Abstract
The promoter sequences directing viral gene expression and genome replication of arenaviruses reside within the 3′ and 5′ termini of each RNA segment. The terminal 19 nucleotides at both ends are highly conserved among all arenavirus species and are almost completely complementary to each other. This study aimed at characterizing the Lassa virus promoter in detail. The relevance of each position in the promoter was studied by site-directed mutagenesis using the Lassa virus minireplicon system. The data indicate that the Lassa virus promoter functions as a duplex, regulates transcription and replication in a coordinated manner, and is composed of two functional elements, a sequence-specific region from residue 1 to 12 and a variable complementary region from residue 13 to 19. The first region appears to interact with the replication complex mainly via base-specific interactions, while in the second region solely base pairing between 3′ and 5′ promoter ends is important for promoter function.
The family Arenaviridae comprises at least 23 virus species (5). Several arenaviruses, such as Lassa virus, Junin virus, Guanarito virus, Machupo virus, and lymphocytic choriomeningitis virus (LCMV), are important human pathogens. Lassa virus persists in the small rodent Mastomys natalensis, which is prevalent in sub-Saharan Africa. Transmission of the virus to humans causes Lassa fever, a life-threatening infection associated with bleeding and organ failure (13).
Arenaviridae belong to the segmented negative-sense RNA viruses. The bisegmented genome consists of a small (S) and a large (L) RNA segment. Each segment contains two viral genes in opposite orientations, an arrangement called the ambisense-coding strategy (1). The S segment encodes the nucleoprotein (NP) and the glycoprotein precursor, which is posttranslationally cleaved into GP-1 and GP-2. The L segment encodes the small zinc-binding matrix protein Z and the large L protein, which contains an RNA-dependent RNA polymerase domain. NP, L protein, and viral RNA form the transcriptionally active unit, the ribonucleoprotein (RNP) complex. Both proteins are the minimal trans-acting factors required for RNA replication and transcription. Minimal cis-acting elements are the 5′ and 3′ noncoding regions (NCR) at the ends of the RNA segments as well as the intergenic region (13, 21, 23).
The promoter sequences directing viral gene expression and genome replication reside within the 3′ and 5′ termini of each RNA segment. The terminal 19 nucleotides at both ends are highly conserved among arenaviruses and are almost completely complementary to each other. They probably hybridize, forming a panhandle structure, with the remaining part of the RNA molecule representing the circumference of the pan. This prediction is supported by electron microscopic studies (29). Recently, functional studies using the LCMV minireplicon system provided experimental evidence that the conserved termini are essential to promote replication and transcription (25). Deletion analysis showed that both 3′ and 5′ termini are required for transcriptional activation, indicating that the promoter indeed functions as a duplex. The promoter elements of several segmented negative-strand RNA viruses, including influenza A virus (2, 6-8, 10, 11, 15, 16, 22, 24, 26), Thogoto virus (19, 20), Bunyamwera virus (3, 4, 17), and Uukuniemi virus (9), have already been characterized in detail, and structural models of the promoter have been proposed. A detailed analysis of the arenavirus promoter is still lacking. Therefore, the present study aimed at establishing a promoter model for Lassa virus. The relevance of each position in the promoter duplex was studied by site-directed mutagenesis using the Lassa virus minireplicon system (13).
