Abstract
The compound 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid (DSB) potently and specifically inhibits human immunodeficiency virus type 1 (HIV-1) replication by delaying the cleavage of the CA-SP1 junction in Gag, leading to impaired maturation of the viral core. In this study, we investigated HIV-1 resistance to DSB by analyzing HIV-1 mutants encoding a variety of individual amino acid substitutions in the CA-SP1 cleavage site. Three of the substitutions were lethal to HIV-1 replication owing to a deleterious effect on particle assembly. The remaining mutants exhibited a range of replication efficiencies; however, each mutant was capable of replicating in the presence of concentrations of DSB that effectively inhibited wild-type HIV-1. Mutations conferring resistance to DSB also led to impaired binding of the compound to immature HIV-1 virions and loss of DSB-mediated inhibition of cleavage of Gag. Surprisingly, two of the DSB-resistant mutants retained an intermediate ability to bind the compound, suggesting that binding of DSB to immature HIV-1 particles may not be sufficient for antiviral activity. Overall, our results indicate that Gag amino acids L363 and A364 are critical for inhibition of HIV-1 replication by DSB and suggest that these residues form key contacts with the drug in the context of the assembling HIV-1 particle. These results have implications for the design of and screening for novel inhibitors of HIV-1 maturation.
We and others have previously described the novel mechanism of action for the small-molecule human immunodeficiency virus type 1 (HIV-1) maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid (DSB) (alternatively referred to as YK-FH312 and PA-457) (reviewed in references 2 and 14). DSB potently inhibits HIV-1 replication by delaying the cleavage of the CA-SP1 junction during HIV-1 particle maturation (8, 9, 17). The impaired cleavage results in decreased infectivity of nascent HIV-1 virions, resulting in a defect reminiscent of that associated with an HIV-1 mutant containing an uncleavable CA-SP1 junction (9). Adaptation of HIV-1 for growth in the presence of DSB has identified a few amino acid substitutions in the residues flanking the CA-SP1 scissile bond that confer resistance to the drug (9, 17). DSB specifically associates with immature HIV-1 particles produced from a protease (PR)-defective HIV-1 mutant, and this association is inhibited by amino acid changes in the target that confer resistance to the compound (16). Collectively, these studies suggest that DSB interferes with the cleavage of the CA-SP1 junction by specifically binding to this region of the Gag protein during assembly and maturation of HIV-1. This hypothesis is also supported by a recent study demonstrating inhibitory activity by DSB toward protease-mediated cleavage of the CA-SP1 junction when present in complexes of a recombinant HIV-1 Gag protein assembled in vitro (11).
In the present study, we generated nine novel HIV-1 mutants encoding various amino acid substitutions in the CA-SP1 cleavage site and analyzed these for particle production, sensitivity to DSB, and the effects of the mutations on the ability of the drug to bind to immature HIV-1 particles. The results indicate that a variety of amino acid substitutions at the P1 and P1′ residues in the CA-SP1 cleavage site can render HIV-1 less susceptible to DSB.
MATERIALS AND METHODS
Viruses and cells.
293T and P4 (HeLa-CD4/LTR-lacZ) cells were cultured in Dulbecco's modification of Eagle's medium (DMEM). CEM cells were cultured in RPMI 1640 medium. All cultures also contained fetal bovine serum (10% final concentration; HyClone), penicillin, and streptomycin and were maintained in a 37°C humidified incubator at 5% CO2.
Viruses in this study were based on the HIV-1 molecular clone pNL4-3. Specific mutations were generated in the pNL4-3 plasmid by PCR using the segment overlap extension strategy (7) to generate mutant DNAs extending from the SpeI to ApaI restriction sites in pNL4-3. The product was inserted into pNL4-3 by using these restriction sites, and the desired mutant proviruses were identified by restriction digestion and by DNA sequencing of the entire SpeI-ApaI region. For immature HIV-1 particles containing the CA-SP1 cleavage site substitutions, PR-defective clones were generated by transferring the SpeI-ApaI fragments containing the CA-SP1 mutations into pNL4-3.PR−, a variant of pNL4-3 containing Ala residues in place of the Asp-Thr-Gly active site residues in PR (16).
