Abstract
The monoclonal antibody 2G2 has been used extensively for detection and quantification of structural changes of human rhinovirus serotype 2 during infection. It recognizes exclusively A and B subviral particles, not native virus. We have elucidated the basis of this selectivity by determining the footprint of 2G2. Since viral escape mutants obviously cannot be obtained, the structures of complexes between Fab fragments of 2G2 and 80S subviral B particles were determined by cryoelectron microscopy. The footprint of the antibody corresponds to the capsid region that we predicted would undergo the most dramatic changes upon RNA release.
Human rhinoviruses (HRVs) belong to the picornavirus family. They are icosahedral with T=1, pseudo-T=3 symmetry, are about 30 nm in diameter, and are composed of 60 copies of each of the proteins VP1 through VP4 that encapsidate a single-stranded positive-sense RNA genome. Twelve of the 99 serotypes, the minor receptor group, bind members of the low-density lipoprotein receptor family for cell entry, whereas the remaining 87 serotypes use intercellular adhesion molecule 1 (ICAM-1) for infection.
As shown for the major group virus HRV serotype 14 (HRV14) and for the minor group virus HRV2, receptor binding is followed by internalization via the clathrin-mediated endocytic pathway (8, 25). Within the endosome, the virion undergoes coordinated structural changes preceding the RNA release into the cytosol. For major group viruses, these changes are catalyzed by the ICAM-1 receptor and possibly aided by low endosomal pH (22). In the case of minor group viruses, the low pH of the late endosomal compartment is the exclusive trigger for these modifications.
HRV2 has been extensively analyzed with respect to these structural changes (9, 11, 12, 14-17, 20, 21). In the first step, the innermost capsid protein VP4 is released, giving rise to subviral particles sedimenting at about 135S, whereas the native virion sediments at 150S. This change in sedimentation behavior is accompanied by changes in antigenicity. The RNA is then released, resulting in an empty capsid with a sedimentation constant of 80S that is finally degraded in lysosomes (24). Using cryoelectron microscopy (cryo-EM) and X-ray structural data, we produced a model for the HRV2 empty capsid after RNA release (9). The capsid was seen to have expanded by 4%, with a relative movement of all capsid proteins. In particular, the viral protein VP1 around the fivefold axes make an iris type of movement to open a 10-Å-diameter channel, which allows the RNA genome to exit.
The monoclonal antibody 2G2, obtained from a mouse injected with purified HRV2, proved extremely useful for the detection of the structural changes occurring upon infection. For example, it was employed to unequivocally demonstrate the low-pH dependency of HRV2 for infection; no subviral particles were formed in the presence of inhibitors of endosomal acidification, such as monensin (18) or the H+-ATPase-specific drug bafilomycin A1 (2, 3, 5, 23). However, although the antibody has been used in many instances for analytical purposes, its binding epitope has remained unknown. We have now solved the structure of complexes between empty capsids of HRV2 and Fab fragments of 2G2 by cryoelectron microscopy. This analysis reveals that the binding epitope of this antibody lies in the region that we have predicted to change the most upon the transition between the native virion and the empty capsid (9).
HRV2 was produced in a spinner culture of HeLa-H1 cells and purified by sucrose density gradient centrifugation, and empty capsids were obtained by incubation for 10 min at 56°C as described previously (9). The monoclonal antibody 2G2 was prepared from the tissue culture supernatant of cells grown in immunoglobulin G (IgG)-free medium by purification over a protein A column using an fast-protein liquid chromatography system, and Fab fragments were prepared and purified by following standard protocols (1). Fab 2G2 and the empty capsids were mixed at a molar ratio of about 120:1, incubated for 30 min at room temperature, and frozen on a holey carbon grid as described for cryo-EM imaging of complexes between native HRV2 and various soluble fragments of human very-low-density lipoprotein receptor (10, 19). Specimens were observed with a Philips CM200 electron microscope (LaB6 gun) operating at 200 kV. Defocus image pairs were obtained with a nominal magnification of ×38,000 at underfocus values ranging from 1.9 μm to 3.2 μm. Image analysis and three-dimensional reconstruction were performed as described in reference 6, using the previously determined map of the empty HRV2 capsid as a starting model (9). The final reconstruction included 858 particles, and the resolution was estimated by Fourier shell correlation of reconstructions from half data sets using the criterion of a 0.5 correlation. The part of the map which corresponds to the capsid (R < 168 Å) has a resolution of 13.8 Å, and the part of the map which corresponds to the Fabs (R > 168 Å) has a resolution of only 19.7 Å.
