Abstract
Acetylation is the most prominent modification on core histones that strongly affects nuclear processes such as DNA replication, DNA repair and transcription. Enzymes responsible for the dynamic equilibrium of histone acetylation are histone acetyltransferases (HATs) and histone deacetylases (HDACs). In this paper we describe the identification of novel HDACs from the filamentous fungi Aspergillus nidulans and the maize pathogen Cochliobolus carbonum. Two of the enzymes are homologs of Saccharomyces cerevisiae HOS3, an enzyme that has not been identified outside of the established yeast systems until now. One of these homologs, HosB, showed intrinsic HDAC activity and remarkable resistance against HDAC inhibitors like trichostatin A (TSA) when recombinant expressed in an Escherichia coli host system. Phylo genetic analysis revealed that HosB, together with other fungal HOS3 orthologs, is a member of a separate group within the classical HDACs. Immunological investigations with partially purified HDAC activities of Aspergillus showed that all classical enzymes are part of high molecular weight complexes and that a TSA sensitive class 2 HDAC constitutes the major part of total HDAC activity of the fungus. However, further biochemical analysis also revealed an NAD+-dependent activity that could be separated from the other activities by different types of chromatography and obviously represents an enzyme of the sirtuin class.
INTRODUCTION
During the past years it has become clear that chromatin represents an important regulatory element that affects nuclear processes such as DNA replication, recombination, DNA repair and transcription by tuning the accessibility of DNA for various factors. Thereby, cells have elaborated a specific machinery to remodel nucleosomes for specific processes occurring in chromatin (1). In particular, core histones are susceptible to a wide range of post-translational modifications, including phosphorylation, methylation, glycosylation and acetylation. Thereby, acetylation/deacetylation processes at the ε-amino groups of highly conserved N-terminal lysine residues of H2A, H2B, H3 and H4 are the most prominent modifications (2,3). Recently, the biological significance of this modification has started to emerge (4–7). Today a large body of evidence indicates that acetylation might play a major role in the regulation of transcription whereby several alternative explanations for acetylation effects are discussed (2,8–11). Enzymes responsible for the acetylation process are histone acetyltransferases (HATs), which usually act as co-activators of transcription and are frequently associated with enhancer-binding proteins or RNA polymerase II (5,12). The dynamic process of histone acetylation is reversed by histone deacetylases (HDACs). The first HDAC to be found was the human HDAC1, which showed striking sequence homology to the already known transcriptional regulator RPD3 (reduced potassium dependency factor) of Saccharomyces cerevisiae confirming a link between transcriptional regulation and histone deacetylation (13). At that time another structurally related protein with HDAC activity, HDA1, was identified in yeast (14). Today RPD3/HDA1 orthologs are reported from yeast to human and are named in a largely random manner (for review see 15–17). However, HDACs now are categorized into classes according to the yeast proteins RPD3 (class 1) and HDA1 (class 2), respectively. Both classes revealed significant sequence similarities predominantly in a large N-terminal domain (18). Among RPD3-type enzymes another two putative members of this class, HOS1 and HOS2 (HDA one similar), were identified in yeast but have not yet been detected as enzymatically active enzymes. However, there are strong indications that these enzymes preferentially affect ribosomal DNA and ribosomal protein genes, respectively (19). Another yeast sequence, HOS3, which is more distantly related to RPD3 and HDA1 was either classified as a class 2 HDAC (20) or was placed between the two classes (21), but does not actually correlate well with either of the two categories. Interestingly, HOS3 orthologs have been found only in yeast systems so far (1,22).