HIV-1 stocks were produced by calcium phosphate transfection of 293T cells with HIV-1 proviral DNA as previously described (1). 293T culture supernatants were collected, clarified by passage through 0.45-μm syringe filters, and frozen in aliquots at −80°C. The concentrated virus stocks were assayed for p24 by an antigen capture enzyme-linked immunosorbent assay (ELISA) (13).
Compounds.
The synthesis, purification, and characterization of DSB were previously described (6). [3H]dihydro-DSB was prepared by reductive hydrogenation of DSB using tritium, as previously described (16).
Single-cycle-infection and virus replication assays.
To quantify single-cycle HIV-1 infectivity, HIV-1 stocks were thawed and diluted into complete DMEM containing DEAE-dextran (20 μg/ml). Virus dilutions (0.125 ml) were added to 9-mm-diameter culture wells (i.e., 48-well plates) in which P4 indicator cells (20,000) were plated the previous day. Cultures were maintained for 48 h, after which cells expressing β-galactosidase were identified by staining them with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), as previously described (3). Blue-stained cells were quantified by capturing images of the wells using a video camera, followed by particle analysis using NIH Image software. Infectivity was reported as the number of blue-stained cells per nanogram of p24 antigen present in the inoculum.
To quantify HIV-1 replication, cultures of the human CEM T-cell line (40,000 cells in 100 μl) were inoculated with various quantities of the wild-type and mutant viruses. Twelve hours later, DSB was added at specific concentrations. The cultures were maintained for up to 30 days and split 1:2 when necessary to maintain cell viability. Every 2 or 3 days, half of the medium was withdrawn and replaced with fresh medium containing DSB at the original concentrations. Virus growth was determined by quantifying reverse transcriptase activity in the culture supernatants, as previously described (15). Radioactivity was quantified using a MATRIX direct beta counter (Perkin-Elmer). Each growth curve analysis was performed in duplicate; the duplicate parallel cultures exhibited similar reverse transcriptase activity levels at each time point.
Pulse-chase assay of Gag processing.
Kinetic analysis of Gag processing was performed with particles released into supernatants of transfected 293T cells, cultured in the presence and absence of DSB (2.5 μg/ml), that were pulse labeled with medium containing [35S]cysteine and [35S]methionine as previously described (15). Following removal of the labeling medium and culture for various periods of time in chase medium, culture supernatants were supplemented with radioimmunoprecipitation assay buffer and the viral lysates were then subjected to immunoprecipitation with the HIV-1 CA-specific monoclonal antibody (183-H12-5C). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Following electrophoresis, the proteins were fixed and the gels were soaked in Fluoro-Hance (Research Products International Corp.) and dried in a Bio-Rad vacuum gel dryer. The radioactive proteins were detected using a Fuji phosphorimager. The relative levels of radioactivity in the CA and CA-SP1 protein bands were quantified using the instrument software.
SPA for binding of DSB to immature HIV-1 particles.
Immature HIV-1 particles (10 to 30 ml) were concentrated by ultracentrifugation, and the pellets were resuspended in complete DMEM (0.2 ml). The concentrated stocks were assayed for Gag protein concentrations by a p24 capture ELISA. Virus particles (2-μg p24 equivalents) were mixed with 0.5 μg/ml [3H]DSB (prepared as previously described [15]) and 1 mg of wheat germ agglutinin-polyvinyltoluene scintillation proximity assay (SPA) beads (GE Healthcare) in 100-μl volumes in a white, 96-well, flat-bottom plate (Optiplate-96; Perkin-Elmer). The reaction mixtures were maintained at room temperature for 24 h, and the scintillation was quantified with a Packard Topcount instrument. Control assays included reactions without virus (for nonproximity effects and nonspecific binding of the compound to the beads) and reactions lacking [3H]DSB (which gave no signal). Additional controls demonstrated that the binding had reached equilibrium under these conditions and that the binding could be competed by unlabeled DSB but not by the inactive compound betulinic acid (J. Zhou and C. Aiken, unpublished observations). The nonproximity signal obtained from the reactions lacking virus particles (typically 10 to 20% of the wild-type signal) was highly reproducible and was dependent on both [3H]DSB and SPA bead concentrations.
