Abstract
Generating broad cellular immune responses against a diversity of viral epitopes is a major goal of current vaccine strategies for human immunodeficiency virus type 1 (HIV-1) and other pathogens. Virus-specific CD8+ T-lymphocyte responses, however, are often highly focused on a very limited number of immunodominant epitopes. For an HIV-1 vaccine, the breadth of CD8+ T-lymphocyte responses may prove to be critical as a result of the need to cover a wide diversity of viral isolates in the population and to limit viral escape from dominant epitope-specific T lymphocytes. Here we show that epitope modification strategies can alter CD8+ T-lymphocyte epitope immunodominance hierarchies elicited by a DNA vaccine in mice. Mice immunized with a DNA vaccine expressing simian immunodeficiency virus Gag lacking the dominant Db-restricted AL11 epitope generated a marked and durable augmentation of responses specific for the subdominant Db-restricted KV9 epitope. Moreover, anatomic separation strategies and heterologous prime-boost regimens generated codominant responses against both epitopes. These data demonstrate that dominant epitopes can dramatically suppress the immunogenicity of subdominant epitopes in the context of gene-based vaccines and that epitope modification strategies can be utilized to enhance responses to subdominant epitopes.
CD8+ T-lymphocyte responses elicited by a viral infection are typically highly focused on a limited number of immunodominant epitopes. Multiple factors likely contribute to the immunodominance of these responses, including antigen expression levels, antigen expression kinetics, antigen processing efficiency, major histocompatibility complex (MHC)/peptide binding affinity, and T-cell receptor repertoire (6, 11, 14, 23, 25). Responses to subdominant epitopes in the context of viral infections have also been reported to be limited by immunodomination, that is, direct suppression of subdominant responses by dominant epitopes (7, 13, 22).
For a human immunodeficiency virus type 1 (HIV-1) vaccine in particular, the breadth of vaccine-elicited cellular immune responses will likely prove critical. An HIV-1 vaccine designed to elicit cellular immune responses will have to protect against a wide diversity of viral sequences present in human populations (9). Moreover, responses against subdominant epitopes will likely be important for controlling the frequency and the consequences of viral escape from dominant CD8+ T-lymphocyte responses in vaccinated individuals who subsequently become infected (1, 4). In addition, recent studies have shown that CD8+ T-lymphocyte responses to subdominant epitopes are associated with improved control of viral replication in HIV-1-infected individuals (8). It has proven difficult, however, to alter the intrinsic CD8+ T-lymphocyte epitope immunodominance hierarchies elicited by infection or vaccination (18).
Studies involving lymphocytic choriomeningitis virus infection in mice have shown that the loss of a dominant epitope or the restricting MHC class I allele can result in enhanced responses to a subdominant epitope (22-24). The extent to which immunodomination occurs in the setting of gene-based vaccines, however, has not previously been investigated. Moreover, the potential of exploiting epitope modification strategies to increase the breadth of vaccine-elicited cellular immune responses has not been explored in detail.
We have recently reported that C57BL/6 mice immunized with gene-based vaccines encoding simian immunodeficiency virus SIVmac239 Gag develop a dominant Db-restricted CD8+ T-lymphocyte response to the AL11 epitope (positions 312 to 322; AAVKNWMTQTL) and a subdominant Db-restricted response to the KV9 epitope (positions 76 to 84; KSLYNTVCV) (5). In this study, we investigate the immunogenicity of a modified DNA vaccine expressing SIV Gag that specifically lacks the AL11 epitope, and we evaluate novel vaccine strategies aimed at expanding the breadth of vaccine-elicited cellular immune responses.
MATERIALS AND METHODS
Mice and immunizations.
Six- to 8-week-old female C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). For DNA immunizations, mice were injected intramuscularly (i.m.) with 50 μg VRC-2000 plasmid (Vaccine Research Center, Bethesda, MD) encoding wild-type SIVmac239 Gag (Gag WT) or AL11-deleted SIVmac239 Gag (Gag dAL11) in 100 μl saline divided between both quadriceps. The VRC-2000 plasmid includes a cytomegalovirus promoter and enhancer element, a bovine growth hormone polyadenylation signal, and a kanamycin resistance gene. The Gag dAL11 plasmid was constructed by mutating the anchor residue of the AL11 epitope required for Db binding by replacing the asparagine (AAC) at position 316 of the SIV Gag protein with an alanine (GCA) by site-directed mutagenesis. The sequence of the Gag dAL11 plasmid was confirmed by double-stranded sequencing. For recombinant adenovirus type 5 (rAd5) immunizations, mice were injected i.m. with 106 viral particles (vp) of rAd5 encoding wild-type SIVmac239 Gag in 100 μl phosphate-buffered saline (PBS) divided between both quadriceps (5). Prime-boost studies involved priming at week 0 and boosting at week 4. All animal studies were approved by our Institutional Animal Use and Care Committee.
Tetramer binding assays.