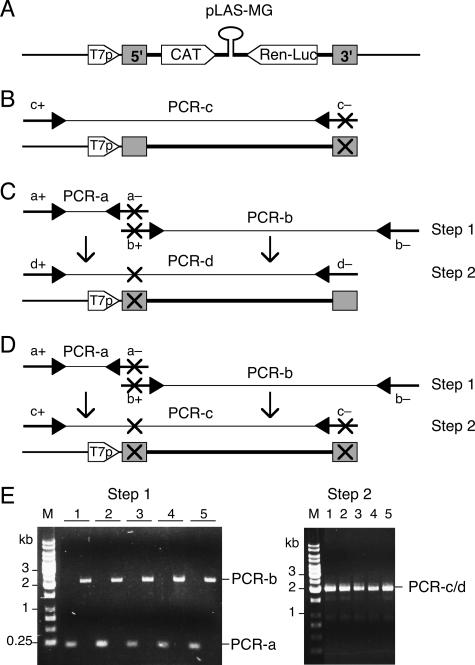
The genomic minigenome pLAS-MG, which is based on the S RNA segment of Lassa virus, served as a template for mutagenesis. It contains 5′ NCR, intergenic region, and 3′ NCR, as well as two reporter genes (encoding chloramphenicol acetyltransferase [CAT] and Renilla luciferase [Ren-Luc]) in place of the viral genes (Fig. 1A). The minigenome is expressed under control of a T7 RNA polymerase promoter and contains a single additional G residue at the 5′ end. In order to facilitate the generation of a large number of promoter mutants, the functional cassette of pLAS-MG (T7 promoter and minigenome) was amplified by mutagenic PCR, and the resulting PCR products were used for transfection without prior cloning. The experimental strategy employs classical PCR mutagenesis (14) and is schematically outlined in Fig. 1. PCR was performed with Phusion DNA polymerase (Finnzymes) and mutagenic primers binding to the 3′ and/or 5′ promoter region of pLAS-MG. Minigenomes with mutations at the 3′ terminus were amplified for 25 cycles with 3 ng linearized pLAS-MG, vector-specific primer pUC-fwd, and mutagenic primer LVS-3400-mut− (CGCACAGTGGATCCTAGGCTATTGGA; the mutagenized region is underlined) (Fig. 1B, PCR-c). Mutations at the 5′ terminus were introduced by a two-step protocol. First, fragments were amplified for 25 cycles with 3 ng linearized pLAS-MG and the primer combination pUC-fwd/LVS-1-mut− (ATGCCTAGGATCCCCGGTGCGCTA; the mutagenized region is underlined) (Fig. 1C and D, PCR-a), as well as the primer combination pUC-rev/LVS-1-mut+ (LVS-1-mut+ is reverse complementary to the corresponding LVS-1-mut− primer) (Fig. 1C and D, PCR-b). Taq polymerase was used for mutagenic PCR if primers contained artificial mutations up to position −5 from the 3′ end to prevent artifacts due to the 3′-5′ exonuclease activity of Phusion DNA polymerase. PCR products were gel purified and fused together in a second PCR with aliquots of both fragments as a template and primers pUC-fwd/LVS-3400-wt− (for 5′ promoter mutants; Fig. 1C, PCR-d) or pUC-fwd/LVS-3400-mut− (for 5′/3′ double mutants; Fig. 1D, PCR-c). Mutant minigenomes were purified using a PCR purification kit (Macherey & Nagel) and quantified spectrophotometrically. The presence of each artificial mutation was ascertained by sequencing. In each mutagenesis PCR, wild-type minigenomes were amplified with unmodified primers in parallel with the mutants and served as a positive control for the transfection experiment.
FIG. 1.
PCR-based mutagenesis of the terminal sequences of the Lassa virus minigenome. (A) Functional elements of minigenome plasmid pLAS-MG (T7p, T7 RNA polymerase promoter; 5′ and 3′, conserved termini forming Lassa virus promoter). (B) Generation of mutants with changes in the 3′ terminus by using a one-step PCR protocol. Arrows indicate primers for PCR. A cross indicates artificial mutation. (C) Generation of mutants with changes in the 5′ terminus by using a two-step PCR protocol. (D) Generation of mutants with changes in the 3′ and 5′ termini by using a two-step PCR protocol. (E) Agarose gel analysis of PCR fragments generated by the two-step PCR protocol. A set of five representative examples is shown. The following primer combinations were used for generation of mutants: PCR-a, pUC-fwd (a+) and mutagenic primer LVS-1-mut− (a−); PCR-b, mutagenic primer LVS-1-mut+ (b+) and pUC-rev (b−); PCR-c, pUC-fwd (c+) and mutagenic primer LVS-3400-mut− (c−); PCR-d, pUC-fwd (d+) and LVS-3400-wt− (d−). Primers LVS-1-mut− (a−) and LVS-1-mut+ (b+) are reverse complementary to each other, facilitating fusion of PCR-a and PCR-b fragments during PCR-c or PCR-d.
