Abstract
Leishmania parasites (order Kinetoplastida, family Trypanosomatidae) cause a spectrum of human diseases ranging from asymptomatic to lethal. The ∼33.6 Mb genome is distributed among 36 chromosome pairs that range in size from ∼0.3 to 2.8 Mb. The complete nucleotide sequence of Leishmania major Friedlin chromosome 1 revealed 79 protein-coding genes organized into two divergent polycistronic gene clusters with the mRNAs transcribed towards the telomeres. We report here the complete nucleotide sequence of chromosome 3 (384 518 bp) and an analysis revealing 95 putative protein-coding ORFs. The ORFs are primarily organized into two large convergent polycistronic gene clusters (i.e. transcribed from the telomeres). In addition, a single gene at the left end is transcribed divergently towards the telomere, and a tRNA gene separates the two convergent gene clusters. Numerous genes have been identified, including those for metabolic enzymes, kinases, transporters, ribosomal proteins, spliceosome components, helicases, an RNA-binding protein and a DNA primase subunit.
INTRODUCTION
The Kinetoplastida are flagellated, primarily parasitic, protozoans found in terrestrial and aquatic environments. Kinetoplastid species cause disease in metazoa ranging from plants to vertebrates, resulting in extensive human suffering and death, as well as considerable economic loss from infection of livestock, wildlife and crops (1,2). Study of these organisms has been valuable for the investigation of fundamental molecular and cellular phenomena, such as RNA editing (3), mRNA trans-splicing (4), GPI anchoring of proteins (5), antigenic variation (6) and telomere organization (7). Their early evolutionary divergence makes comparison of their DNA and protein sequence with those of other eukaryotes, as well as prokaryotes, useful for the identification of ancient conserved motifs and their protein sequences may be a useful source of diversity for protein engineering.
Leishmania parasites belong to the family Trypano somatidae and the numerous human-infective Leishmania spp. are responsible for a wide spectrum of human diseases with pathologies that range from asymptomatic to lethal (8). The World Health Organization (WHO) has estimated that there are over two million new cases of leishmaniasis each year in 88 countries, with 367 million people at risk (http://www.who.int/health-topics/leishmaniasis.htm). Whilst a correlation exists between disease manifestation and parasite species (8), host factors also play an important role in disease pathology (9). Leishmaniasis/HIV co-infection remains a significant problem (10). Diagnosis of Leishmania infection remains problematic; existing methods are labor-intensive and have sensitivity and specificity issues; current chemotherapeutic agents are unsuitable because of their high toxicity; and there are no approved vaccines (11,12). For these reasons, a greater knowledge of Leishmania biochemistry and genetics is sorely required.
The Leishmania karyotype is conserved among species (albeit with considerable size polymorphism among species), except that the Old World species (including Leishmania major) have 36 chromosomes (13) and the New World species are reported to have 35 (Leishmania braziliensis complex) or 34 (Leishmania mexicana complex) (14). More modest chromosome size polymorphisms exist within species amongst the various Leishmania strains. Despite this often high degree of polymorphism, gene order is conserved between strains and species.
In 1994, the Leishmania Genome Network (LGN) (http://www.ebi.ac.uk/parasites/leish.html) was set up under the auspices of the WHO to initiate a Leishmania genome sequencing project, and L.major MHOM/IL/81/Friedlin (LmjF) was selected as the reference strain. This genome is ∼33.6 Mb in size, with chromosomes ranging from ∼0.3 to 2.8 Mb (15). A cosmid library of 9216 clones (∼9-fold genome coverage), constructed in the shuttle vector cLHYG (16), was used for a first generation cosmid contig map of the entire genome (17). The sequence of the smallest Leishmania chromosome, chromosome 1 (285 kb) revealed 79 potential protein-coding genes (1). Remarkably, the genes on this chromosome were organized into two large polycistronic units, with the first 29 genes on one DNA strand and the remaining 50 genes on the other, oriented such that their mRNAs are transcribed divergently towards the telomeres. The 257 kb informational region of chromosome 1 was flanked by telomeric and sub-telomeric sequences differing significantly in size (∼29 kb) between the chromosome 1 homologs (18).
We report here the complete nucleotide sequence and analysis of the third smallest chromosome in L.major Friedlin, chromosome 3, which has a size of 384.5 kb and contains 95 putative protein-coding genes.
MATERIALS AND METHODS
Cosmid mapping
A genomic library was constructed in the shuttle cosmid cLHYG (16) using partial Sau3AI-digested genomic DNA from L.major MHOM/IL/81/Friedlin and 9216 clones were picked, arrayed and transferred to nylon membranes (17). These filters were first hybridized with a chromosome 3/2b DNA probe obtained from clamped homogeneous electric field gel electrophoresis and subsequently with a chromosome 2a DNA probe. Hybridizations were done in 1 M NaCl, 1% SDS, 100 µg/ml denatured salmon sperm DNA for 18 h at 65°C. Filters were washed twice for 20 min in 2× SSC, 1% SDS at 65°C and twice for 20 min in 0.1× SSC, 0.2% SDS at 65°C. Clones that hybridized with the chromosome 3/2b probe but not with the chromosome 2a probe were deemed to be likely to contain sequence from chromosome 3 and were initially mapped by cosmid fingerprinting (17). The precise map locations of these clones were subsequently determined by comparison of their end sequences with the consensus sequence obtained from selected cosmids that were sequenced using the shotgun method described below.
Cosmid shotgun sequencing
Cosmid DNA (from L952, L7535, L4625, L622, L6202, L5204, L1559, L3561, L5801, L509, L6910, L6290, L505, L7472, L712, L7234 and L3223) was sheared by sonication, repaired using the Klenow fragment of Escherichia coli DNA polymerase and T4 polynucleotide kinase and fractionated by agarose gel electrophoresis. Fragments of 0.8–2.0 kb were purified and ligated to SmaI-digested, phosphatase-treated M13mp18 RF DNA. Clones obtained from each ligation were screened for the presence of insert (white color in the presence of isopropyl β-d-thiogalactoside). DNA was prepared by PEG precipitation and templates sequenced using dyeTerminator and dyePrimer chemistry with primer –21M13 (5′-GTAAAA CGACGGCCAGT). Cosmid end sequences were obtained from cosmid DNA using dyeTerminator chemistry with primer HygT3 (5′-CCGTTGCGCCGTAGAAG) or HygT7a (5′-CGATGATAAGCTGTCAAACATG). Generally 600–1000 sequencing reactions were performed during the shotgun phase before assembly using the phred/phrap/consed software suite (19). Gaps and regions of poor sequence quality were finished by sequencing existing clones with different chemistries and/or different primers or by sequencing PCR products amplified from the cosmid DNA. Sequences from several clones containing telomeric and sub-telomeric sequences, obtained by using a PCR-based method (20), overlapped with both ends of the chromosome 3 sequence and were included in the final chromosome 3 consensus. The entire sequence has been deposited in the GenBank sequence database with accession no. AC125735.