Tetrameric H-2Db complexes folded around the immunodominant SIV Gag AL11 epitope (AAVKNWMTQTL) (5) were prepared and utilized to stain peptide-specific CD8+ T lymphocytes as described previously (2). One hundred microliters of mouse blood was collected retro-orbitally in RPMI 1640 containing 40 U/ml heparin. Following lysis of the red blood cells, 0.1 μg of phycoerythrin-labeled Db/AL11 tetramer in conjunction with allophycocyanin-labeled anti-CD8α monoclonal antibody (MAb) (Ly-2; Caltag, San Francisco, CA) was utilized to stain AL11-specific CD8+ T lymphocytes. The cells were washed in PBS containing 2% fetal bovine serum (FBS) and fixed in 0.5 ml PBS containing 1.5% paraformaldehyde. Samples were analyzed by two-color flow cytometry on a FACSArray (BD PharMingen, San Diego, CA). Gated CD8+ T lymphocytes were examined for staining with the Db/AL11 tetramer. CD8+ T lymphocytes from naïve mice were utilized as negative controls and exhibited <0.1% tetramer staining.
ELISPOT assays.
Gag-specific cellular immune responses were assessed by gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays as described previously (5, 10). Murine splenocytes were assessed for responses to individual Gag epitope peptides or a pool of overlapping 15-amino-acid peptides covering the entire SIVmac239 Gag protein (NIH AIDS Research and Reference Reagent Program, Bethesda, MD). Ninety-six-well multiscreen plates (Millipore, Bedford, MA) coated overnight with 100 μl/well of 10 μg/ml rat anti-mouse IFN-γ (BD PharMingen, San Diego, CA) were washed three times with endotoxin-free Dulbecco's PBS (Life Technologies, Gaithersburg, MD) containing 0.25% Tween 20 and blocked with PBS containing 5% FBS for 2 h at 37°C. The plates were washed three times with Dulbecco's PBS containing 0.25% Tween 20, rinsed with RPMI 1640 containing 10% FBS, and incubated in triplicate with 5 × 105 splenocytes per well in a 100-μl reaction volume containing 1 μg/ml peptide. Following an 18-h incubation, the plates were washed nine times with PBS containing 0.25% Tween 20 and once with distilled water. The plates were then incubated for 2 h with 75 μl/well of 5-μg/ml biotinylated rat anti-mouse IFN-γ (BD PharMingen, San Diego, CA), washed six times with PBS containing 0.25% Tween 20, and incubated for 2 h with a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Birmingham, AL). Following five washes with Coulter Wash and one with PBS, the plates were developed with nitroblue tetrazolium-5-bromo-4-chloro-3-indolyl-phosphate chromogen (Pierce, Rockford, IL), the reaction was stopped by washing with tap water, and the plates were air dried and read using an ELISPOT reader (Cellular Technology Ltd., Cleveland, OH).
ICS assays.
Intracellular cytokine staining (ICS) assays were utilized to assess epitope-specific CD4+ and CD8+ T-lymphocyte responses. Mouse blood was collected in RPMI 1640 containing 40 U/ml heparin. For ICS assays involving multiple peptide stimulation conditions, peripheral blood from four similarly injected mice was pooled. All ICS reagents and MAbs were obtained commercially (BD PharMingen, San Diego, CA). Following lysis of the red blood cells, 106 peripheral blood mononuclear cells (PBMC) were stimulated at 37°C in 200 μl medium containing 4 μg/ml of peptide. After 2 h, 50 μl of medium containing 100 μg/ml GolgiStop was added, and the cells were cultured for an additional 4 h at 37°C. Cells were then stained with fluorescein isothiocyanate-conjugated anti-CD3 (145-2C11) and peridinin chlorophyll protein-conjugated anti-CD8 (53-6.7) MAbs for 30 min at 4°C, washed twice with cold PBS containing 2% FBS, and fixed and permeabilized with Cytofix/Cytoperm. After two washes with the Perm/Wash solution, cells were incubated with phycoerythrin-conjugated anti-interleukin-2 (JES6-5H4) and/or allophycocyanin-conjugated anti-IFN-γ (XMG1.2) MAbs for 30 min at 4°C. Cells were then washed twice with Perm/Wash and once with PBS and then were resuspended in PBS containing 1.5% formaldehyde. Samples were analyzed using a FACSCaliber and analyzed with FlowJo software.
Recombinant vaccinia virus-Gag challenge.
Vaccinated mice were challenged intraperitoneally (i.p.) with 5 × 106 PFU of replication-competent recombinant vaccinia virus expressing SIVmac239 Gag (Therion Biologics, Cambridge, MA) in 100 μl sterile PBS as described previously (21). On day 6 following challenge, ovaries were harvested and homogenized by three freeze-thaw cycles and vigorous vortexing. The homogenate was trypsinized for 30 min at 37°C. COS-7 cells were plated in six-well plates at a density of 5 × 105 cells/well and incubated overnight. Cell monolayers were then infected with log dilutions of the homogenate in medium. After 2 days, vaccinia virus plaques were visualized by staining with 0.1% crystal violet and 20% ethanol.