Recently, a third class of HDACs with homology to yeast SIR2 (silent information regulator) was established. HDAC activity of this enzyme class was demonstrated not only for the yeast enzyme but also for a mouse SIR2 ortholog (23). Furthermore, SIR2 was shown to possess ADP-ribosyltransferase activity (24,25), which is separable from HDAC activity, although deacetylation of histones is dependent on NAD+ (25,26). This NAD+ dependence might provide a link between cellular metabolism and chromatin structure and maybe is involved in the aging of cells (for review see 27). In contrast to the activity of class 1 and class 2 HDACs, SIR2-like proteins (sirtuins) cannot be inhibited by known HDAC inhibitors, such as trichostatin A (TSA) or HC toxin. However, they are conserved among species from bacteria to human (28), but so far little information is available concerning the functional role and the targets of these homologs.
Another separate class of HDACs, the HD2 enzymes are named after the founding member, the maize HD2 (29,30). Silencing of AtHD2A expression, a HD2 ortholog in Arabidopsis thaliana, resulted in aborted seed development in transgenic Arabidopsis plants, suggesting that the AtHD2A gene product is important in the reproductive development of A.thaliana (31). However, enzymes of the HD2 class have been found only in plants so far.
Recently, we have characterized RpdA and HosA, two class 1 enzymes of Aspergillus nidulans, which were the first HDACs identified in a filamentous fungus (32). Shortly thereafter we also found orthologs of these enzymes in the plant pathogenic fungus Cochliobolus carbonum (33,34). Interestingly, engineered Cochliobolus mutants of one of these genes had smaller conidia, exhibited a ∼50% reduction in total HDAC activity, and showed a reduction in growth on certain carbohydrates (33). Very recently, another fungal class 1 HDAC was identified in the corn smut fungus Ustilago maydis, which can substitute for RPD3 in S.cerevisiae. In an U.maydis knock out strain the proliferation to mature teliospores was blocked upon karyogamy (35).
Now we report the identification and molecular characterization of another four fungal HDAC sequences of A.nidulans and C.carbonum. With high sequence similarity to yeast HDA1, two of the deduced enzymes, the A.nidulans HdaA and the C.carbonum HDC3, are typical class 2 enzymes. The other two, HosB and HDC4, are the first HOS3 orthologs found in multicellular eukaryotes. Comprehensive alignments of these fungal HDAC sequences together with the well known yeast sequences resulted in a novel phylogenetic HDAC tree. Further investigations of the Aspergillus enzymes revealed that HosB has intrinsic HDAC activity and is resistant against HDAC inhibitors even when expressed in a prokaryotic expression system. However, it is not HosB but HdaA that seems to play the major role in the deacetylation process of histones in A.nidulans.
MATERIALS AND METHODS
Strains and growth media
Generally, A.nidulans strain A4 (Glasgow wild type) provided from the Fungal Genetic Stock Center (FGSC, Kansas City, KS) and C.carbonum strain SB111 provided from the American Type Culture Collection (90305) were used. Aspergillus was grown in minimal medium according to (36) for 24 h at 37°C in shake culture; Cochliobolus was grown in HMT medium according to (37) for 2 days at 25°C in shake culture.
Identification and characterization of the HDA1 orthologs hdaA and HDC3
Total A.nidulans and C.carbonum RNA was used to prepare cDNA employing Superscript reverse transcriptase (Life Technologies, Bethesda, MD). cDNA was used as template for the amplification of specific fragments of hdaA and HDC3. Two mixed degenerated 17mer oligodeoxynucleotides were used as primers for polymerase chain reaction (PCR). Forward primer HDA1 was 5′-CCNCCNGGNCAYCAYGC-3′ based on the amino acid sequence PPGHHA, and reversed primer HDA2 was 5′-GTNCCRTTNCCRTGRTG-3′ based on HHGNGT. Both amino acid sequences descend from highly conserved regions of class 2 HDACs (Fig. 1). Amplification products of ∼150 bp were isolated, cloned into a pGEM T-Easy vector (Promega) and sequenced.
