Abstract
GB virus type C (GBV-C) is a human flavivirus that may cause persistent infection, although most infected individuals clear viremia and develop antibodies to the envelope glycoprotein E2. To study GBV-C E2 antigenicity and cell binding, murine anti-E2 monoclonal antibodies (MAbs) were evaluated to topologically map immunogenic sites on GBV-C E2 and for the ability to detect or block recombinant E2 binding to various cell lines. Five competition groups of MAbs were identified. Groups I and II did not compete with each other. Group III competed with both groups I and II. Group IV did not compete with group I, II, or III. One MAb competed with all of the other MAbs, suggesting that the epitopes bound by these MAbs are intimately related. Individually, none of the MAbs competed extensively with polyclonal human convalescent antibody (PcAb); however, combinations of all five MAb groups completely blocked PcAb binding to E2, suggesting that the epitopes bound by these MAbs form a single, immunodominant antigenic site. Only group I and III MAbs detected purified recombinant E2 bound to cells in binding assays. In contrast, group II MAbs neutralized the binding of E2 to cells. Both PcAb and MAbs were conformation dependent, with the exception of one group II MAb (M6). M6 bound to a five-amino-acid sequence on E2 if the peptide included four C-terminal or eight N-terminal residues, suggesting that the GBV-C E2 protein contains a single immunodominant antigenic site which includes a complex epitope that is involved in specific cellular binding.
GB virus type C (GBV-C) is a positive-sense, single-stranded RNA virus that was independently discovered by two laboratory groups in 1995 (15, 27). On the basis of the nucleotide and deduced amino acid sequences, GBV-C was classified as a member of the family Flaviviridae (14, 15, 17, 27). Because it was initially identified in the serum of humans with chronic non-A, non-B, non-C hepatitis and shared 30% amino acid identity with hepatitis C virus (HCV), one of the groups named the virus hepatitis G virus (HGV) (15). The other group called the virus GBV-C because of its close phylogenetic relationship to previously discovered primate viruses GBV-A and GBV-B (27). The genome includes a 5′ nontranslated region that contains an internal ribosomal entry site (26) and is followed by a long open reading frame that encodes a predicted polyprotein of ∼ 3,000 amino acids (14). The polyprotein is predicted to be cleaved at the amino terminus into two envelope glycoproteins (E1 and E2), whereas the nonstructural proteins are processed by viral proteases (2, 14). On the basis of comparisons with HCV, the GBV-C E2 protein is predicted to form a heterodimer with E1 on the endoplasmic reticulum membrane, has a C-terminal hydrophobic transmembrane domain, and is glycosylated (14, 28).
Many well-controlled epidemiological studies have failed to show any association between GBV-C and either acute or chronic hepatitis; thus, most investigators do not refer to the virus as HGV (1, 21, 29). The virus has a worldwide distribution and is very common in humans, with approximately 2% of healthy U.S. blood donors actively viremic at the time of donation (1, 15, 29). However, because no disease state has been associated with GBV-C in controlled studies, the Food and Drug Administration has not implemented donor screening for GBV-C (1). The virus is transmitted by sexual, parenteral, and vertical routes (21), and thus it is not surprising that the prevalence of GBV-C is significantly higher among people with sexually transmitted or blood-borne infections compared to the general population. For example, up to 42% of human immunodeficiency virus (HIV)-positive people are GBV-C viremic in cross-sectional studies (22, 29, 36, 39, 43). Although persistent infection occurs in some individuals, the majority of infected people who are immune competent clear GBV-C within 2 years following acquisition (24, 30, 31, 32). Clearance is associated with the development of E2 antibodies (3, 30, 31, 32, 34), which appear to provide some protection against reinfection (9, 35), suggesting that these antibodies are neutralizing.
Because of poor replication characteristics and lack of pathogenicity in humans, studies of GBV-C replication have been limited. However, there has been increased interest in GBV-C because GBV-C viremia was found to be associated with significantly prolonged survival of HIV-infected people in several, though not all, studies (reviewed in reference 29). A meta-analysis of published studies comprising 1,294 HIV-infected people found that persistent viremia with GBV-C was associated with a 59% reduction in mortality (relative hazard, 0.41; 95% confidence interval, 0.23 to 0.69) (43). GBV-C replicates in vitro in peripheral blood mononuclear cells (PBMCs) and in CD4+ and CD8+ T-cell subsets (4-6, 38, 39, 41, 42). Infection induces chemokines and downregulates chemokine receptors (CCR5), and coinfection of PBMCs with GBV-C and HIV results in inhibition of HIV replication (10, 39, 41, 42).
GBV-C replicates in B and T lymphocytes in vitro (5), and most evidence suggests that lymphocytes or bone marrow progenitor cells are the primary site of replication (13, 18). Like those of HCV, the interactions of GBV-C with cellular receptors are predicted to involve envelope glycoprotein E2. Nattermann et al. found that exposure of PBMCs to E2 induced the release of RANTES and downregulated surface expression of CCR5 (16). Furthermore, Jung et al. demonstrated that cells transfected with either infectious RNA or a deletion mutant that expressed the N-terminal third of the polyprotein (including the E1 and E2 coding regions) resulted in inhibition of HIV replication, increased release of chemokines, and decreased surface expression of CCR5 compared to cells transfected with antisense GBV-C RNA, indicating that the E2 protein may be involved in the inhibition of HIV replication (10).
To characterize antigenic sites on GBV-C E2 protein and to determine the relationship between these sites and E2 binding to cells, we studied a panel of previously described monoclonal antibodies (MAbs) (25) and developed an E2 cell binding assay. Competition studies suggest the existence of a single, conformation-dependent, immunodominant antigenic site on the GBV-C E2 protein that is composed of three distinct epitopes. Two antibodies neutralized E2 binding to cells, and one of these MAbs reacted with a complex linear peptide present on E2.
MATERIALS AND METHODS
Recombinant proteins and synthetic peptides.
