Abstract
RNA interference with viral vectors that express short hairpin RNAs (shRNAs) has emerged as a powerful tool for functional genomics and therapeutic purposes. However, little is known about shRNA in vivo processing, accumulation, functional kinetics, and side effects related to shRNA saturation of the cellular gene silencing machinery. Therefore, we constructed first-generation recombinant adenoviruses encoding different shRNAs against murine ATP-binding cassette multidrug resistance protein 2 (Abcc2), which is involved in liver transport of bilirubin to bile, and analyzed Abcc2 silencing kinetics. C57/BL6 mice injected with these viruses showed significant impairment of Abcc2 function for up to 3 weeks, as reflected by increased serum bilirubin levels. The lack of Abcc2 function correlated with a specific reduction of Abcc2 mRNA and with high levels of processed shRNAs targeting Abcc2. Inhibition was lost at longer times postinfection, correlating with a decrease in the accumulation of processed shRNAs. This finding suggests that a minimal amount of processed shRNAs is required for efficient silencing in vivo. This system was also used to evaluate the effect of shRNA expression on the saturation of silencing factors. Saturation of the cellular silencing processing machinery alters the accumulation and functionality of endogenous microRNAs (miRNAs) and pre-miRNAs. However, expression of functional exogenous shRNAs did not change the levels of endogenous miRNAs or their precursors. In summary, this work shows that adenoviral vectors can deliver sufficient shRNAs to mediate inhibition of gene expression without saturating the silencing machinery.
RNA interference (RNAi) is a sequence-specific posttranscriptional gene-silencing process originally described in Caenorhabditis elegans and plants (53). Subsequently, RNAi has been described in many organisms where regulation of gene expression is achieved by inducing DNA heterochromatinization, mRNA cleavage, translation inhibition, or gene deletion (29). It has been calculated that RNAi controls the expression of 30% of human genes, some of which are involved in developmental timing, neuronal and hematopoietic cell fate, stem cell division, and cell death and proliferation (35, 47). Moreover, a clear connection between cancer and RNAi has been recently shown (8, 19, 38, 43, 46).
MicroRNAs (miRNAs) are short, ∼22-nucleotide (nt)-long, noncoding RNAs that function as silencing effector molecules (29). miRNAs are transcribed as long primary transcripts (∼160 nt long or longer, called pri-miRNAs) that undergo a sequential maturation process. First, pri-miRNAs are cleaved in the nucleus by an RNase III called Drosha into imperfectly pairing stem-loop precursors of ∼70 nt called pre-miRNAs (29, 34). The pre-miRNAs are then exported to the cytoplasm by a Ran-GTP-dependent process mediated by exportin 5 (Exp-5) (5, 16, 30, 40, 59). Once in the cytoplasm, Dicer processes pre-miRNAs, rendering mature double-stranded miRNAs (see Fig. 2A) that interact with the RNA-induced silencing complex (RISC) (4, 29, 53). The antisense strand of the miRNA must be incorporated into RISC to mediate sequence-specific mRNA regulation (53).
FIG. 2.
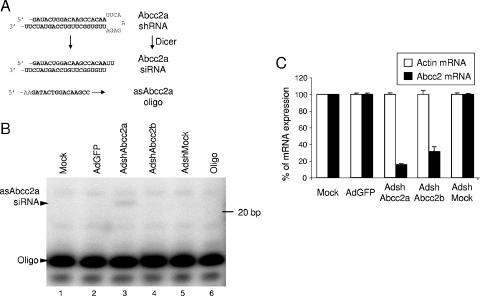
Hepa 1-6 cells infected with AdshAbcc2 show decreased Abcc2 mRNA levels. (A) Schematic representation of shRNA processing detected by primer extension. An shRNA targeting Abcc2 mRNA (Abcc2a shRNA) can be processed by Dicer to an siRNA (Abcc2a siRNA). The accumulation of the antisense strand of Abcc2a siRNA can be detected by hybridization or extension from oligonucleotide asAbcc2a. (B) Detection of the antisense strand of Abcc2a siRNA in AdshAbcc2a-infected cells. Buffer only (Oligo) or RNA isolated from Hepa 1-6 cells (Mock) or cells infected for 72 h with the AdGFP, AdshMock, or AdshAbcc2 virus was subjected to primer extension with oligonucleotide asAbcc2a. The positions of the oligonucleotide and of the antisense strand of Abcc2a siRNA (asAbcc2a siRNA) are indicated to the left. The position of the 20-bp marker is indicated to the right. (C) Abcc2 mRNA levels are decreased in AdshAbcc2-infected cells. Total RNA described in panel B was used to quantify Abcc2 and actin mRNA by LightCycler RT-PCR. The values shown are percentages of Abcc2 and actin mRNA expression compared to those of mock-infected cells. Error bars indicate standard deviations obtained after quantification of three different experiments.
RNAi-based technology is a powerful tool for functional genomic studies and constitutes a promising source for new therapeutic approaches (17). These goals can be achieved by molecules similar to pre-miRNAs and miRNAs called short hairpin RNAs (shRNAs) and small interfering RNAs (siRNAs), respectively. shRNAs are similar in structure to pre-miRNAs and mature following the same Exp-5/Dicer pathway into siRNAs (16, 30, 31, 59). siRNAs are ∼21-nt-long RNA duplexes that interact with RISC to mediate gene silencing similarly to what has been described above for miRNAs (53). However, pairing between siRNAs and target sequences is generally designed to be perfect, activating a RISC-mediated cleavage of the target mRNA, while in most cases the functional binding size between the miRNA and the target mRNA is only 6 to 8 nt, inducing a RISC-mediated inhibition of mRNA translation or a decrease in mRNA stability (53).
Delivery of siRNAs and shRNAs into mammalian cells has been achieved by several means. In cell culture, the simplest method is transfection of chemically synthesized siRNAs or plasmids expressing shRNAs (6, 11). In vivo silencing has been obtained by administration of synthetic siRNAs or by viral vectors expressing shRNAs such as lentiviruses, retroviruses, adeno-associated viruses (AAV), or adenoviruses (21, 36, 42, 49, 52, 55, 57). Viral vectors offer several advantages for delivery of shRNAs, including longer expressions and better transduction efficiencies than siRNA administration (21, 57). Several reports have described inhibition of liver messengers with siRNAs and shRNAs, making the liver a suitable target organ for RNAi approaches (36, 52, 55, 57). Adenoviruses are excellent vectors for liver gene delivery because of their high liver tropism and infection efficiency that allows massive hepatic transduction with a short-term expression profile (51, 56). However, little is known about adenovirus-delivered kinetics of functional shRNAs. Also, RNAi amplification or cell-to-cell transmission, as happens in C. elegans, has never been evaluated in the liver. Finally, undesirable side effects of shRNA expression in the liver have not been studied in detail. Side effects related to shRNA delivery could be associated with off-target gene silencing, induction of interferon response, or saturation of the endogenous RNAi machinery (23, 24, 58).
