Abstract
Membrane penetration by nonenveloped reoviruses is mediated by the outer-capsid protein, μ1 (76 kDa). Previous evidence has suggested that an autolytic cleavage in μ1 allows the release of its N-terminally myristoylated peptide, μ1N (4 kDa), which probably then interacts with the target-cell membrane. A substantial rearrangement of the remaining portion of μ1, μ1C (72 kDa), must also have occurred for μ1N to be released, and some regions in μ1C may make additional contacts with the membrane. We describe here a particle-free system to study conformational rearrangements of μ1. We show that removal of the protector protein σ3 is not sufficient to trigger rearrangement of free μ1 trimer and that free μ1 trimer undergoes conformational changes similar to those of particle-associated μ1 when induced by similar conditions. The μ1 rearrangements require separation of the μ1 trimer head domains but not the μ1N/C autocleavage. We have also obtained a relatively homogeneous form of the structurally rearranged μ1 (μ1*) in solution. It is an elongated monomer and retains substantial α-helix content. We have identified a protease-resistant ∼23-kDa fragment of μ1*, which contains the largely α-helical regions designated domains I and II in the conformation of μ1 prior to rearrangement. We propose that the μ1 conformational changes preceding membrane penetration or disruption during cell entry involve (i) separation of the β-barrel head domains in the μ1 trimer, (ii) autolytic cleavage at the μ1N/C junction, associated with partial unfolding of μ1C and release of μ1N, and (iii) refolding of the N-terminal helical domains of μ1C, with which μ1N was previously complexed, accompanied by dissociation of the μ1 trimer.
Mammalian reoviruses have a multishelled architecture that segregates distinct functions into specific protein layers (20). The inner-capsid particle, or core, contains the complete viral transcription machinery. It enters the cell during infection but does not disassemble further. Rather, it transcribes, caps, and exports mRNA into the cytoplasm from each of the 10 viral double-stranded RNA genome segments. The core, approximately 700 Å in diameter, has an icosahedrally symmetric structure, based on 120 copies of a principal shell protein, λ1, and 12 pentameric turrets of the multidomain capping enzyme λ2 (8, 26). In virions, proteins μ1 and σ3, associated as heterohexamers (μ13σ33), coat the outer surface of the core by forming a layer intercalated between the λ2 turrets. The role of this outer layer is to mediate membrane penetration or disruption and to deposit cores into the cytoplasm (3, 13, 16, 19, 21, 22, 24, 25).
The initial step in reovirus penetration is proteolytic removal of σ3 (2, 9, 21, 22, 27). The proteolysis, which primes μ1 for its role in membrane translocation of the core, can occur either in the intestinal lumen, prior to receptor binding, or in endocytic vesicles. It yields an infectious subviral particle (ISVP). ISVPs can also be produced in vitro by treatment of virions with chymotrypsin. A transient yet distinctive particle, designated ISVP*, appears to be an essential intermediate in the subsequent penetration process (3, 25). Its properties include release of the cell-attachment protein σ1 (which projects from the λ2 turrets on ISVPs), rearrangement of μ1 into a protease-sensitive conformation, release of the N-terminal myristoylated peptide μ1N (see below), and derepression of core-particle transcriptional activity.
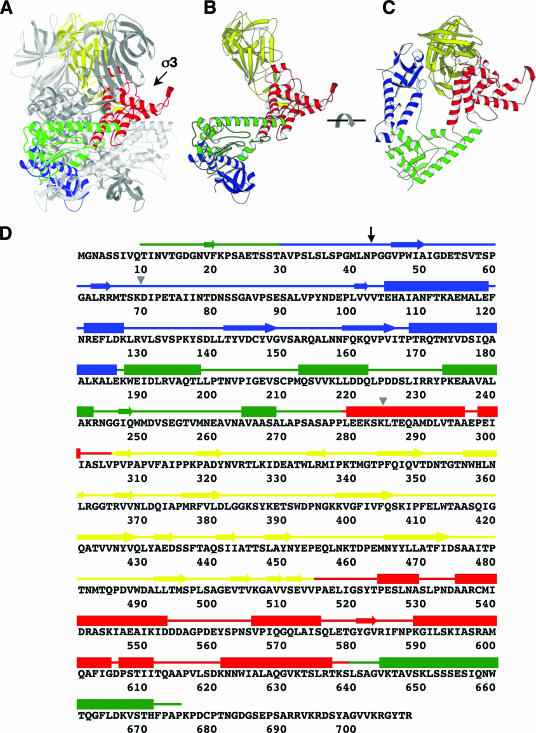
Analysis of the molecular mechanism of reovirus core translocation requires an understanding of the structural details of μ1 in the various conformations that it adopts during cell entry. The μ1 protein itself is a 76-kDa polypeptide, myristoylated at its N terminus (24). An autolytic cleavage at some stage between viral assembly and entry creates fragments μ1N (residues 2 to 42, plus the myristoyl group) and μ1C (residues 43 to 708) (24). The fragments remain associated with each other and with the ISVP (15, 24). Mutations in μ1 that prevent autocleavage also block membrane penetration (25). The crystal structure of μ13σ33 shows that μ1 is a tightly associated trimer, with the three σ3 subunits projecting axially from its periphery (Fig. 1) (15). The central segment of each μ1 polypeptide chain folds into a jelly-roll β-barrel; the N- and C-terminal parts of the chain fold together and associate with the corresponding parts of the other two chains to form a largely α-helical pedestal, which supports the apically clustered barrel domains (15). The myristoyl group at the N terminus of μ1N is probably tucked into an elongated, hydrophobic pocket on a lateral face of this pedestal; the autocleavage site at the C terminus of μ1N is buried within the pedestal interior (15, 29). It is necessary for infectivity (of cores recoated with μ13σ33) that μ1N is cleaved from μ1C; it is therefore plausible to suppose that productive infection requires release of μ1N for membrane targeting through its myristoyl group, and such release of μ1N from the ISVP* has indeed been demonstrated (25). Proximity of the three myristoyl groups and the three autocleavage sites, in the conformation of μ1 found on the virion surface as well as in the crystal structure, further suggests that cleavage, release of μ1N, and membrane targeting of μ1N are all related events.
FIG. 1.