GBV-C E2 protein truncated at the C terminus to remove the hydrophobic transmembrane domain (nucleotides 1166 to 2162, encoding Ala205 to Asn536 [numbering based on GenBank accession number AF121950 with nucleotides 556 to 558 as the AUG start codon]) (38) was expressed in Chinese hamster ovary (CHO) cells. The construct featured a secretory leader, and antigenic protein was efficiently released into the culture supernatant. Recombinant GBV-C E2 protein was purified by anion-exchange chromatography (pH 9), followed by cation-exchange chromatography (pH 6) with a linear salt gradient (0 to 0.5 M). Recombinant HCV E2 protein, Ala384 to Lys715, expressed in CHO cells was obtained from Austral Biologicals (San Ramon, CA). A panel of synthetic peptides (13-mers) spanning the putative mature E2 protein sequence were synthesized as previously described (25). Each peptide overlapped the adjacent peptide by nine residues and featured an N-terminal biotinylated ɛ-lysyl-β-alanyl-ɛ-aminocaproyl-β-alanine spacer. Additional synthetic peptides were kindly provided by Opendra Sharma of the NIH AIDS Research and Reference Reagent Program. Peptide enzyme-linked immunosorbent assay (ELISA) was performed by applying E2 protein or synthetic peptides directly to wells in borate buffer (pH 8.3) unless otherwise noted. Furthermore, the full-length sequence of the GBV-C E2 coding region was amplified from a clinical virus isolate (38), cloned, and expressed with the Escherichia coli pRSET vector (Invitrogen, Carlsbad, CA) featuring an N-terminal Xpress epitope. Deletion mutants were made by using convenient SphI and XmaI restriction sites. Expression and purification of recombinant GBV-C E2 proteins were evaluated by ELISA, polyacrylamide gel electrophoresis, and Western blotting, whereas peptides were analyzed by ELISA.
Antibodies.
GBV-C E2 antibodies in human sera or plasma were detected with the μPlate anti-HGenv ELISA (Roche Diagnostics GmbH) as previously described (31). Immunoglobulin G (IgG) was purified from human GBV-C E2-positive and E2-negative sera by protein G chromatography. Previously described murine MAbs to GBV-C E2 protein generated by DNA immunization were used in these studies (M11, M17 [both IgG1], M6 [IgG2a], M3, M5, M13, M19, and M30 [IgG2b]) (25). Mouse isotype control antibodies IgG1, IgG2a(κ), and IgG2b(κ) (BD Biosciences Pharmingen, San Diego, CA) and an HCV anti-E2 MAb that recognizes HCV E2 bound to MOLT-4 cells (7) served as control antibodies. Written informed consent was obtained from all blood donors, and the University of Iowa Institutional Review Board approved these studies.
Cell lines.
Human CD4+ T (MOLT-4, Jurkat), embryonic kidney (HEK 293), cervical adenocarcinoma (HeLa), murine fibroblast (3T3), and CHO cell lines were obtained from the American Type Culture Collection (Manassas, VA). MOLT-4 and Jurkat cells were grown in RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with 10% heat-inactivated fetal calf serum (FCS) and Pen/Strep/l-glut (100 U of penicillin per ml, 100 mg of streptomycin sulfate per ml, and 2 mM l-glutamine). CHO K1 and HeLa cells were grown in Ham's F12 (Invitrogen) supplemented with 10% FCS and Pen/Strep/l-glut, and HeLa medium contained 1.5 g/liter sodium bicarbonate. 3T3 cells were grown in Dulbecco modified Eagle medium with Earle's salts (Mediatech) containing 10% FCS and Pen/Strep/l-glut. HEK 293 cells were grown in DMEM-ES supplemented with 10% FCS and Pen/Strep/l-glut.
GBV-C and HCV E2 cell binding assay.
Cells (1 × 106) were cooled to 4°C prior to incubation with HCV or GBV-C E2 protein at various concentrations for 1 h as previously described (37, 40). Cells were then washed with cold phosphate-buffered saline (PBS) and incubated with antibodies to HGV E2 or control antibodies for 1 h at 4°C. Cells were washed again with PBS and incubated with isotype-specific, R-phycoerythrin (PE)-conjugated goat anti-mouse IgG2a(γ) and IgG2b(γ) antibodies (Southern Biotech Inc., Birmingham, AL) or Alexa Fluor 488 goat anti-human IgG (Molecular Probes, Eugene, OR) for 45 min at 4°C. Cells were again washed and analyzed by flow cytometry (BD, Franklin Lakes, NJ) as previously described (37). For neutralization-of-binding studies, antibodies (100 μg/ml) were preincubated with HGV E2 for 30 min at 4°C prior to being added to cells, and detection anti-E2 MAbs of different isotypes were used to detect E2 bound to cells as previously described (23, 37).
Statistics.
All statistical analyses were performed with SigmaStat software V2.03S (Jandel Scientific, Chicago, IL). All comparisons were of two samples and utilized t tests.
RESULTS
GBV-C E2 binding to cultured cell lines.
To determine if GBV-C E2 protein binds specifically to cultured cell lines, a direct E2 cell binding assay was developed. GBV-C E2 protein lacking the C-terminal transmembrane domain in order to facilitate purification (Ala205-to-Asn536 protein fragment) was fused to an Fc secretory leader sequence for expression in CHO cells. The E2 protein was purified from culture supernatant and demonstrated an apparent relative molecular mass of 45 kDa (Fig. 1A). E2 is glycosylated (20); thus, the apparent molecular mass is greater than predicted (35.6 kDa). For cell binding experiments, GBV-C E2 was applied to microtiter plate wells directly or after the wells had been coated with capture antibodies (8). All eight GBV-C MAbs reacted with GBV-C E2 by ELISA and did not bind HCV E2, and none of the control antibodies reacted with either GBV-C or HCV E2 protein (data not shown).
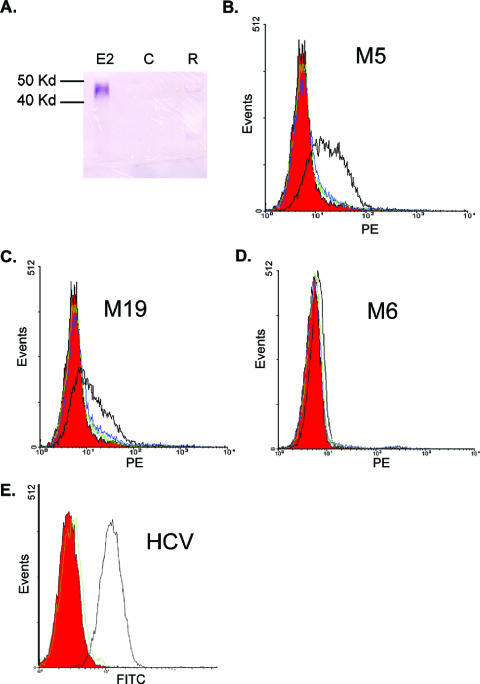
FIG. 1.