Several reports have shown that the endogenous RNAi machinery is limiting. Thus, Exp-5 has been saturated in tissue culture cells expressing high levels of shRNAs and overexpression of Exp-5 has been shown to increase RNAi efficiency (58). Saturation of RISC has also been demonstrated in tissue culture cells (23). Also, Dicer function has been inhibited in the presence of double-stranded RNA-binding proteins or by incubation with double-stranded RNAs, such as virus-associated (VA) RNAs (2, 37, 39). VA RNAs are ∼160-nt-long double-stranded RNA molecules expressed in adenovirus-infected cells (41). Thus, adenovirus expressing shRNAs could saturate the cellular silencing machinery by expression of exogenous shRNAs and VA RNAs, as well as expression of adenovirus miRNAs, as has been recently described (2, 3, 48). Also, wild-type adenovirus (AdWT) infection has been shown to inactivate RISC in tissue culture (2). Inhibition of the silencing machinery would decrease the level and functionality of endogenous miRNAs and therefore alter the expression of target cellular genes. This altered expression in the cell could lead to toxicity by different mechanisms. In fact, the expression of miRNAs is lower in cancer cells and it has been suggested that reduced miRNA expression leads to a cancer-specific block of expression that interferes with the normal development of the cells (38).
To study adenovirus-delivered shRNA kinetics and cellular effects, we constructed first-generation recombinant adenoviruses encoding different shRNAs to silence the murine ATP-binding cassette subfamily C member 2 multidrug resistance protein 2 (Abcc2/MRP2/cMOAT) (AdshAbcc2). Abcc2 is a liver export pump located in the canalicular membrane of hepatocytes responsible for secretion of conjugated bilirubin to bile (7, 27, 28). Mutations in the gene for Abcc2 are responsible for Dubin-Johnson syndrome, a hereditary disease characterized by conjugated hyperbilirubinemia (25, 26, 32, 54). We monitored Abcc2 silencing after intravenous administration of AdshAbcc2 in C57/BL6 mice for up to 150 days. By quantitative reverse transcription (RT)-PCR and measurement of serum bilirubin accumulation, we demonstrated Abcc2 knockdown and functional inhibition for 3 weeks. Abcc2 inhibition correlated with higher levels of processed antisense shRNAs targeting Abcc2, while lower nonfunctional accumulation was detected for up to 150 days. Moreover, adenovirus-mediated expression of functional shRNAs did not affect the accumulation or processing of endogenous miRNAs such as let-7, miR16, or miR21. These results indicate that adenovirus vectors can be used to express a sufficient level of shRNAs in mouse liver capable of silencing target genes without inhibition of the cellular RNAi machinery.
MATERIALS AND METHODS
Plasmids.
Enhanced green fluorescent protein (GFP) cDNA was digested with NotI, filled with Klenow, excised with PstI from pEGFP (Promega), and cloned into PstI-SmaI-digested pAdLox (kind gift from S. Hardy) downstream of the cytomegalovirus promoter (18, 45) to generate pAdLoxGFP. We used pSuper (kindly given by R. Agami) to construct the plasmids that express shRNAs (6). Proper oligonucleotide sequences (Sigma) were cloned into the BglII and HindIII sites of pSuper to obtain plasmids expressing shAbcc2a and shAbcc2b (Table 1; Fig. 1A). Similarly, oligonucleotides containing Abcc2-unrelated sequences were used to obtain a plasmid expressing shMock. These plasmids were digested with EcoRI and XhoI, and fragments containing shRNA sequences downstream of the H1 promoter were cloned in the orientation opposite to that of the adenovirus E1 gene into XhoI-EcoRI-opened pAdlox. This generated pAdLoxshAbcc2a, pAdLoxshAbcc2b, and pAdLoxshMock. Positive clones were verified by DNA sequencing (ABI Prism 310 genetic analyzer from Perkin-Elmer).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence | Usea |
|---|---|---|
| pSAbcc2AS | GATCCCCGATACTGGACAAGCCACAATTCAAGAGATTGTGGCTTGTCCAGTATCTTTTTGGAAA | Cloning of pSUPERshAbcc2a |
| pSAbcc2aAS | AGCTTTTCCAAAAAGATACTGGACAAGCCACAATCTCTTGAATTGTGGCTTGTCCAGTATCGGG | Cloning of pSUPERshAbcc2a |
| pSAbcc2bS | GATCCCCTTCTCTACCTATGCACTTGTTCAAGAGACAAGTGCATAGGTAGAGAATTTTTGGAAA | Cloning of pSUPERshAbcc2b |
| pSAbcc2bAS | AGCTTTTCCAAAAATTCTCTACCTATGCACTTGTCTCTTGAACAAGTGCATAGGTAGAGAAGGG | Cloning of pSUPERshAbcc2b |
| Abcc2S197 | CCAAGCAGGTGTTCGTTGTGT | qPCR of Abcc2 mRNA |
| Abcc2AS628 | TGTCATACCAACTAAACGTAACGC | qPCR of Abcc2 mRNA |
| mActin5′ | ACTGCGCTTCTTGCCGC | qPCR of murine actin mRNA |
| mActin3′ | CATGACGCCCTGGTGTC | qPCR of murine actin mRNA |
| asAbcc2a | AAGATACTGGACAAGCC | PE of shAbcc2a, Northern blotting |
| asAbcc2b | AATTCTCTACCTATGCA | PE of shAbcc2b, Northern blotting |
| Let-7 | AAAACTATACAACCTACTAC | PE of Let-7, Northern blotting |
| miR16 | AACGCCAATATTTACGTGC | PE of miR16, Northern blotting |
| miR21 | AGTCAACATCAGTCTGATAA | PE of miR21, Northern blotting |
| pre-miR16 | AGAATCTTAACGCCAATATT | PE of pre-miR16, Northern blotting |
| pre-miR21 | GATTCAACAGTCAACATCAG | PE of pre-miR21, Northern blotting |
| U6 snRNA | TGCTAATCTTCTCTGTATCGT | PE of U6 snRNA, Northern blotting |
| Let-7Arrestor | UGAGGUAGUAGACGCAG | Invader quantification of Let-7 |
| Let-7Probe | CCGTCGCTGCGTCTACTACCTCAGGCUUCGGCC | Invader quantification of Let-7 |
| Let-7Invader | GGCUUCGGCCAACTATACAACT | Invader quantification of Let-7 |
| miR21Arrestor | UAGCUUAUCAGAGUGCGC | Invader quantification of miR21 |
| miR21Probe | AACGAGGCGCACTCTGATAAGCTAGGCUUCGGCC | Invader quantification of miR21 |
| miR21Invader | GGCUUCGGCCTCAACATCAGG | Invader quantification of miR21 |
qPCR, quantitative PCR; PE, primer extension.
FIG. 1.
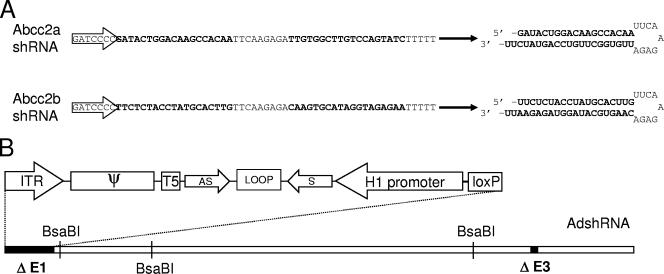
Schematic representation of shRNAs targeting Abcc2 mRNA (Abcc2 shRNAs) and adenovirus expressing Abcc2 shRNAs (AdshAbcc2). (A) Sequence and structure of Abcc2 shRNAs. Two constructs were designed that express shRNAs targeting Abcc2 mRNA sequences. The linear sequence and the predicted folded hairpin are shown for each Abcc2 shRNA. Transcription initiation is indicated by an arrow. Sense and antisense sequences are shown in bold. (B) A first-generation adenovirus genome with E1 and E3 deleted (black boxes) is shown, and the relative positions of BsaBI restriction sites are indicated. The 5′ end of the genome is highlighted above, including the 5′ internal terminal repeat (ITR), the packaging signal (ψ), the loxP site, and, in the opposite direction, the H1 promoter driving the expression of the shRNA formed by sense (s) and antisense (as) sequences separated by a loop.