Statistical analyses.
Statistical analyses were performed with GraphPad Prism version 4.01 (GraphPad Software, Inc., 2004). Immunologic and virologic data are presented as means with standard errors. Data comparisons among groups were performed by analyses of variance with Bonferroni adjustments to account for multiple comparisons. In all cases, P values of less than 0.05 were considered significant.
RESULTS
Construction of the SIV Gag dAL11 DNA vaccine.
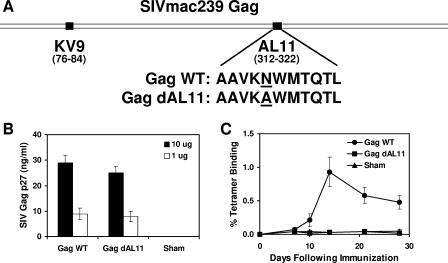
We initiated studies by constructing a plasmid DNA vaccine expressing a mutated form of SIVmac239 Gag that specifically lacked the immunodominant Db-restricted AL11 epitope (AAVKNWMTQTL) (5). As depicted in Fig. 1A, we deleted the AL11 epitope by introducing a point mutation that changed the position 5 anchor residue of the AL11 epitope required for Db binding from asparagine (N) to alanine (A). To confirm that this mutation did not abrogate protein expression, we transiently transfected 293 cells with 10 μg or 1 μg of DNA vaccine expressing either wild-type SIV Gag (Gag WT) or AL11-deleted SIV Gag (Gag dAL11) and analyzed culture supernatants after 48 h for SIV Gag p27 expression by enzyme-linked immunosorbent assay. As shown in Fig. 1B, the two constructs expressed comparable levels of SIV Gag.
FIG. 1.
Construction of the Gag dAL11 DNA vaccine. (A) The Gag dAL11 DNA vaccine was produced by mutating the position 5 anchor residue of the AL11 epitope from asparagine (N) to alanine (A). (B) Expression of Gag WT and Gag dAL11 DNA vaccines was assessed in transiently transfected 293 cells by enzyme-linked immunosorbent assay. (C) AL11-specific CD8+ T-lymphocyte responses were evaluated by Db/AL11 tetramer binding assays of C57BL/6 mice immunized with Gag WT and Gag dAL11 DNA vaccines. Error bars indicate standard errors.
We next evaluated whether the Gag WT and Gag dAL11 DNA vaccines elicited AL11-specific CD8+ T-lymphocyte responses in mice. Groups of C57BL/6 mice (n = 4/group) were immunized with 50 μg of each of these DNA vaccines or a sham plasmid, and AL11-specific CD8+ T-lymphocyte responses were assessed by Db/AL11 tetramer binding assays. As shown in Fig. 1C, the Gag dAL11 DNA vaccine did not elicit detectable AL11-specific responses, as expected, whereas the Gag WT plasmid induced high-frequency AL11-specific responses. IFN-γ ELISPOT assays confirmed that splenocytes from mice immunized with the Gag dAL11 DNA vaccine were unable to recognize the AL11 epitope AAVKNWMTQTL as well as the mutated peptide AAVKAWMTQTL (data not shown). Competitive peptide binding assays further confirmed that the mutated peptide AAVKAWMTQTL was unable to bind the class I molecule Db, whereas the wild-type AL11 and KV9 epitopes bound Db with comparable affinities (data not shown). These data demonstrate that the Gag dAL11 DNA vaccine expressed wild-type levels of SIV Gag protein but specifically lacked the immunodominant Db-restricted AL11 epitope.
Immunogenicity of Gag WT and Gag dAL11 DNA vaccines.