Southern analysis
Genomic DNA preparation and Southern blot analyses were performed as described previously (21). LmjF genomic DNA was digested with restriction enzymes, separated by 0.8% agarose gel electrophoresis and transferred to Hybond N+ membranes (Amersham) by capillary action. For use as a probe, a 560 bp fragment containing part of the ‘left-most’ gene on chromosome 3 (Chr3_0010) was PCR amplified with oligonucleotides Chr3_0010-1 (5′-ATTCGAGAGAGGATGCGACTGGCAC) and Chr3_0010-2 (5′-TCTGGAGGTGCTGGACATTGGAGGA). A 996 bp fragment corresponding to the ‘right-most’ gene on chromosome 3 (Chr3_0890) was amplified with the oligonucleotides Chr3_0890-1 (5′-TCCTTCAGCTCTGTATTAGTCCG) and Chr3_0890-2 (5′-CAATAACGAGCGTCCAGAGTGGA). Fragments were labeled with [α-32P]dCTP using the High Prime labeling system (Amersham).
Clamped homogeneous electric field analysis
Agarose blocks containing 107 promastigotes from LmjF were prepared as previously described (22). Chromosome-sized DNA was separated from these blocks by clamped homogeneous electric field (CHEF) gel electrophoresis (Bio-Rad Mapper system) in a 1% SeaKem LE agarose gel with 1× TBE buffer. Conditions to separate DNA in the range of 150–600 kb utilized a linear switch time gradient of 15.02–44.48 s at 6 V/cm for 23 h 22 min at 14°C. The gel was transferred to a Hybond N+ membrane (Amersham) by capillary action. The Chr3_0010 fragment described above was used as a probe.
Nuclear run-on assays
These experiments were performed as described elsewhere (23). Briefly, nuclei were isolated from 2 × 108 LmjF promastigotes by washing twice in PBS, resuspending in 10 ml ice-cold lysis buffer (10 mM Tris–HCl pH 7.5, 3 mM CaCl2, 2 mM MgCl2) and adding NP-40 to a final concentration of 0.5%. Cells were transferred to a Dounce homogenizer and broken with 40 strokes; the nuclei were collected by centrifugation (1400 g) and washed once with lysis buffer. To elongate nascent RNA, nuclei were resuspended in 100 µl of 100 mM Tris–HCl (pH 7.5), 25% glycerol, 0.15 mM spermine, 0.5 mM spermidine, 2 mM DTT, 40 U RNasin (Promega), 2 mM MgCl2, 4 mM MnCl2, 50 mM NaCl, 50 mM KCl, 2 mM ATP, 2 mM GTP, 2 mM UTP, 10 µM CTP and 300 µCi of [α-32P]CTP (3000 Ci/mmol; Amersham). The incubation was carried out for 6 min at 26°C, after which DNase I (10 U) was added. Incubation was continued for 5 min at 37°C, then stopped by the addition of 100 µl of 10 mM Tris–HCl (pH 7.5), 10 mM EDTA, 1% SDS and 100 µg/ml proteinase K. After 5 min incubation at 37°C, RNA was extracted with phenol-chloroform and separated from free nucleotides by G-50 Sephadex chromatography. Labeled nascent RNA was hybridized to Hybond filters (Amersham) containing dots of 1 µg of single-stranded M13 DNA. Hybridization was performed for 48 h at 50°C in 50% formamide, 5× SSC, 0.2% SDS, 4× Denhardt’s reagent and 100 µg/ml salmon sperm DNA. Post-hybridization washes were carried out in 0.1× SSC and 0.1% SDS at 65°C. In the assays carried out in the presence of UV light, promastigotes (in a total volume of 15 ml) were irradiated in Petri dishes, with agitation, in a Stratalinker UV crosslinker (Stratagene). After irradiation, cells were incubated for at least 1.5 h at 28°C to allow the clearing of RNA polymerases engaged prior to irradiation.
Sequence analysis
The G+C content of the entire chromosome 3 sequence was calculated using the sequence and annotation viewer software Artemis, version 4 (24), as was the overall G+C content of coding sequences. The RepeatMasker program (A.F.A. Smit and P. Green, unpublished data) was used to search for simple repeats and low complexity regions within the sequence. The GCG programs Window and StatPlot (25) were used to identify polypyrimidine tracts within the chromosome 3 sequence. The results were then parsed into a suitable format for import into Artemis, where their locations with respect to predicted protein-coding regions were identified.
The percentage of cumulative GC skew for the whole chromosome sequence was calculated with window length (L) = 10 kb and sliding window (SL) = 1 kb using the expression:
%GC skew = Σi(δG – δC) × 100/(δG + δC),
where δG and δC are the counts of nucleotides G and C in the given window length.
Similarly, purine excess for the whole chromosome was calculated using the expression:
Pexcess = Σi(δpurine – δpyrimidine),
where δpurine and δpyrimidine are the sum of nucleotides (A,G) and (C,T) with L and SL as above.
The consensus sequence for chromosome 3 was examined for putative protein-coding open reading frames (ORFs) using a combination of gene prediction techniques, combined in our Machine Automated Gene Identification (Magi) software (25a). The methods included Glimmer 2.0 (26) (trained with previously identified protein-coding genes from LmjF chromosomes 1 and 4), in-house adaptations of Testcode (27), CodonUsage (28) and Genescan (29). Putative genes predicted by at least three of these four methods, along with graphical representations of the Testcode and Genescan statistics, were exported from Magi and imported into the Artemis annotation platform (24) for visual inspection and manual editing. The sequences was searched for tRNA genes using tRNAscan (30).
The amino acid sequence predicted from each putative gene was used as a query sequence for local blastp and tblastn (31) searches (through Artemis) of the GenBank non- redundant protein and a kinetoplastid-specific nucleotide database, respectively. These databases were created from sequence collections downloaded by ftp from ftp.ncbi.nih.gov/blast/db/ and ftp.ebi.ac.uk/pub/databases/parasites/blastdbs/. Generally, hits with blast scores of >50 and e-values of <1 × e–6 were considered potentially significant, although some exceptions were made upon visual inspection of the alignments. Each protein sequence was then searched against numerous collections of protein motifs and families (SWISS_PROT release 39.27, ProDom version 2001.2, SMART version 3.3, PROSITE release 16.46, Pfam version 6.6, PRINTS release 31.0, Domo version 2.0 and BLOCKs release 13.0). These searches were carried out using various search algorithms (http://blocks.fhcrc.org/blocks/blocks_search.html; http://hits.isb-sib.ch/cgi-bin/PFSCAN; http://dna.stanford.edu/emotif/emotif-search.html; http://www.sanger.ac.uk/Software/Pfam/). BLAST searching of the database of Clusters of Orthologous Groups of Proteins (COGs) (http://www.ncbi.nlm.nih.gov/COG/cog99nitor.html) (32) was performed simultaneously, along with identification of putative protein localization sites and transmembrane spanning regions using PSORT version II (http://psort.nibb.ac.jp/form2.html) (33) and TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (34). Protein domains with distinct evolutionary origins and functions were identified within the predicted genes using the NCBI Conserved Domain Database and Search Service, version 1.54 (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (35). Gene ontology (GO) terms (36) were assigned to the predicted proteins, based on their top matches to proteins with GO annotations from SwissProt and TrEMBL. This analysis was carried out through the GOblet server running at the Max Planck Institute (MPI) for Molecular Genetics (http://goblet.molgen.mpg.de/cgi-bin/GOblet.cgi).