Cell lines.
Human embryonic kidney 293 cells (adenovirus E1 transformed) and murine hepatoma cell line Hepa 1-6 were obtained from the American Type Culture Collection. The CRE8 selective cell line was kindly provided by S. Hardy. All cell lines were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 100 U/ml streptomycin-penicillin. Cell lines were cultured at 37°C in a 5% CO2 atmosphere. All cell culture reagents were obtained from GIBCO BRL/Life Technologies.
Adenovirus construction and cell infection.
Recombinant adenoviruses expressing GFP (AdGFP) and mock shRNA or shRNAs against murine Abcc2 (AdshAbcc2a and AdshAbcc2b) were constructed with the Cre-lox recombination system (18, 45). Briefly, CRE8 cells were cotransfected by using calcium phosphate with Ψ5 DNA and SfiI-predigested pAdLoxGFP, pAdLoxshMock, pAdLoxshAbcc2a, or pAdLoxshAbcc2b. After 7 to 10 days, lysates from cotransfected cells were used to infect CRE8 cells in order to remove the Ψ5 helper virus contamination. Four passages were performed to ensure efficient ψ5 removal (data not shown). Lysates from the last passages were used to infect 293 cells to amplify adenoviruses. Packaged viral DNA was purified as previously described (18) and analyzed in agarose gels after BsaBI digestion (Fig. 1B). These adenoviruses and an adenovirus that expresses luciferase (AdLuc; kindly provided by J. Wilson) were amplified in 293 cells, purified by double cesium chloride gradient ultracentrifugation, and desalted by dialysis overnight at 4°C against phosphate-buffered saline, pH 7.2 (45). Adenovirus infective unit titers were determined by limiting dilution. Hepa 1-6 cells were infected with adenovirus as previously described, with a multiplicity of infection (MOI) of 600 (45). This MOI was sufficient to infect 99% of the cells as determined by fluorescence-activated cell sorter analysis (data not shown).
Animal treatments and serum analysis.
Five- to 6-week-old C57/BL6 mice (Harlan) received 2 × 109 infective units of adenovirus in 200 μl of saline by intravenous injection into the tail vein. Blood and liver samples were collected at the indicated times postinfection. Mouse blood samples were drawn from the retroorbital sinus and immediately stored in darkness. After 30 to 60 min of incubation at room temperature, samples were centrifuged at 10,000 rpm in a table microcentrifuge and serum samples were stored at −20°C. Bilirubin levels were determined with a total bilirubin kit (ABX Diagnostics) in a Hitachi autoanalyzer (Roche Diagnostics) by following the supplier's recommendations. When indicated, animals received an intravenous injection in the tail vein of 100 μl of 100 mM NaCl and 50 mM Na2CO3 with 10 mg/kg bilirubin (Sigma) 40 min before blood was drawn (22). All experiments with animals were performed by following guidelines from the institutional ethical commission.
Protein analysis.
For liver histological analysis, mice were intravenously injected by the tail vein with 150 μl of AdGFP containing 2 × 109 or 109 infective units. At 7 and 24 days after adenovirus administration, the animals were sacrificed and their livers were excised and fixed overnight in 3.7% paraformaldehyde in CSK buffer (14). Samples were then embedded in Tissue-Tek OCT (Sakura), snap-frozen in liquid nitrogen, and stored at −80°C. For GFP analysis, samples were sliced, dried overnight at room temperature in the dark, mounted with Vectashield 4′,6′-diamidino-2-phenylindole (DAPI) solution (Vector Laboratories), and analyzed with a Nikon fluorescence microscope.
Preparation and analysis of RNA.
Total RNA from Hepa 1-6 cells was purified by the guanidinium method as previously described (9). Frozen livers were homogenized in denaturing solution (9) with an Ultraturrax (Ika-Werke) at 4°C, and total RNA was purified by the guanidinium method.
Total RNA was repurified with an RNeasy Minikit (QIAGEN) for real-time quantitative RT-PCR. The RT reaction was done with Moloney murine leukemia virus reverse transcriptase (Promega) for 1 h at 37°C. Real-time quantitative PCR was performed with a LightCycler system (Roche Diagnostics) with LightCycler Faststart Master DNA SYBR green I (Roche) by following the supplier's instructions. Sequences of forward and reverse primers for Abcc2 mRNA and murine actin mRNA quantification by LightCycler RT-PCR are indicated in Table 1. The amplification program consisted of 1 cycle of 95°C for 1 min, followed by 40 cycles of 95°C for 20 s, 59°C (Abcc2) or 64°C (actin) for 5 s, and 72°C for 20 s (Abcc2) or 15 s (actin). Melting curve analysis was performed at 83°C (Abcc2) or 91°C (actin). The average of triplicate data of each sample was used to calculate the relative change in gene expression after normalization to actin mRNA.
Primer extension analysis of shRNAs, miRNAs, and U6 snRNA was performed with total RNA isolated from Hepa 1-6 cells or from homogenized frozen liver extracts, as previously described (3, 12). Primer sequences are shown in Table 1. Primer extension conditions were set up to work in the linear range. Northern blot assays were done as previously described, with oligonucleotides in Table 1 (3, 13). Invader quantification of let-7 and miR21 was done by following the recommendations of the supplier (Third Wave) (1). U6 snRNA quantification was done in the same experiments as an internal control. The oligonucleotides used are listed in Table 1.
Statistical analysis.
Results are shown as the arithmetic mean ± the standard error of the mean. Statistical comparisons were made by analysis of variance. Differences were deemed significant for a real alpha of 0.05. All statistical analyses were carried out with SPSS v11.0 (SPSS, Inc.).
RESULTS
Adenovirus-mediated Abcc2 silencing in murine hepatoma cells.
Two different shRNAs, targeting Abcc2 mRNA nt +265 to +283 and +311 to +329 (Abcc2a and -b shRNAs, respectively), were designed according to recommended properties (10) and mFold-predicted secondary structure for Abcc2 mRNA (60) (data not shown; Fig. 1A). Abcc2 shRNA sequences were cloned into pSuper (6) and then inserted into the genome of recombinant adenovirus defective in the E1 and E3 early genes (18). This generated adenovirus expressing shRNAs against Abcc2 mRNA (AdshAbcc2a and -b) transcribed from the H1 promoter in the direction opposite to that of the adenovirus 5′ internal terminal repeat (Fig. 1B). A similar adenovirus was generated which expresses an shRNA unrelated to Abcc2 (AdshMock).
In AdshAbcc2-infected cells, the corresponding shRNA should be expressed and processed by Dicer into siRNAs (Fig. 2A). However, only one of the strands of a single siRNA will remain attached to the RISC complex (53). Target Abcc2 mRNA expression should be inhibited if enough RISC complexes incorporate the antisense strand of Abcc2 siRNA. Thus, the presence of this antisense strand was analyzed by primer extension of total RNA isolated from Hepa 1-6 cells infected with the AdshAbcc2 viruses at 600 infective units/cell. This MOI guarantees that all cells have been infected (data not shown). As shown in Fig. 2B, a 21-nt band was detected by primer extension with a specific oligonucleotide that hybridizes to the antisense strand of Abcc2a siRNA (oligonucleotide asAbcc2a, Fig. 2A and Table 1). This band was specific for AdshAbcc2a-infected cells (Fig. 2B, lane 3), as it was not detected in mock-infected cells or in cells infected with AdshAbcc2b, AdshMock, or AdGFP (Fig. 2B, lanes 1 and 2 and lanes 4 and 5). Similar experiments confirmed the presence of the antisense strand of Abcc2b siRNA and the sense strand of Abcc2a and Abcc2b siRNAs (data not shown). However, we were unable to detect unprocessed Abcc2 shRNAs by Northern blot assay or primer extension, indicating efficient processing by Dicer (data not shown).