Purification of GBV-C E2 protein and development of a cell binding assay. (A) Immunoblot assay of culture supernatant from CHO cells expressing E2 (lane E2), CHO cells without E2 (lane C), or Roche cell lysates containing E2 (lane R). E2 detected with anti-E2 MAb M6. Kd, kilodaltons. (B to D) GBV-C E2 protein (10 μg/ml) was incubated with MOLT-4 cells, and cell-bound E2 was detected by GBV-C anti-E2 MAbs M5 (B), M19 (C), and M6 (D) (all with open black lines). Controls (isotype control antibody and anti-HCV E2 MAb, respectively) are shown in red and green. Cells incubated with HCV E2 (10 μg/ml) and detected with M5 (B), M19 (C), and M6 (D) are shown in blue. Panel E demonstrates HCV E2 binding to MOLT-4 cells including isotype control antibody (red), HCV MAb (black), or GBV-C M5 (green).
GBV-C E2 or HCV E2 (10 μg/ml) was incubated with the CD4+ T-lymphocyte cell line MOLT-4 for 1 h at 4°C, and binding was assessed with the GBV-C anti-E2 MAbs, isotype control antibodies, or the anti-HCV E2 MAb. GBV-C E2 binding was detected with GBV-C MAbs M5 and M19; however, no binding was detected with anti-GBV-C E2 MAb M6 or with either the isotype control antibodies or the HCV MAb (Fig. 1B to D). GBV-C E2 incubated with MOLT-4 cells was also detected by MAbs M17 and M13 but not by MAb M11 (data not shown). In contrast, HCV E2 protein bound to cells was detected by the HCV E2 MAb but not by GBV-C E2 MAb M5 (Fig. 1E), further demonstrating the specificity of binding. Nonspecific binding to cells was seen with MAbs M3 and M30. Consequently, these antibodies could not be used to identify E2 cell binding.
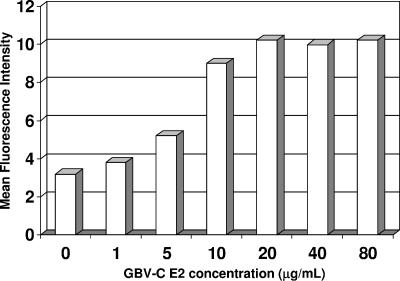
MOLT-4 cells were incubated with a series of concentrations of GBV-C E2 (Fig. 2), and the estimated affinity (Kd) of recombinant E2 for its putative receptor was estimated as previously described (23). The mean fluorescence intensity of each E2 concentration was calculated by subtracting the mean fluorescence intensity obtained with the isotype control antibody and PE-labeled isotype-specific anti-mouse antibody without E2 from that obtained with E2 as described by others (19, 23). Isotype-specific antibodies were maintained in excess. Plotting these values, the Kd was ∼10−7.5 μg/ml with M5 as the detection MAb (Fig. 2), similar to that previously demonstrated for HCV E2 binding to MOLT-4 cells (23). Comparable results were obtained with M19, and all isotype-specific probes had similar reactivities (data not shown).
FIG. 2.
E2 binding affinity. GBV-C E2 binding saturation of MOLT-4 cells consistently occurred at concentrations between 10 and 40 μg/ml. The estimated binding Kd, calculated as described in Results, was ∼10−7.5 μg/ml.
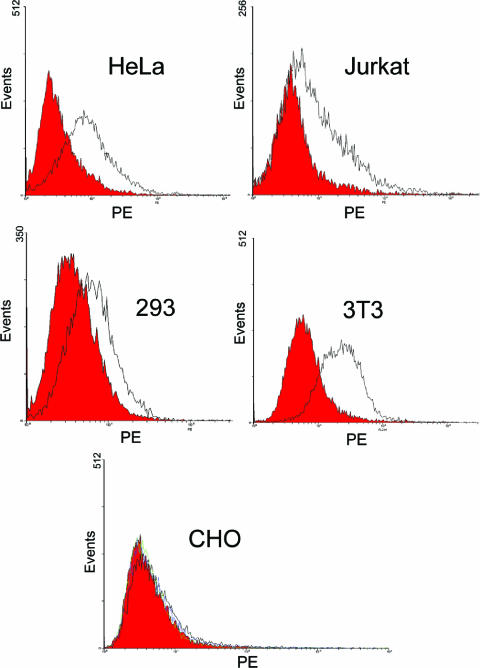
To determine if GBV-C E2 binds to other human cell types or cells of nonhuman origin, E2 binding to HeLa, Jurkat, HEK 293, murine 3T3, and CHO cells was assessed. GBV-C E2 bound to all three human (HeLa, Jurkat, and 293) and murine (3T3) cell lines (Fig. 3A to D) but not to CHO cells (Fig. 3E).
FIG. 3.
Cell specificity of GBV-C E2 binding. GBV-C E2 protein bound to HeLa, Jurkat, HEK 293, and murine 3T3 cells but not to CHO cells, as detected by MAb M5 (black) and isotype control antibody (red).
E2 MAbs: competition and neutralization of binding.
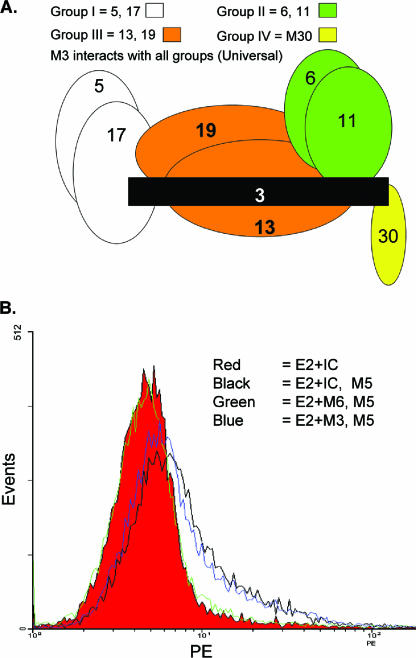
Previous competition immunoassays demonstrated that discrete GBV-C E2 binding patterns existed for the eight MAbs studied (25). The competition pattern reported by Schmolke et al. (25) was confirmed, and Fig. 4A illustrates the competition binding pattern of the different MAbs. For example, MAbs M5 and M17 competed almost completely with each other (group I) but not at all with M6 or M11 (group II). The group III MAbs competed weakly with both groups I and II, whereas M30 (group IV) did not compete with group I, II, or III MAbs. These antibodies appear to be topologically related into a single antigenic site, as M3 (“universal” MAb) competed with all of the other antibodies (25).