μ1 prior to structural rearrangement. (A) Side view of a μ1 trimer (as in the crystal structure of the μ13σ33 heterohexamer) without bound σ3, with one subunit highlighted (domain I, blue; domain II, green; domain III, red; domain IV, yellow). The other two μ1 subunits are in gray. One σ3 binding site is indicated by an arrow. (B) Side view of an isolated μ1 subunit, colored as described for panel A. (C) Top view of an isolated μ1 subunit, colored as described for panel A. (D) Amino acid sequence and secondary structure elements of reovirus μ1. The amino acid sequence of μ1 from reovirus serotype T1L is shown (5). Secondary structure elements, derived from the crystal structure of the μ13σ33 heterohexamer (15), are presented as tubes for α-helices and arrows for β-stands. The four domains of μ1 are colored as described above. The black arrow indicates the position of the autolytic cleavage, and the gray triangles indicate the starting and ending positions of the proteolysis-resistant fragment of μ1*.
Inspection of the folded structure of the μ1 trimer pedestal shows that, should release of μ1N from μ1C indeed be a critical step, major conformational rearrangements of μ1 are likely to ensue and that these rearrangements are probably fundamental aspects of the penetration mechanism. The N-terminal segment of μ1 is tightly wound into the pedestal structure, which would have to unfold to some extent in order to release μ1N and which might then refold into what is likely to be a very different conformation (15). This sort of reorganization underlies how the membrane fusion proteins of enveloped viruses facilitate entry (12), and it also appears to be an essential property of the rotavirus penetration protein VP4 (7). In order to determine a rearranged structure, we have established a particle-free method to promote the conformational change in μ1, by using conditions similar to those found to induce the ISVP→ISVP* transition (3). We find that μ1 treated in this manner has properties similar to those of ISVP*-associated μ1 and that the rearrangement requires separation of the β-barrel head domains but not the μ1N/C autocleavage. We have, moreover, obtained a relatively homogeneous form of the rearranged μ1 for biophysical characterization in solution.
MATERIALS AND METHODS
Cloning, protein expression, and purification of μ13σ33 heterohexamers.
Wild-type μ13σ33 heterohexamer was prepared by coexpression of the type 1 Lang (T1L) μ1 protein and σ3 protein in insect cells infected with a recombinant baculovirus carrying the T1L M2 and S4 genes, which encode μ1 and σ3 proteins, respectively, as previously described (5, 15). In brief, sf21 cells were grown in Hink's TNM-FH insect medium (JRH) supplemented with 10% (vol/vol) fetal bovine serum (Sigma), infected at one PFU per cell, and harvested at 72 h postinfection. The cells were resuspended by lysis buffer (20 mM Tris-Cl, pH 8.5, 2 mM MgCl2, 150 mM KCl, 5 mM dithiothreitol [DTT], 2 mM benzamidine, 2 μg/ml pepstatin A, 2 μg/ml leupeptin, and 0.2 mg/ml ethanolic phenylmethylsulfonyl fluoride [PMSF], supplemented with complete EDTA-free protease inhibitor cocktail [Roche]) at 1 × 108 cells/ml lysis buffer and disrupted by sonication on ice. Cell debris was removed by ultracentrifugation (180 kg; 1 h at 4°C). The supernatant was applied to a Q-Sepharose high-performance (HP) column (Amersham Biosciences) that was equilibrated with buffer A (20 mM Tris-Cl, pH 8.5, 2 mM MgCl2, 5 mM DTT) and 150 mM NaCl. μ13σ33 heterohexamer was eluted with a linear salt gradient from 150 mM to 450 mM NaCl. Then, 3 M (NH4)2SO4 was added dropwise to the fractions containing μ13σ33 to achieve a final concentration of 0.7 M. The sample was loaded onto a phenyl-Sepharose HP column (Amersham Biosciences) equilibrated in buffer A and 0.7 M (NH4)2SO4, and eluted with a linear gradient from 0.7 M to 0 M (NH4)2SO4. The eluted protein was diluted 1:3 with buffer A, applied to a Mono Q column (Amersham Biosciences) equilibrated with buffer A and 200 mM NaCl, and eluted with a linear salt gradient from 200 mM to 350 mM NaCl. The protein complex was further purified by gel filtration chromatography using a Superdex 200 column (Amersham Biosciences) in a buffer containing 20 mM Bicine-HCl, pH 9.0, 2 mM MgCl2, 10 mM DTT, 100 mM NaCl, and 0.02% NaN3. The purified protein can be concentrated to 3 mg/ml and stored at −80°C. The (μ1-N42A)3σ33 heterohexamer was also prepared as described above.
Amino acid changes of μ1 Ser385→Cys and Ser436→Cys for mutant DS1, as well as Thr325→Cys and Thr445→Cys for mutant DS2, were introduced into cDNA copies of the T1L M2 gene by site-directed mutagenesis using the QuickChange method, according to the manufacturer's instructions (Stratagene). Bsu36I-MluI restriction fragments conferring the desired mutations were subcloned into the shuttle plasmid pFastbacDUAL-M2L-S4L for recombinant baculovirus production using the Bac-to-Bac system (Invitrogen) (5, 6). The disulfide mutant (μ1-DS1)3σ33 and (μ1-DS2)3σ33 heterohexamers were purified similarly to the method described above except that DTT was absent from all the buffers.
Conformational changes of μ1 in solution.
To remove σ3 protein from μ13σ33 heterohexamer, the purified wild-type or mutant protein complex (0.3 mg/ml) was incubated with various amounts of chymotrypsin in a 20-μl reaction mix containing 20 mM Tris-Cl, pH 8.5, and 100 mM NaCl or CsCl, at room temperature for 30 min. The protease digestions were quenched with the addition of 5 mM PMSF (final concentration). To trigger conformational changes in μ1, the reaction mixes were incubated at 42°C for 30 min and moved onto ice. For the trypsin sensitivity assay, the reaction mixes were divided into two parts, and one part (10 μl) was treated with trypsin at 0.1 mg/ml on ice for 1 h and quenched with the addition of 0.3 mg/ml soybean trypsin inhibitor (Worthington). The samples were disrupted by boiling for 2 to 5 min in gel loading buffer and subjected to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Immunoprecipitation.