The detection of the antisense siRNAs correlated with a decrease in Abcc2 mRNA. The levels of Abcc2 mRNA were quantified by LightCycler RT-PCR of RNA isolated at 72 h postinfection from Hepa 1-6 cells mock infected or infected with AdGFP or the AdshAbcc2 viruses. Abcc2 mRNA levels of cells infected with AdshAbcc2a or AdshAbcc2b were 15% and 31%, respectively, of the values observed in AdGFP- or mock-infected cells (Fig. 2C). However, similar amounts of the nontargeted actin mRNA were found in all cases. This ensures integrity of the RNA samples and specificity of Abcc2 mRNA inhibition.
Intravenous administration of the AdshAbcc2 viruses to mice causes hyperbilirubinemia and a decrease in Abcc2 mRNA.
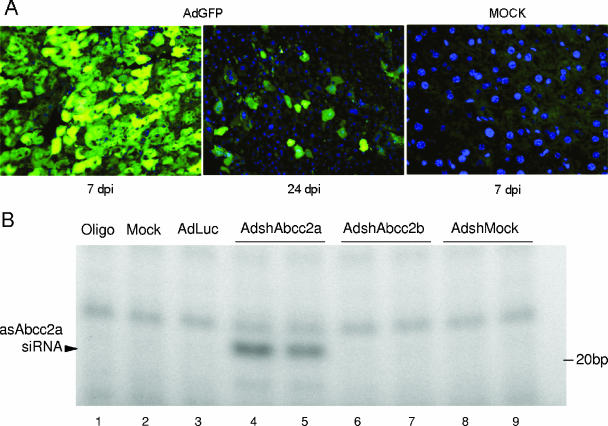
Adenoviruses have been proven to be valuable gene delivery vectors in vivo with special tropism for the liver (51). To analyze if AdshAbcc2 viruses could inhibit Abcc2 mRNA expression in mouse liver, we first set up conditions to infect most liver cells. Thus, increasing amounts of AdGFP were administered by the tail vein to three to five C57/BL6 mice per group (data not shown). Animals were sacrificed 7 to 24 days postinfection (dpi) for histological analysis of liver samples by fluorescence microscopy. Under our experimental conditions, 2 × 109 infective units/mouse is the minimal dose of AdGFP needed to detect GFP expression in more than 95% of the hepatocytes after 7 dpi (Fig. 3A). According to the adenovirus short-term expression properties, less than 5% of the hepatocytes expressed GFP 24 days after AdGFP administration (Fig. 3A) (56). In agreement with these results, a strong light emission signal was observed with a charge-coupled device camera in the liver area of mice injected with AdLuc at 7 dpi but not at 24 dpi (data not shown). These experiments also showed that adenovirus transgene expression increases up to 7 dpi with the vector in C57/BL6 mice (data not shown; 56).
FIG. 3.
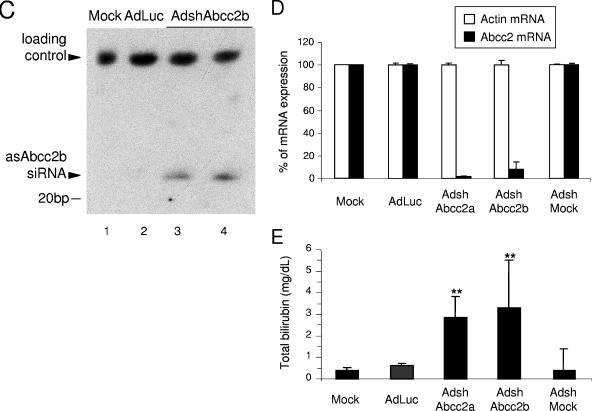
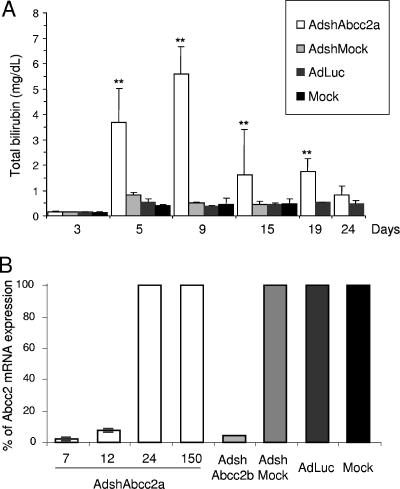
AdshAbcc2 administration in mice affects Abcc2 functionality. (A) Dose required to infect more than 95% of hepatocytes. Mice were mock infected or infected with 2 × 109 infective units of AdGFP/mouse, and liver samples were processed at 7 or 24 dpi. Histological analysis of liver samples stained with DAPI (blue) was visualized in a fluorescence microscope. GFP expression is shown in green. (B, C) The antisense strand of Abcc2 siRNAs is detected in AdshAbcc2-infected animals. Mice were mock infected or infected with AdLuc or AdshMock as controls or infected with AdshAbcc2a or AdshAbcc2b. At 7 days posttreatment, serum and liver samples were analyzed. Buffer only (oligo) or total liver RNA was analyzed by primer extension with oligonucleotide asAbcc2a (B) or by Northern blot assay with oligonucleotide asAbcc2b (C). The position of the antisense strand of Abcc2a siRNA or Abcc2b siRNA (asAbcc2 siRNA) is indicated to the left. The position of the 20-bp marker is indicated to the right. (D) Abcc2 mRNA levels are decreased in AdshAbcc2-infected animals. Total RNA described in panel B was used to quantify Abcc2 and actin mRNAs by LightCycler RT-PCR. Results were plotted as indicated in Fig. 2C. (E) AdshAbcc2 administration to mice causes hyperbilirubinemia. Mice described in panel B were injected with synthetic bilirubin, and 40 min later, serum samples were collected to quantify bilirubin levels. Error bars indicate standard deviations obtained with five to seven animals.
Thus, five to seven C57/BL6 mice per group were injected intravenously with 2 × 109 infective units of AdLuc, AdshAbcc2a, AdshAbcc2b, or AdshMock. A group of mice receiving the vehicle alone (saline) was also included as a negative control. At 7 dpi, the animals were sacrificed for collection of serum and liver samples. RNA isolated from liver extracts was analyzed by primer extension or Northern blot assay. As was observed in Hepa 1-6 cells (Fig. 2B), a 21-nt specific band was extended from oligonucleotide asAbcc2a (Table 1) incubated with liver RNA from AdshAbcc2a-infected animals (Fig. 3B, lanes 4 and 5). A similar band was observed when RNA from AdshAbcc2b-infected animals was hybridized with oligonucleotide asAbcc2b (Table 1; Fig. 3C, lanes 3 and 4). These bands were specific since they were not detected in mock-infected animals or in animals infected with the other adenoviruses tested (Fig. 3B, lanes 2 and 3 and lanes 6 to 9, and C, lanes 1 and 2). The presence of the antisense strand of Abcc2 siRNAs correlated with a decrease in Abcc2 mRNA accumulation. Quantification of Abcc2 mRNA levels by quantitative RT-PCR from liver RNA samples showed that Abcc2 mRNA amounts were 5% and 12% of the levels of the control groups in AdshAbcc2a- and AdshAbcc2b-receiving mice, respectively (Fig. 3D). This decrease was specific, as similar levels of the nontargeted actin mRNA were found in all cases (Fig. 3D).