FIG. 4.
Epitope mapping of anti-GBV-C MAbs. (A) Patterns of competition between GBV-C E2 MAbs for binding to E2 are illustrated. Group I MAbs (M5 and M17) competed extensively with each other but not with group II or IV. Group III competed weakly with groups I and II, and universal M3 competed with all four groups. (B) GBV-C E2 (10 μg/ml) was incubated with either isotype control antibody (IC), M6 (green), or M3 (blue) prior to addition to MOLT-4 cells. Bound E2 was detected by MAb M5 and PE-conjugated isotype-specific species. M3 or IC did not block E2 binding to cells, whereas M6 neutralized E2 binding to cells.
Group I and III MAbs detected E2 bound to cells, whereas group II MAbs did not (Fig. 1B to D). To determine if group II antibodies neutralized E2 binding to cells, a neutralization-of-binding assay was used as previously described for HCV E2 (23, 37). GBV-C E2 was incubated with M5 (group I), with M6 or M11 (group II), with universal M3, or with the respective isotype control antibodies for 30 min prior to adding the mixture to MOLT-4 cells. Bound E2 was detected with MAb M5. Figure 4B demonstrates that preincubation of E2 with M6 resulted in loss of E2 binding to MOLT-4 cells, while M3 did not inhibit E2 binding. M11 similarly blocked E2 binding, whereas preincubation of E2 with M5 did not alter E2 binding patterns (data not shown). Thus, E2 bound to cells was detected by group I and III antibodies, whereas group II antibodies neutralized E2 cellular binding.
The commercial ELISA was modified to test MAbs individually or in combination for the ability to inhibit the binding of human polyclonal anti-GBV-C E2 antibodies (PcAb) to recombinant E2 protein. Briefly, biotinylated MAb M5 was used to bind E2 protein in solution and the complex was applied to streptavidin-coated microtiter plate wells. Human serum or purified human IgG was added to the wells, and E2-specific antibodies were detected with alkaline phosphatase-conjugated anti-human antibodies. For competition studies, the murine MAbs (100 μg/ml) were added to wells prior to the addition of doubling dilutions of human IgG from E2-positive or -negative subjects. None of the absorbance values for groups II (M6, M11), III (M13, M19), and IV (M30) or the M3 universal MAb were significantly lower than the control lacking antibody when tested individually for competition with PcAb. However, group I MAbs (M5 and M17) significantly competed with anti-GBV-C E2 PcAb (P < 0.01 for both [t test]; Fig. 5). Combining two groups of antibodies (excluding group I), no competition was observed; however, combining groups II, III, and IV resulted in significant inhibition (P = 0.02) and addition of the M3 universal antibody to these three groups increased inhibition (P < 0.01; Fig. 5). These combinations were not significantly greater than group I alone, and all of these values remained in the positive absorbance range, indicating that they did not completely block PcAb binding to E2. However, the combination of all four of the overlapping groups of antibodies resulted in complete inhibition of human PcAb in this assay (P < 0.01) and the reduction in absorbance was significantly greater than that of group I antibodies alone (compared to M5, P < 0.01; compared to M17, P = 0.04). Thus, these data suggest that the MAbs against the overlapping epitopes within this antigenic site represent the vast majority of the anti-E2 antibodies elicited during natural human infection (Fig. 5), and therefore, this antigenic site appears to be immunodominant. The group I epitope appears to be the most immunogenic within this antigenic site. Together, the data presented in Fig. 4 and 5 and summarized in Table 1 suggest that the epitopes recognized by these eight MAbs form a closely spaced, immunodominant cluster on E2, although alternative interpretations such as inhibition of antibody binding due to induced changes in E2 conformation cannot be excluded.
FIG. 5.
Competition of anti-GBV-C E2 MAbs and human polyclonal anti-E2 serum for binding to E2. Absorbance values for human polyclonal anti-E2 antibody incubated in the absence of a competing MAb (No Ab) or in the presence of individual anti-E2 MAbs or combinations of anti-E2 MAbs are shown. The dashed line demonstrates the cutoff value for a positive test. Symbols: *, significantly different from no-antibody control (P < 0.01); **, P = 0.02; †, P < 0.01 compared to group I absorbance values (M5 and M17). All other comparisons were not significant.
TABLE 1.
GBV-C MAbs used in this study
| Antibody | Competition group | Isotype | E2 bindinga | Polyclonal competition |
|---|---|---|---|---|
| M3 | Universal (U) | IgG2b(κ) | NS | No |
| M5 | I | IgG2b(λ) | D | Yes |
| M6 | II | IgG2a(κ) | NB | No |
| M11 | II | IgG1(κ) | NB | No |
| M13 | III | IgG2b(κ) | D | No |
| M17 | I | IgG1(κ) | D | Yes |
| M19 | III | IgG2b(κ) | D | No |
| M30 | IV | IgG2b(κ) | NS | No |
NS, nonspecific background; D, detects E2 bound to cells; NB, neutralizes binding of E2 to cells.
Defining a linear epitope on GBV-C E2.