Monolayers of BSR T7/5 cells (1 × 105 cells per well of a 24-well plate) were transfected with 250 ng amplified minigenome, 250 ng pCITE-NP and 250 ng pCITE-L (expression constructs for NP and L protein, respectively), and 10 ng pCITE-FF-luc (expression construct for firefly luciferase) as a transfection control. Each mutant was transfected in duplicate. One day after transfection, cells were lysed in 100 μl passive lysis buffer (Promega) per well, and 20 μl of the lysate was assayed for firefly luciferase and Ren-Luc activities by using the dual-luciferase reporter assay system (Promega) as described by the manufacturer. Ren-Luc levels were corrected with respect to the firefly luciferase levels (standardized relative light units). For measuring CAT expression, cells were lysed 24 h after transfection in 200 μl CAT lysis buffer. CAT was assayed using a commercial CAT enzyme-linked immunosorbent assay (Roche) as described by the manufacturer. CAT levels were calculated using a standard curve and corrected with respect to the firefly luciferase levels (standardized CAT levels).
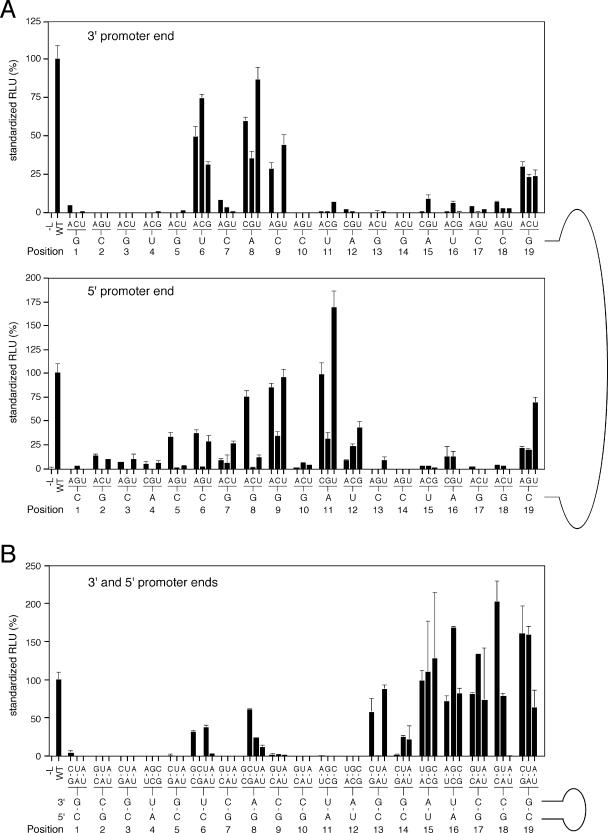
In the first set of experiments, the relevance of every nucleotide of the conserved termini was determined. Each of the 19 positions at the 3′ and 5′ termini was individually changed into the three remaining bases (Fig. 1B and C), and the transcriptional activity of the mutant minigenome was analyzed in a replicon system. Most point mutations resulted in a dramatic loss of Ren-Luc activity (Fig. 2A). Only a few mutants remained functional. At the 3′ end, mutations at positions 6 and 8 (numbering starts at the 3′ end) were tolerated, while at the 5′ end, mutations at position 8, 9, and 11 (numbering starts at the 5′ end) were tolerated. Reduced though still significant activity was seen at positions 9 and 19 of 3′ end and at positions 5, 6, 7, 12, and 19 of the 5′ end. Structural flexibility at positions 6 and 8 is consistent with the existence of natural variability at these positions. Overall, both the central part of the promoter from position 6 to 12 and position 19 were less susceptible to single mutations than the remaining positions. Differences between the 3′ and 5′ ends were most prominent at positions 11 and 12. At some positions, specific mutations abolished replicon activity while others were tolerated. For example, at position 3′-9C, bases A and U were tolerated, while a G resulted in complete loss of function. Similar effects were observed at position 5′-5C (A tolerated), 5′-6C (A and U tolerated), and 5′-8G (A and U tolerated). Thus, it appears that A and U are more likely to be tolerated at positions showing G or C in the natural context.