RESULTS AND DISCUSSION
Design and testing of HIV-1 mutants containing substitutions in the CA-SP1 cleavage site.
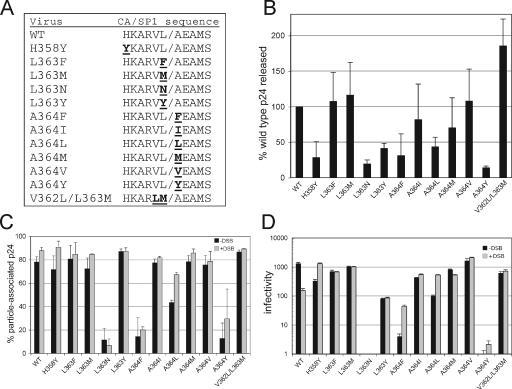
To analyze the spectrum of mutations capable of conferring resistance to DSB, we constructed a panel of 12 HIV-1 mutants containing specific amino acid substitutions within the CA-SP1 cleavage site. Three of these (L363F, A364V, and V362L/L363M) were previously characterized as viruses that were partially or totally resistant to the inhibitory action of the compound (9, 15, 17). We chose additional substitutions that are represented at these positions in various natural substrates for PR (12) and thus were likely to be recognized by the enzyme (Fig. 1A). Previous studies showed that some mutations in SP1 result in impaired assembly of HIV-1 particles (4, 5, 10). To test whether the mutations that we constructed are competent for assembly, virus particles were produced by transfection and the resulting supernatants assayed for levels of HIV-1 antigen by a p24 capture ELISA. Four of the mutations (H358Y, L363N, A364F, and A364Y) resulted in a 69 to 86% decrease the quantity of p24 released from the transfected cells (Fig. 1B). For three of these mutants (L363N, A364F, and A364Y), only a small fraction (11%, 14%, and 13%, respectively) of the p24 in the culture supernatants could be pelleted by ultracentrifugation (Fig. 1C), suggesting a defect in particle formation. The A364L mutant gave an intermediate value, with 43% of the p24 antigen released from cells in pelletable form. Similar effects of these mutations were observed in constructs containing inactivating mutations in the viral protease, indicating that the impaired particle production was independent of particle maturation (data not shown). The remaining mutants efficiently produced particles (72 to 87% of the p24 released in pelletable form), thus permitting studies of their sensitivity to DSB.
FIG. 1.
HIV-1 mutants used in this study. (A) Altered amino acids are indicated in underlined bold type. (B) Release of p24 from 293T cells transfected with the mutant proviruses. (C) Analysis of particle-associated p24 antigen. Virus stocks were produced by transfection of 293T cells with proviral DNA, and the supernatants were harvested and assayed for p24 by an ELISA. One-milliliter samples were then subjected to ultracentrifugation in a Beckman TLA-45 rotor at 100,000 × g for 30 min at 4°C. The supernatants were recovered and assayed for p24. The results were expressed as fractions of the total p24 that were depleted from the supernatants upon ultracentrifugation. The gray shaded bars represent the results for viruses that were harvested from cells cultured in the presence of DSB (2.5 μg/ml). In panels B and C, the values shown are the means for three independent experiments, with error bars representing 1 standard deviation. (D) Infectivities of mutant viruses produced in the presence and absence of DSB. Shown are the mean values from triplicate determinations, with error bars representing 1 standard deviation. The results are representative of two independent experiments. WT, wild type.
Substitutions in the CA-SP1 cleavage site confer resistance of HIV-1 to DSB in single-cycle-infection assays.
To evaluate the effects of DSB on the infectivities of the mutants, we harvested virus particles released from transfected 293T cells that were cultured in the presence and absence of a fixed concentration of DSB (2.5 μg/ml) and titrated the virus stocks on HeLa-CD4/LTR-lacZ (P4) indicator target cells. DSB inhibited the infectivity of the wild-type virus approximately ninefold (Fig. 1D), a magnitude consistent with the reported effect of DSB on HIV-1 in single-cycle infections. By contrast, none of the mutant viruses was substantially inhibited by DSB. The infectivities of several of the poorly infectious mutants (H358Y, A364F, and A364L) were actually stimulated 4- to 10-fold when the viruses were produced in the presence of the compound (Fig. 1D). Two of these mutants, H358Y and A364L, exhibited enhanced particle stability when produced in the presence of DSB (Fig. 1C), suggesting that the compound may enhance the infectivities of these mutants by stabilizing the particles. Collectively, these results indicate that a wide variety of substitutions at the CA-SP1 junction can confer HIV-1 resistance to DSB in single-cycle infection assays.