RESULTS
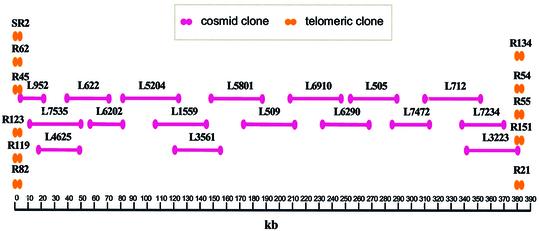
LmjF chromosome 3 was sequenced from a set of overlapping cosmid and smaller clones. A contig map of these cosmids was constructed using a combination of cosmid fingerprinting, hybridization with chromosome 3-specific probes and cosmid end sequencing, as previously described (21). Seventeen cosmid clones, representing a tile-path for most of chromosome 3, were selected for shotgun sequencing (Fig. 1). In addition, 11 clones containing telomeric and sub-telomeric sequences were prepared using a PCR-based method (20), since these sequences were not present in this cosmid library. The sequences obtained from the cosmid clones and telomeric clones were assembled into a final consensus sequence of 384 518 bp.
Figure 1.
Leishmania major Friedlin chromosome 3 cosmid map. The locations of the sequenced cosmids and telomeric PCR clones are shown relative to the final consensus sequence. The scale is shown at the bottom in kb.
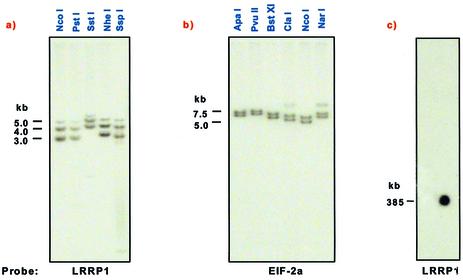
The consensus sequence from the ‘left’ end of chromosome 3 has 1165 bp 5′ to the first protein-coding gene. The distance from the last protein-coding gene at the ‘right’ end to the end of the available sequence is 2283 bp. The consensus sequence terminated in 40 nt of telomere hexamer repeat (THR) sequence (CCCTAA) at the ‘left’ end and 520 nt of the THR repeat at the ‘right’ end. This sequence is found at the telomeres of all Leishmania chromosomes (21,37). No other telomere-associated (TAS) or sub-telomeric (LST) repeats were found adjacent to the THR repeats in chromosome 3, unlike in other Leishmania chromosomes (18). Genomic Southern analysis using a number of different restriction enzymes and probes from each telomere detected two major fragments in each case (Fig. 2a and b). The presence of minor bands in some digests at the ‘right’ telomere is most likely due to incomplete digestions. The different fragment sizes observed presumably reflect a small size difference between chromosome homologs in the size of the telomeric regions, and indicate that a further 800 or 1710 bp (depending upon the homolog) of unsequenced DNA is located at the ‘left’ end of the chromosome 3 sequence. It is likely that this additional sequence is composed largely of THR repeats, although the presence of TAS-A and/or TAS-B sequences (18) cannot be ruled out. Similarly, a further 1180 or 2280 bp of unsequenced DNA is located at the ‘right’ end. The presence of a third, less intense, band with the Chr3_0010-1 probe, which is not due to incomplete digestion, suggests further heterogeneity in the length of the telomeric sequences at the ‘left’ end of one of the chromosome 3 homologs. Southern blot analysis of CHEF gel-separated chromosomal DNA with this probe showed hybridization only to chromosome 3 (Fig. 2c), indicating that the additional restriction fragment is not likely due to cross-hybridization with a fragment elsewhere in the genome. These results indicate that LmjF chromosome 3 has the smallest combined telomeric and sub-telomeric capping region identified in any trypanosomatid species to date (18,20,38–40). Thus, the total size of the chromosome 3 homologs ranges from 387.1 to 389.5 kb. This is consistent with measurements from PFGE separations of LmjF chromosomes, where both chromosome 3 homologs co-migrate together with chromosome 2b, indicating that the size difference between the chromosome 3 homologs is small (15).
Figure 2.
Southern blot analysis of the telomeric ends of LmjF chromosome 3. Chromosome 3 genomic DNA was digested with the restriction enzymes indicated and probed with (a) a probe for Chr3_0010 (the ‘left-most’ gene on chromosome 3) and (b) a probe for Chr3_0890 (the ‘right-most’ gene on chromosome 3). (c) CHEF gel separated LmjF chromosomal DNA was probed with Chr3_0010. Size markers (in kb) are shown to the left of each panel.
Analysis of the chromosome 3 sequence revealed 1154 ORFs larger than 225 nt (75 amino acids) in length. A single gene encoding a tRNALysCUU was identified using tRNAscan. Putative protein-coding ORFs were identified using a combination of gene prediction techniques (Glimmer 2.0, CodonUsage, Testcode and Genescan) as well as manual inspection in Artemis. These analyses identified 95 putative protein-coding ORFs (Fig. 3). The gene density is 1 gene per 4.0 kb and the mean size of these predicted protein-coding ORFs is 2517 bp, with a range of 318 (Chr3_0870) to 9342 bp (Chr3_0690); the predicted protein-coding sequence accounts for ∼60.8% of chromosome 3. No intron sequences were detected in these genes, as is typical for the Trypano somatidae. The average G+C content for sequence predicted to be coding is 65.13%, which is somewhat higher than that of the genome in general (62.5%).
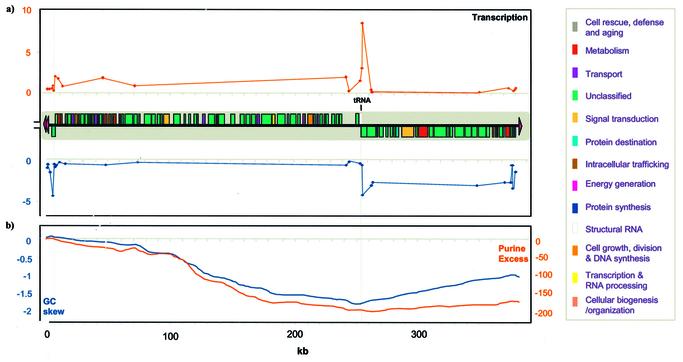
Figure 3.
Gene organization of LmjF chromosome 3. (a) The location and coding strand of the 95 protein-coding ORFs and the one tRNA gene are indicated by the boxes, which are color-coded according to functional category, as indicated at the side. Telomeric hexamer repeat regions are indicated by red arrowheads. The results from a nuclear run-on experiment are plotted against the position of each single-stranded M13 DNA on chromosome 3, with the signal from probes on the top strand shown above the gene map and those from the bottom strand shown below. The x-axis scale indicates distance in kb. The y-axes represent relative signal intensity (in arbitrary units × 103). (b) Plots of cumulative GC skew (red) and purine excess (blue) are shown in sliding 10 kb windows across chromosome 3, with units indicated on the left and right, respectively.