FIG. 2.
Analysis of promoter mutants by using the Lassa virus minireplicon system. Mutant minigenomes, pCITE-NP, and pCITE-L were transfected into BSR T7/5 cells, and Ren-Luc activity was measured 1 day after transfection. Ren-Luc levels were corrected with respect to the firefly luciferase levels (standardized relative light units [RLU]). Means (± ranges) from duplicate transfection experiments are shown relative to that for the wild-type (WT) promoter. (A) Mutants with single exchanges at either the 3′ or 5′ promoter end. The wild-type sequence is shown below the diagrams. Above the wild-type sequence, the introduced mutations are indicated. Each position, at either the 3′ or 5′ end, was changed into the three remaining bases and tested independently. (B) Double mutants with complementary exchanges in the 3′ and 5′ promoter ends. Each base pair formed between the 3′ and 5′ promoter ends was changed into the remaining three possible base pairs. The mutant base pairs are shown above the wild-type base pairs. At positions 6 and 8 with a mismatch in the natural promoter, a perfect match was introduced by changing the sequence into all four possible base pairs.
The minireplicon had been engineered to reflect the natural situation of ambisense gene expression by insertion of two marker genes, those for CAT and Ren-Luc (Fig. 3A). Consequently, replicative intermediates and transcripts can be easily distinguished by length in Northern blots. To test whether promoter mutations selectively interfere with either replication or transcription, BSR T7/5 cells were transfected with selected promoter mutants and total RNA was purified with RNeasy columns (QIAGEN, Hilden, Germany). RNA was resolved in a 1.5% agarose-formaldehyde gel and blotted onto a Hybond N+ membrane (Amersham Pharmacia Biotech). Antigenomic RNA and Ren-Luc mRNA were detected using a 32P-labeled riboprobe hybridizing to the Ren-Luc gene. In agreement with the levels of Ren-Luc enzyme activity, the mutants showed strong reductions of both antigenome and Ren-Luc mRNA levels, except mutants with changes at positions 6 and 19 of the 3′ end (Fig. 3B). There was no indication of selective reduction of either antigenome or Ren-Luc mRNA levels, suggesting that the promoter does not differently regulate replication and transcription.
FIG. 3.
Influence of selected mutations on replication and transcription of the minigenome. (A) Functional elements of the minireplicon (5′ and 3′, conserved termini forming the Lassa virus promoter). Arrows indicate the two RNA species expressed by the replicon. The mRNAs terminate in the intergenic region, while the antigenome terminates at the 5′ end of genomic RNA. (B) Northern blot analysis of RNAs expressed by mutant minigenomes. Antigenomic RNA and Ren-Luc mRNA were detected using a riboprobe hybridizing to the Ren-Luc gene. Mutations in the promoter are indicated above the blot. WT, wild-type; NC, negative control cells lacking all replicon components. Negative control cells expressing the minigenome and NP but lacking L protein did not produce a background signal (data not shown). (C) Influence of mutations at the 3′ promoter end of genomic RNA on CAT expression level. Mutant minigenomes, pCITE-NP, and pCITE-L were transfected into BSR T7/5 cells, and CAT activity was measured 1 day after transfection. Means (± ranges) from duplicate transfection experiments are shown relative to that for the wild-type (WT) promoter.