Monoclonal antibody (MAb) 4A3, specific for structurally rearranged μ1, was expressed and purified as described previously (28). Protein A-conjugated beads (100 μl) (Pierce) were incubated with 10 μg antibody 4A3 and 200 μl immunoprecipitation (IP) buffer (containing 10 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 1% NP-40) at room temperature for 2 h and washed with 500 μl IP buffer three times. Then, 20-μl samples were added to the antibody-bound beads with 180 μl ice-cold IP buffer. The reaction mixes were incubated with rotary mixing at 4°C for 1 h and washed with 500 μl ice-cold IP buffer six times. Proteins were released from the beads by boiling in gel loading buffer for 2 to 5 min and then subjected to SDS-PAGE analysis.
Generation of recoated cores containing WT μ1 and μ1-DS1.
Recoated cores containing wild-type (WT) or μ1-DS1 together with the wild-type σ3 protein were generated from purified T1L cores and insect cell lysates containing recombinant baculovirus-expressed μ13σ33 as described previously (5, 6). Recoated cores (μ1-DS1) containing Cys385-Cys436 disulfide bonds were purified from insect cell lysates by centrifugation on CsCl gradients and dialysis against virion buffer (10 mM Tris-Cl, pH 7.5, 140 mM NaCl, 1.5 mM MgCl2) as described previously (5, 6). Recoated cores (μ1-DS1) lacking Cys385-Cys436 disulfide bonds were purified in a similar manner, except that CsCl gradients and dialysis buffer were supplemented to contain 40 mM β-mercaptoethanol immediately before use. Recoated cores (WT μ1) used in the experiments whose results can be seen in Fig. 3 were purified in parallel with recoated cores (μ1-DS1) as described above.
FIG. 3.
An engineered interchain disulfide bond arrests μ1 conformational change and virus-induced hemolysis. (A) Surface representation of μ1 trimer, viewed from the outside relative to the orientation in virus particles. Black bars represent approximate positions of residues 385 and 436, which form an interchain disulfide bond in μ1-DS1. (B) Disulfide bond formation in μ1-DS1. ISVP-like particles generated by chymotryptic digestion of recoated cores and containing either wild-type μ1 (μ1-WT) or μ1-DS1 (5 × 1010 particles per lane) were subjected to nonreducing SDS-PAGE. Positions of viral proteins are marked. (C) Disulfide-bond-dependent arrest of hemolysis mediated by ISVP-like particles (μ1-DS1). Recoated cores of (μ1-WT) or (μ1-DS1), purified in the absence or presence of 40 mM β-mercaptoethanol (β-ME), were converted to ISVP-like particles by treatment with chymotrypsin. The ISVP-like particles (5 × 1012 particles per ml) were then incubated with bovine RBCs (3% [vol/vol]) at 37°C for 20 min in the absence or presence of 40 mM β-ME). The extent of hemolysis was determined as described in Materials and Methods. (D) Status of μ1 conformational change in hemolysis reactions described for panel C. Trypsin sensitivity assays were carried out to test whether wild-type or mutant μ1 had undergone a conformational change; 10-μl aliquots from each sample were left untreated or incubated with trypsin (100 μg/ml) for 30 min on ice and then subjected to reducing SDS-PAGE.
Generation of ISVP-like particles from recoated cores.
Nonpurified ISVP-like particles were obtained by digesting recoated cores in virion buffer at a concentration of 5 × 1012 to 1 × 1013 particles/ml with Nα-p-tosyl-l-lysine chloromethyl ketone-treated chymotrypsin (200 μg/ml) for 10 to 20 min at 37°C. Digestion was stopped by the addition of PMSF (2 to 5 mM) at 4°C.
Hemolysis assay.
The capacities of nonpurified ISVP-like particles obtained from recoated cores to mediate hemolysis were determined as described previously (4). Briefly, washed citrated bovine calf red blood cells (RBCs; 3% [vol/vol]) were incubated with viral particles at the indicated concentrations in virion buffer containing 300 mM CsCl for 1 h at 37°C, and the extent of hemoglobin release into the supernatant was measured relative to that of controls containing only virion buffer and RBCs (0%) or water and RBCs (100%).
Purification of μ1*.
To ensure complete digestion of σ3 and minimal cleavage of μ1, μ13σ33 heterohexamer was incubated with chymotrypsin at a mass ratio of 1:4 (chymotrypsin to μ13σ33 heterohexamer) in a 3-ml reaction mix containing 20 mM Tris-Cl, pH 8.5, 100 mM CsCl, and 5% foscholine-16 (Anatrace) on ice for 30 min, and protease digestion was quenched by the addition of 5 mM PMSF. Conformational rearrangement of μ1 was triggered by incubation of the reaction mix at 42°C for 30 min. The sample was immediately applied to a size exclusion column (Superdex 200; Amersham Biosciences) in a buffer containing 50 mM Bicine-HCl, pH 9.0, 10 mM DTT, 100 mM NaCl, and 0.01% foscholine-16. The purified protein can be concentrated up to 10 mg/ml and stored at −80°C. The purified protein was analyzed by matrix-assisted laser desorption ionization mass spectrometry (MS) and N-terminal sequencing (HHMI mass spectrometry laboratory).
Light scattering.
The static light=scattering system consists of an 18-angle light-scattering detector (Dawn EOS; Wyatt Technology) in combination with a refractive index detector (Optilab Rex; Wyatt Technology). Light-scattering data were collected from about 50 μg of purified μ1* at room temperature using a Superdex 200 column (Amersham Biosciences) in a running buffer containing 50 mM Bicine-HCl, pH 9.0, 10 mM DTT, 100 mM NaCl, and 0.01% foscholine-16. Data analysis was carried out using ASTRA 5 software (Wyatt Technology) (11) after calibration and normalization with monomeric bovine serum albumin (Sigma) in the same running buffer.
CD spectrometry.
The circular dichroism (CD) spectrum of purified μ1* at a 0.5-mg/ml concentration was measured between wavelengths of 200 and 260 nm at 25°C in a buffer containing 20 mM sodium phosphate, pH 8.0, 100 mM NaCl, 0.5 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride], and 0.01% foscholine-16. A secondary-structure prediction program was used to estimate the amounts of different secondary-structure motifs (1). For the melting experiments, the CD spectrum of μ1* at wavelength 222 nm was collected from 25°C to 95°C.