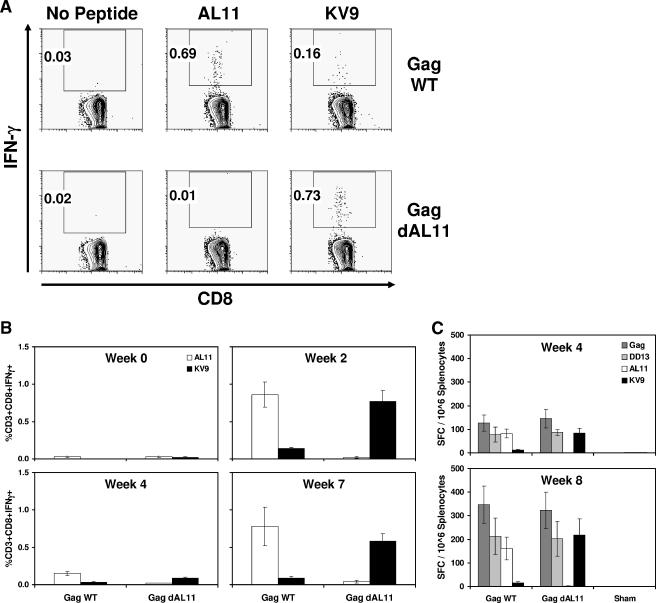
We next assessed the magnitudes of dominant and subdominant epitope-specific CD8+ T-lymphocyte responses elicited by the Gag WT or Gag dAL11 DNA vaccine. C57BL/6 mice were immunized i.m. with 50 μg of these DNA vaccines, and AL11- and KV9-specific CD8+ T-lymphocyte responses were assessed by IFN-γ ICS assays using PBMC obtained at week 2 following vaccination. Figure 2A depicts epitope-specific CD8+ T-lymphocyte responses in representative mice. The Gag WT DNA vaccine elicited dominant AL11-specific responses that were over fourfold higher than subdominant KV9-specific responses, consistent with our previous studies (5, 10, 12). In contrast, the Gag dAL11 DNA vaccine failed to elicit detectable AL11-specific immune responses but generated markedly enhanced KV9-specific immune responses. In fact, the KV9-specific CD8+ T-lymphocyte responses elicited by the Gag dAL11 DNA vaccine (0.73% of CD3+ CD8+ lymphocytes) were comparable in magnitude to the AL11-specific CD8+ T-lymphocyte responses elicited by the Gag WT DNA vaccine (0.69% of CD3+ CD8+ lymphocytes).
FIG. 2.
Cellular immune responses elicited by Gag WT and Gag dAL11 DNA vaccines. Groups of C57BL/6 mice (n = 12/group) were immunized i.m. at week 0 and week 4 with 50 μg Gag WT or Gag dAL11 DNA vaccine. AL11- and KV9-specific CD8+ T-lymphocyte responses were assessed by IFN-γ ICS assays. (A) Representative mice at week 2. (B) Mean responses in both groups at multiple time points following immunization. (C) IFN-γ ELISPOT assays in response to a pool of overlapping Gag peptides, the CD4+ T-lymphocyte epitope DD13, and the CD8+ T-lymphocyte epitopes AL11 and KV9 at week 4 and week 8. SFC, spot-forming cells. Error bars indicate standard errors.
To confirm these results and to evaluate the kinetics of epitope-specific CD8+ T-lymphocyte responses, groups of C57BL/6 mice (n = 12/group) were immunized i.m. with 50 μg of the Gag WT or Gag dAL11 DNA vaccine at week 0 and week 4. ICS assays were performed at multiple time points following immunization. As shown in Fig. 2B, responses after the initial immunization peaked at week 2 but then declined by week 4. Following the homologous boost immunization, both AL11- and KV9-specific responses increased, but the relative epitope immunodominance hierarchies remained comparable with the patterns observed following the initial immunization. Thus, deletion of the AL11 epitope resulted in a dramatic and sustained augmentation of responses to the KV9 epitope. These data suggest that KV9-specific responses are suppressed by the dominant AL11 epitope in the Gag WT DNA vaccine.
We performed IFN-γ ELISPOT assays using splenocytes from these mice at week 4 and week 8 in response to a pool of overlapping SIV Gag peptides, the CD8+ T-lymphocyte epitopes AL11 and KV9, and a newly identified CD4+ T-lymphocyte epitope DD13 (positions 299 to 311; DRFYKSLRAEQTD). Fine mapping of the DD13 epitope is depicted in Table 1. As demonstrated in Fig. 2C, mice immunized with the Gag WT and Gag dAL11 DNA vaccines elicited comparable responses to the Gag peptide pool, indicating that deletion of the dominant AL11 epitope did not substantially reduce the overall magnitude of responses to SIV Gag. Consistent with the ICS assays, the Gag WT DNA vaccine induced dominant AL11-specific ELISPOT responses and subdominant KV9-specific ELISPOT responses, whereas the Gag dAL11 DNA vaccine generated no detectable AL11-specific responses but markedly enhanced KV9-specific responses. The Gag WT and Gag dAL11 DNA vaccines elicited comparable responses to the CD4+ T-lymphocyte epitope DD13, demonstrating that deletion of the Db-restricted AL11 epitope selectively enhanced immune responses to the Db-restricted KV9 epitope but did not modulate responses to the MHC class II-restricted DD13 epitope.
TABLE 1.