Pyrimidine-rich tracts provide part of the processing signals for the addition of the 39 nt spliced leader (or mini exon) sequence at the 5′ end of all mRNAs and for 3′ polyadenylation (41). Tracts of 10 pyrimidines or more were identified in all but two intergenic regions on chromosome 3 (data not shown). The size distribution in these regions was between 9 and 29 nt (mean length 24). Pyrimidine tracts of this size range were rare within the predicted protein-coding regions (fewer than half of the ORFs contained tracts of 10 nt or longer). The mean tract size in coding regions was 13 pyrimidines (maximum 24).
Interspersed and simple tandem repeats in the finished sequence were masked and a combination of blastx/blastp/tblastn database searches carried out in order to assign each of the 95 putative protein-coding genes on chromosome 3 to 13 different categories based on their putative biological function (Table 1). In parallel, a number of databases containing collections of biologically significant sites and patterns were queried with the sequence of each putative protein. Protein localization signals and probable transmembrane regions were also identified. Out of the 95 putative protein-coding genes identified on chromosome 3, 57 were predicted to be soluble, 17 were predicted to have a single transmembrane domain and 20 were predicted to have two or more transmembrane domains. Of this latter group, 12 were predicted to be localized to the plasma membrane and five to the endoplasmic reticulum.
Table 1. Functional classification of L.major chromosome 3 genes.
| Category | n | |
|---|---|---|
| Cell growth, division and DNA synthesis | 5 | |
| Putative DNA primase large subunit dp58 (Chr3_0070) | ||
| Helicase 1 (Chr3_0490) | ||
| Helicase 2 (Chr3_0610) | ||
| Serine-threonine protein kinase 4 (Chr3_0170) | ||
| RNA-binding protein (Chr3_0430) | ||
| Cell rescue, defense, death and aging | 0 | |
| Cellular organization/biogenesis | 1 | |
| Ankyrin repeat protein 1 (Chr3_0390) | ||
| Energy generation | 1 | |
| Quinone oxidoreductase (Chr3_0480) | ||
| Intracellular trafficking | 0 | |
| Metabolism | 6 | |
| Long chain fatty acyl CoA synthetase (Chr3_0180) | ||
| 2-Aminoethylphosphonate:pyruvate aminotransferase (Chr3_0030) | ||
| Inorganic pyrophosphatase (Chr3_0820) | ||
| δ-1-Pyroline-5-carboxylate dehydrogenase (Chr3_0160) | ||
| d-3-Phosphoglycerate dehydrogenase (Chr3_0020) | ||
| Multicopper oxidase 1 (Chr3_0860) | ||
| Protein destination | 3 | |
| 26S Protease regulatory ATPase subunit (Chr3_0450) | ||
| Protein arginine methyltransferase (Chr3_0500) | ||
| Cytochrome c oxidase assembly protein (Chr3_0080) | ||
| Protein synthesis | 4 | |
| Elongation initiation factor 2, α subunit (Chr3_0890) | ||
| Ribosomal protein L38 (Chr3_0200) | ||
| 60S Acidic ribosomal protein P1_2 (Chr3_0350) | ||
| 60S Acidic ribosomal protein P1_1 (Chr3_0345) | ||
| Signal transduction | 2 | |
| Serine-threonine protein kinase 2 (Chr3_0280) | ||
| Serine-threonine protein kinase 3 (Chr3_0690) | ||
| Structural RNA | 1 | |
| tRNALysCUU (Chr3_tRNA_Lys01) | ||
| Transcription and RNA processing | 1 | |
| U2AF23/25 spliceosome component (Chr3_0155) | ||
| Transport | 6 | |
| Lysine-arginine-ornithine transport system kinase 1 (Chr3_0510) | ||
| Leishmania unknown permease 1 (Chr3_0330) | ||
| Phosphate-repressible phosphate permease 1 (Chr3_0410) | ||
| ABC transporter protein 1 (Chr3_0130) | ||
| Choline transporter protein 1 (Chr3_0060) | ||
| Tubulin-specific chaperone (Chr3_0680) | ||
| Unclassified | 65 | |
| No homology/motifs | 14 | |
| Chr3_0115; Chr3_0140; Chr3_0150; Chr3_0205; Chr3_0220; Chr3_0290; Chr3_0360; Chr3_0370; Chr3_0470; Chr3_0640; Chr3_0650; Chr3_0720; Chr3_0770; Chr3_0850. | ||
| Uninformative blast homology/motifs | 32 | |
| Chr3_0010; Chr3_0015; Chr3_0040; Chr3_0100; Chr3_0110; Chr3_0210; Chr3_0230; Chr3_0240; Chr3_0270; Chr3_0300; Chr3_0310; Chr3_0312; Chr3_0315; Chr3_0340; Chr3_0400; Chr3_0420; Chr3_0460; Chr3_0510; Chr3_0590; Chr3_0600; Chr3_0630; Chr3_0670; Chr3_0700; Chr3_0710; Chr3_0730; Chr3_0760; Chr3_0780; Chr3_0790; Chr3_0800; Chr3_0830; Chr3_0840; Chr3_0880. | ||
| Shared with other Kinetoplastidae | 20 | |
| Chr3_0050; Chr3_0090; Chr3_0120; Chr3_0175; Chr3_0180; Chr3_0250; Chr3_0260; Chr3_0380; Chr3_0440; Chr3_0485; Chr3_0520; Chr3_0540; Chr3_0550; Chr3_0560; Chr3_0580; Chr3_0620; Chr3_0660; Chr3_0740; Chr3_0750; Chr3_0870. | ||
| Total for chromosome 3 | 96a |
aIncluding the structural RNA Chr3_tRNA_Lys01
The predicted amino acid sequences from 29 of the chromosome 3 genes (30.5%) showed sufficiently high sequence similarity to previously identified proteins to be assigned putative specific functions. The remaining 69.5% of genes remain unclassified. This percentage of unclassified genes is similar to that currently found in the whole LmjF genome project (70%). Within this category, 47% of the genes are predicted to encode proteins with similarity to proteins with unknown function in other organisms or show insufficient similarity (i.e. only short motifs) to allow functional assignment, 31% show similarity only to genes in other trypanosomatid species and 22% show no similarity to previously identified protein sequences.
Most of the functional protein classifications identified by the LGN are represented on chromosome 3, with the exception of proteins predicted to be involved in intracellular trafficking or cell rescue, defense, death and aging (Table 1). The numbers of genes predicted to encode for proteins with these functions within the whole genome are low in general. Of the 29 proteins whose functions have been determined, six are homologous to proteins involved in transport and a further six are homologous to proteins involved in metabolism, three of these being involved in amino acid metabolism. Proteins involved in cell growth, division and DNA synthesis and in protein synthesis are also relatively common on this chromosome (accounting in total for another nine genes). Two DEAD/DEAH box helicases have been identified, as have two unrelated permeases and three ribosomal proteins.