Limited proteolysis.
For limited proteolysis of structurally rearranged μ1, we incubated 10 μg of purified μ1* with various amounts of trypsin in a 20-μl reaction mix containing 50 mM Bicine-HCl, pH 9.0, 10 mM DTT, 100 mM NaCl, and 0.01% foscholine-16 on ice for 1 h. The reactions were quenched with the addition of 1 mg/ml soybean trypsin inhibitor and divided in two parts. One set of samples was subjected to SDS-PAGE followed by standard GelCode Blue (Pierce) staining and visual inspection. Another set of samples was applied to an SDS-PAGE gel and transferred to polyvinylidene difluoride membrane in a buffer containing 50 mM CAPS, pH 10.0, and 10% methanol (MeOH). The polyvinylidene difluoride membrane was stained with 0.025% Coomassie brilliant blue R-250 (Sigma) in 40% MeOH until samples were seen and then destained with 50% MeOH and air dried. Protein bands were cut from the dried membrane and sent for N-terminal sequencing (Tufts Core Facility). Limited proteolysis samples were also applied to matrix-assisted laser desorption ionization mass spectrometry (Tufts Core Facility).
RESULTS
Removal of σ3 is necessary but not sufficient to trigger μ1 rearrangement in solution.
To enable in vitro analysis of μ1 structural rearrangements and the ensuing membrane penetration, we sought to acquire the penetration-inducing form of μ1 (rearranged μ1, or μ1*) free of a reovirus particle. To obtain soluble protein from insect cells using a recombinant baculovirus vector, μ1 must be coexpressed with σ3, and the resulting free heterohexamer (μ13σ33, μ1 trimer plus three σ3 monomers) is then stable in solution (5, 15). Although σ3 functions as a “molecular clamp” to inhibit conformational change in the μ1 trimer (15), it appears that removal of σ3 does not directly induce μ1 rearrangements on a virus particle but rather renders the μ1 trimers metastable, i.e., prone to undergoing conformational changes when suitably promoted (3). Interactions between adjacent μ1 trimers in the viral surface lattice, as well as those between the trimers and the viral core proteins λ2 and σ2 (29), may contribute to determining this metastability, and we anticipated that free μ1 trimer (μ13), after removal of σ3 from free μ13σ33 heterohexamer, might be unstable.
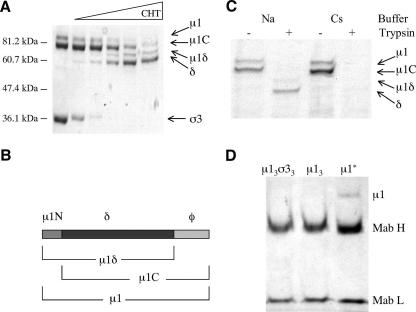
To test whether removal of σ3 is sufficient to trigger μ1 conformational changes in solution, we expressed and purified wild-type free μ13σ33 as described previously (15). Limited proteolysis by chymotrypsin led to complete digestion of σ3 while leaving μ1 essentially intact (Fig. 2A, lanes 3 and 4). Note that two different kinds of cleavage occur in μ1, chymotryptic proteolysis in a loop between residues 581 and 582 (left exposed by the loss of σ3), creating the fragments μ1δ and φ (Fig. 1A and D and 2B) (22), and an autocleavage between residues 42 and 43, which occurs during sample preparation for SDS-PAGE (15, 23) (Fig. 2B). We therefore observed four large μ1 fragments on the gel: intact μ1, μ1C, μ1δ, and δ (Fig. 2A and B). The smaller fragments μ1N and φ were not observed on the gel, due to its low percentage of cross-linking and an inability to resolve species with a molecular mass less than about 15 kDa. Moreover, the free μ13 appeared to be stable in vitro after proteolytic removal of σ3, as indicated by its overall insensitivity to proteolytic cleavage (Fig. 2C, lane 2) (3). The gel-filtration chromatographic profile of the μ1 trimer indicates that its three subunits remain associated with each other (L. Zhang and S. C. Harrison, unpublished data). The μ1 trimer is poorly soluble, however, and tends to aggregate over a wide range of pHs, ionic strengths, and temperatures (L. Zhang and S. C. Harrison, unpublished data). Nondenaturing detergents, such as foscholine (used in the experiments noted above whose results are unpublished to help solubilize rearranged μ1) or β-octyl glucoside, did not inhibit aggregation of the μ1 trimer (L. Zhang and S. C. Harrison, unpublished data).
FIG. 2.
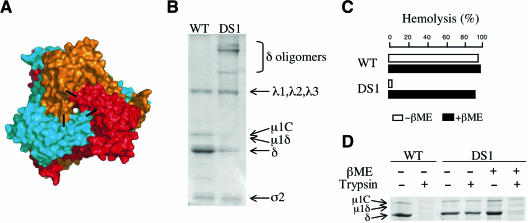
Removal of σ3 and conformational change of μ1 in solution. (A) Removal of σ3 by digestion with chymotrypsin (CHT). The μ13σ33 heterohexamer is treated with increasing amounts of chymotrypsin (first lane from left, untreated μ13σ33). (B) Diagram of μ1 and its fragments. The autocleavage (between residues 42 and 43), which occurs during sample preparation, and the δ/φ cleavage by chymotrypsin (between residues 581 and 582) create three larger μ1 cleavage products, μ1C, μ1δ, and δ, and two smaller fragments, μ1N and φ. (C) In vitro induction of a conformational change in μ1. Free μ1 trimers, prepared as described in Materials and Methods, were incubated at 42°C for 30 min in different buffers containing either NaCl (Na) or CsCl (Cs). Each sample was treated with trypsin to detect the conformational change. (D) Structurally rearranged μ1 interacts with MAb 4A3 (28), a conformation-specific antibody. Antibody pull-down assays of wild-type μ13σ33 heterohexamer, μ1 trimer (μ13), and rearranged μ1 (μ1*). The bands for the heavy (H) and light (L) chains of MAb 4A3 are indicated.
A conformational change in free μ13 similar to one undergone by particle-associated μ13 in the ISVP→ISVP* transition.