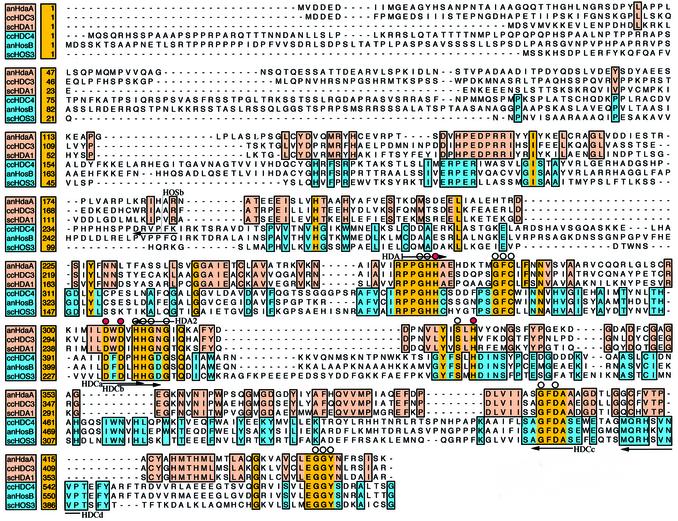
Figure 1.
Multiple sequence alignment of HDA1-type HDACs and HOS3-type HDACs of A.nidulans, C.carbonum and S.cerevisiae. The alignment shows only the conserved N-terminal parts of the enzymes. Identical amino acids within a class are boxed in pink (HDA1 class) or blue (HOS3 class), identical amino acids between both classes are boxed in yellow. Gaps are indicated by dashes introduced for alignment purpose. Numbers indicate the amino acid position of each protein. Small transparent circles represent amino acids that are invariant in all known HDACs of C.carbonum, A.nidulans and S.cerevisiae. Red circles indicate invariant amino acids identified to play an essential role in HDAC activity. Selected primers used for PCR approaches and cloning of the enzymes derived from amino acids sequences are marked by arrows.
Full-length genomic sequences of hdaA and HDC3 were obtained by using the amplified fragments as probes for the screening of an A.nidulans chromosome-specific cosmid library constructed in pWE15 (38) and pLORIST2 (39) and a λEMBL3 genomic library of C.carbonum DNA (40), respectively. To determine number and location of prospected introns, genomic sequences were compared with the corresponding cDNA sequences generated by reverse transcribed (RT)–PCR. The nucleotide and deduced amino acid sequences of HdaA and HDC3 have been deposited in the GenBank database (accession numbers AF306859 and AF307341, respectively).
Identification and characterization of the HOS3 orthologs hosB and HDC4
Blast searches with the chromosome IV-specific genomic database (http://aspergillus-genomics.org/) of A.nidulans and the HOS3 sequence of S.cerevisiae revealed a sequence of 45 amino acids with 39% identity to the query. Based on the according contig (g2000Jan142006_4209) two specific oligodeoxynucleotides, HOSa (5′-CCATCTCCTGTTTCCCCC-3′, a non-coding sequence of the promoter region of hosB), and reversed primer HOSb (5′-GGAACGGCGGAACAGGG-3′) based on amino acid sequence PVPPFQ (Fig. 1) were designed and used for the amplification of a ∼900 bp hosB fragment. The amplification product subsequently was used to screen the chromosome-specific A.nidulans cosmid library (as described above) reduced to clones representing sequences of chromosome IV (41) that led to the full-length sequence of hosB. Sequence information of hosB and yeast HOS3 allowed the construction of another two degenerated forward primers HDCa (5′-GAYTTYGAYYTNCAYCA-3′) and HDCb (5′-GAYYTNCAYCAYGGNGA-3′) based on amino acid sequences DFDLHH and DLHHGD and two degenerated reversed primers HDCc (5′-SWNGCRTCRAANCCNGC-3′) and HDCd (5′-ACRTTNACYTTRTGNCKYTGCAT-3′) based on sequences AGFDAS and MQRHKVNVP (Fig. 1). Primers were used in different combinations for the amplification of a 400 bp HDC4 cDNA fragment of C.carbonum, which was used for the screening of the λEMBL3 genomic library of C.carbonum (as described above) and led to the full-length sequence of HDC4. The nucleotide and deduced amino acid sequences of HosB and HDC4 have been deposited in the GenBank database (accession numbers AF537125 and AF537126, respectively).