CAT mRNA can be transcribed only from newly synthesized antigenomic RNA (Fig. 3A). To ascertain that the antigenome is actually used as a template for transcription, CAT levels were measured for the wild-type replicon and the set of 3′ promoter mutants that had been tested by Northern blotting. Only the wild type and the three mutants that showed detectable levels of antigenome in Northern blots expressed CAT (mutants 6U-A, 17C-U, and 19G-U; compare Fig. 3B and C), indicating the antigenome functions as transcriptional template. This is consistent with the accumulation of genomic RNA synthesized by the Lassa virus polymerase 24 h after transfection as shown previously by Northern blot analysis (13).
The testing of single mutants also confirmed that both termini are required for transcriptional activity of the replicon. This raises the question of whether a specific base or base pairing between the 3′ and 5′ ends is relevant for promoter activity. Therefore, a second set of experiments was performed to test whether activity could be rescued by restoring complementarity between the 3′ and 5′ termini. To this end, 3′ and 5′ double mutants were generated (Fig. 1D). Each of the 19 positions of the 3′ terminus was changed into the three remaining bases, whereas a complementary change was introduced at the corresponding position of the 5′ terminus to maintain base pairing. All double changes at positions 1 to 12 resulted in complete loss of activity, except for some changes at positions 6 and 8 (Fig. 2B). This shows that base pairing cannot rescue promoter activity in this region and strongly suggests that a specific base at a certain position is required for promoter activity. The complete loss of function due to double mutations compared to the single mutations indicates an additive or even synergistic effect of the mutations and supports the view that the activity of the promoter depends on the specific sequence between positions 1 and 12 at both termini.
In contrast to the case for residues 1 to 12, compensatory mutations at positions 13 to 19 efficiently rescued transcriptional activity, indicating that base pairing rather than a specific sequence is essential in this part of the promoter. There were only three double mutants which did not show transcriptional activity (base pairs are given 3′ to 5′): at position 13, G-C to U-A; at position 14, G-C to C-G; and at position 18, C-G to A-U. It might be that at these positions the presence of a specific base at either the 3′ or 5′ end interferes with replicon activity. Residues 13 to 19 are referred to as the variable complementary region since primarily base pairing is important for promoter strength in this region.
The rescue experiments did not exclude intrastrand interactions in the region between positions 1 and 12. Therefore, the secondary structure was predicted separately for the 5′ and 3′ ends of the promoter by using mfold (version 3.2) (30). This analysis revealed possible intrastrand interactions between positions 1 and 9, positions 2 and 8, and positions 3 and 7 of the 3′ end and between positions 1 and 7 and positions 2 and 6 of the 5′ end, as shown in Fig. 4. To test the relevance of intrastrand interaction for promoter activity, each predicted base pair was inverted by mutagenesis as described above. This led to a change in sequence, but the base pairing was maintained. None of these double mutants showed transcriptional activity in the replicon system (Fig. 4). Residues 1 to 12 are referred to as the sequence-specific region, since restoring predicted inter- and intrastrand interactions by compensatory mutations could not rescue promoter activity.
FIG. 4.
Predicted intrastrand interaction and experimental evaluation of the structure. Intrastrand base pairs were predicted by mfold. Each predicted base pair was inverted separately by mutagenesis at either the 3′ or 5′ end. The activities of promoter mutants were calculated as standardized relative light units and are shown relative to the activity of the wild-type (WT) promoter.
In conclusion, the presented data suggest that the Lassa virus promoter (i) functions as a duplex; (ii) is composed of two functional elements, a sequence-specific region from residues 1 to 12 and a variable complementary region from residues 13 to 19 where solely base pairing between the 3′ and 5′ promoter ends is important; and (iii) regulates transcription and replication in a coordinated manner.