One characteristic of the reovirus ISVP→ISVP* transition is an increase in μ1 protease sensitivity, resulting from the generation of a new conformer of μ1 by structural reorganization (μ1→ μ1*) (3). μ1 trimers on the surface of reovirus T1L ISVPs undergo a conformational change at elevated temperatures (32°C and above) in the presence of K+, Rb+, or Cs+ but not Li+ or Na+ ions (3, 19; K. S. Myers, M. A. Agosto, J. K. Middleton, J. Yin, and M. L. Nibert, unpublished data). To investigate whether free μ13 undergoes structural rearrangements under similar triggering conditions, we carried out trypsin sensitivity assays on the free trimers. Incubation of free μ13 in the presence of Cs+ ions at elevated temperatures indeed yielded a protease-sensitive μ1 conformer (μ1*) (Fig. 2C). Moreover, this species interacted with a conformation-specific MAb that recognizes rearranged μ1 on the surface of an ISVP* (3). Neither free μ13σ33 nor unheated μ13 bound this MAb (Fig. 2D). These results suggest that the conformational change in free μ13 heated gently in the presence of Cs+ ions resembles the change that μ1 undergoes on the ISVP surface when treated similarly.
The conformational changes in μ13 require separation of μ1 head domains.
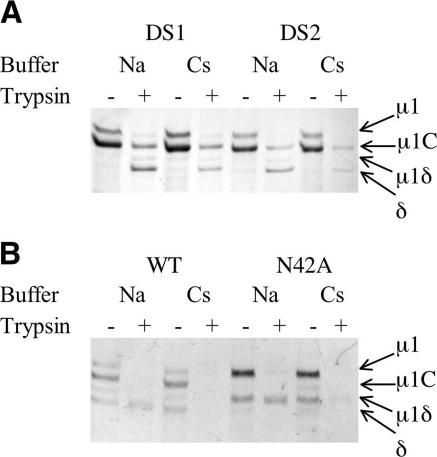
Based on analysis of the crystal structure (15), two different Cys double mutations have been introduced into μ1 at positions 385/436 (μ1-DS1) and 325/445 (μ1-DS2) (Fig. 1A and D and 3A). Both double mutants involve the introduction of a pair of Cys residues that can form disulfide bonds under oxidizing conditions and link adjacent β-barrel head domains (domain IV) (Fig. 3A). These μ1 disulfide-bond mutants have been used to examine whether separation of the μ1 head domains is required for the heat- and Cs+-induced conformational changes. ISVP-like particles derived from reovirus cores recoated with μ1-DS1 and maintained in an oxidizing environment in which the disulfide bonds are formed (Fig. 3B) show no increase in μ1 protease sensitivity and fail to disrupt membranes, as determined by hemolysis (Fig. 3C and D). In contrast, ISVP-like particles derived from reovirus cores recoated with μ1-DS1 and maintained in a reducing environment in which the disulfide bonds are not formed exhibit μ1 protease sensitivity and hemolysis comparable to those exhibited by ISVPs containing wild-type μ1 (Fig. 3C and D). We therefore prepared μ13σ33 containing μ1-DS1 and μ1-DS2, using the same protocol as for wild-type heterohexamers but omitting reducing agents. Protease sensitivity assays showed that these two disulfide bonds inhibit the conformational changes in free μ13 (Fig. 4A), just as μ1-DS1 inhibits the ISVP→ISVP* transition when incorporated into μ1 on the surface of a recoated particle. The similarity of the requirements for the conformational changes in solution and on the ISVP surface supports our conclusion that μ1 undergoes the same structural transition under both circumstances.
FIG. 4.
Requirements for the μ1 conformational change in solution. Trypsin sensitivity assays were carried out to test whether free mutant μ1 had undergone a conformational change. Free wild-type and mutant μ1 trimers [μ13, (μ1-DS1)3, (μ1-DS2)3, and (μ1-N42A)3], prepared as described in Materials and Methods, were incubated at 42°C for 30 min in different buffers containing either NaCl (Na) or CsCl (Cs). Each sample was treated with trypsin to detect the conformational change. (A) The μ1 conformational change requires separation of the μ1 head domains. (B) The μ1 conformational change does not require autocleavage at the μ1N/C junction. Note that the δ/φ cleavage is more extensive here than in the experiment whose results are shown in Fig. 2C, probably because of slightly different chymotrypsin to μ13σ33 heterohexamer ratios.
μ1 conformational change does not require autolytic cleavage at the μ1N/C junction.
Like its wild-type counterpart, the autocleavage-inactive μ1-N42A trimer, derived by chymotryptic removal of σ3 from the (μ1-N42A)3σ33 heterohexamer, undergoes a heat- and Cs+-induced conformational change (Fig. 4B). This result shows that structural rearrangements in μ1 do not require autocleavage at the μ1N/C junction. It also affords a further parallel between studies of functional properties of recoated cores and conformational properties of free μ1 trimer, as reovirus cores recoated with the (μ1-N42A)3σ33 heterohexamer fail to enter cells but do exhibit hallmarks of the ISVP→ISVP* transition (25). The crystal structure of μ13σ33 has shown that autocleavage can occur (perhaps slowly) without conformational change and even without removal of σ3 (15). Thus, structural rearrangement and autocleavage of μ1 are not strictly dependent on each other, although the former may facilitate the latter (23).
Purification of μ1*.