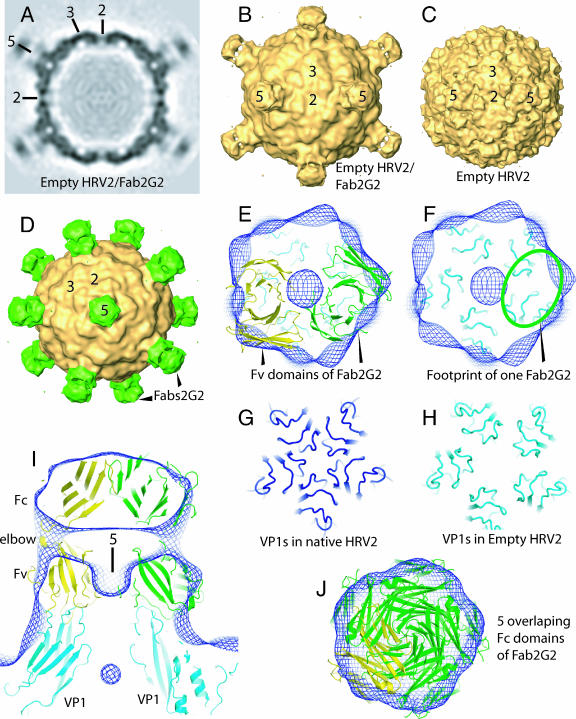
The reconstruction of the empty 80S B particles of HRV2 in complex with Fab fragments of 2G2 (Fig. 1A and B) reveals extra density on the fivefold axes compared with the 80S particles alone (Fig. 1C). This extra density represents the antibody fragments (Fig. 1D). A comparison of the densities of the viral capsid and the Fabs reveals that the occupancy of the Fabs is only about 40%. The position of the Fab in the map is well determined by the position of the narrow “pillars” of density, which link the annular region of density (Fv modules) in contact with the capsid with the distal disk of density (Fc modules) (Fig. 1B, D, and I). The “pillars” of density correspond to the Fab “elbow,” i.e., the peptide chains linking the Fv and Fc modules of the Fab. The footprints of the antibody on the virus surface overlap, so a maximum of two out of five possible copies of the Fab fragment can attach to each star-like mesa at the fivefold axes (Fig. 1E and F). Thus, the partial occupancy of Fabs can be accounted for by steric hindrance. Also, the footprint of each Fab overlaps two neighboring VP1s. For comparison, the VP1 surface loops around the fivefold axis on the native capsid and on the 80S particle as shown in Fig. 1G and H. This is the region of the capsid that displays the most dramatic change upon conversion of the native virus to the empty capsid. It is clear that the epitope recognized by 2G2 is absent from the native virion due to the extensive structural modification. The VP1s have moved relative to each other. It is notable that the star-like mesa is also the binding site of module 3 of human very-low-density lipoprotein receptor, as shown by cryo-EM (10, 19) and essentially confirmed by X-ray analysis (27).
FIG. 1.