The bases of the sequence-specific region probably interact with a promoter-binding protein(s), most likely with the L protein (Fig. 5). Residues at positions 1, 2, 3, 4, 5, 7, 10, 11, and 12 of the 3′ end and at positions 1, 2, 3, 4, and 10 of the 5′ end are particularly invariable, indicating very specific interactions. Other positions are more flexible, especially positions 6 and 8, suggesting a lack of interaction or binding of L protein to the RNA backbone or common structural groups of the bases. The latter hypothesis might also explain why A and/or U bases were tolerated to some degree at positions 3′-9C, 5′-5C, and 5′-6C, while G completely blocked transcriptional activity. Bases U and A are structurally more closely related to C than G is. The data do not exclude the existence of base pairing in the sequence-specific part of the promoter. The fact that there is complementarity in this region suggests that base pairing is important, perhaps at an early stage of promoter-polymerase interaction. However, if the sequence may not be changed without affecting promoter function, it is difficult to determine the relevance of base pairing experimentally.
FIG. 5.
Model of the promoter of genomic S RNA of Lassa virus. The model assumes that the promoter functions as a duplex, in agreement with previous studies with LCMV (25). The unbound promoter may form a complete duplex. Upon binding to the replication complex, residues 1 to 12 seem to interact in a sequence-specific manner with the polymerase or another protein of the complex. Particularly invariable sites are shown in boldface, with dots indicating base-specific interactions. Residues 13 to 19 do not appear to form base-specific interactions with the replication complex. However, complementarity between the 3′ and 5′ ends of the promoter is required for activity.
In contrast to the case for the sequence-specific part, there was clear evidence that base pairing is important for promoter function in the variable complementary region. This part of the promoter might not be involved in interactions with the polymerase at all, or the interaction may involve ribose and phosphate groups rather than the base. However, some base-specific interactions at positions 13, 14, and 18, where only certain base pairs were tolerated, seem to exist. The high level of sequence flexibility of the complementary part of the promoter is somewhat surprising in view of the complete conservation of its sequence among the arenavirus family. This discrepancy may point to the existence of overlapping functions in the terminal sequence which could not be measured with the replicon system. For example, the termini of influenza A virus and Bunyamwera virus RNAs are involved in incorporation of RNA into virions (12, 18, 27).
On the other hand, the effects observed in the replicon system may not be exclusively related to promoter activity. Although this assumption is purely speculative, interference of the artificial mutations with other functions, e.g., RNP assembly, might have contributed to the experimental outcome. The availability in the future of in vitro assays with purified, enzymatically active L protein may provide further insight into L-protein-promoter interaction.
Testing a selected range of mutations at the 3′ and 5′ ends of the promoter did not reveal evidence for differential regulation of antigenome and mRNA synthesis by the promoter, which is in agreement with findings obtained with the LCMV minigenome (25). The ratio between the RNA species may be regulated subsequent to promoter binding, e.g., during initiation, elongation, or termination.
A comparison of our findings with data on promoter sequences of other segmented negative-strand RNA viruses reveals similarities but also differences. All promoters analyzed thus far function as duplexes and are composed of two functional elements. Relevant examples are the promoter models for influenza A virus (2, 6-8, 10, 11, 15, 16, 22, 24, 26), Thogoto virus (19, 20), Bunyamwera virus (3, 4, 17), and Uukuniemi virus (9). Great similarity in structure and function is seen between the Lassa virus, Uukuniemi virus, and Bunyamwera virus promoters. In all three viruses, the terminal sequences of RNA segments are almost completely complementary to each other. The sequence-specific part ranges from position 1 to 12 (Lassa virus), 1 to 11 (Bunyamwera virus), or 1 to 10 (Uukuniemi virus), while complementarity per se is the main structural feature of the adjacent variable complementary region. The major difference between Lassa virus and Bunyamwera virus lies in the sequence variability of the latter region. The S, M, and L RNA segments of the Bunyamwera virus share only the 11 sequence-specific residues, while the sequence of the variable complementary region is segment specific. Thus, this region exhibits natural variability, indicating that the sequence of this part is less important. In contrast, this region is completely conserved among the L and S segments of arenaviruses, suggesting an additional function(s) as discussed above.