Northern blot analysis
The expression of hdaA and hosB was analyzed in northern hybridization experiments according to (42). Fungi were grown for 24 h, northern analysis was done as described earlier (32). Hybridizations were performed with 32P-labeled PCR fragments of hdaA and hosB. Results were corrected for loading and blotting of mRNA by using the γ-actin gene of A.nidulans (43) as an internal control.
Expression and purification of recombinant HDACs of A.nidulans in E.coli, production of polyclonal antibodies
The coding sequences of HdaA and HosB, but also the sequences of already identified class 1 HDACs RpdA and HosA (32) were amplified from fungal cDNA. Recognition sites for restriction enzymes were fused to the PCR primers in order to clone the amplified products into a pQE 60 or pQE9 expression vector (Qiagen), respectively. Subsequently, Escherichia coli M15 cells were transformed with the generated recombinant expression constructs. Induction with IPTG led to the recombinant proteins with a 6× His affinity tag on the C-(pQE 60) or the N-terminus (pQE 9) allowing purification of the fragments on an immobilized metal-ion affinity resin. Eluted products were detected at a wavelength of 280 nm and purified fractions were analyzed by SDS–PAGE and Coomassie brilliant blue staining (Fig. 2A and B). 100 µg of purified recombinant proteins were used for each of a total of four subcutaneous injections in rabbit. After the final bleed, blood was allowed to clot and serum was separated by centrifugation at 10 000 g for 10 min at 4°C. Antibodies were purified from antiserum by protein G–Sepharose affinity chromatography (prepacked HiTrap affinity columns, Amersham Pharmacia Biotech) following the manufacturers instructions. Specificity of the antisera was tested by immunoblotting using crude protein extracts of induced E.coli expression strains. Each antiserum reacted with the corresponding recombinant fragment; no cross reactivity between antisera and E.coli proteins or the other recombinant HDAC fragment was detected.
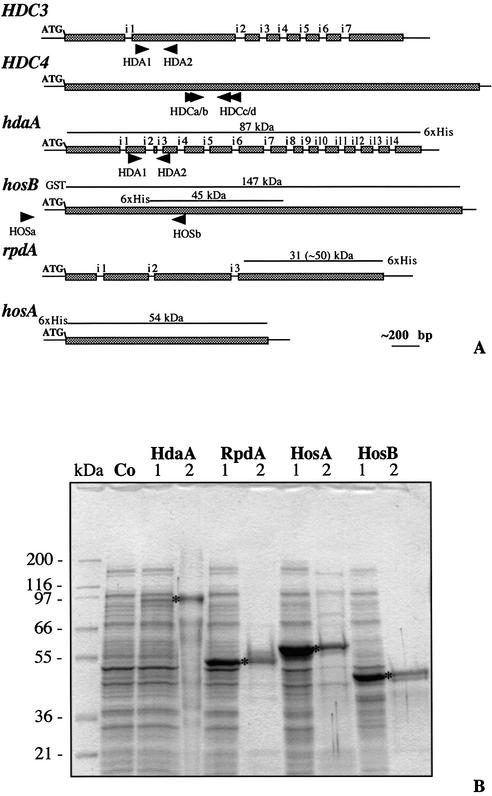
Figure 2.