Fine mapping of the DD13 epitope
| Sequence | Peptide | IFN-γ ELISPOT assay result (spot-forming cells/106 PBMC)
|
||
|---|---|---|---|---|
| Total | CD4 depleted | CD8 depleted | ||
| YVDRFYKSLRAEQTD | YD15 | 130 | ||
| VDRFYKSLRAEQTD | VD14 | 114 | ||
| DRFYKSLRAEQTD | DD13 | 120 | ||
| RFYKSLRAEQTD | RD12 | 22 | ||
| FYKSLRAEQTD | FD11 | 1 | ||
| YKSLRAEQTD | YD10 | 10 | ||
| YVDRFYKSLRAEQTD | YD15 | 180 | ||
| DRFYKSLRAEQTD | DD13 | 158 | ||
| DRFYKSLRAEQT | DT12 | 60 | ||
| DRFYKSLRAEQ | DQ11 | 2 | ||
| DRFYKSLRAE | DE10 | 2 | ||
| DRFYKSLRAEQTD | DD13 | 193 | 0 | 218 |
Splenocytes from C57BL/6 mice immunized with a DNA vaccine expressing SIV Gag demonstrated reactivity to peptide 75 (YD15) in matrix IFN-γ ELISPOT assays. Fine mapping was performed using single amino- and carboxy-terminal amino acid deletions and revealed that the minimal epitope was DD13. Unfractionated, CD4-depleted, and CD8-depleted IFN-γ ELISPOT assays confirmed DD13 as a CD4+ T-lymphocyte epitope.
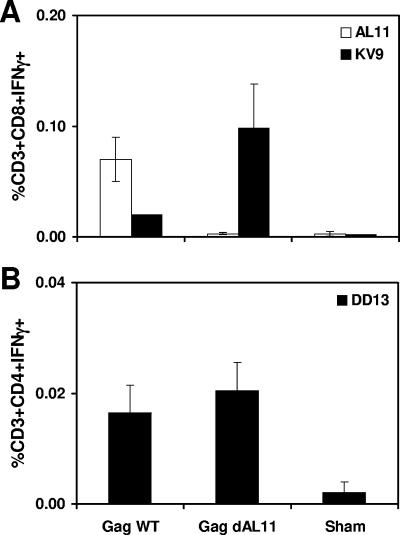
To investigate the durability of the observed epitope hierarchies, C57BL/6 mice (n = 4/group) were immunized i.m. with 50 μg of the Gag WT or Gag dAL11 DNA vaccine, and epitope-specific cellular immune responses were evaluated after 6 months. As shown in Fig. 3, AL11-, KV9-, and DD13-specific responses elicited by both vaccines diminished in magnitude compared with those depicted in Fig. 2, but they exhibited stable long-term relative epitope immunodominance hierarchies.
FIG. 3.
Durability of epitope-specific cellular immune responses. Groups of C57BL/6 mice (n = 4/group) were immunized i.m. once with 50 μg Gag WT or Gag dAL11 DNA vaccine. AL11- and KV9-specific CD8+ T-lymphocyte responses (A) and DD13-specific CD4+ T-lymphocyte responses (B) were assessed by IFN-γ ICS assays 6 months after immunization. Error bars indicate standard errors.
Anatomic separation strategies.
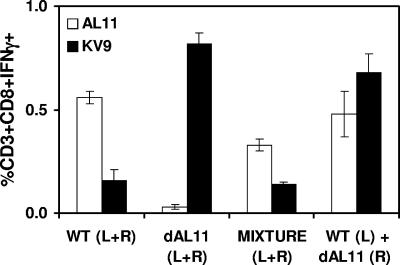
It would be highly desirable to develop a gene-based vaccine strategy that could augment responses to subdominant epitopes without losing potentially important responses to dominant epitopes, thus expanding the breadth of vaccine-elicited cellular immune responses. We reasoned that this might be possible by administering vaccines expressing Gag WT and Gag dAL11 either at separate anatomic sites or in heterologous prime-boost regimens. To explore the first possibility, we evaluated the immunogenicities of the Gag WT and Gag dAL11 DNA vaccines when mixed together in the same syringe or when administered at the same time but at separate anatomic sites. Groups of C57BL/6 mice (n = 8/group) were immunized once i.m. with the following DNA vaccine regimens: 50 μg Gag WT (half the dose in each leg), 50 μg Gag dAL11 (half the dose in each leg), 25 μg Gag WT and 25 μg Gag dAL11 mixed together in the same syringe (half the dose in each leg), or 25 μg Gag WT (left leg) and 25 μg Gag dAL11 (right leg). AL11- and KV9-specific CD8+ T-lymphocyte responses were assessed by ICS assays at week 2 following immunization. As shown in Fig. 4, mixing the Gag WT and Gag dAL11 DNA vaccines together failed to enhance KV9-specific responses. These data suggest that the AL11 epitope from the Gag WT DNA vaccine suppressed KV9-specific responses elicited by the Gag dAL11 DNA vaccine when the plasmids were mixed together. In contrast, administration of the Gag WT and Gag dAL11 plasmids at the same time but at separate anatomic sites resulted in high-frequency responses against both epitopes. These findings indicate that AL11 immunodomination over KV9 involves primarily local rather than systemic mechanisms. Thus, anatomic separation of plasmids can enhance responses to subdominant epitopes without substantial loss of responses to dominant epitopes.
FIG. 4.