Abcc2 transports serum bilirubin to bile in hepatocytes. Alterations in the Abcc2 gene are the cause of Dubin-Johnson syndrome, which is characterized by hyperbilirubinemia and jaundice (7, 25, 26, 32). It could then be expected that Abcc2 mRNA silencing would cause a decrease in Abcc2 expression, reduced transport of bilirubin to bile, and hyperbilirubinemia. To assess whether AdshAbcc2a and AdshAbcc2b lead to Abcc2 functional inhibition in vivo, bilirubin was measured in serum samples collected from three to five mice per group infected as described above. We observed a significant increase in serum bilirubin in mice receiving AdshAbcc2a or AdshAbcc2b (0.90 ± 0.66 mg/dl) compared to control animals (0.11 ± 0.05 mg/dl). Serum bilirubin forms as a by-product of erythrocyte turnover, which, under normal conditions, does not saturate the machinery that transports bilirubin to bile. To study the effect of AdshAbcc2 viruses with saturating bilirubin, five to seven mice per group infected as described above received 10 mg/kg of synthetic bilirubin intravenously and blood was collected 40 min later (22). After this time, synthetic bilirubin was completely cleared in control mice, while mice injected with AdshAbcc2a or AdshAbcc2b showed marked hyperbilirubinemia (Fig. 3E).
AdshAbcc2 liver transduction impairs bilirubin clearance for up to 3 weeks.
Once we detected Abcc2 functional inhibition and bilirubin clearance impairment 7 days after AdshAbcc2 injection, we analyzed in vivo Abcc2 silencing kinetics following AdshAbcc2 administration. Five to seven C57/BL6 mice per group were mock injected or injected with 2 × 109 infective units of AdshAbcc2a or with AdshMock or AdLuc as negative controls. Liver and serum samples were collected on different days postinfection. Serum bilirubin was quantified 40 min after synthetic bilirubin injection on days −1, 3, 5, 9, 19, 24, and 150 post virus administration. We found normal levels of serum bilirubin in all animals at −1 and 3 dpi (Fig. 4A and data not shown). This was expected since expression from adenovirus is observed from 48 h posttransduction (56) and some time should be required from Abcc2 mRNA degradation to functional impairment of Abcc2 protein. Remarkably, we detected high bilirubin values from day 5 until day 19 postinjection of AdshAbcc2a (Fig. 4A). In these animals, the increase in bilirubin was maximal at 9 dpi and was no longer significant at day 24 after AdshAbcc2a administration (Fig. 4A). In control animals or in all animals analyzed at 150 dpi, serum bilirubin showed values similar to baseline levels (Fig. 4A and data not shown).
FIG. 4.
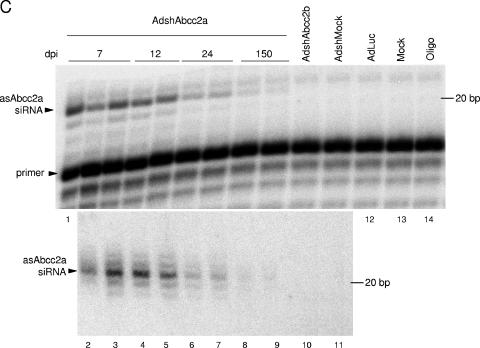
AdshAbcc2 liver transduction affects Abcc2 functionality for up to 3 weeks. (A) AdshAbcc2-infected animals show hyperbilirubinemia for 19 days. Mice were mock injected or injected with 2 × 109 infective units of AdshMock or AdLuc as negative controls or with AdshAbcc2a. Bilirubin was quantified in serum samples collected on days 3, 5, 9, 15, 19, and 24 post virus administration and 40 min after synthetic bilirubin injection. Error bars indicate standard deviations. Asterisks highlight significant differences. (B) Abcc2 mRNA is decreased in AdshAbcc2-treated animals for up to 12 days. Liver RNA was isolated from animals treated as described in panel A, at 7, 12, 24, and 150 dpi. Liver RNA was also isolated from animals transduced with AdshAbcc2b for 7 days. Liver RNA was used to quantify Abcc2 mRNA by LightCycler RT-PCR. Results are plotted as indicated in Fig. 2C. (C) Antisense siRNAs against Abcc2 mRNA were detected at 150 dpi with AdshAbcc2a. Liver RNA described in panel B or buffer (oligo) was analyzed by primer extension with oligonucleotide Abcc2aAS (top). Some of the samples were also analyzed by Northern blot assay hybridized to oligonucleotide asAbcc2a (bottom). The positions of the oligonucleotide and the antisense Abcc2a siRNA (asAbcc2a siRNA) are indicated to the left. The position of the 20-bp marker is indicated to the right.
RNA was isolated from liver samples taken from the animals described above at 7, 12, 24, and 150 dpi. Liver RNA was also isolated at 7 dpi from three mice transduced with AdshAbcc2b. Abcc2 mRNA levels quantified by real-time RT-PCR showed a significant reduction in mice infected with AdshAbcc2b or with AdshAbcc2a for 7 or 12 days (Fig. 4B). This result is in agreement with the functional inhibition of Abcc2 shown above. Control animals and all animals analyzed after 24 or 150 dpi showed normal levels of Abcc2 mRNA (Fig. 4B and data not shown). Also, the inhibition was specific for Abcc2 mRNA, as normal levels of actin mRNA were found in all cases.
The accumulation of small RNAs against Abcc2 mRNA was analyzed in the liver RNA samples described above by Northern blot assay and primer extension with oligonucleotide asAbcc2a. Both techniques detected high levels of antisense Abcc2a siRNAs in mice transduced with AdshAbcc2a at 7 and 12 dpi (Fig. 4C, lanes 1 to 5). Thus, the decrease in Abcc2 mRNA and the impairment of bilirubin clearance correlated with a strong accumulation of siRNAs against Abcc2 mRNA. Small RNAs were not detected with oligonucleotide asAbcc2a in control samples (Fig. 4C, lanes 10 to 14). Interestingly, we were able to detect a weak signal corresponding to antisense Abcc2a siRNA at day 24, and even day 150, postinfection (Fig. 4C, lanes 6 to 9). However, these levels of small RNA were not sufficient to silence Abcc2 mRNA since bilirubin clearance and Abcc2 mRNA values were similar to those of controls (Fig. 4A and B).
Expression of functional shRNAs does not affect processing or accumulation of endogenous liver miRNAs.
