work was supported by NIH grants AI048722 and GM063872 and by American Cancer Society Research Scholar Award RSG-02-049-01 (to C.M.H.).
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 4.Gresser, I., M. Aguet, L. Morel-Maroger, D. Woodrow, F. Puvion-Dutilleul, J. C. Guillon, and C. Maury. 1981. Electrophoretically pure mouse interferon inhibits growth, induces liver and kidney lesions, and kills suckling mice. Am. J. Pathol. 102:396-402. [PMC free article] [PubMed] [Google Scholar]
- 5.Gresser, I., M. G. Tovey, C. Maury, and I. Chouroulinkov. 1975. Lethality of interferon preparations for newborn mice. Nature 258:76-78. [DOI] [PubMed] [Google Scholar]
- 6.Hemmi, H., O. Takeuchi, S. Sato, M. Yamamoto, T. Kaisho, H. Sanjo, T. Kawai, K. Hoshino, K. Takeda, and S. Akira. 2004. The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath, C. M. 2004. The Jak-STAT pathway stimulated by interferon alpha or interferon beta. Sci. STKE 2004:tr10. [DOI] [PubMed] [Google Scholar]
- 8.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 9.Inohara, N., M. Chamaillard, C. McDonald, and G. Nunez. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74:355-383. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 11.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 175:10968-10977. [DOI] [PubMed] [Google Scholar]
- 12.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 13.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 14.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, K. S., and R. A. Flavell. 2004. Shielding the double-edged sword: negative regulation of the innate immune system. J. Leukoc. Biol. 75:428-433. [DOI] [PubMed] [Google Scholar]
- 16.Komuro, A., M. Nagai, N. E. Navin, and M. Sudol. 2003. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 278:33334-33341. [DOI] [PubMed] [Google Scholar]
- 17.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14:432-436. [DOI] [PubMed] [Google Scholar]
- 18.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinon, F., and J. Tschopp. 2005. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 26:447-454. [DOI] [PubMed] [Google Scholar]
- 20.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzhitov, R., and C. A. Janeway, Jr. 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91:295-298. [DOI] [PubMed] [Google Scholar]
- 22.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1171. [DOI] [PubMed] [Google Scholar]
- 23.Perry, A. K., E. K. Chow, J. B. Goodnough, W. C. Yeh, and G. Cheng. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 199:1651-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260-5268. [DOI] [PubMed] [Google Scholar]
- 25.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 26.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 27.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 29.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-Box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 31.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]