Three leucine-rich repeat (LRR) proteins, which show no significant identity to one another at the protein level, are encoded on chromosome 3 (Chr3_0010, Chr3_0100 and Chr3_0780). The functions of these proteins are unknown, but proteins containing these characteristic LRR motifs have been identified in a wide variety of species and are associated with widely differing functions (42). It has been suggested that LRRs may mediate protein–protein interactions, as well as cellular adhesion (43). Other identified functions of LRR-containing proteins include binding to enzymes (44) and vascular repair (45). Two glycoproteins of Leishmania, the parasite surface antigen-2/gp46 (PSA-2) and proteophosphoglycan (PPG1) are known to contain LRRs ((46). These findings have led to the recent suggestion that proteins containing these motifs may be involved in interactions with the mammalian host cell and/or insect vector (47). One LRR-containing protein from chromosome 3 (Chr3_0010) also contains a RING finger motif and shows greatest similarity to ESAG8/T-LR, which is encoded in several VSG gene expression sites of Trypanosoma brucei (48). While the function of ESAG8 is unknown, it has recently been shown to interact with a protein with a possible role in regulation of mRNA stability (49).
A number of gene duplication events are apparent on chromosome 3. It is estimated that between 5 and 10% of the genes in L.major Friedlin occur in more than one copy, either as gene duplications or as members of multigene families (50). The neighboring genes, Chr3_0300, Chr3_0310, Chr3_0312 and Chr3_0315, show varying degrees of similarity at both the DNA and amino acid levels. At the amino acid level, Chr3_0315 and Chr3_0310 share 67% identity, whilst Chr3_0312 and Chr3_0310 share 39%; the remaining pairings have identities of ∼20%, with the exception of Chr3_0315 and Chr3_0300, which show only 9% identity. Thus, it appears that two separate duplication events occurred to give rise to these four genes. Interestingly, whilst all of these proteins are predicted to belong to a group of uncharacterized, hydrophobic, evolutionarily related membrane proteins (GPR1/FUN34/YAAH) from fungi, bacteria and archaea (51,52), only Chr3_0300, Chr3_0310 and Chr3_0315 were predicted to contain multiple transmembrane domains. Ribosomal protein P1 copies 1 and 2 (Chr3_0345 and Chr3_0350) are identical at the amino acid level (only a single synonymous nucleotide substitution separates them at the DNA level), suggesting that the duplication event that gave rise to them may have occurred relatively recently. The level of identity at both the protein (26%) and nucleotide (13%) level is lower for Chr3_0560 and Chr3_0580, perhaps suggesting that this duplication is more ancient and that these two proteins may now have divergent functions.
The 95 putative protein-coding genes are organized into two large polycistronic clusters of 65 and 29 genes, plus a single gene located at one telomere (Fig. 3). The two clusters are organized convergently, i.e. transcription of the mRNAs is orientated away from each telomere. A tRNA gene is located within the convergent strand switch region at the boundary between the clusters. A single gene at the ‘left’ telomere is transcribed towards the telomere, i.e. divergently from the neighboring gene cluster. Nuclear run-on studies of chromosome 3 (Fig. 3a) indicate that transcription of chromosome 3 mirrors the gene organization pattern, since the hybridization signal for the gene-coding strand is ∼10-fold higher than that for the non-coding strand. Thus, transcription of chromosome 3 appears to be polycistronic and initiates upstream of each gene cluster, as seem for LmjF chromosome 1 (23).
RepeatMasker, a search program for simple repeats and low complexity regions, identified 257 mono- to hexanucleotide microsatellite sequences, including the telomeric hexamer repeats, comprising ∼3% of chromosome 3. Of the microsatellite sequences identified, 44% were GC-rich, 4% were AT-rich and 52% contained equal numbers of C/G and A/T nucleotides. CA·TG dinucleotide repeats were the most abundant class of microsatellites, accounting for 1% of the chromosome. A further 0.5% of chromosome 3 was GC-rich low complexity sequence and 2% was made up of other classes of tandemly repetitive DNA, including minisatellite sequences (repeats with motifs of 6–18 nt). No putative transposable elements were identified in this LmjF sequence.
GC skew is a statistical method to measure the strand-specific over-representation of guanine and the inflection point on a plot of GC skew versus nucleotide position corresponds to the position of the putative origin or terminus of DNA replication in a number of organisms (53). A non-random distribution of nucleotide bias was found for chromosome 3 using analyses similar to that previously described (54). The maximum value of cumulative GC skew identified coincides with the region between the single gene at the ‘left’ end of the chromosome and the polycistronic cluster of neighboring genes (Fig. 3b). Purine excess analysis has also been used to identify the putative origin or terminus of DNA replication in a number of bacterial species (55). The maximum value for purine excess is also located at this strand-switch point. The minimum GC skew and purine excess values coincide with the strand-switch region between the two polycistronic gene clusters, where the tRNA is located. A second maximum for GC skew purine excess is located upstream of the THR sequences at the ‘right’ end of chromosome 3.
DISCUSSION
This paper reports the complete sequence of LmjF chromosome 3. This sequence adds to the published large-scale contiguous finished sequence from protozoan parasite genomes, which currently consists of a draft sequence of the entire Plasmodium falciparum genome (56), one chromosome from L.major (21) and a region of chromosome 3 from Trypanosoma cruzi (57). Ninety-five protein-coding genes were identified on this chromosome, including proteins belonging to families as yet unidentified in this species. The gene density of LmjF chromosome 3 is 1 gene every 4.0 kb, which is slightly lower than the 1 every 3.3 kb observed for the informational region of LmjF chromosome 1 (21), but consistent with that observed for those LmjF chromosomes with large sections of completed sequence (chromosomes 1–6, 12–15, 19, 21–25, 27, 29, 31 and 34–36) (data not shown). In contrast, the gene density for a 75 kb portion of chromosome 21 is 1 gene every 2.4 kb (58), but the genes within this region are significantly smaller on average than those on chromosome 1, and probably reflect a cluster of smaller genes. Clusters of larger, more dispersed genes are also seen elsewhere in the LmjF genome. The gene density is also comparable to that obtained from T.cruzi (1 per 4.5 kb) (57) and P.falciparum (1 per 4 kb) (59), but is somewhat higher than that of Caenorhabditis elegans (1 gene per 5 kb) (60) and lower than that of Saccharomyces cerevisiae (1 gene per 2 kb) (61).
As first reported for chromosome 1 (21), the genes on LmjF chromosome 3 are organized into large polycistronic clusters. Similar organization of genes in T.cruzi and T.brucei (57) indicates that this pattern is common in trypanosomatids. However, in contrast to chromosome 1, chromosome 3 contains two large convergent polycistronic gene clusters and a single divergently transcribed gene. Whilst genes with related functions are clustered in polycistronic transcription units in many prokaryotic species, thus facilitating co-regulation (62), no such organization is the case in LmjF chromosome 3 or chromosome 1. The polycistronic units of LmjF are much larger than those found in these bacterial operons and contain genes with apparently unrelated functions. This suggests that the basis for this organization into large polycistronic clusters is unrelated to gene function.