Proteolytic removal of σ3 appears to result in exposure of hydrophobic surfaces on μ1 (3). We found that the μ1 trimer aggregates over time in a variety of buffers, spanning a range of pHs and ionic strengths (L. Zhang and S. C. Harrison, unpublished). It is thus inefficient to purify μ13 prior to the transition to μ1*, trigger the transition in solution, and repurify the rearranged μ1. We therefore attempted first to trigger the μ1 rearrangement without purifying μ13 from the limited chymotryptic digestion mixture and then to use gel-filtration chromatography to isolate μ1* directly. We could obtain a relatively soluble and homogeneous form of μ1* by using foscholine, a lipid-like detergent, to protect its hydrophobic surface. We used a minimal amount of protease to remove σ3 so that the proteolytic cleavage site at the δ/φ junction of μ1 was mostly intact, triggered the μ1 conformational rearrangement in the presence of 5% foscholine (wt/vol) immediately after removal of σ3, and recovered about one-third of the μ1 as a rearranged species by using gel-filtration chromatography to purify the product (in a buffer containing foscholine-12 or foscholine-16 at a concentration of 1.5 times its critical micelle concentration or higher). Purified μ1* appears to be relatively stable in detergent solution, as shown by gel-filtration chromatography after storage at 4°C for about a week (Fig. 5A). Like μ1* on the surface of ISVP* particles (3) and unpurified μ1* in solution (Fig. 2C), purified μ1* is protease sensitive and binds the rearrangement-specific MAb described above (Fig. 5B). These results support our conclusion that we have indeed obtained in solution a μ1 conformer that has the properties of μ1* on ISVP* particles. Moreover, N-terminal sequencing and mass spectrometry show that purified μ1* contains a polypeptide of residues 43 to 691 (Fig. 1D). These data suggest that the autocleavage has occurred at the μ1N/C junction in purified μ1*, and μ1N has likely been released from purified μ1* (25), although our results described above show that the autocleavage is not required for the conformational rearrangement. The extreme C terminus of μ1 has also been cleaved in a disordered region (residue 691) (Fig. 1D) (15), consistent with previous findings (18, 21). It is not clear whether φ, if produced by chymotrypsin during σ3 removal, still associates with the δ fragment in the rearranged conformation of μ1.
FIG. 5.
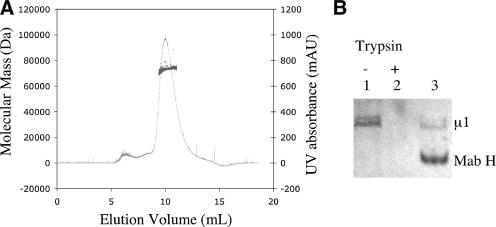
Purification of rearranged μ1 (μ1*). (A) Rearranged μ1 is a monomer in solution. Size exclusion chromatographic profile of purified μ1* is shown by its UV absorbance (black curve), and the estimated native molecular mass of μ1* calculated by the coupled static light-scattering system is also shown (gray dots). AU, absorbance units. (B) Purified μ1* is trypsin sensitive and interacts with a conformation-specific antibody. Lane 1 and 2, trypsin sensitivity assay of purified μ1*. Lane 3, antibody pull-down assay of purified μ1*. The band for the heavy chain (H) of MAb 4A3 is indicated.
μ1* is a monomer in solution.
We used multiangle light scattering (11) to measure the molecular mass of μ1* in a buffer containing 0.01% foscholine-16 (Fig. 5A). The result, 73.4 kDa, lies between the expected molecular masses of a μ1 monomer (74.4 kDa, ending at residue 691) and a μ1C monomer (70.1 kDa, ending at residue 691). By contrast, its apparent mass estimated from the elution volume on gel-filtration chromatography is much larger, about 800 kDa (Fig. 5A). Indeed, μ1* elutes in an even smaller volume than the μ13σ33 heterohexamer (330 kDa) in the same buffer (Zhang and Harrison, unpublished). We conclude that μ1* is a very elongated molecule.
μ1* retains substantial α-helix content.
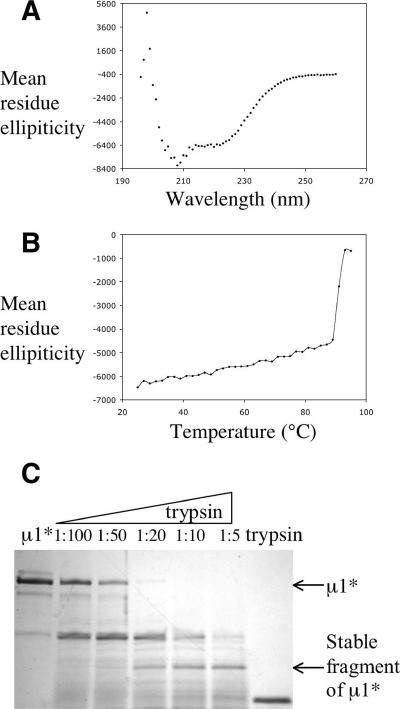
The CD spectrum of purified μ1* has strong negative peaks at 208 nm and 222 nm (Fig. 6A). We estimate from the CD spectrum, using the program of Andrade et al. (1), that μ1* contains about 21% α-helix structure and about 23% β-strand structure. In the heterohexamer, μ1 has three helical domains (I to III), with contributions from both the N- and C-terminal parts of the polypeptide chain at its base and a β-barrel domain (IV) from the middle of the chain, containing about 200 amino acids, at its apex (Fig. 1D) (15). About 27% of the μ1 residues are in α-helices, and 18% are in β-strands. Inspection of the structure suggests that the β-barrel domain is unlikely to unfold or rearrange but that the helical domains might be prone to do so. Indeed, domains I and II incorporate μ1N in a tightly folded pattern, and the release of μ1N would require unfolding (Fig. 1A). The percent β-strand calculated from the CD spectrum is in accord with these predictions. Moreover, a substantial fraction of α-helix present in μ1 appears to remain in μ1*. The ratio of ellipticities at 222 nm and 208 nm can be used as an index of the presence of a coiled-coil conformation (14): the CD spectrum of μ1* has a θ222/θ208 ratio close to 0.8 (Fig. 6A), as expected for distributed helices rather than oligomeric coiled-coils. This is the anticipated result for a monomeric protein. The temperature dependence of the CD spectrum of μ1* (Fig. 6B) shows that the helical domains unfold at about 90°C, the temperature at which θ222 decreases abruptly. The helical domains are therefore very thermostable.
FIG. 6.
Rearranged μ1 is a helical protein. (A) CD spectrum of rearranged μ1 at room temperature. (B) CD melting curve of rearranged μ1 at 222 nm. (C) Limited proteolysis of rearranged μ1. μ1* was treated with increasing amounts of trypsin; the mass ratio of trypsin to μ1* is indicated for each lane (first lane from left, untreated μ1*; last lane from left, trypsin alone). Rearranged μ1 and a relatively stable fragment of μ1* are indicated by arrows.
A stable μ1* fragment.