Substitutions at the P1 and P1′ positions of the CA-SP1 cleavage site confer the ability of HIV-1 to replicate in the presence of high concentrations of DSB.
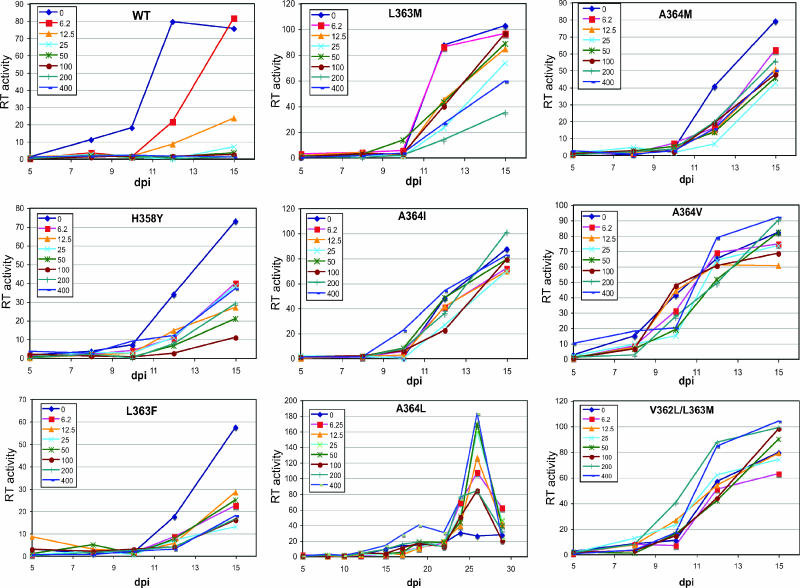
To determine the effects of the mutations on sensitivity to DSB in a continuous-replication assay, we inoculated cultures of human CEM T cells with fixed quantities of the wild-type and mutant viruses and monitored the spread of the viruses in cultures containing various concentrations of DSB by quantifying the accumulation of p24 antigen in the culture media. As previously reported, wild-type HIV-1 was highly sensitive to the compound, exhibiting a 90% reduction in p24 accumulation at day 12 (the peak for the control culture lacking DSB) at a DSB concentration of 12.5 ng/ml (Fig. 2, WT). Four mutants (L363N, L363Y, A364F, and A364Y) were markedly impaired for replication and therefore were not studied further (data not shown). By contrast, all of the remaining mutants were capable of replicating in the presence of elevated concentrations of DSB. The mutants fell into two general groups: one group, including H358Y, L363F, and L363M, exhibited intermediate sensitivities to DSB. Viruses in the second group, including A364I, A364M, A364V, and V362L/L363M, were strongly resistant, exhibiting robust replication in the presence of DSB concentrations of up to 400 ng/ml. The replications of the A364V and V362L/L363M mutants were actually slightly stimulated at the highest concentrations of DSB. The replication of mutant A364L was relatively inefficient, requiring a prolonged culture period to reach maximal p24 production. Nonetheless, A364L replication was resistant to DSB and was markedly stimulated at the highest dose of the compound (400 ng/ml). These results indicate that a variety of substitutions at the CA-SP1 junction can allow HIV-1 to replicate at elevated concentrations of DSB.
FIG. 2.
Replication of wild-type (WT) and mutant HIV-1 viruses in the presence of DSB. Cultures of CEM cells were inoculated with the indicated viruses (1 ng p24), and DSB was added at the indicated concentrations 12 h after infection. HIV-1 replication was monitored by quantifying the accumulation of p24 in the culture supernatants. The data shown are the mean values for duplicate parallel cultures, which agreed well. RT, reverse transcriptase.
Mutations conferring resistance to DSB prevent the delay in CA-SP1 processing induced by the compound.