Recombinant expression of HDACs for the production of polyclonal antibodies in rabbit. (A) Schematic representation of HDC3, HDC4, hdaA, hosB, rpdA and hosA. Bars illustrate the open reading frames interrupted by introns (i). The positions of primers used for amplification of gene fragments are shown by triangles. Lines above the bars indicate the fragments with the predicted molecular weight used for over-expression of the Aspergillus enzymes in E.coli (see Fig. 3B and 4A). The numbers in parentheses give the molecular mass of recombinant RpdA-fragment as it actually migrates in SDS–PAGE. (B) Fifteen microliters of E.coli extracts containing recombinant fragments of RpdA and HosB or the full-length proteins HdaA and HosA (1), or aliquots of the corresponding purified proteins (2) were subjected to SDS–10% PAGE and stained with Coomassie blue. Molecular mass of marker proteins is indicated; corresponding recombinant products are marked by asterisks. Purified recombinant products subsequently were used for the immunization of rabbits for the production of specific antibodies. The HdaA expression strain grown under non-induced conditions served as negative control (Co).
For the production of active recombinant HosB, the coding sequence of hosB was cloned into a pGEX-5X-1 expression vector (Amersham Pharmacia Biotech). Escherichia coli BL21 were used as host cells for the expression of HosB. Induction with 1 mM IPTG led to a soluble, active HosB fused to the C-terminus of glutathione S-transferase (GST) of Schistosoma japonicum (44) (Fig. 3A). Aliquots of the supernatant of briefly sonicated host cells were assayed for HDAC activity with and without different concentrations of TSA (Fig. 3B and C). A strain expressing the GST tag only served as negative control.
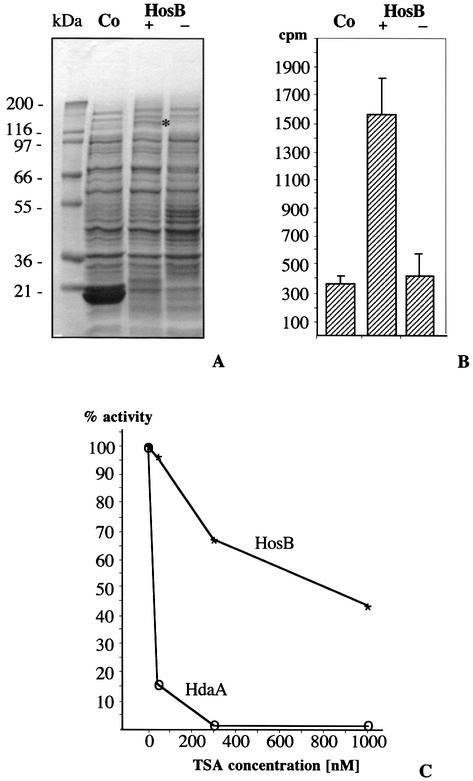
Figure 3.
Expression of recombinant HosB and analysis of HDAC activity. (A) Enzymatically active HosB was expressed as a GST-fusion protein. Coomassie staining of IPTG-induced E.coli GST-HosB cell extracts revealed the presence of the recombinant HosB fusion protein in the expected length (+). The recombinant protein is marked with an asterisk. No recombinant product could be detected under non-induced growth conditions (–). A strain expressing the GST-tag served as control for the activity assay (Co). (B) HDAC activity in the supernatant of the corresponding lysed E.coli strains. Supernatant of the induced strain (+) versus not induced cells (–) and the induced control strain (Co). (C) Inhibition of recombinant HosB and purified HdaA obtained from the peak fraction of Figure 4B. Inhibition of both enzymes was tested under various concentrations of TSA. Inhibition was calculated as a ratio of enzyme reactions containing the corresponding inhibitor versus no inhibitor.
Protein extraction
Mycelia of Aspergillus were collected and washed in a sintered glass filter, thoroughly dried with filter paper and immediately frozen in liquid nitrogen. Fifteen grams of frozen mycelia were ground to powder in an IKA grinding machine and the powder was suspended in 60 ml of buffer B [15 mM Tris–HCl, pH 8.0, 0.25 mM EDTA, 1 mM 2-mercaptoethanol, 10% (v/v) glycerol]. The mixture was stirred on ice for 10 min and centrifuged for 30 min at 37 000 g at 4°C. The supernatant was used for SourceQ-anion exchange-chromatography.