Anatomic separation of Gag WT and Gag dAL11 DNA vaccines. Groups of C57BL/6 mice (n = 8/group) were immunized once i.m. with the following DNA vaccine regimens: 50 μg Gag WT (half the dose in each leg), 50 μg Gag dAL11 (half the dose in each leg), 25 μg Gag WT and 25 μg Gag dAL11 mixed together in the same syringe (half the dose in each leg), or 25 μg Gag WT (left leg) and 25 μg Gag dAL11 (right leg). AL11- and KV9-specific CD8+ T-lymphocyte responses were assessed by IFN-γ ICS assays at week 2 following immunization. Error bars indicate standard errors.
Heterologous prime-boost strategies.
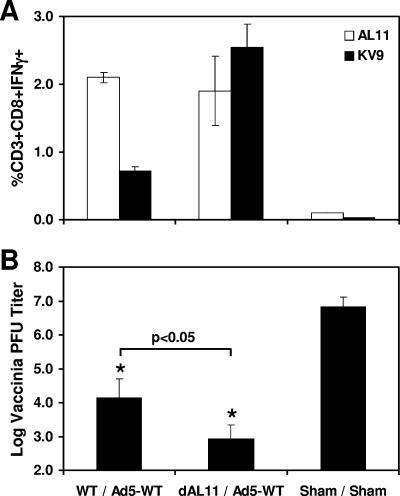
We next investigated whether heterologous prime-boost strategies could similarly elicit codominant responses against both epitopes. Priming with a DNA vaccine followed by boosting with a rAd5 vector expressing the same antigen has proven particularly effective at eliciting high-frequency cellular immune responses (16, 19, 20). We hypothesized that potent responses against both dominant and subdominant epitopes could potentially be generated by selectively deleting the dominant epitope in the DNA vaccine prime in the context of a heterologous DNA-rAd5 prime-boost regimen. Groups of mice (n = 8/group) were primed with 50 μg Gag WT or Gag dAL11 DNA vaccine at week 0, and both groups were boosted with 106 vp of rAd5-Gag WT at week 4. Control mice were primed with sham DNA and boosted with an empty rAd5 vector. As shown in Fig. 5A, mice that were primed with the Gag dAL11 DNA vaccine and boosted with rAd5-Gag WT generated high-frequency, codominant responses against both the KV9 and AL11 epitopes.
FIG. 5.
Immunogenicities and protective efficacies of heterologous prime-boost regimens. Groups of C57BL/6 mice (n = 8/group) were immunized i.m. at week 0 with 50 μg Gag WT or Gag dAL11 DNA vaccine and boosted at week 4 with 106 vp of rAd5 expressing Gag WT. Control mice received a sham plasmid and an empty rAd5 vector. (A) AL11- and KV9-specific CD8+ T-lymphocyte responses by IFN-γ ICS assays 2 weeks after the boost immunization at week 6. (B) Mice were challenged i.p. at week 10 with 5 × 106 PFU of vaccinia virus-Gag, and vaccinia virus titers in ovaries were assessed on day 6 after challenge. Virus titers in both vaccinated groups (*) were significantly lower than those in the sham control group (P < 0.001). Virus titers were lower in mice primed with the Gag dAL11 DNA vaccine than in mice primed with the Gag WT DNA vaccine (P < 0.05). Error bars indicate standard errors.
To assess the protective efficacy of these vaccine regimens, vaccinated mice were challenged i.p. at week 10 with 5 × 106 PFU of recombinant vaccinia virus expressing SIVmac239 Gag. Vaccinia virus titers in ovaries were assessed on day 6 following challenge. As demonstrated in Fig. 5B, mice primed with the Gag WT and Gag dAL11 DNA vaccines exhibited highly significant 2.4- and 3.9-log reductions, respectively, of median virus titers compared with sham-vaccinated mice (P < 0.001). Moreover, mice primed with the Gag dAL11 DNA vaccine had significantly lower virus titers than mice primed with the Gag WT DNA vaccine (P < 0.05), presumably as a result of the augmented KV9-specific responses with only minimal loss of AL11-specific responses. These data suggest that expanding the breadth of vaccine-elicited cellular immune responses improves protective efficacy in this recombinant vaccinia virus challenge model.
DISCUSSION
Vaccine-elicited CD8+ T-lymphocyte responses are typically highly focused on a very limited number of immunodominant epitopes. In this study, we demonstrate that epitope modification strategies substantially enhanced responses to a subdominant CD8+ T-lymphocyte epitope elicited by a DNA vaccine in mice. In particular, deletion of the dominant AL11 epitope in an SIV Gag DNA vaccine resulted in a dramatic and sustained augmentation of responses to the subdominant KV9 epitope. These data demonstrate that the presence of a dominant epitope can dramatically suppress CD8+ T-lymphocyte responses to a subdominant epitope in gene-based vaccines.
Previous studies involving a lymphocytic choriomeningitis virus infection model in mice showed that responses to a subdominant glycoprotein epitope were enhanced by mutating the dominant nucleoprotein epitope (23), by making mice tolerant to the dominant epitope (24), or by deleting the restricting MHC class I allele (22). Here we extend these observations by showing that epitope modification strategies augment cellular immune responses to a subdominant epitope in the context of DNA vaccines, which may have substantially different immunodominance constraints compared with viral infections (3, 15). We also show that anatomic separation strategies and heterologous prime-boost regimens utilizing epitope-modified DNA vaccines can be utilized to expand the breadth of vaccine-elicited cellular immune responses.