Several studies on the influenza A virus promoter have provided evidence for a so-called corkscrew structure (6-8). Similar structures have also been proposed for Thogoto virus (19, 20, 28). These structures involve intrastrand base pairings in the 5′- and 3′-terminal sequences of the RNA. Similar intrastrand interactions were also predicted for the Lassa virus promoter (Fig. 4). However, we did not obtain experimental evidence for the existence of these structures. This does not exclude their formation at a certain step of promoter binding. As discussed above for possible interstrand interactions, rescue experiments may not be informative if residues are invariable due to base-specific interaction with the replication complex.
Acknowledgments
This work was supported by grant E/B41G/1G309/1A403 from the Bundesamt für Wehrtechnik und Beschaffung. The Bernhard-Nocht-Institut is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Auperin, D. D., and J. B. McCormick. 1989. Nucleotide sequence of the Lassa virus (Josiah strain) S genome RNA and amino acid sequence comparison of the N and GPC proteins to other arenaviruses. Virology 168:421-425. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. H., H. K. Cheong, J. H. Lee, C. Cheong, M. Kainosho, and B. S. Choi. 2001. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. USA 98:10602-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, J. N., and G. W. Wertz. 2004. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J. Virol. 78:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., J. W. Rodgers, and G. W. Wertz. 2005. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J. Virol. 79:12602-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charrel, R. N., and X. de Lamballerie. 2003. Arenaviruses other than Lassa virus. Antiviral Res. 57:89-100. [DOI] [PubMed] [Google Scholar]
- 6.Crow, M., T. Deng, M. Addley, and G. G. Brownlee. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 8.Flick, R., and G. Hobom. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80:2565-2572. [DOI] [PubMed] [Google Scholar]
- 9.Flick, R., F. Elgh, and R. F. Pettersson. 2002. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J. Virol. 76:10849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 100:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hass, M., U. Golnitz, S. Muller, B. Becker-Ziaja, and S. Gunther. 2004. Replicon system for Lassa virus. J. Virol. 78:13793-13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, M. T., J. D. Parvin, S. Gupta, M. Krystal, and P. Palese. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. USA 84:8140-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. J., E. Fodor, G. G. Brownlee, and B. L. Seong. 1997. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78:353-357. [DOI] [PubMed] [Google Scholar]
- 17.Kohl, A., E. F. Dunn, A. C. Lowen, and R. M. Elliott. 2004. Complementarity, sequence and structural elements within the 3′ and 5′ non-coding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J. Gen. Virol. 85:3269-3278. [DOI] [PubMed] [Google Scholar]
- 18.Kohl, A., A. C. Lowen, V. H. Leonard, and R. M. Elliott. 2006. Genetic elements regulating packaging of the Bunyamwera orthobunyavirus genome. J. Gen. Virol. 87:177-187. [DOI] [PubMed] [Google Scholar]
- 19.Leahy, M. B., J. T. Dessens, and P. A. Nuttall. 1997. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J. Virol. 71:8352-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leahy, M. B., J. T. Dessens, D. C. Pritlove, and P. A. Nuttall. 1998. The Thogoto orthomyxovirus cRNA promoter functions as a panhandle but does not stimulate cap snatching in vitro. J. Gen. Virol. 79:457-460. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torreqq. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., and P. Palese. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J. Virol. 66:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parvin, J. D., P. Palese, A. Honda, A. Ishihama, and M. Krystal. 1989. Promoter analysis of influenza virus RNA polymerase. J. Virol. 63:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez, M., and J. C. de la Torre. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 77:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccone, M. E., A. Fernandez-Sesma, and P. Palese. 1993. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 28:99-112. [DOI] [PubMed] [Google Scholar]
- 27.Tchatalbachev, S., R. Flick, and G. Hobom. 2001. The packaging signal of influenza viral RNA molecules. RNA 7:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber, F., O. Haller, and G. Kochs. 1997. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch. Virol. 142:1029-1033. [DOI] [PubMed] [Google Scholar]
- 29.Young, P. R., and C. R. Howard. 1983. Fine structure analysis of Pichinde virus nucleocapsids. J. Gen. Virol. 64:833-842. [DOI] [PubMed] [Google Scholar]
- 30.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]