LmjF chromosomes 1 (54) and 3 have GC skew and purine excess patterns that mirror the polycistronic gene clusters. The effects of selective pressure on events associated with replication, transcription and DNA repair are thought to influence base composition (63,64). Replication-associated mutations tend to skew gene locations to the lagging DNA strand (65). In addition, errors associated with transcription may also skew the base composition of the different DNA strands (55). However, the nucleotide bias in LmjF is the opposite of that seen in bacteria, where there is a positive correlation between gene orientation and GC skew, AT skew or purine excess (66). Hence, the organization of genes in clusters and the patterns of base skew and base composition seen in Leishmania may reflect replication and/or transcription processes. This implies that regions upstream of the clusters may contain replication origins and/or promoters.
Little is known about transcription of protein-coding genes by RNA polymerases II and III in LmjF and in other trypanosomatid species. Most protein-coding genes in these organisms are transcribed polycistronically (4) and regulation of their gene expression is believed to be primarily post-transcriptional, occurring at the levels of trans-splicing, polyadenylation, mRNA stability, translation or protein stability (4), suggesting that trypanosomatids may have relatively few polymerase II promoters. Transcriptional analysis of LmjF chromosome 1 using strand-specific nuclear run-on assays shows that transcription preferentially initiates within a <100 bp region in the strand-switch region between the two gene clusters, from which transcription appears to proceed bi-directionally towards each telomere (23). Nuclear run-on analysis of chromosome 3 indicates that transcription is similarly consistent with the pattern of gene organization (Fig. 3a). The results obtained using UV-irradiated nuclei suggest that transcription initiates in three different regions: between the divergently organized left-most gene and large left gene cluster, telomeric to the right-most gene cluster and upstream of the tRNA gene that separates the two gene clusters (Martinez-Calvillo et al., submitted for publication). RNA polymerase II appears to mediate transcription at the first two sites, while the last is carried out by polymerase III. Transcription of both strands appears to terminate within the tRNA gene region (Martinez-Calvillo et al., submitted for publication).
The sequence of the region between the two polycistronic clusters on chromosome 3 predicts significant curvature and stem–loop structures (58). Curvature has been associated with transcription promoter activity (67) as well as replication (68) and centromere functions (69). No centromeric or replication origin consensus sequence of any kinetoplastid has been identified to date. In a recent study, targeted replacement of the strand-switch region on two copies of chromosome 1 did not prevent normal replication and segregation of these chromosomes (70). This may be taken to indicate that it does not constitute an origin of replication or centromere, but it is possible that trans-complementation from the strand-switch region on the remaining chromosome 1 copy could be maintaining the other chromosomes lacking origins.
The structure of telomeric and sub-telomeric regions in a wide variety of organisms is highly plastic, often varying significantly in both size and composition between chromosomes within a single genome (71). Large differences are seen in the size of the sub-telomeric sequence between LmjF chromosome 1 homologs (18), due to the differences in the number of copies of simple sequence repeats. It had previously been shown that at least one telomere of chromosome 3 lacked the sub-telomeric repeats found in other LmjF chromosomes (72). The complete sequencing of chromosome 3 shows that there are only modest (1–2 kb) differences in the size of the telomeric sequences between homologs and that both ends lack sub-telomeric repeats. The role of sub-telomeric repeats in Leishmania is an open question. They are likely to play a role in the maintenance of genome stability by protecting the non-repetitive chromosomal termini from degradation, end-to-end fusion and rearrangement (73,74). Rearrangements in sub-telomeric repeat sequences have been linked to immune evasion through alteration of immunogenic epitopes on the cell surface in other organisms (75,76), though this has not yet been observed in Leishmania. It has recently been suggested that the 272 bp repeats in the sub-telomeric region of chromosome 1a have centromeric function, based upon analysis of a version of chromosome 1a in which all sequence other than the ‘right-hand’ sub-telomeric and telomeric repeats was deleted through homologous recombination (77). Similarly, analysis of chromosome fragmentation experiments on amplified small linear chromosome in Leishmania donovani suggested that the region responsible for mitotic stability contained homopolymer tracts (78). The absence of sub-telomeric repeats on LmjF chromosome 3 indicates that they are not a requirement for chromosome stability, re-opening the question of their biological function in other chromosomes.
Only 31% of the chromosome 3 genes could be assigned function on the basis of homology to proteins from other organisms. These proteins span a wide range of functional classes, as detailed in Table 1. Approximately 33% show homology to unclassified proteins in other species. Substantial numbers (36%) of chromosome 3 genes show no sequence similarity to proteins from organisms other than related trypanosomatid species. These genes may be specific to the trypanosomatids or may have diverged sufficiently as to have no significant sequence similarity to their functional homologs in other species. Since the total number of genes in LmjF is likely to be about 8600 (79), it is reasonable to infer that completion of the entire Leishmania genome sequence may identify approximately 3000 genes with potentially parasite-specific functions.
Completion of the sequence of chromosome 3 increases knowledge of the molecular biology of these parasites, contributing to the overall understanding of genome structure and comparative biology/evolution of this and related species. It has led to the identification of 95 new genes with key cellular functions that may be potential drug targets, vaccine candidates or useful for diagnostics or for vaccine development. In addition, study of LmjF chromosomes is valuable for future analysis of many aspects of basic molecular biology.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a PHS grant (R01 AI40599) from the National Institutes of Health to K.D.S. and post-doctoral fellowships from the International Training and Research in Emerging Infectious Diseases (ITREID) program to S.M.-C. and G.A.