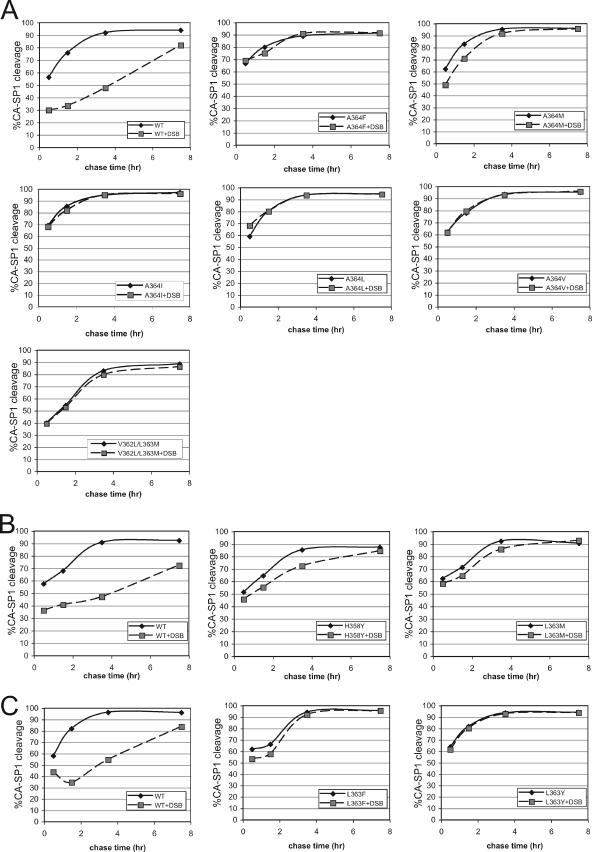
To analyze the effects of the CA-SP1 mutations on DSB-induced inhibition of CA-SP1 cleavage, we performed pulse-chase analysis of Gag processing in nascent virions. The images of the phosphorimager scans of the gels are shown in Fig. S1 in the supplemental material. Quantitation of the relative radioactivity levels in the CA and CA-SP1 bands revealed that DSB induced a marked delay in the cleavage of the CA-SP1 site in wild-type HIV-1 (Fig. 3A to C). By contrast, this effect was either markedly reduced or eliminated by the mutations conferring resistance to DSB. Comparison of the results for the wild-type and mutant viruses in the absence of DSB revealed differences in the CA-SP1 processing rates induced by the mutations, with several mutants exhibiting slight (H358Y) to pronounced (L363F and V362L/L363M) delays in the cleavage of this site relative to wild-type HIV-1. Collectively, these results indicate that the mutations render HIV-1 resistant to DSB by blocking the ability of the compound to delay the cleavage of the CA-SP1 junction.
FIG. 3.
Pulse-chase analysis of the effects of DSB on CA-SP1 cleavage in HIV-1 particles. Cultures of transfected 293T cells were cultured in the presence and absence of DSB (2.5 μg/ml) and were pulsed with 35S-labeled amino acids for 20 min. Supernatants were harvested from the cultures at the indicated chase times. Following lysis with detergent, the supernatants were subjected to immunoprecipitation with an antibody specific for CA. Immunoprecipitated proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the radioactive protein bands were detected and quantified using a phosphorimager. The values shown are the extents of cleavage as determined by quantifying the radioactivity signals in the CA-SP1 band as percentages of the sum of the signals in the CA and CA-SP1 bands. The results shown in panels A, B, and C were obtained in separate experiments, each of which included wild-type (WT) HIV-1 as a control. The results are representative of a minimum of two independent analyses.
Mutations at the CA-SP1 junction that confer resistance to DSB also reduce the binding of the compound to immature HIV-1 particles.