Anatomic separation of the Gag WT and Gag dAL11 DNA vaccines resulted in high-frequency responses against both the KV9 and AL11 epitopes (Fig. 4). These data suggest that the suppression of KV9-specific responses by the AL11 epitope involves a local rather than a systemic mechanism. We speculate that the mechanism of action likely involves epitope competition for MHC class I binding in professional antigen-presenting cells in local draining lymph nodes. Although direct data supporting this model are not yet available, we did observe that deletion of the dominant Db-restricted AL11 epitope enhanced responses to the Db-restricted KV9 epitope but did not modulate responses to the MHC class II-restricted DD13 epitope (Fig. 2 and 3). These data suggest that immunodomination may be most relevant for epitopes that share a common MHC restricting allele, which is consistent with our epitope competition model.
We also explored the utility of heterologous prime-boost regimens to elicit codominant responses against both the KV9 and AL11 epitopes (Fig. 5). Priming mice with the Gag dAL11 DNA vaccine and boosting with the rAd5-Gag WT vector elicited high-frequency responses against both AL11 and KV9 epitopes, presumably as a result of combining primary AL11-specific responses with secondary KV9-specific responses that were primed in the absence of the AL11 epitope. Importantly, the Gag dAL11 DNA prime, rAd5-Gag WT boost regimen was more effective than the Gag WT DNA prime, rAd5-Gag WT boost regimen in protecting against a recombinant vaccinia virus-Gag challenge, demonstrating the functional relevance of the enhanced KV9-specific immune responses generated by the epitope-modified DNA vaccine. Taken together, these studies suggest that subdominant epitope-specific responses should optimally be primed in the absence of suppressive dominant epitopes to optimize the breadth of vaccine-elicited cellular immune responses.
Our results contrast with those from a prior study in which simultaneous, but not sequential, immunization of mice with mutant OVA peptide variants resulted in a diverse array of T-cell responses (17). These differences are likely explained by the fact that the mutant OVA peptides in this prior study were highly related competitive agonist and antagonist peptides, and simultaneous administration of these peptides may have avoided the “original antigenic sin” associated with priming with one of these peptides (17). In contrast, the KV9 and AL11 peptides in the present study are independent epitopes that presumably interact with distinct populations of T cells.
This study is limited to the evaluation of dominant and subdominant epitope-specific responses in the defined system of SIV Gag DNA vaccination of C57BL/6 mice. The extent to which immunodomination limits the breadth of cellular immune responses elicited by more complex gene-based vaccines in nonhuman primates and humans, however, has not yet been determined. A variety of factors in addition to immunodomination have been shown to determine epitope immunodominance hierarchies, including the level and kinetics of antigen production, the efficiency of antigen processing, the affinity of MHC-peptide binding, and the extent of the T-cell receptor repertoire. Nevertheless, it is clear from the present study that immunodomination can limit responses to subdominant epitopes in certain settings. These data suggest that further studies should be conducted to evaluate the extent that immunodomination may limit the breadth of cellular immune responses elicited by the multi-antigen gene-based vaccines that are currently under development.
Acknowledgments
We thank Menzo Havenga, Jaap Goudsmit, Norman Letvin, Brianne Barker, Diana Truitt, Darci Gorgone, Michelle Lifton, Gary Nabel, and Dennis Panicali for generous advice, assistance, and reagents. The SIV Gag overlapping peptides were obtained from the NIH AIDS Research and Reference Reagent Program.