Three-dimensional reconstructions of the empty capsid of HRV2 alone and in complex with Fab fragments of 2G2. Shown are a central section (A) and a surface representation (B) of the map of the complex viewed down a twofold axis. (C) Comparable surface view of the empty HRV2 capsid (9). (D) View down a fivefold axis of the complex with the 2G2 Fabs shaded in green. Representative two-, three-, and fivefold icosahedral axes are marked. The structure of the Fab 2G2 is not determined and so was replaced for docking by Fab 4C4 (28). A view down a fivefold axis of a thick section shows two Fab molecules docked in the complex map (E), and the footprint of one Fab which overlaps two VP1 molecules is outlined in green (F). One Fab is shaded in yellow, and the other is shaded in green. The Cα chains of VP1 in the empty capsid are depicted in cyan. The density map of the complex is represented by a blue mesh. Docking was performed manually using the program O. The disposition of the VP1s around a fivefold axis for the native capsid shaded in dark blue (26) (G) and the empty capsid shaded in cyan (9) (H) shows that each VP1 has moved with respect to its neighbor, thus modifying any epitope that covers two VP1s. (I) A thick section parallel to a fivefold axis showing the fit of the Fab in the density of the complex. (J) The fit of the constant domains of the Fab. As for the variable domains, steric hindrance is evident and will prevent more than two Fabs from binding on each fivefold axis. Panels A to D were produced using Amira (Mercury Computer Systems, Courtaboeuf, France), and panels E to J were made with PyMOL (http://pymol.sourceforge.net/).
It is interesting to compare this HRV2/Fab complex with that of a cucumber mosaic virus (CMV)/Fab complex (4). In the case of CMV, the antibodies bound only to the pentons of the T=3 virus, not to the hexons, most probably because of the relative differences in the distances between the subunits at these axes. In both cases, the difference in binding is apparently determined by the relative position of neighboring capsid proteins, but for HRV2 the difference arises because of a movement of the capsid proteins to allow exit of the RNA and for CMV the difference arises because of different symmetry positions on the capsid, i.e., fivefold versus pseudosixfold symmetry.
Complexes between HRV2 80S particles and 2G2 IgG have previously been analyzed by capillary electrophoresis. It is interesting that, with a high excess of antibody as used in these experiments, no aggregates appeared, and a well-defined complex migrating as a single small peak was formed; this complex was well separated from residual IgG and empty particles. However, lowering the concentration of the antibody also resulted in aggregation (13). This is typical of classical antibody-mediated aggregation due to monomeric binding of the IgGs. Such high antibody concentrations are detrimental to the contrast in cryo-EM and thus could not be employed in our present EM analysis; therefore, we had to make use of Fab fragments.
Using cryo-EM, we identified the footprint of a nonneutralizing antibody that was highly specific for the empty capsid. This antibody binds to a region that was shown in a previous cryo-EM study to present the greatest differences between the native particle and the empty capsid. The footprint of this antibody thus helps to confirm the cryo-EM model of empty HRV2 and explains the high specificity of the antibody. It is interesting that in the only other structure determination of an empty picornavirus after uncoating, i.e., poliovirus by cryo-EM (7), the VP1 on the fivefold axis are not seen to make a similar large scale movement to open a channel on the fivefold axis. This result implies that there are differences in the uncoating mechanism of these picornaviruses.
Acknowledgments
We thank F. Metoz for assistance in running the computers, D. Belnap, J. F. Conway, and B. Heymann for help with the PFT2 and EM3DR2 programs, T. S. Baker for the PFT and ROBEM programs employed, and I. Goesler for preparing HRV2 and 2G2.