We used limited proteolysis with trypsin, followed by N-terminal sequencing and MS, to identify stable fragments of μ1*. A fragment with a molecular mass of about 23.4 kDa detected by MS extends from residue 70 to residue 284 (Fig. 1D and 6C). Thus, it derives primarily from domains I and II (Fig. 1). The size of this stable fragment is consistent with the CD measurements, as it could account for much of the estimated α-helix content of μ1*. Although the μ1* monomer is more protease sensitive than the unrearranged μ1 trimer, this fragment of μ1* is still relatively protease resistant, also consistent with our conclusion from the data in Fig. 6B that it has a well-ordered conformation.
DISCUSSION
In this report, we describe characteristics of the conformational rearrangements in the reovirus penetration protein, μ1. We demonstrate that under similar priming and promoting (or triggering) conditions, free μ1 in solution undergoes conformational changes similar to the μ1 rearrangements during the ISVP→ISVP* transition. Removal of the σ3 protector protein primes μ1 for structural rearrangement, i.e., proteolysis of σ3 protein is necessary but not sufficient to trigger μ1 rearrangement in solution (Fig. 7). The metastability of the μ1 trimer after σ3 removal (that is, its retention in the conformation present in μ13σ33 until triggering) was thought to be contributed by the interactions between adjacent μ1 trimers on the virion lattice and between μ1 trimers and the viral core proteins (15). Our results suggest instead that interactions within the μ1 trimer itself contribute substantially to its metastability.
FIG. 7.
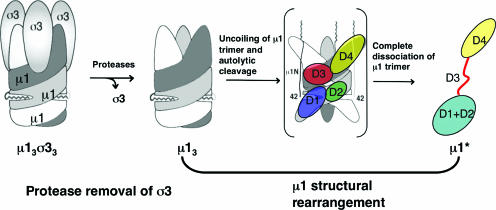
Model of μ1 conformational change during reovirus entry. Proteolytic removal of σ3 primes trimeric μ1 to undergo a conformational change in the endosome. The structural rearrangements involve uncoiling of the μ1 trimer and an autolytic cleavage, resulting in release of the N-terminally myristoylated μ1N peptide, which can then associate with the endosomal membrane. μ1C must unfold, at least in part, to release μ1N; it may, in the process, dissociate from the other two subunits of the trimer and refold as a monomer. Evidence in this paper suggests that the top β-barrel domain (domain IV) remains in the same conformation as in the μ13σ33 heterohexamer, that the middle domain (domain III) unfolds and become flexible, and that the bottom domains, domains I and II, refold into a monomeric, largely helical structure. The color scheme for domains I to IV matches that described in the legend to Fig. 1. Note that for a clearer view of the proposed intermediate state of μ1 during the structural rearrangements, positions of domains I and II are labeled for one subunit of μ1 (in light gray) and those of domains III and IV are labeled for another subunit (in dark gray).
It is not clear what promotes or triggers μ1 conformational changes during infection in vivo. Exposure of reovirus ISVPs to elevated temperature in the presence of larger monovalent cations promotes the ISVP→ISVP* transition in vitro (3). Our results confirm that under these in vitro conditions, we can produce in solution a distinct μ1 conformer (μ1*) that mimics the structurally rearranged μ1 conformer on the ISVP* surface. A host-factor interaction by μ1N is unlikely to be the principal trigger of the transition because the crystal structure of μ13σ33 heterohexamer shows that μ1N is tightly coiled into the base of the μ1 subunit and that its myristoyl group cannot reach to the surface of the virion without release of μ1N, i.e., without some type of conformational change having already occurred (15). Membrane interaction through some part of the μ1 protein other than the myristoylated μ1N peptide, such as the anion binding site identified in the crystal structure of the μ13σ33 heterohexamer, is another candidate for a triggering event (15). K+ ions, which promote the ISVP→ISVP* transition in vitro in the same way as do Cs+ ions (3; Myers et al., unpublished), might also have a role. The μ1 structural rearrangements might also be initiated by other reovirus surface proteins in vivo. For example, binding of σ1 protein to the cellular receptor might induce opening of the λ2 turret, and the conformational change of λ2 might then propagate to μ1 (10).
The conformational transition of μ1 involves two major changes: cleavage and release of the myristoylated μ1N peptide and substantial structural rearrangements in μ1C (Fig. 7) (3, 15, 25). Either the released myristoylated μ1N peptide or the structurally rearranged μ1C, or both, might be responsible for direct membrane interaction. Reovirus cores recoated with a mutant μ1 N42A protein, which cannot undergo autocleavage and which is inactive in mediating cell entry of those particles, still exhibit a number of the characteristics of the ISVP→ISVP* transition in vitro (25). Our results with μ1-N42A trimers confirm that the μ1 conformational change in solution does not require cleavage at the μ1N/C junction. The μ1N peptide cannot be released and reach the cellular membrane, however, without substantial changes in μ1 and possibly dissociation of the μ1 trimer (15). Autocleavage and μ1 conformational change are thus likely to be coupled events on the surface of the ISVP and during the infectious process. Moreover, although the crystal structure of the μ13σ33 heterohexamer shows that autocleavage can occur in vitro even without proteolytic removal of σ3 (15), recent electron microscope data suggest that the majority of the μ1 subunits on the virus surface contain an uncleaved peptide bond at the μ1N/C junction (29). Biochemical analysis of μ1 both in solution (L. Zhang, S. C. Harrison, and M. L. Nibert, unpublished data) and on the ISVP surface (23) suggest that autocleavage takes place during the ISVP→ISVP* transition, as part of the larger-scale structural changes. Although purified μ1* likely consists of a cleaved form of μ1 and previous study has suggested that the μ1N peptide is indeed released from μ1C during the ISVP→ISVP* transition (25), we cannot conclude yet whether the cleaved μ1N peptide remains associated with the rest of the molecule in solution, due to the low resolution of the molecular mass estimation obtained by light scattering.