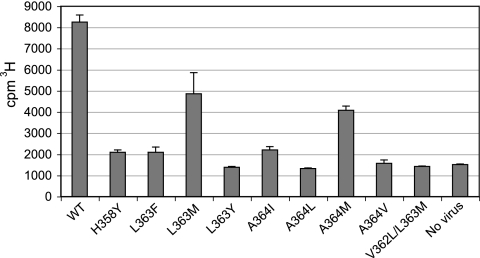
DSB binds specifically to immature HIV-1 particles (16). To facilitate studies of the association of [3H]DSB with multiple HIV-1 mutants, we established a system employing wheat germ agglutinin-coated SPA beads to which immature HIV-1 particles were bound. The assay was designed to detect DSB only when bound to the immobilized virions by virtue of the close proximity of the radiolabeled drug to the SPA beads, which contain a scintillant. This strategy overcomes the requirement for a washing step to remove unbound ligand, thus permitting a “mix-and-read” equilibrium assay format. Immature virus particles were produced by transfection, and the particles were concentrated by ultracentrifugation, resuspended, and normalized by a p24 capture ELISA. [3H]DSB was added to the particles at a concentration of 0.5 μg/ml, followed by addition of the scintillation proximity beads. This DSB concentration was previously determined to be subsaturating for binding to immature HIV-1 particles (16). The reaction mixtures were incubated for 24 h at room temperature, and the scintillation signals were then quantified. Using this approach, we observed specific binding of DSB to immature HIV-1 particles containing wild-type Gag (Fig. 4). The assay signal was approximately five times greater than that from the control sample lacking virus particles. Control experiments demonstrated that the binding had reached equilibrium under these conditions and that the signal was minimal in assays in which virus particles were omitted (data not shown). To determine whether the signal detects specific binding, we tested the ability of [3H]DSB to bind to particles containing both V362L and L363M substitutions. We previously demonstrated that these mutations render HIV-1 highly resistant to DSB and prevent the binding of the compound to immature HIV-1 particles (15). No significant binding of DSB to the immature V362L/L363M mutant particles was observed using the assay, thus confirming the ability of the assay to discriminate specific from nonspecific binding (Fig. 4). In additional control studies, we observed that the mutations at the CA-SP1 junction did not interfere with the binding of PR-defective virions to the SPA beads (J. Zhou and C. Aiken, unpublished results). The assay could therefore be used to determine the effects of Gag mutations on the binding of the drug to immature HIV-1 particles.
FIG. 4.
Association of [3H]DSB with immature HIV-1 particles containing substitutions at the CA-SP1 cleavage site. The reactions were performed in duplicate; shown are the averages for the duplicates, with error bars indicating the ranges of the values. Results shown are from one of three independent experiments. WT, wild type; No virus, control sample lacking virus particles.
To test the consequences of the CA-SP1 mutations on the binding of DSB to immature HIV-1 particles, we constructed PR-defective HIV-1 proviruses encoding the CA-SP1 mutations. Virus stocks were produced by transfection, concentrated by ultracentrifugation, and assayed for [3H]DSB binding by using the scintillation proximity assay. All of the DSB-resistant mutants exhibited a reduction in DSB binding relative to the wild-type control (Fig. 4). However, the impairment in binding differed among the mutants, with the L363Y, A364L, and A364V mutants exhibiting the lowest signals (<10% of those from control wild-type virions). We could not quantify the binding of DSB to the L363N, A364F, and A364Y mutants, since these particles were unstable and sufficient quantities were not recovered for the assay. The H358Y, L363F, and A364I mutants exhibited approximately 10% of the wild-type DSB binding signal, while the L363M and A364M mutants exhibited 50% and 30%, respectively, of the wild-type DSB binding signal. Thus, resistance to DSB was associated with a reduced capacity of immature HIV-1 particles to bind the drug. Interestingly, the last two mutants retained a significant capacity to bind DSB despite exhibiting resistance to the compound, suggesting the possibility that binding of DSB to its target may not be sufficient to inhibit infectivity.
Resistance to DSB in vitro is conferred by a variety of CA-SP1 mutations: implications for the antiviral mechanism and drug development.
The results of this study indicate that single amino acid substitutions in the CA-SP1 cleavage site confer resistance to DSB in culture by reducing the ability of the compound to associate with Gag during particle assembly. Although several of the mutations resulted in moderate to severely impaired HIV-1 replication in the absence of the compound, others resulted in only slight delays relative to what was found for the wild type. These findings suggest that the emergence of drug-resistant variants may occur readily when DSB is administered to HIV-infected individuals. Nonetheless, the highly conserved sequence of the CA-SP1 cleavage site suggests that DSB resistance in vivo may be constrained by impaired viral fitness. Ultimately, these studies will be addressed by identification and analysis of DSB-resistant variants that emerge in patients undergoing DSB monotherapy.