SourceQ-chromatography
The supernatant was incubated batchwise with 10 ml of Source 15Q anion exchange medium (Amersham Pharmacia Biotech) for 5 h at 4°C, equilibrated with buffer B. The matrix was pelleted by centrifugation and transferred to an FPLC column (1.6 × 15 cm). Elution of proteins was performed with 50 ml of a linear gradient from 10 to 500 mM NaCl in buffer B at a flow rate of 1 ml/min. Fractions of 1.5 ml were collected and assayed for HDAC activity. Fractions with HDAC activity were pooled and concentrated to a final volume of 1 ml by centrifugation (2500 g, 4°C) using Centriprep YM-10 (Amicon).
Size exclusion chromatography (Superdex 200)
Pooled and concentrated fractions from SourceQ-chromatography were applied onto a Superdex 200 FPLC column (1.6 × 60 cm, 120 ml, Amersham Pharmacia Biotech), equilibrated with 200 mM NaCl in buffer B. The flow rate was maintained at 1 ml/min, and fractions of 1.5 ml were collected and assayed for HDAC activity. For estimation of the molecular weight of the native enzymes the Superdex 200 column was calibrated with proteins of known molecular weight.
Histone deacetylase assay
HDAC activity was determined as described (45) using [3H]acetate pre-labeled chicken reticulocyte histones as substrate. Fifty microliter samples of the chromatography fractions were mixed with 10 µl of total [3H]acetate pre-labeled chicken reticulocyte histones (4 mg/ml). After incubation for 1 h at 25°C the reaction was stopped by addition of 50 µl of 1 M HCl/0.4 M acetate and 0.8 ml ethylacetate. After centrifugation at 10 000 g for 5 min an aliquot of 600 µl of the upper phase was counted for radioactivity in 3 ml liquid scintillation cocktail. For the detection of NAD+-dependent HDAC activity, NAD+ was added to the mixture to a final concentration of 0.5 mM. The incubation temperature was 30°C for 1 h.
Immunoprecipitation
For immunoprecipitation (IP) experiments 50 µl of partially purified HDAC fractions of A.nidulans (S200-chromatography) were mixed with 2 µg of affinity purified HDAC antibodies, 30 µl of protein A–Sepharose, equilibrated with buffer B, and were incubated for 2 h by permanent shaking at 4°C. To avoid unspecific binding the mixture was adjusted to 300 mM NaCl. After centrifugation for 2 min at 3000 g the supernatant was saved for HDAC assay and protein blotting. Pellets were washed three times with 500 µl of buffer B + 300 mM NaCl and finally resuspended in 20 µl of buffer B. Entire pellets and 50 µl of supernatants were used for standard HDAC assay. Immunoblotting precipitates were mixed with SDS sample buffer, boiled, centrifuged for 5 min at 10 000 g and the resulting supernatant was used for SDS–PAGE.
Immunoblotting
Enzyme fractions of IP experiments and fractions from the S200-chromatography were electrophoresed in precast 10% SDS Tris–Glycin polyacrylamide gels (Invitrogen, USA) at 35 mA for 1.5 h at room temperature. Gels were blotted onto Hybond nitrocellulose membrane (Amersham Pharmacia Biotech) at 25 V for 2 h and membranes were blocked with 2% (w/v) skim milk in phosphate-buffered saline (PBS, 20 mM at pH 7.4) for 2 h. Membrane strips were incubated with 1:1000 diluted affinity purified antibodies against RpdA, HdaA, HosA and HosB in 2% skim milk in PBS at 4°C overnight. After washing, strips were incubated for 2 h with alkaline phosphatase conjugated secondary anti-rabbit Ig (Sigma) and immunodetection was performed using the BCIP/NBT Color Substrate (Promega) following the manufacturer’s instructions.