Our results allow us to outline some of the structural features of μ1*, the product of the conformational changes (Fig. 7). It is a monomer in solution and exposes substantial hydrophobic surface. Mutations that cross-link the head domains of μ1 trimers inhibit the conformational changes in μ1 (Fig. 3 and 4A), and many of the known mutations in μ1 that confer enhanced thermostability on virions have mapped to subunit interfaces within a trimer (13). Both of these observations suggest that dissociation of trimer interfaces occurs during the ISVP→ISVP* transition and not just during conformational changes in isolated μ1. The μ1 head domain, a jelly-roll β-barrel, appears likely to fold independently of the rest of the subunit and is not likely to undergo a large structural rearrangement. The amount of β-strand in μ1*, as estimated from its CD spectrum, is consistent with this suggestion. Domains I to III of μ1 are mainly α-helical and intertwined with helical domains from other subunits of the trimer. We believe that it is these domains that refold during trimer dissociation. Moreover, these helical domains contain several hydrophobic sequences and long amphipathic helices, which might be responsible for the hydrophobicity and probable membrane interaction of μ1*.
The hydrodynamic properties of μ1* suggest that the helical domains adopt a substantially more elongated structure than they do in the μ1 trimer. The total helix content of μ1 appears to decrease somewhat during the transition from μ1 to μ1*, but a reasonable fraction remains. The stable fragment of μ1* produced by limited proteolysis contains most of domains I and II, and if the residues in their constituent helices remain helical, those segments could account for most of the α-helical portions of μ1*. We therefore suggest that μ1* contains much of domains I and II in an elongated alignment, with a flexible (and protease-sensitive) link, created by the unfolding of domain III, to the β-barrel (Fig. 7). The long α-helix near the C terminus of μ1 contributes to domain II, anchoring the polypeptide chain within this domain upon its return from the β-barrel. It is possible that this segment, which would not have been easily detected in our analysis of limited proteolysis data, also contributes to the elongated, monomeric μ1*. Limited proteolysis of ISVP* in the absence of membrane generates a different protease-resistant fragment of μ1*, which contains the β-barrel domain (21); interaction between this domain and other proteins of the virus may protect it from protease cleavage. The enhanced stability of domains I and II of μ1* in the presence of foscholine suggests that the refolded helical portions of μ1* participate in membrane interaction.
The structural protein VP6 of rotavirus has an overall structure similar to that of μ1 (its conformation in the μ13σ33 heterohexamer), suggesting an evolutionary relationship between these two proteins. VP6 does not, however, mediate membrane penetration. Like μ1, VP6 forms a tight trimer, but its folded structure consists of only two distinct domains: a distal β-barrel, which is analogous to domain IV of μ1, and a proximal α-helical domain, which interacts with the inner layer of the virion (17). Consistent with our data, structural comparison of μ1 and VP6 also suggests that the additional α-helical elements of μ1 at the base of the trimer, mostly from domains I and II, might be the components that mediate the membrane-penetration function of μ1.
The penetration machineries of nonenveloped viruses undergo primed and triggered conformational rearrangements to facilitate cell entry, just as the fusion machineries of enveloped viruses do. A number of nonenveloped viruses, exemplified by reoviruses, picornaviruses, and probably polyomaviruses, have an N-terminally myristoylated peptide cleaved from or associated with a jelly-roll β-barrel subunit. Proteolytic cleavage is also a critical priming step in the case of rotavirus VP4, which has neither an N-terminal myristoyl group nor a standard jelly-roll β-barrel but does have a surface likely to associate with a lipid bilayer when exposed by the cleavage (7). It remains a puzzle whether membrane penetration by nonenveloped viruses involves a defined penetration pore or a more generalized lysis of an endosomal membrane. In the case of reovirus μ1, a higher-order oligomer of the conformationally rearranged protein has not yet been identified. We anticipate that the results described here can lead us to a strategy for obtaining a high-resolution structure of μ1*.
Acknowledgments
We thank A. L. Odegard for constructing the recombinant baculovirus carrying genes encoding the mutant (μ1-N42A)3σ33 protein; D. S. King at the HHMI mass spectrometry laboratory and the Tufts Core Facility for mass spectrometry and N-terminal sequencing; J. Zimmer for help with the static light-scattering experiment; and members of the Harrison and Nibert laboratories for helpful discussions.
This work was supported in part by NIH grants R01 CA13202 to S.C.H. and R01 AI46440 to M.L.N. L.Z. acknowledges a Helen Hay Whitney postdoctoral fellowship, and K.C. acknowledges a Bernard N. Fields postdoctoral fellowship.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Bodkin, D. K., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran, K., J. S. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The delta region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandran, K., X. Zhang, N. H. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 75:5335-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dormitzer, P. R., E. B. Nason, B. V. Prasad, and S. C. Harrison. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, J., D. Tang, G. Leone, and P. W. Lee. 1994. Binding of reovirus to receptor leads to conformational changes in viral capsid proteins that are reversible upon virus detachment. J. Biol. Chem. 269:17043-17047. [PubMed] [Google Scholar]
- 11.Folta-Stogniew, E., and K. R. Williams. 1999. Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10:51-63. [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, S. C. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 64:231-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, S. Y., A. K. Taneja, and R. S. Hodges. 1984. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 259:13253-13261. [PubMed] [Google Scholar]
- 15.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu, M., I. Petitpas, J. Navaza, J. Lepault, E. Kohli, P. Pothier, B. V. Prasad, J. Cohen, and F. A. Rey. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 20:1485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendez, I. I., Y. M. She, W. Ens, and K. M. Coombs. 2003. Digestion pattern of reovirus outer capsid protein σ3 determined by mass spectrometry. Virology 311:289-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton, J. K., T. F. Severson, K. Chandran, A. L. Gillian, J. Yin, and M. L. Nibert. 2002. Thermostability of reovirus disassembly intermediates (ISVPs) correlates with genetic, biochemical, and thermodynamic properties of major surface protein μ1. J. Virol. 76:1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibert, M., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 21.Nibert, M. L. 1989. Ph.D. thesis. Harvard University, Boston, Mass.
- 22.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nibert, M. L., A. L. Odegard, M. A. Agosto, K. Chandran, and L. A. Schiff. 2005. Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345:461-474. [DOI] [PubMed] [Google Scholar]
- 24.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odegard, A. L., K. Chandran, X. Zhang, J. S. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 27.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Virgin, H. W., IV, and K. L. Tyler. 1991. Role of immune cells in protection against and control of reovirus infection in neonatal mice. J. Virol. 65:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, X., Y. Ji, L. Zhang, S. C. Harrison, D. C. Marinescu, M. L. Nibert, and T. S. Baker. 2005. Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 angstrom resolution. Structure (Cambridge) 13:1545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]