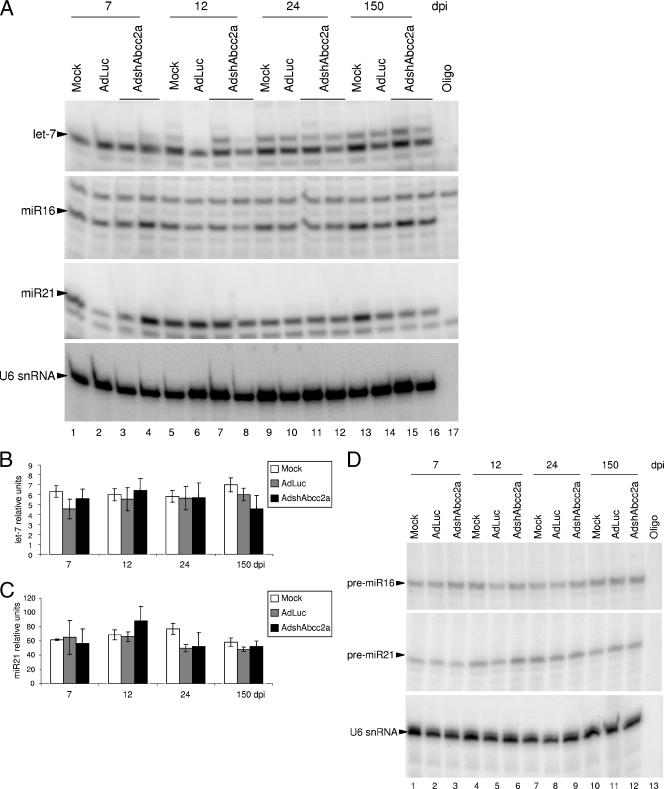
Because shRNAs and miRNAs share the same processing and nuclear export machinery (30, 31), we explored whether functional expression of shRNAs in the liver could modify endogenous miRNA processing or accumulation. Thus, miRNAs were quantified in liver RNA samples isolated from mice mock transduced or transduced with 2 × 109 infective units of AdLuc or AdshAbcc2a at 7, 12, 24, or 150 dpi. These RNAs were normalized to U6 snRNA, and accumulation of several endogenous miRNAs was analyzed. Out of seven cellular miRNAs chosen, only let-7, miR16, miR21, and liver-specific miR122 were expressed to detectable amounts in mouse liver cells (33). Accumulation of these miRNAs was analyzed by primer extension and Northern blot assay with specific oligonucleotides (Fig. 5A; Table 1; data not shown). Quantification of the results obtained with primer extensions and Northern blot assays indicated variability in the amount of endogenous miRNAs in different animals. However, statistical analysis revealed nonsignificant differences between liver extracts from AdLuc- or mock-treated mice and AdshAbcc2a-injected mice (data not shown). Also, there were no differences among controls and mice treated with AdshAbcc2b and analyzed at 7, 12, or 24 days posttreatment (data not shown). The Invader technique to quantify endogenous miRNAs was also used to reduce the mouse-to-mouse variability observed by Northern blot assay or primer extension (1). When quantification of let-7 or miR21 was carried out with Invader assays, similar results were obtained but with a lower variability. No differences were observed between controls or AdshAbcc2a-infected animals analyzed at 7, 12, 24, or 150 days posttreatment (Fig. 5B and C).
FIG. 5.
Expression of functional shRNAs does not affect processing or accumulation of endogenous liver miRNAs. (A) Accumulation of miRNAs is not altered in AdshAbcc2-infected cells as detected by primer extension. Buffer (oligo) or liver RNA described in Fig. 5 was analyzed by primer extension with oligonucleotides that hybridize to let-7, miR16, miR21, or U6 snRNA (as a loading control). (B, C) let-7 (B) and miR21 (C) accumulation is not altered in AdshAbcc2-infected cells as detected by Invader assay. RNAs described in panel A were analyzed by Invader assay and plotted as relative units. Error bars indicate standard deviations obtained with five to seven animals. (D) Abnormal accumulation of precursor miRNAs is not detected in AdshAbcc2-infected cells. Buffer (oligo) or liver RNA, as described in panel A, was analyzed by primer extension with oligonucleotides that hybridize to pre-miR16, pre-miR21, or U6 snRNA (as a loading control).
Competition of functional shRNAs for the silencing processing machinery could still induce the accumulation of miRNA precursors (pre-miRNAs). Thus, a blockade of Exp-5 or Dicer should alter the localization and accumulation of pre-miRNAs. Therefore, accumulation of pre-miR16 and pre-miR21 was analyzed by primer extension (Fig. 5D) and Northern blot assay (data not shown) with specific antisense oligonucleotides (Table 1). Again, quantification of pre-miR16 or pre-miR21 did not reveal significant differences between liver extracts from control mice or AdshAbcc2a-transduced mice analyzed at 7, 12, 24, or 150 days posttreatment (data not shown).
DISCUSSION
One of the major obstacles to using siRNAs in vivo is their efficient delivery into the target tissue or organ. The experience accumulated in recent years in the gene therapy field has led to the optimization of several viruses as gene delivery vectors. Thus, viral vectors expressing shRNAs have proven to be valuable vehicles to achieve RNAi (21, 49, 55, 57). However, detailed studies analyzing the kinetics of inhibition or the side effects related to functional shRNA expression in vivo have not been carried out. To address these questions, we have chosen first-generation adenoviruses that silence the hepatocellular Abcc2 transporter in mouse liver. With these vectors, we were able to induce Abcc2 silencing in tissue cultured liver cells (Fig. 2C) and in murine liver (Fig. 3D and 4B), whereas other approaches such as hydrodynamic administration of plasmid coding for shAbcc2 had failed (data not shown).
AdshAbcc2 caused Abcc2 silencing from day 5 up to day 19 after vector administration (Fig. 4A). As Abcc2 transports bilirubin into bile, silencing of Abcc2 was detected by hyperbilirubinemia (Fig. 3D and 4A). Hyperbilirubinemia is also detected in patients with Dubin-Johnson syndrome, in whom Abcc2 function is impaired (25, 26, 32, 54). Thus, our AdshAbcc2 adenoviruses could be administered in mice to generate transient knockdowns that could serve as mouse models for Dubin-Johnson syndrome and for the study of the pathophysiological consequences derived from Abcc2 dysfunction. Moreover, Abcc2, among other ATP-binding cassette transporters, has important clinical implications since it is responsible for excretion of drugs and xenotoxins and for resistance to multiple chemotherapeutic agents (20). Thus, our system could enable a better definition of the implication of this transporter in specific metabolic functions and may serve to enhance the sensitivity of liver cancer cells to chemotherapeutic agents.
Impairment of Abcc2 function correlates with a decrease in liver Abcc2 mRNA as detected by RT-PCR (Fig. 3D and 4B). Abcc2 mRNA levels decreased at days 7 and 12 after adenovirus injection but were back to normal at day 24, when the increase in blood bilirubin was not significant (Fig. 4A and B). This kinetics of inhibition follows a classical short-term expression pattern of first-generation adenoviral vectors (Fig. 3A). Silencing is mediated by the antisense strand of processed shRNAs. These antisense small RNA mediators were detected by Northern blot assay or primer extension up to 150 days after AdshAbcc2 administration in mice (Fig. 4C). This suggests the existence of an siRNA reservoir and/or the persistence of few copies of adenovirus genomes in liver cells. We favor the latter hypothesis since we were able to detect viral genomes by quantitative PCR in DNA extracts from murine livers 150 days after injection with AdshAbcc2a (data not shown). However, the low levels of antisense Abcc2 siRNA detected 24 or 150 days post AdshAbcc2 administration were not sufficient to silence Abcc2 (Fig. 4). Only the levels detected at days 7 and 12 were efficient inhibitors of Abcc2 (Fig. 4). These results indicate that there is a threshold amount of siRNAs required for efficient silencing. Also, these functional levels need to be expressed in the majority of cells. Thus, efficient silencing was not detected when we used fewer than 2 × 109 infective units of AdshAbcc2/mouse (data not shown). These data also show that mouse liver cells do not support efficient amplification or spread of siRNA signals, as has been shown in worms or plants (53).