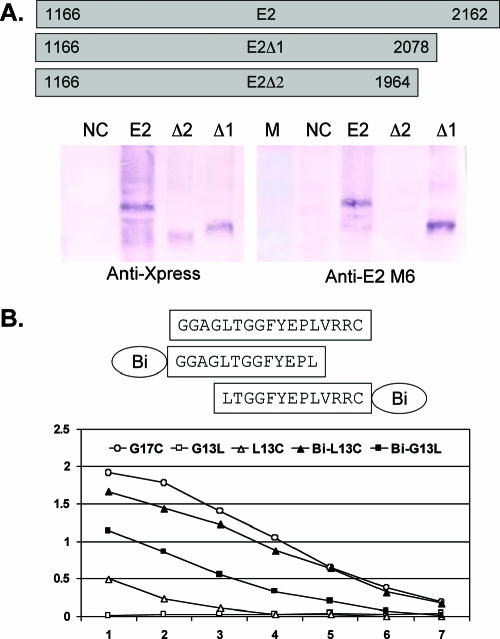
Human anti-E2-positive IgG did not react with GBV-C E2 expressed by baculovirus (40), nor did it react with synthetic peptides spanning the putative E2 protein (25) or with E2 expressed by CHO cells or E. coli in Western blot assays (data not shown), suggesting that naturally acquired GBV-C E2 antibodies are strictly conformation dependent. To further assess the reactivity of denatured E2 protein with anti-E2 MAbs, immunoblot analysis was performed with the GBV-C E2 Ala205-to-Asn536 fragment and C-terminal Ala205-to-Cys508 (E2Δ1) and Ala205-to-Met470 (E2 Δ2) deletion mutant forms expressed by E. coli (Fig. 6A). Of the eight MAbs, only M6 reacted with E2 or E2Δ1 constructs in the immunoblot studies. M6 did not react with E2Δ2, which lacks the 33 C-terminal residues of E2Δ1. E2Δ2 expression was confirmed by anti-Xpress antibody (Fig. 6A). These data are consistent with the previous characterization of these eight MAbs and polyclonal E2 antibody, which demonstrated that only M6 reacted with two overlapping synthetic peptides sharing amino acids Gly481 to Cys497 of the E2 linear sequence (25). Synthetic peptides (13-mers overlapping by nine amino acids) originally used to map M6 included an N-linked biotinylation adaptor (ɛ-lysyl-β-alanyl-ɛ-aminocaproyl-β-alanine) coupled to biotin (Bi) (25). These two peptides, the 17-mer overlapping sequence (G481AGGLTGGFYEPLVRRC497), plus the same two peptides lacking the biotinylation adaptor were tested for reactivity with M6 by ELISA. The 17-mer overlapping peptide consistently reacted most strongly, and as reported, both biotinylated peptides reacted well with M6 (Fig. 6B). However, the upstream 13-mer peptide (G13L; GAGGLTGGFYEPL) did not react at all with M6 when it lacked the biotinylation linker, and the downstream 13-mer (L13C; LTGGFYEPLVRRC) reacted significantly less well than did the same peptide with the adaptor (Bi-G13L and Bi-L13C, respectively), suggesting that there is both a sequence and size requirement for optimal interaction with M6.
FIG. 6.
Characterization of MAb M6 binding to linear epitopes on the GBV-C E2 protein. (A) The E2 coding sequences were expressed in E. coli, and protein expression was detected by Western blot assay with anti-Xpress or M6 antibodies as shown. NC, negative control (E. coli without GBV-C E2 protein); M, marker. (B) ELISA detection of M6 binding to overlapping 13-mer peptides with or without the biotinylation adaptor (Bi) and a 17-mer peptide containing the entire epitope. Removal of Bi resulted in decreased binding to L13C and loss of binding of G13L.
To assess this further, a series of peptides were synthesized representing amino acids within a 21-amino-acid region on E2 (Fig. 7A). M6 binding to G17C always had the highest absorbance value in the ELISA, and at a concentration of 0.25 μg/ml, only Bi-L13C and Bi-G13L had absorbance values >50% of this value. Deletions from both the C- and N-terminal ends of G17C revealed that reactivity could be maintained if the FYEPL sequence was present and at least four amino acids were present on the C terminus or eight amino acids and the Bi adaptor were present on the N terminus (Fig. 7B). A 17-mer peptide starting at Ser477 and extending to Leu493 did not react at a concentration of 0.25 μg/ml; however, at higher concentrations, this peptide was recognized by M6 in the ELISA (Fig. 7A). C-terminal Cys497 could be removed from the 17-mer if at least six residues were present upstream of Phe489.
FIG. 7.
Peptide mapping of M6 epitope. (A) A series of peptides within the 21-amino-acid region shown were synthesized, and their reactivity (0.25 μg/ml) with anti-GBV-C E2 MAb M6 was determined. The 17-mer (G17C) had the highest reactivity (100%), and the relative absorbance values (%) of M6 binding to the peptides compared to the 17-mer (G17C) is shown. Peptides (Pep) presented as solid bars reacted with M6, whereas open-bar peptides did not. The gray bar (S17L) did not react at 0.25 μg/ml; however, it did react with M6 at all higher concentrations in a dose-dependent manner. The minimal peptides reacting with M6 with and without the biotinylation adaptor (Bi) are shown in panel B.
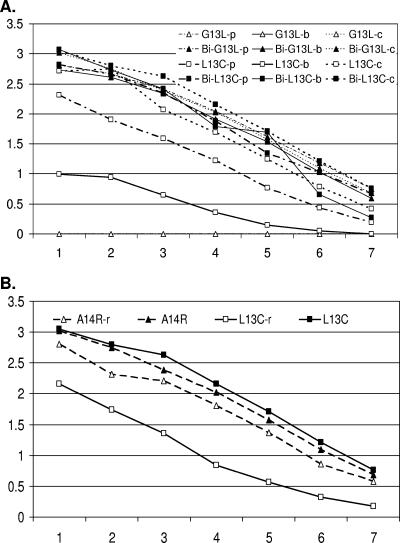
To determine if pH altered the binding of the synthetic peptides to M6, the G13L and L13C peptides (with and without the biotin adaptor Bi) were applied directly to ELISA plates with PBS, borate, or carbonate buffer (pH 7.0, 8.0, or 9.6, respectively). Alteration of pH did not lead to M6 recognition of G13L without the Bi adaptor, nor did it alter M6 recognition of Bi-G13L and Bi-L13C (Fig. 8A). However, pH altered the binding of L13C without the biotinylated spacer. In order to determine whether reducing conditions might alter intra- or interpeptide interactions, a peptide with (L13C) or lacking C-terminal Cys497 (A14R) was incubated in a 5% β-mercaptoethanol solution and tested by ELISA. No difference in reactivity was seen in the A14R peptide; however, reducing the peptide containing C-terminal Cys497 (L13C) resulted in significant loss of reactivity compared to nonreduced (L13C) (Fig. 8B).
FIG. 8.