DDBJ/EMBL/GenBank accession no. AC125735
REFERENCES
- 1.Camargo E.P. (1999) Phytomonas and other trypanosomatid parasites of plants and fruit. Adv. Parasitol., 42, 29–112. [DOI] [PubMed] [Google Scholar]
- 2.Zambrano-Villa S., Rosales-Borjas,D., Carrero,J.C. and Ortiz-Ortiz,L. (2002) How protozoan parasites evade the immune response. Trends Parasitol., 18, 272–278. [DOI] [PubMed] [Google Scholar]
- 3.Stuart K. (1991) RNA editing in trypanosomatid mitochondria. Annu. Rev. Microbiol., 45, 327–344. [DOI] [PubMed] [Google Scholar]
- 4.Perry K. and Agabian,N. (1991) mRNA processing in the Trypanosomatidae. Experientia, 47, 118–128. [DOI] [PubMed] [Google Scholar]
- 5.Krakow J.L., Hereld,D., Bangs,J.D., Hart,G.W. and Englund,P.T. (1986) Identification of a glycolipid precursor of the Trypanosoma brucei variant surface glycoprotein. J. Biol. Chem., 261, 12147–12153. [PubMed] [Google Scholar]
- 6.Borst P. and Rudenko,G. (1994) Antigenic variation in African trypanosomes. Science, 264, 1872–1873. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 8.Ashford R.W. (2000) The leishmaniases as emerging and reemerging zoonoses. Int. J. Parasitol., 30, 1269–1281. [DOI] [PubMed] [Google Scholar]
- 9.Fowell D.J. and Locksley,R.M. (1999) Leishmania major infection of inbred mice unmasking genetic determinants of infectious diseases. Bioessays, 21, 510–518. [DOI] [PubMed] [Google Scholar]
- 10.Alvar J., Canavate,C., Gutierrez-Solar,B., Jimenez,M., Laguna,F., Lopez-Velez,R., Molina,R. and Moreno,J. (1997) Leishmania and human immunodeficiency virus coinfection: the first 10 years. Clin. Microbiol. Rev., 10, 298–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder A., Greif,G. and Haberkorn,A. (2001) Chemotherapeutic approaches to protozoa: kinetoplastida—current level of knowledge and outlook. Parasitol. Res., 87, 778–780. [DOI] [PubMed] [Google Scholar]
- 12.Reed S.G. (2001) Leishmaniasis vaccination: targeting the source of infection. J. Exp. Med., 194, F7–F9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wincker P., Ravel,C., Blaineau,C., Pages,M., Jauffret,Y., Dedet,J., Bastien,P. and Dedet,J.P. (1996) The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res., 24, 1688–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britto C., Ravel,C., Bastien,P., Blaineau,C., Pagès,M., Dedet,J.P. and Wincker,P. (1998) Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New World Leishmania genomes. Gene, 222, 107–117. [DOI] [PubMed] [Google Scholar]
- 15.Bastien P., Blaineau,C., Britto,C., Dedet,J.-P., Dubessay,P., Pagès,M., Ravel,C., Winker,P., Blackwell,J.M., Leech,V. et al. (1998) The complete chromosomal organization of the reference strain of the Leishmania genome project, L. major ‘Friedlin’. Parasitol. Today, 14, 301–303. [DOI] [PubMed] [Google Scholar]
- 16.Ryan K.A., Dasgupta,S. and Beverley,S.M. (1993) Shuttle cosmid vectors for the trypanosomatid parasite Leishmania. Gene, 131, 145–150. [DOI] [PubMed] [Google Scholar]
- 17.Ivens A.C., Lewis,S.M., Bagherzadeh,A., Zhang,L., Chang,H.M. and Smith,D.F. (1998) A physical map of the Leishmania major Friedlin genome. Genome Res., 8, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunkin S.M., Kiser,P., Myler,P.J. and Stuart,K.D. (2000) The size difference between Leishmania major Friedlin chromosome one homologues is localized to sub-telomeric repeats at one chromosomal end. Mol. Biochem. Parasitol., 109, 1–15. [DOI] [PubMed] [Google Scholar]
- 19.Wilson R., Ainscough,R., Anderson,K., Baynes,C., Berks,M., Bonfield,J., Burton,J., Connell,M., Copsey,T., Cooper,J. et al. (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- 20.Fu G. and Barker,D.C. (1998) Rapid cloning of telomere-associated sequence using primer-tagged amplification. Biotechnology, 24, 386–390. [DOI] [PubMed] [Google Scholar]
- 21.Myler P.J., Audleman,L., deVos,T., Hixson,G., Kiser,P., Lemley,C., Magness,C., Rickell,E., Sisk,E., Sunkin,S. et al. (1999) Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA, 96, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholler J.K., Reed,S.G. and Stuart,K. (1986) Molecular karyotype of species and subspecies of Leishmania. Mol. Biochem. Parasitol., 20, 279–293. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Calvillo S., Yan,S., Nguyen,D., Fox,M., Stuart,K.D. and Myler,P.J. (2003) Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell, 11, 1291–1299. [DOI] [PubMed] [Google Scholar]
- 24.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.-A. and Barrell,B. (2000) Artemis: sequence visualisation and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 25.Womble D.D. (2000) GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol., 132, 3–22. [DOI] [PubMed] [Google Scholar]
- 25a.Aggarwal G., Worthey,E.A., McDonagh,P.D. and Myler,P.J. (2003) Importing statistical measures into Artemis enhances gene identification in the Leishmania genome project. BMC Bioinformatics, 4, 23 (7 June 2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzberg S.L., Pertea,M., Delcher,A.L., Gardner,M.J. and Tettelin,H. (1999) Interpolated Markov models for eukaryotic gene finding. Genomics, 59, 24–31. [DOI] [PubMed] [Google Scholar]
- 27.Fickett J.W. (1982) Recognition of protein coding regions in DNA sequences. Nucleic Acids Res., 10, 5303–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bibb M.J., Findlay,P.R. and Johnson,M.W. (1984) The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene, 30, 157–166. [DOI] [PubMed] [Google Scholar]
- 29.Tiwari S., Ramachandran,S., Bhattacharya,A., Bhattacharya,S. and Ramaswamy,R. (1997) Prediction of probable genes by Fourier analysis of genomic sequences. Comput. Appl. Biosci., 13, 263–270. [DOI] [PubMed] [Google Scholar]
- 30.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 32.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 34.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 35.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu G. and Barker,D.C. (1998) Characterization of Leishmania telomeres reveal unusual telomeric repeats and conserved telomere-associated sequence. Nucleic Acids Res., 26, 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiurillo M.A., Beck,A., deVos,T., Myler,P.J., Stuart,K. and Ramirez,J.L. (2000) Cloning and characterization of Leishmania donovani telomeres. Exp. Parasitol., 94, 248–258. [DOI] [PubMed] [Google Scholar]
- 39.Rudenko G. (2000) The polymorphic telomeres of the African trypanosome Trypanosoma brucei. Biochem. Soc. Trans., 28, 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horn D., Spence,C. and Ingram,A.K. (2000) Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J., 19, 2332–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramamoorthy R., Donelson,J.E. and Wilson,M.E. (1996) 5′ Sequences essential for trans-splicing of msp (gp63) RNAs in Leishmania chagasi. Mol. Biochem. Parasitol., 77, 65–76. [DOI] [PubMed] [Google Scholar]
- 42.Miao E.A., Scherer,C.A., Tsolis,R.M., Kingsley,R.A., Adams,L.G., Baumler,A.J. and Miller,S.I. (1999) Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol., 34, 850–864. [DOI] [PubMed] [Google Scholar]