Application of RNAi to gene therapy approaches has first to face the challenge of avoiding side effects. RNAi-induced toxicity has been associated with three major causes: off-target inhibition, activation of the interferon response, and saturation of the cellular silencing machinery. Several authors have questioned the specificity of RNAi, as expression of some nontarget genes may be decreased (24). We have not conducted detailed experiments to rule out off-target inhibition in our system. However, the expression of specific cellular genes such as that for murine actin was not altered by expression of Abcc2 shRNAs (Fig. 2C and 3D; data not shown). Also, all mice survived throughout the study and showed normal behavior without apparent loss of weight or other signs of illness. Liver functionality was not affected, as ascites, hepatomegaly, hepatocyte necrosis, or liver fibrosis was not apparent after autopsies were carried out. In addition, bilirubin levels were not altered in mice treated with adenovirus expressing siRNAs other than the specific inhibitors of Abcc2 (Fig. 3E and 4A). Finally, some sequences of siRNAs and shRNAs or high doses of these small RNAs can induce the interferon response (24, 44). However, we could not detect induction of the interferon pathway by quantitative RT-PCR of 2′5′oligoadenylate synthetase mRNA with RNA isolated from Hepa 1-6 cells or murine livers infected with AdshAbcc2 (data not shown).
Numerous authors have pointed out the need for more information about shRNA toxicity by saturation of the cellular silencing machinery (17, 29, 58). Their worries are based on the saturability of silencing factors such as Exp-5, Dicer, and RISC (23, 39, 58). Processing and function of shRNAs require the cellular silencing machinery. Thus, overexpression of shRNAs can saturate Exp-5 in tissue culture cells and in vivo (15, 58). During the process of writing this report, a study was published showing that high levels of AAV-delivered shRNAs induce liver injury in mice (15). The authors showed that overexpressed shRNAs saturate Exp-5, leading to a decrease in the accumulation and functionality of liver miRNAs. We hypothesized that a similar result could be obtained with adenovirus-delivered shRNAs. Besides, adenovirus infection could affect the silencing machinery by additional mechanisms. On the one hand, first-generation adenoviruses express VA RNAs that could saturate Exp-5 (5, 16, 50; data not shown). On the other hand, VA RNAs can inhibit Dicer function and can be processed to functional viral miRNAs that could help in saturation of the silencing machinery (2, 3, 39, 48). Finally, other adenovirus miRNAs could be expressed from other sequences in the adenovirus genome (2, 3, 39, 48).
To address the saturation of the silencing machinery, we analyzed the processing and accumulation of cellular miRNAs in murine livers infected with first-generation adenovirus vectors that express shRNAs. The cellular miRNAs chosen were let-7, miR16, miR21, and miR122, as they are well expressed in mouse liver. These miRNAs and their pre-miRNAs were analyzed by Northern blot assay, primer extension, and Invader (Fig. 5 and data not shown). Even when adenovirus-mediated shRNA delivery had inhibited the expression of the target gene for 1 week (from day 5 to day 12, when miRNAs were analyzed), no differences were observed in the accumulation of let-7, miR16, and miR21 miRNAs or pre-miRNAs. Also, we did not observe alterations in the expression of miRNA-regulated genes such as that for Ras, Notch, or RelA (data not shown). These experiments were performed under conditions in which more than 95% of the cells were expressing adenovirus-delivered shRNAs. This result indicates that efficient silencing in vivo can be achieved without altering miRNA biogenesis or accumulation or the functionality of the RNAi machinery.
A similar result was described by Grimm et al. when they used minimal doses of AAV expressing shRNAs (AAV/shRNA) or, in most cases, when they delivered higher doses of these vectors but the stem length of the shRNAs was 19 nt. These conditions are identical to our experimental settings. However, Grimm et al. also reported that doses of AAV/shRNA fivefold higher than the dose required for complete liver transduction (1012 particles of AAV/shRNA) led to toxicity in 73% of the animals because of oversaturation of the cellular miRNA pathway. It would be of interest to analyze if higher levels of adenovirus expressing shRNAs can also induce toxicity in our system. To address this question, we have tried to produce high titers of AdshAbcc2a and -b. However, all of our attempts ended in production of adenovirus enriched in infective particles that had lost expression of Abcc2 shRNA, probably because these sequences have been lost by recombination (data not shown). Also, in our experiments, doses of AdGFP or AdWT higher than 4 × 109 infective units per mouse led to toxicity that was unrelated to inhibition of the silencing pathway (data not shown). Therefore, we evaluated the effect of high levels of adenovirus expressing shRNAs in 293 cells, which allow replication of first-generation adenovirus vectors. In uninfected cells, the shRNA-mediated inhibition of a luciferase reporter was fivefold whereas the inhibition dropped to two- to threefold in cells infected for 3 days with AdWT, AdGFP, or AdshAbcc2a (data not shown). Similar results have been obtained by others with AdWT (2, 39). These results indicate that adenovirus-infected cells may have impaired shRNA processing or function. However, in the same cells we did not detect an alteration of the processing or function of endogenous miRNAs (data not shown). We believe that longer times postinfection may be required to affect these miRNAs. In fact, a decrease in mouse liver miRNAs has only been detected after 2 weeks of infection with AAV overexpressing shRNAs (15).
In summary, we show that adenoviral vectors are suitable for delivery of shRNAs and allow the silencing of a liver transporter for up to 3 weeks. The silencing of the Abcc2 RNA correlated with inhibition of the bilirubin export function and with the presence of high levels of shRNAs expressed from the adenoviral vectors. Moreover, we showed the existence of a threshold in the levels of processed antisense shRNA required for efficient target silencing. Importantly, the data presented in this work indicate that adenovirus expressing functional shRNAs may efficiently inhibit target genes without alteration or saturation of the miRNA silencing machinery.
Acknowledgments
This work was supported by CICYT (SAF2003-01804), FIS (01/1310 and 01/0843), Instituto Carlos III (C03/02 and PI051098), and the Education and Health Department of the Navarra Government and through the agreement between FIMA and the UTE project CIMA. O.A. is a holder of an FPI fellowship from the Spanish Ministry of Education. I.N. and M.V. are recipients of an FIMA postdoctoral fellowship.