Effects of pH and β-mercaptoethanol on reactivity of the GBV-C anti-E2 MAb with synthetic peptides. (A) Synthetic 13-mers with and without a biotinylation adaptor (Bi) were applied to microtiter plate wells in PBS (p; pH 7.0), borate (b; pH 8.3), or carbonate (c; pH 9.6) buffer. The 13-mer (GGAGLTGGFYEPL) lacking Bi did not react at all (open triangles), whereas the same peptide with Bi (solid triangles) reacted well. pH altered the binding of the 13-mer featuring C-terminal Cys497 and lacking Bi (open squares) but not that of the same peptide featuring Bi (solid squares). (B) M6 reactivity with the C-terminally truncated peptide A14R was not altered by reducing (r) with β-mercaptoethanol. However, the reduced peptide containing C-terminal Cys497 (L13C-r) demonstrated significantly less reactivity with M6 than nonreduced L13C did.
DISCUSSION
GBV-C E2 protein was shown to specifically bind to cell lines with a reasonably high affinity (Kd, ∼10−7.5 μg/ml), similar to that previously demonstrated for HCV E2 protein binding to CD81 (Kd, ∼10−8 μg/ml) (23). The specificity of both the GBV-C E2 protein and E2 MAbs was demonstrated by using HCV E2 and HCV E2 MAb in the same system (Fig. 2), by demonstrating that binding was concentration dependent, and finally by demonstrating that GBV-C did not bind to CHO cells. Of eight previously described MAbs generated against E2 by DNA immunization, only four were able to detect E2 bound to cells (groups I and III). E2 bound to the four human cell lines tested (MOLT-4, Jurkat, HeLa, and 293) and also to murine 3T3 cells.
Competition immunoassays previously demonstrated that group I, II, and IV MAbs recognize discrete epitopes that are topologically closely localized, as evidenced by competition between group III and both groups I and II and competition among all four groups by universal M3 (Fig. 4A) (25). The E2 cell binding assay showed functional differences in the different epitope groups as well, as the MAbs in groups I and III detected E2 bound to cells while group II antibodies neutralized E2 binding to cells. The group IV MAbs and M3 demonstrated high background fluorescence in the cell binding assay and thus could not be used to assess E2 binding.
Unlike HCV, the presence of antibody to GBV-C is not commonly observed in viremic individuals (3, 30-32). However, immunocompetent people infected with GBV-C who clear viremia develop E2 antibodies (3, 34) and these antibodies appear to be partly protective against reinfection following liver transplantation, suggesting neutralizing activity (9, 35). Only group I GBV-C E2 MAbs inhibited PcAb independently, and none of the MAbs tested demonstrated more than 50% inhibition when competing with human polyclonal E2 antibody individually. However, when groups II, III, and IV were combined, they were able to significantly inhibit PcAb binding to E2 (Fig. 5). Complete competition for PcAb binding was seen when all groups (I, II, III, IV and universal MAb M3) were combined. Thus, the E2 antibodies elicited during natural infection appear to be topologically related and to reside in an antigenic site composed of the cluster of epitopes recognized by these MAb groups. It is possible that the antibodies are not strictly topologically related but that they alter the conformation of E2 upon binding, thereby reducing the binding of PcAb. However, this interpretation is unlikely, as group I, III, and IV antibodies did not compete with each other in immunoassays. Furthermore, the additive effect of several combinations of antibodies in blocking the binding of PcAb (Table 1) is difficult to explain on the basis of conformational changes because this would require the existence of multiple conformation states of E2 during natural infection. The existence of an immunodominant antigenic site on E2 is further suggested by the fact that there do not appear to be antigenic differences in the E2 antibody reactivity elicited during GBV-C infection by different genotypes occurring in diverse regions of the world (31, 33).
Of the eight MAbs tested, only M6 (of group II) reacted with E. coli-expressed GBV-C E2 and synthetic peptides, indicating that all of the other antibodies recognize conformation-dependent epitopes on E2. Interestingly, both M6 and M11 neutralized E2 binding to cells and these antibodies competed extensively with each other for binding to E2, suggesting that the M11 conformational epitope shares features of the M6 epitope. Human polyclonal anti-E2 also recognized only conformational epitopes on E2, and M6 did not compete with polyclonal human E2 antibody, suggesting that the M6 epitope is not exposed to the human immune system during natural infection but was generated because of the presentation of E2 to the mouse immune system during DNA immunization (25).
Although previous studies found that M6 reacted with two overlapping 13-mer peptides defining a 17-amino-acid linear epitope (25), M6 did not recognize one of these peptides when synthesized without a biotinylation adaptor (Fig. 6B). Mapping of the M6 epitope demonstrated a minimal epitope of nine residues including FYEPLVRRC. However, removal of up to four C-terminal residues (VRRC) from the 17-mer peptide did not abolish reactivity as long as the peptide contained the biotin adaptor. These data suggest that there is both a size and a sequence requirement for M6 reactivity and that there may be a conformational component to the peptide involved in binding. This was further suggested by the finding that altering either the pH or reducing the peptides influenced the binding of M6 in the solid-phase ELISA (Fig. 8). However, it is possible that the high pH leads to either better plate binding or possibly a stronger MAb interaction due to the alpha amino acid pK values of this sequence.
The neutralization of E2 binding to cells by M6 indicates that the complex linear epitope that reacts with M6 in ELISAs is involved in GBV-C E2 cell binding. Recent data demonstrated that a synthetic GBV-C E2 peptide including the M6 epitope did not have structure by circular-dichroism measurements in aqueous solution. However, addition of lipids to the buffer resulted in the stabilization of a more ordered secondary structure characteristic of an α-helix (11, 12). These peptides were shown to perturb lipid bilayers, suggesting that the peptides are involved in membrane fusion (11). The finding that M6 neutralizes binding of E2 to cells is consistent with the hypothesis that this region of E2 is involved in cell binding and/or fusion and suggests that structural changes associated with the addition of lipids to E2 protein may be involved in the formation of the antigenic site.
In summary, these data indicate that GBV-C E2 protein specifically bound to transformed cell lines and that the binding was not strictly species specific. GBV-C E2 protein appears to have a conformation-dependent, immunodominant antigenic site composed of discrete epitopes with different functional properties. Group II MAbs neutralized binding of E2 to MOLT-4 cells, and one of these MAbs (M6) recognized a complex linear epitope on E2 previously suggested to be involved in virus-cell fusion (11). The linear epitope of M6 does not appear to be exposed during natural human GBV-C infection but was immunogenic during DNA immunization (25). The cell binding assay should prove useful for identifying a GBV-C cellular receptor and for further studying GBV-C antigenic structure.