We acknowledge support from NIH grants AI058727 (to D.H.B.), AI066305 (to D.H.B.), and P30 AI060354.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., A. Craiu, S. Santra, M. A. Egan, J. E. Schmitz, M. J. Kuroda, T. M. Fu, J. H. Nam, L. S. Wyatt, M. A. Lifton, G. R. Krivulka, C. E. Nickerson, C. I. Lord, B. Moss, M. G. Lewis, V. M. Hirsch, J. W. Shiver, and N. L. Letvin. 2001. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J. Virol. 75:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., M. G. Pau, J. H. Custers, W. Koudstaal, S. Kostense, M. J. Havenga, D. M. Truitt, S. M. Sumida, M. G. Kishko, J. C. Arthur, B. Korioth-Schmitz, M. H. Newberg, D. A. Gorgone, M. A. Lifton, D. L. Panicali, G. J. Nabel, N. L. Letvin, and J. Goudsmit. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172:6290-6297. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, M. A., A. K. Pinto, K. A. Daniels, J. P. Schneck, R. M. Welsh, and L. K. Selin. 2002. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat. Immunol. 3:627-634. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., K. Pang, K. A. Masterman, G. Kennedy, S. Basta, N. Dimopoulos, F. Hornung, M. Smyth, J. R. Bennink, and J. W. Yewdell. 2004. Reversal in the immunodominance hierarchy in secondary CD8+ T cell responses to influenza A virus: roles for cross-presentation and lysis-independent immunodomination. J. Immunol. 173:5021-5027. [DOI] [PubMed] [Google Scholar]
- 8.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. St John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173-178. [DOI] [PubMed] [Google Scholar]
- 9.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 10.Lemckert, A. A., S. M. Sumida, L. Holterman, R. Vogels, D. M. Truitt, D. M. Lynch, A. Nanda, B. A. Ewald, D. A. Gorgone, M. A. Lifton, J. Goudsmit, M. J. Havenga, and D. H. Barouch. 2005. Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-Ad5 immunity. J. Virol. 79:9694-9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo, A. X., S. F. van Lelyveld, A. Craiu, and K. L. Rock. 2000. Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol. 164:4003-4010. [DOI] [PubMed] [Google Scholar]
- 12.Nanda, A., D. M. Lynch, J. Goudsmit, A. A. Lemckert, B. A. Ewald, S. M. Sumida, D. M. Truitt, P. Abbink, M. G. Kishko, D. A. Gorgone, M. A. Lifton, L. Shen, A. Carville, K. G. Mansfield, M. J. Havenga, and D. H. Barouch. 2005. Immunogenicity of recombinant fiber-chimeric adenovirus serotype 35 vector-based vaccines in mice and rhesus monkeys. J. Virol. 79:14161-14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newberg, M. H., K. J. McEvers, D. A. Gorgone, M. A. Lifton, S. H. Baumeister, R. S. Veazey, J. E. Schmitz, and N. L. Letvin. 2006. Immunodomination in the evolution of dominant epitope-specific CD8+ T lymphocyte responses in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 176:319-328. [DOI] [PubMed] [Google Scholar]
- 14.Probst, H. C., K. Tschannen, A. Gallimore, M. Martinic, M. Basler, T. Dumrese, E. Jones, and M. F. van den Broek. 2003. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 171:5415-5422. [DOI] [PubMed] [Google Scholar]
- 15.Santra, S., D. H. Barouch, M. J. Kuroda, J. E. Schmitz, G. R. Krivulka, K. Beaudry, C. I. Lord, M. A. Lifton, L. S. Wyatt, B. Moss, V. M. Hirsch, and N. L. Letvin. 2002. Prior vaccination increases the epitopic breadth of the cytotoxic T-lymphocyte response that evolves in rhesus monkeys following a simian-human immunodeficiency virus infection. J. Virol. 76:6376-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 17.Singh, R. A., J. R. Rodgers, and M. A. Barry. 2002. The role of T cell antagonism and original antigenic sin in genetic immunization. J. Immunol. 169:6779-6786. [DOI] [PubMed] [Google Scholar]
- 18.Subbramanian, R. A., M. J. Kuroda, W. A. Charini, D. H. Barouch, C. Costantino, S. Santra, J. E. Schmitz, K. L. Martin, M. A. Lifton, D. A. Gorgone, J. W. Shiver, and N. L. Letvin. 2003. Magnitude and diversity of cytotoxic-T-lymphocyte responses elicited by multiepitope DNA vaccination in rhesus monkeys. J. Virol. 77:10113-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan, N. J., T. W. Geisbert, J. B. Geisbert, L. Xu, Z. Y. Yang, M. Roederer, R. A. Koup, P. B. Jahrling, and G. J. Nabel. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 21.Sumida, S. M., P. F. McKay, D. M. Truitt, M. G. Kishko, J. C. Arthur, M. S. Seaman, S. S. Jackson, D. A. Gorgone, M. A. Lifton, N. L. Letvin, and D. H. Barouch. 2004. Recruitment and expansion of dendritic cells in vivo potentiate the immunogenicity of plasmid DNA vaccines. J. Clin. Investig. 114:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Most, R. G., R. J. Concepcion, C. Oseroff, J. Alexander, S. Southwood, J. Sidney, R. W. Chesnut, R. Ahmed, and A. Sette. 1997. Uncovering subdominant cytotoxic T-lymphocyte responses in lymphocytic choriomeningitis virus-infected BALB/c mice. J. Virol. 71:5110-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Most, R. G., K. Murali-Krishna, J. G. Lanier, E. J. Wherry, M. T. Puglielli, J. N. Blattman, A. Sette, and R. Ahmed. 2003. Changing immunodominance patterns in antiviral CD8 T-cell responses after loss of epitope presentation or chronic antigenic stimulation. Virology 315:93-102. [DOI] [PubMed] [Google Scholar]
- 24.von Herrath, M. G., J. Dockter, M. Nerenberg, J. E. Gairin, and M. B. Oldstone. 1994. Thymic selection and adaptability of cytotoxic T lymphocyte responses in transgenic mice expressing a viral protein in the thymus. J Exp. Med. 180:1901-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]