We are grateful to S. Hardy for the adenovirus Cre-lox System, R. Agami for pSuper, and J. Wilson for AdLuc. We thank M. Zaratiegui for helpful scientific discussions and help with statistical analysis, E. Casales for technical assistance, L. Martínez and A. Vales for bilirubin quantifications, P. Alzuguren for serum sample collection support, and P. Garcés for tissue sample processing. We are also grateful to C. Smerdou, R. Hernández-Alcoceba, M. D. Weitzman, J. Suh, and P. Gastaminza for critical readings of the manuscript.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Allawi, H. T., J. E. Dahlberg, S. Olson, E. Lund, M. Olson, W. P. Ma, T. Takova, B. P. Neri, and V. I. Lyamichev. 2004. Quantitation of microRNAs using a modified Invader assay. RNA 10:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, M. G., P. C. Haasnoot, N. Xu, S. Berenjian, B. Berkhout, and G. Akusjarvi. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79:9556-9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, O., N. Razquin, M. Zaratiegui, I. Narvaiza, and P. Fortes. 2006. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 80:1376-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 5.Bohnsack, M. T., K. Czaplinski, and D. Gorlich. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Büchler, M., J. Konig, M. Brom, J. Kartenbeck, H. Spring, T. Horie, and D. Keppler. 1996. cDNA cloning of the hepatocyte canalicular isoform of the multidrug resistance protein, cMrp, reveals a novel conjugate export pump deficient in hyperbilirubinemic mutant rats. J. Biol. Chem. 271:15091-15098. [DOI] [PubMed] [Google Scholar]
- 8.Caldas, C., and J. D. Brenton. 2005. Sizing up miRNAs as cancer genes. Nat. Med. 11:712-714. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 12.Fortes, P., D. Bilbao-Cortes, M. Fornerod, G. Rigaut, W. Raymond, B. Seraphin, and I. W. Mattaj. 1999. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 13:2425-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortes, P., Y. Cuevas, F. Guan, P. Liu, S. Pentlicky, S. P. Jung, M. L. Martinez-Chantar, J. Prieto, D. Rowe, and S. I. Gunderson. 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl. Acad. Sci. USA 100:8264-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes, P., A. I. Lamond, and J. Ortin. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 76(Pt. 4):1001-1007. [DOI] [PubMed] [Google Scholar]
- 15.Grimm, D., K. L. Streetz, C. L. Jopling, T. A. Storm, K. Pandey, C. R. Davis, P. Marion, F. Salazar, and M. A. Kay. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537-541. [DOI] [PubMed] [Google Scholar]
- 16.Gwizdek, C., B. Ossareh-Nazari, A. M. Brownawell, A. Doglio, E. Bertrand, I. G. Macara, and C. Dargemont. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278:5505-5508. [DOI] [PubMed] [Google Scholar]
- 17.Hannon, G. J., and J. J. Rossi. 2004. Unlocking the potential of the human genome with RNA interference. Nature 431:371-378. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, S., M. Kitamura, T. Harris-Stansil, Y. Dai, and M. L. Phipps. 1997. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann, U., and H. K. Kroemer. 2004. The ABC transporters MDR1 and MRP2: multiple functions in disposition of xenobiotics and drug resistance. Drug Metab. Rev. 36:669-701. [DOI] [PubMed] [Google Scholar]
- 21.Hommel, J. D., R. M. Sears, D. Georgescu, D. L. Simmons, and R. J. DiLeone. 2003. Local gene knockdown in the brain using viral-mediated RNA interference. Nat. Med. 9:1539-1544. [DOI] [PubMed] [Google Scholar]
- 22.Huang, W., J. Zhang, S. S. Chua, M. Qatanani, Y. Han, R. Granata, and D. D. Moore. 2003. Induction of bilirubin clearance by the constitutive androstane receptor (CAR). Proc. Natl. Acad. Sci. USA 100:4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutvágner, G., M. J. Simard, C. C. Mello, and P. D. Zamore. 2004. Sequence-specific inhibition of small RNA function. PLoS. Biol. 2:E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, A. L., and P. S. Linsley. 2004. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 20:521-524. [DOI] [PubMed] [Google Scholar]
- 25.Kartenbeck, J., U. Leuschner, R. Mayer, and D. Keppler. 1996. Absence of the canalicular isoform of the MRP gene-encoded conjugate export pump from the hepatocytes in Dubin-Johnson syndrome. Hepatology 23:1061-1066. [DOI] [PubMed] [Google Scholar]
- 26.Keitel, V., J. Kartenbeck, A. T. Nies, H. Spring, M. Brom, and D. Keppler. 2000. Impaired protein maturation of the conjugate export pump multidrug resistance protein 2 as a consequence of a deletion mutation in Dubin-Johnson syndrome. Hepatology 32:1317-1328. [DOI] [PubMed] [Google Scholar]
- 27.Keppler, D., and J. Konig. 1997. Hepatic canalicular membrane 5: expression and localization of the conjugate export pump encoded by the MRP2 (cMRP/cMOAT) gene in liver. FASEB J. 11:509-516. [DOI] [PubMed] [Google Scholar]
- 28.Keppler, D., J. Konig, and M. Buchler. 1997. The canalicular multidrug resistance protein, cMRP/MRP2, a novel conjugate export pump expressed in the apical membrane of hepatocytes. Adv. Enzyme Regul. 37:321-333. [DOI] [PubMed] [Google Scholar]
- 29.Kim, V. N. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6:376-385. [DOI] [PubMed] [Google Scholar]
- 30.Kim, V. N. 2004. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 14:156-159. [DOI] [PubMed] [Google Scholar]
- 31.Kim, V. N. 2005. Small RNAs: classification, biogenesis, and function. Mol. Cells 19:1-15. [PubMed] [Google Scholar]
- 32.König, J., A. T. Nies, Y. Cui, I. Leier, and D. Keppler. 1999. Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochim. Biophys. Acta 1461:377-394. [DOI] [PubMed] [Google Scholar]
- 33.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 34.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, D. L., J. E. Hagstrom, A. G. Loomis, J. A. Wolff, and H. Herweijer. 2002. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 32:107-108. [DOI] [PubMed] [Google Scholar]
- 37.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834-838. [DOI] [PubMed] [Google Scholar]
- 39.Lu, S., and B. R. Cullen. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 78:12868-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 303:95-98. [DOI] [PubMed] [Google Scholar]
- 41.Ma, Y., and M. B. Mathews. 1996. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J. Virol. 70:5083-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 43.Meltzer, P. S. 2005. Cancer genomics: small RNAs with big impacts. Nature 435:745-746. [DOI] [PubMed] [Google Scholar]
- 44.Moss, E. G., and J. M. Taylor. 2003. Small-interfering RNAs in the radar of the interferon system. Nat. Cell Biol. 5:771-772. [DOI] [PubMed] [Google Scholar]
- 45.Narvaiza, I., G. Mazzolini, M. Barajas, M. Duarte, M. Zaratiegui, C. Qian, I. Melero, and J. Prieto. 2000. Intratumoral coinjection of two adenoviruses, one encoding the chemokine IFN-γ-inducible protein-10 and another encoding IL-12, results in marked antitumoral synergy. J. Immunol. 164:3112-3122. [DOI] [PubMed] [Google Scholar]
- 46.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 47.Pasquinelli, A. E., S. Hunter, and J. Bracht. 2005. MicroRNAs: a developing story. Curr. Opin. Genet. Dev. 15:200-205. [DOI] [PubMed] [Google Scholar]
- 48.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 49.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 50.Sano, M., Y. Kato, and K. Taira. 2006. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 580:1553-1564. [DOI] [PubMed] [Google Scholar]
- 51.Smith, T., N. Idamakanti, H. Kylefjord, M. Rollence, L. King, M. Kaloss, M. Kaleko, and S. C. Stevenson. 2002. In vivo hepatic adenoviral gene delivery occurs independently of the coxsackievirus-adenovirus receptor. Mol. Ther. 5:770-779. [DOI] [PubMed] [Google Scholar]
- 52.Soutschek, J., A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan, and H. P. Vornlocher. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432:173-178. [DOI] [PubMed] [Google Scholar]
- 53.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19:517-529. [DOI] [PubMed] [Google Scholar]
- 54.Tsujii, H., J. Konig, D. Rost, B. Stockel, U. Leuschner, and D. Keppler. 1999. Exon-intron organization of the human multidrug-resistance protein 2 (MRP2) gene mutated in Dubin-Johnson syndrome. Gastroenterology 117:653-660. [DOI] [PubMed] [Google Scholar]
- 55.Uprichard, S. L., B. Boyd, A. Althage, and F. V. Chisari. 2005. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. USA 102:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolff, G., S. Worgall, N. van Rooijen, W. R. Song, B. G. Harvey, and R. G. Crystal. 1997. Enhancement of in vivo adenovirus-mediated gene transfer and expression by prior depletion of tissue macrophages in the target organ. J. Virol. 71:624-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia, H., Q. Mao, H. L. Paulson, and B. L. Davidson. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20:1006-1010. [DOI] [PubMed] [Google Scholar]
- 58.Yi, R., B. P. Doehle, Y. Qin, I. G. Macara, and B. R. Cullen. 2005. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]