Acknowledgments
We thank Carl W. Dieffenbach, Carolyn Williams (NIH Division of AIDS), and Opendra Sharma (NIH AIDS Research and Reference Reagent Program) for helpful discussions and providing synthetic peptides and Dietmar Zdunek for providing anti-E2 diagnostic test reagents.
This work was supported by VA Merit Review grants (J.T.S. and J.X.) and Public Health Service grant AI58740 from the National Institute of Allergy and Infectious Diseases (J.T.S.).
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Alter, H. J. 1997. G-pers creepers, where'd you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion 37:569-572. [DOI] [PubMed] [Google Scholar]
- 2.Belyaev, A. S., S. Chong, A. Novikov, A. Kongpachith, F. R. Masiarz, M. Lim, and J. P. Kim. 1998. Hepatitis G virus encodes protease activities which can effect processing of the virus putative nonstructural proteins. J. Virol. 72:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feucht, H. H., B. Zollner, S. Polywka, B. Knodler, M. Schroter, H. Nolte, and R. Laufs. 1997. Distribution of hepatitis G viremia and antibody response to recombinant proteins with special regard to risk factors in 709 patients. Hepatology 26:491-494. [DOI] [PubMed] [Google Scholar]
- 4.Fogeda, M., S. Navas, J. Martin, M. Casqueiro, E. Rodriguez, C. Arocena, and V. Carreno. 1999. In vitro infection of human peripheral blood mononuclear cells by GB virus C/hepatitis G virus. J. Virol. 73:4052-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George, S. L., D. Varmaz, and J. T. Stapleton. 2006. GB virus C replicates in primary T and B lymphocytes. J. Infect. Dis. 193:451-454. [DOI] [PubMed] [Google Scholar]
- 6.George, S. L., J. Xiang, and J. T. Stapleton. 2003. Clinical isolates of GB virus type C vary in their ability to persist and replicate in peripheral blood mononuclear cell cultures. Virology 316:191-201. [DOI] [PubMed] [Google Scholar]
- 7.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrignani, C. Wychowski, I. Nakano, C. Trepo, C. Desgranges, and G. Inchauspe. 1998. Characterization of human monoclonal antibodies specific to the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 249:32-41. [DOI] [PubMed] [Google Scholar]
- 8.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. H. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassoba, H. M., M. G. Pessoa, N. A. Terrault, N. J. Lewis, M. Hayden, J. C. Hunt, X. Qiu, S. C. Lou, and T. L. Wright. 1998. Antienvelope antibodies are protective against GBV-C reinfection: evidence from the liver transplant model. J. Med. Virol. 56:253-258. [PubMed] [Google Scholar]
- 10.Jung, S., O. Knauer, N. Donhauser, M. Eichenmuller, M. Helm, B. Fleckenstein, and H. Reil. 2005. Inhibition of HIV strains by GB virus C in cell culture can be mediated by CD4 and CD8 T-lymphocyte derived soluble factors. AIDS 19:1267-1272. [DOI] [PubMed] [Google Scholar]
- 11.Larios, C., J. Casas, M. A. Alsina, C. Mestres, M. J. Gomara, and I. Haro. 2005. Characterization of a putative fusogenic sequence in the E2 hepatitis G protein. Arch. Biochem. Biophys. 442:149-159. [DOI] [PubMed] [Google Scholar]
- 12.Larios, C., B. Christiaens, M. J. Gomara, M. A. Alsina, and I. Hara. 2005. Interaction of synthetic peptides corresponding to hepatitis G virus (HGV/GBV-C) E2 structural protein with phospholipid vesicles. FEBS J. 272:2456-2466. [DOI] [PubMed] [Google Scholar]
- 13.Laskus, T., M. Radkowski, L. F. Wang, H. Vargas, and J. Rakela. 1998. Detection of hepatitis G virus replication sites by using highly strand-specific Tth-based reverse transcriptase PCR. J. Virol. 72:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leary, T. P., A. S. Muerhoff, J. N. Simons, T. J. Pilot-Matias, J. C. Erker, M. L. Chalmers, G. G. Schlauder, G. J. Dawson, S. M. Desai, and I. K. Mushahwar. 1996. Sequence and genomic organization of GBV-C: a novel member of the Flaviviridae associated with human non-A-E hepatitis. J. Med. Virol. 48:60-67. [DOI] [PubMed] [Google Scholar]
- 15.Linnen, J., J. Wages, Z.-Y. Zhang-Keck, K. E. Fry, K. Z. Krawczynski, H. Alter, E. Koonin, M. Gallagher, M. Alter, S. Hadziyannis, P. Karayiannis, K. Fung, Y. Nakatsuji, J. W. K. Shih, M. Piatak, C. Hoover, J. Fernandez, S. Chen, J.-C. Zou, T. Morris, K. C. Hyams, S. Ismay, J. D. Lifson, G. Hess, S. K. H. Foung, H. Thomas, D. Bradley, H. Margolis, and J. P. Kim. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505-508. [DOI] [PubMed] [Google Scholar]
- 16.Nattermann, J., H. D. Nischalke, B. Kupfer, J. Rockstroh, L. Hess, T. Sauerbruch, and U. Spengler. 2003. Regulation of CC chemokine receptor 5 in hepatitis G virus infection. AIDS 17:1457-1462. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto, H., H. Nakao, T. Inoue, M. Fukuda, J. Kishimoto, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1997. The entire nucleotide sequence of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J. Gen. Virol. 78:737-745. [DOI] [PubMed] [Google Scholar]
- 18.Pessoa, M. G., N. A. Terrault, J. Detmer, J. Kolberg, M. Colllins, H. M. Hassoba, and T. L. Wright. 1998. Quantitation of hepatitis G and C viruses in the liver: evidence that hepatitis G virus is not hepatotropic. Hepatology 27:877-880. [DOI] [PubMed] [Google Scholar]
- 19.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Brugio, E. Di Stasio, B. Girardina, M. Houghton, S. Abrignani, and G. Gransi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilot-Matias, T. J., R. J. Carrick, P. F. Coleman, T. P. Leary, T. K. Surowy, J. N. Simons, A. S. Muerhoff, S. L. Buijk, M. L. Chalmers, G. J. Dawson, S. M. Desai, and I. K. Mushahwar. 1996. Expression of the GB virus C E2 glycoprotein using the Semliki Forest virus vector system and its utility as a serologic marker. Virology 225:282-292. [DOI] [PubMed] [Google Scholar]