Although the molecular target of DSB is likely the Gag protein within assembled immature HIV-1 particles, the structure of the binding site has yet to be determined due to the poorly behaved nature of the C-terminal end of CA in protein crystals and in solution. Our finding that a variety of amino acid substitutions at the P1 and P1′ positions of the CA-SP1 cleavage site confer resistance to DSB and inhibit the binding of the compound suggest that the DSB binding site exhibits tight molecular constraints due to the presence of the specific side chains at these positions. These substitutions could directly disrupt molecular contacts between Gag and DSB or may affect binding indirectly by perturbing the secondary structure of this region of the Gag protein.
Interestingly, the L363M and A364M mutants retained substantial capacities to bind DSB yet were markedly resistant to the compound in single-cycle and continuous-replication assays. These observations suggest the possibility that binding of DSB may not be sufficient for antiviral activity. If correct, this would suggest that DSB inhibits the cleavage of the CA-SP1 junction by altering the conformation of the cleavage site rather than by steric interference with the access of the protease.
Studies of the DSB antiviral mechanism have revealed a novel target for antiviral therapy: the HIV-1 Gag protein. To facilitate the present study, we developed a novel scintillation proximity assay to quantify the binding of radiolabeled DSB to immature HIV-1 particles. This approach is simple, rapid, and highly specific. Because binding of DSB to immature HIV-1 particles is saturable and can be specifically competed by unlabeled DSB (16), a potential approach for identifying novel maturation inhibitors is to screen libraries of compounds for molecules that competitively inhibit the binding of DSB to immature HIV-1 virus-like particles. Because such inhibitors may occupy the binding site of DSB, it is expected that some will also interfere with HIV-1 maturation.
Supplementary Material
Acknowledgments
We thank members of the Aiken laboratory for helpful suggestions and Sebastian Joyce for use of the Packard Topcount instrument. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: hybridoma 183-H12-5C from Bruce Chesebro.
This work was supported by NIH grant AI062452.
Footnotes
Published ahead of print on 11 October 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C., and C. H. Chen. 2005. Betulinic acid derivatives as HIV-1 antivirals. Trends Mol. Med. 11:31-36. [DOI] [PubMed] [Google Scholar]
- 3.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, X., A. Roldan, J. Hu, M. A. Wainberg, and C. Liang. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo, X., B. B. Roy, J. Hu, A. Roldan, M. A. Wainberg, and C. Liang. 2005. The R362A mutation at the C-terminus of CA inhibits packaging of human immunodeficiency virus type 1 RNA. Virology 343:190-200. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto, F., Y. Kashiwada, L. M. Cosentino, C. H. Chen, P. E. Garrett, and K. H. Lee. 1997. Anti-AIDS agents-XXVII. Synthesis and anti-HIV activity of betulinic acid and dihydrobetulinic acid derivatives. Bioorg. Med. Chem. 5:2133-2143. [DOI] [PubMed] [Google Scholar]
- 7.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 8.Kanamoto, T., Y. Kashiwada, K. Kanbara, K. Gotoh, M. Yoshimori, T. Goto, K. Sano, and H. Nakashima. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob. Agents Chemother. 45:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakalian, M., C. P. McMurtrey, F. J. Deeg, C. W. Maloy, F. Li, C. T. Wild, and K. Salzwedel. 2006. 3-O-(3′,3′-dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 Gag precursor assembled in vitro. J. Virol. 80:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 13.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 14.Yu, D., C. T. Wild, D. E. Martin, S. L. Morris-Natschke, C. H. Chen, G. P. Allaway, and K. H. Lee. 2005. The discovery of a class of novel HIV-1 maturation inhibitors and their potential in the therapy of HIV. Expert Opin. Investig. Drugs 14:681-693. [DOI] [PubMed] [Google Scholar]
- 15.Zhou, J., C. H. Chen, and C. Aiken. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 280:42149-42155. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, J., X. Yuan, D. Dismuke, B. M. Forshey, C. Lundquist, K. H. Lee, C. Aiken, and C. H. Chen. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.