This work was supported by Austrian Science Foundation grant P17516-B10.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Andrew, S. M., and J. A. Titus. 2000. Fragmentation of immunoglobulin G, p. 16.4.1-16.4.6. In J. S. Bonifacino, M. Dasso, J. B. Hartford, J. Lippincott-Schwartz, and M. Yamada (ed.), Current protocols in cell biology, 1st ed., vol. 2. John Wiley and Sons, Inc., New York, N.Y. [Google Scholar]
- 2.Bayer, N., E. Prchla, M. Schwab, D. Blaas, and R. Fuchs. 1999. Human rhinovirus HRV14 uncoats from early endosomes in the presence of bafilomycin. FEBS Lett. 463:175-178. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, N., D. Schober, E. Prchla, R. F. Murphy, D. Blaas, and R. Fuchs. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645-9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, V. D., E. S. Chase, A. W. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabec, M., G. Baravalle, D. Blaas, and R. Fuchs. 2003. Conformational changes, plasma membrane penetration, and infection by human rhinovirus type 2: role of receptors and low pH. J. Virol. 77:5370-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruemmer, A., F. Scholari, M. Lopez-Ferber, J. F. Conway, and E. A. Hewat. 2005. Structure of an insect parvovirus (Junonia coenia densovirus) determined by cryo-electron microscopy. J. Mol. Biol. 347:791-801. [DOI] [PubMed] [Google Scholar]
- 7.Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeTulleo, L., and T. Kirchhausen. 1998. The clathrin endocytic pathway in viral infection. EMBO J. 17:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewat, E., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317-326. [DOI] [PubMed] [Google Scholar]
- 10.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korant, B. D., K. Lonberg-Holm, J. Noble, and J. T. Stasny. 1972. Naturally occurring and artificially produced components of three rhinoviruses. Virology 48:71-86. [DOI] [PubMed] [Google Scholar]
- 12.Korant, B. D., K. Lonberg-Holm, F. H. Yin, and J. Noble-Harvey. 1975. Fractionation of biologically active and inactive populations of human rhinovirus type 2. Virology 63:384-394. [DOI] [PubMed] [Google Scholar]
- 13.Kremser, L., M. Petsch, D. Blaas, and E. Kenndler. 2006. Capillary electrophoresis of affinity complexes between subviral 80S particles of human rhinovirus and monoclonal antibody 2G2. Electrophoresis 27:2630-2637. [DOI] [PubMed] [Google Scholar]
- 14.Lonberg-Holm, K., L. B. Gosser, and E. J. Shimshick. 1976. Interaction of liposomes with subviral particles of poliovirus type 2 and rhinovirus type 2. J. Virol. 19:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonberg-Holm, K., and J. Noble-Harvey. 1973. Comparison of in vitro and cell-mediated alteration of a human rhinovirus and its inhibition by sodium dodecyl sulfate. J. Virol. 12:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonberg-Holm, K., and B. D. Korant. 1972. Early interaction of rhinoviruses with host cells. J. Virol. 9:29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonberg-Holm, K., and F. H. Yin. 1973. Antigenic determinants of infective and inactivated human rhinovirus type 2. J. Virol. 12:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neubauer, C., L. Frasel, E. Kuechler, and D. Blaas. 1987. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology 158:255-258. [DOI] [PubMed] [Google Scholar]
- 19.Neumann, E., R. Moser, L. Snyers, D. Blaas, and E. A. Hewat. 2003. A cellular receptor of human rhinovirus type 2, the very-low-density lipoprotein receptor, binds to two neighboring proteins of the viral capsid. J. Virol. 77:8504-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble, J. N., and K. Lonberg-Holm. 1973. Interactions of components of human rhinovirus type 2 with HeLa cells. Virology 51:270-278. [DOI] [PubMed] [Google Scholar]
- 21.Noble-Harvey, J., and K. Lonberg-Holm. 1974. Sequential steps in attachment of human rhinovirus type 2 to HeLa cells. J. Gen. Virol. 25:83-91. [DOI] [PubMed] [Google Scholar]
- 22.Nurani, G., B. Lindqvist, and J. M. Casasnovas. 2003. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J. Virol. 77:11985-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prchla, E., E. Kuechler, D. Blaas, and R. Fuchs. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schober, D., P. Kronenberger, E. Prchla, D. Blaas, and R. Fuchs. 1998. Major and minor-receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol. 72:1354-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyers, L., H. Zwickl, and D. Blaas. 2003. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J. Virol. 77:5360-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1179-1194. [DOI] [PubMed] [Google Scholar]
- 27.Verdaguer, N., I. Fita, M. Reithmayer, R. Moser, and D. Blaas. 2004. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11:429-434. [DOI] [PubMed] [Google Scholar]
- 28.Verdaguer, N., G. Schoehn, W. F. Ochoa, I. Fita, S. Brookes, A. King, E. Domingo, M. G. Mateu, D. Stuart, and E. A. Hewat. 1999. Flexibility of the major antigenic loop of foot-and-mouth disease virus bound to a Fab fragment of a neutralising antibody: structure and neutralisation. Virology 255:260-268. [DOI] [PubMed] [Google Scholar]