- 21.Polgreen, P. M., J. Xiang, Q. Chang, and J. T. Stapleton. 2003. GB virus type C/hepatitis G virus: a nonpathogenic flavivirus associated with prolonged survival in HIV-infected individuals. Microbes Infect. 5:1255-1261. [DOI] [PubMed] [Google Scholar]
- 22.Rey, D., J. Vidinic-Moularde, P. Meyer, C. Schmitt, S. Fritsch, J. M. Lang, and F. Stoll-Keller. 2000. High prevalence of GB virus C/hepatitis G virus RNA and antibodies in patients infected with human immunodeficiency virus type 1. Eur. J. Clin. Microbiol. Infect. Dis. 19:721-724. [DOI] [PubMed] [Google Scholar]
- 23.Rosa, D., S. Campagnoli, Q.-L. Choo, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauleda, S., J. I. Esteban, J. M. Hernandez, H. Reesink, D. Castella, J. Quer, G. Hess, R. Esteban, and J. Guardia. 1999. Evaluation of RNA and E2 antibodies in prospectively followed recipients of hepatitis G virus-infected blood. Transfusion 39:633-638. [DOI] [PubMed] [Google Scholar]
- 25.Schmolke, S., M. Tacke, U. Schmitt, A. M. Engel, and B. Ofenloch-Haehnle. 1998. Identification of hepatitis G virus particles in human serum by E2-specific monoclonal antibodies generated by DNA immunization. J. Virol. 72:4541-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons, J. N., S. M. Desai, D. E. Schultz, S. M. Lemon, and I. K. Mushahwar. 1996. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genomic organization. J. Virol. 70:6126-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons, J. N., T. P. Leary, G. J. Dawson, T. J. Pilot-Matias, A. S. Muerhoff, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1:564-569. [DOI] [PubMed] [Google Scholar]
- 28.Stapleton, J. T. 2003. GB virus type C/hepatitis G virus. Semin. Liver Dis. 23:137-148. [DOI] [PubMed] [Google Scholar]
- 29.Stapleton, J. T., C. F. Williams, and J. Xiang. 2004. GB virus C: a beneficial infection? J. Clin. Microbiol. 42:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacke, M., K. Kiyosawa, K. Stark, V. Schlueter, B. Ofenloch-Haehnle, G. Hess, and A. M. Engel. 1997. Detection of antibodies to a putative hepatitis G virus envelope protein. Lancet 349:318-320. [DOI] [PubMed] [Google Scholar]
- 31.Tacke, M., S. Schmolke, V. Schlueter, S. Sauleda, J. I. Esteban, E. Tanaka, K. Kiyosawa, H. J. Alter, U. Schmitt, G. Hess, B. Ofenloch-Haehnle, and A. M. Engel. 1997. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology 26:1626-1633. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, E., K. Kiyosawa, K. Shimoda, K. Hino, M. Tacke, S. Schmolke, A. M. Engel, and G. Hess. 1998. Evolution of hepatitis G virus infection and antibody response to envelope protein in patients with transfusion-associated non-A, non-B hepatitis. J. Viral Hepatitis 5:153-159. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, E., M. Tacke, M. Kobayashi, Y. Nakatsuji, K. Kiyosawa, S. Schmolke, A. M. Engel, G. Hess, and H. J. Alter. 1998. Past and present hepatitis G virus infections in areas where hepatitis C is highly endemic and those where it is not endemic. J. Clin. Microbiol. 36:110-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, D. L., D. Vlahov, H. J. Alter, R. Marshall, J. Astemborski, and K. E. Nelson. 1998. Association of antibody to GB virus C (hepatitis G virus) with viral clearance and protection from reinfection. J. Infect. Dis. 177:539-542. [DOI] [PubMed] [Google Scholar]
- 35.Tillmann, H. L., S. Heringlake, C. Trauwein, D. Meissner, B. Nashan, H. J. Schlitt, J. Kratochvil, J. Hunt, X. Qiu, C. Lou, R. Pichlmayr, and M. P. Manns. 1998. Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology 28:379-384. [DOI] [PubMed] [Google Scholar]
- 36.Williams, C. F., D. Klinzman, T. E. Yamashita, J. Xiang, P. M. Polgreen, C. Rinaldo, C. Liu, J. Phair, J. B. Margolick, D. Zdunek, G. Hess, and J. T. Stapleton. 2004. Persistent GB virus C infection and survival in HIV-infected men. N. Engl. J. Med. 350:981-990. [DOI] [PubMed] [Google Scholar]
- 37.Wünschmann, S., D. Klinzman, J. Medh, D. Klinzman, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang, J., S. Wunschmann, W. N. Schmidt, J. Shao, and J. T. Stapleton. 2000. Full-length GB virus C (hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J. Virol. 74:9125-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang, J., S. Wunschmann, D. J. Diekema, D. Klinzman, K. D. Patrick, S. L. George, and J. T. Stapleton. 2001. Effect of coinfection with GB virus C (hepatitis G virus) on survival among patients with HIV infection. N. Engl. J. Med. 345:707-714. [DOI] [PubMed] [Google Scholar]
- 40.Xiang, J., S. Wunschmann, D. Klinzman, S. L. George, W. N. Schmidt, D. R. LaBrecque, and J. T. Stapleton. 2002. Recombinant hepatitis C virus-like particles expressed by baculovirus: utility in cell-binding and antibody detection assays. J. Med. Virol. 68:537-543. [DOI] [PubMed] [Google Scholar]
- 41.Xiang, J., S. L. George, S. Wunschmann, Q. Chang, D. Klinzman, and J. T. Stapleton. 2004. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1α, MIP-1β, and SDF-1. Lancet 363:2040-2046. [DOI] [PubMed] [Google Scholar]
- 42.Xiang, J., M. A. Sathar, J. H. McLinden, D. Klinzman, Q. Chang, and J. T. Stapleton. 2005. South African GB virus C isolates: interactions between genotypes 1 and 5 GBV-C isolates and the human immunodeficiency virus. J. Infect. Dis. 192:2147-2151. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, W., K. Chaloner, H. L. Tillmann, C. F. Williams, and J. T. Stapleton. 2006. Effect of early and late GBV-C viremia on survival of HIV infected individuals: a meta-analysis. HIV Med. 7:173-180. [DOI] [PubMed] [Google Scholar]