- 43.Kobe B. and Kajava,A.V. (2001) The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol., 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 44.Tian S.S. and Zinn,K. (1994) An adhesion molecule-like protein that interacts with and is a substrate for a Drosophila receptor-linked protein tyrosine phosphatase. J. Biol. Chem., 269, 28478–28486. [PubMed] [Google Scholar]
- 45.Hickey M.J., Williams,S.A. and Roth,G.J. (1989) Human platelet glycoprotein IX: an adhesive prototype of leucine-rich glycoproteins with flank-center-flank structures. Proc. Natl Acad. Sci. USA, 86, 6773–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilg T., Montgomery,J., Stierhof,Y.-D. and Handman,E. (1999) Molecular cloning and characterization of a novel repeat-containing Leishmania major gene, ppg1, that encodes a membrane-associated form of proteophosphoglycan with a putative glycosylphosphatidylinositol anchor. J. Biol. Chem., 274, 31410–31420. [DOI] [PubMed] [Google Scholar]
- 47.Montgomery J., Ilg,T., Thompson,J.K., Kobe,B. and Handman,E. (2000) Identification and predicted structure of a leucine-rich repeat motif shared by Leishmania major proteophosphoglycan and Parasite Surface Antigen 2. Mol. Biochem. Parasitol., 107, 289–295. [DOI] [PubMed] [Google Scholar]
- 48.Revelard P., Lips,S. and Pays,E. (1990) A gene from the VSG expression site of Trypanosoma brucei encodes a protein with both leucine-rich repeats and a putative zinc finger. Nucleic Acids Res., 18, 7299–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoek M., Zanders,T. and Cross,G.A.M. (2002) Trypanosoma brucei expression-site-associated-gene-8 protein interacts with a Pumilio family protein. Mol. Biochem. Parasitol., 120, 269–283. [DOI] [PubMed] [Google Scholar]
- 50.Myler P.J., Beverley,S.M., Cruz,A.K., Dobson,D.E., Ivens,A.C., McDonagh,P.D., Madhubala,R., Martinez-Calvillo,S., Ruiz,J.C., Saxena,A. et al. (2001) The Leishmania genome project: new insights into gene organization and function. Med. Microbiol. Immunol., 190, 9–12. [DOI] [PubMed] [Google Scholar]
- 51.Lalo D., Stettler,S., Mariotte,S., Slonimski,P.P. and Thuriaux,P. (1993) Two yeast chromosomes are related by a fossil duplication of their centromeric regions. C. R. Acad. Sci. III, 316, 367–373. [PubMed] [Google Scholar]
- 52.Yun C.W., Tamaki,H., Nakayama,R., Yamamoto,K. and Kumagai,H. (1997) G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 240, 287–292. [DOI] [PubMed] [Google Scholar]
- 53.McLean M.J., Wolfe,K.H. and Devine,K.M. (1998) Base composition skews, replication orientation and gene orientation in 12 prokaryote genomes. J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 54.McDonagh P.D., Myler,P.J. and Stuart,K.D. (2000) The unusual gene organization of Leishmania major chromosome 1 may reflect novel transcription processes. Nucleic Acids Res., 28, 2800–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freeman J.M., Plasterer,T.N., Smith,T.F. and Mohr,S.C. (1998) Patterns of genome organization in bacteria. Science, 279, 1827a. [Google Scholar]
- 56.Gardner M.J., Hall,N., Fung,E., White,O., Berriman,M., Hyman,R.W., Carlton,J.M., Pain,A., Nelson,K.E., Bowman,S. et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson B., Åslund,L., Tammi,M., Tran,A.-N., Hoheisel,J.D. and Pettersson,U. (1998) Complete sequence of an 93.4 kb contig from chromosome 3 of Trypanosoma cruzi containing a strand switch region. Genome Res., 8, 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tosato V., Ciarloni,L., Ivens,A.C., Rajandream,M.-A., Barrell,B.G. and Bruschi,C. (2001) Secondary DNA structure analysis of the coding strand switch regions of five Leishmania major Friedlin chromosomes. Curr. Genet., 40, 186–194. [DOI] [PubMed] [Google Scholar]
- 59.Gardner M.J., Tettelin,H., Carucci,D.J., Cummings,L.M., Aravind,L., Koonin,E.V., Shallom,S., Mason,T., Yu,K., Fujii,C. et al. (1998) Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science, 282, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 60.The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 61.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes [see comments]. Science, 274, 546, 563–567. [DOI] [PubMed] [Google Scholar]
- 62.Glansdorff N. (1999) On the origin of operons and their possible role in evolution toward thermophily. J. Mol. Evol., 49, 432–438. [DOI] [PubMed] [Google Scholar]
- 63.Muto A. and Osawa,S. (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl Acad. Sci. USA, 84, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 65.Francino M.P. and Ochman,H. (1997) Strand asymmetries in DNA evolution. Trends Genet., 13, 240–245. [DOI] [PubMed] [Google Scholar]
- 66.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 67.Aiyar S.E., Gourse,R.L. and Ross,W. (1998) Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc. Natl Acad. Sci. USA, 95, 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lavigne M., Roux,P., Buc,H. and Schaeffer,F. (1997) DNA curvature controls termination of plus strand DNA synthesis at the centre of HIV-1 genome. J. Mol. Biol., 266, 507–524. [DOI] [PubMed] [Google Scholar]
- 69.Vig B.K. (1995) The centromere:kinetochore complex. Southeast Asian J. Trop. Med. Public Health, 26 (suppl. 1), 68–76. [PubMed] [Google Scholar]
- 70.Dubessay P., Ravel,C., Bastien,P., Crobu,L., Dedet,J.P., Pages,M. and Blaineau,C. (2002) The switch region on Leishmania major chromosome 1 is not required for mitotic stability or gene expression, but appears to be essential. Nucleic Acids Res., 30, 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mefford H.C. and Trask,B.J. (2002) The complex structure and dynamic evolution of human subtelomeres. Nature Rev. Genet., 3, 91–102. [DOI] [PubMed] [Google Scholar]
- 72.Pedrosa A.L., Ruiz,J.C., Tosi,L.R. and Cruz,A.K. (2001) Characterization of three chromosomal ends of Leishmania major reveals transcriptional activity across arrays of reiterated and unique sequences. Mol. Biochem. Parasitol., 114, 71–80. [DOI] [PubMed] [Google Scholar]
- 73.Pryde F.E. and Louis,E.J. (1997) Saccharomyces cerevisiae telomeres. A review. Biochemistry, 62, 1232–1241. [PubMed] [Google Scholar]
- 74.Cano M.I. (2001) Telomere biology of trypanosomatids: more questions than answers. Trends Parasitol., 17, 425–429. [DOI] [PubMed] [Google Scholar]
- 75.Pays E. and Steinert,M. (1988) Control of antigen gene expression in African trypanosomes. Annu. Rev. Genet., 22, 107–126. [DOI] [PubMed] [Google Scholar]
- 76.Marshall V.M., Coppel,R.L., Martin,R.K., Oduola,A.M.J., Anders,R.F. and Kemp,D.J. (1991) A Plasmodium falciparum MSA-2 gene apparently generated by intragenic recombination between the two allelic families. Mol. Biochem. Parasitol., 45, 349–352. [DOI] [PubMed] [Google Scholar]
- 77.Dubessay P., Ravel,C., Bastien,P., Stuart,K., Dedet,J., Blaineau,C. and Pages,M. (2002) Mitotic stability of a coding DNA sequence-free verison of Leishmania major chromosome 1 generated by targeted chromosome fragmentation. Gene, 289, 151–159. [DOI] [PubMed] [Google Scholar]
- 78.Dubessay P., Ravel,C., Bastien,P., Lignon,M.F., Ullman,B., Pages,M. and Blaineau,C. (2001) Effect of large targeted deletions on the mitotic stability of an extra chromosome mediating drug resistance in Leishmania. Nucleic Acids Res., 29, 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Myler P.J. and Stuart,K.D. (2000) Recent developments from the Leishmania genome project. Curr. Opin. Microbiol., 3, 412–416. [DOI] [PubMed] [Google Scholar]