Computational methods
WUBLASTP and BLASTX were used for the identification of protein coding regions by database similarity search. Multiple alignments were produced using CLUSTALW 1.8 (46), the graphic representation of the alignment was done with SeqVu 1.1 (Garven Institute of Medical Research, Sydney, Australia) for the Macintosh Computer. Phylogenetic trees were generated based on the alignment in CLUSTALW using PAUP* (version 4.0b10, Swofford/2002; Sinauer Associates, Inc., Sunderland, UK). Preliminary sequence data from Aspergillus fumigatus were obtained from the Institute for Genomic Research website at http://www.tigr.org. Sequence data of Neurospora crassa were obtained from the Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research website at http://www-genome.wi.mit.edu.
RESULTS
Identification and characterization of new HDACs of A.nidulans and C.carbonum
Fragments of hdaA and HDC3, both members of the HDA1 class, and hosB and HDC4, two HOS3 homologs, were amplified from cDNA of A.nidulans and C.carbonum as described in Materials and Methods.
Comparison of genomic and cDNA sequences revealed open reading frames of 2346 (hdaA), 2541 (HDC3), 3375 (hosB) and 3579 bp (HDC4), equivalent to enzymes of a predicted molecular mass of 87, 95, 121 and 128 kDa, respectively. Multiple sequence alignment of the deduced amino acid sequences with HDACs of S.cerevisiae (Fig. 1) and Schizosaccharomyces pombe (data not shown) confirmed HdaA and HDC3 as class 2 enzymes and HosB and HDC4 as members of the HOS3 group, respectively.
The coding sequences of the HDA1 orthologs HDC3 and hdaA are interrupted by 7 and 14 introns (Fig. 2A), respectively, with exon/intron splice sites and intron sizes typical for filamentous fungi. Comparison between the cDNA and the genomic sequence revealed that the shortest exon of hdaA encodes for not more than four amino acids. In contrast, no introns are present in the HOS3 orthologs hosB and HDC4. Sequences have been deposited in the GenBank database (accession numbers are specified in Table 1). The chromosome specificity of the Aspergillus cosmid library allowed the chromosomal localization of hdaA on chromosome II, and hosB on chromosome IV, respectively.
Table 1. Histone deacetylases of A.nidulans and C.carbonum.
| Name | Class | Chromosome | Introns | GenBank Accession number | Reference | Highest identity to | ||
|---|---|---|---|---|---|---|---|---|
| S.cerevisiae | A.fumigatus | N.crassa | ||||||
| RpdA | 1 | III | 3 | AF163862 | (32) | RPD3 | Contig 322 | Contig 1.91 |
| HosA | 1 | II | 0 | AF164342 | (32) | HOS2 | Contig 22 | Contig 1.1059 |
| HdaA | 2 | II | 14 | AF306859 | This paper | HDA1 | Contig 610 | Contig 1.17 |
| HosB | – | IV | 0 | AF537125 | This paper | HOS3 | Contig 143 | Contig 1.451 |
| HDC1 | 1 | ND | 0 | AF306507 | (33) | HOS2 | Contig 22 | Contig 1.1059 |
| HDC2 | 1 | ND | 1 | AF349677 | [J.D. Walton, unpublished] | RPD3 | Contig 322 | Contig 1.91 |
| HDC3 | 2 | ND | 7 | AF307341 | This paper | HDA1 | Contig 610 | Contig 1.17 |
| HDC4 | – | ND | 0 | AF537126 | This paper | HOS3 | Contig 143 | Contig 1.451 |
Chromosomal location and intron information of the corresponding genes are given. Sequence information of the deduced proteins and the identity to HDACs of S.cerevisiae and putative HDAC sequences of N.crassa and A.fumigatus are shown on the right columns of the table. The first four enzymes are from A.nidulans, the latter are from C.carbonum. (ND) not determined.
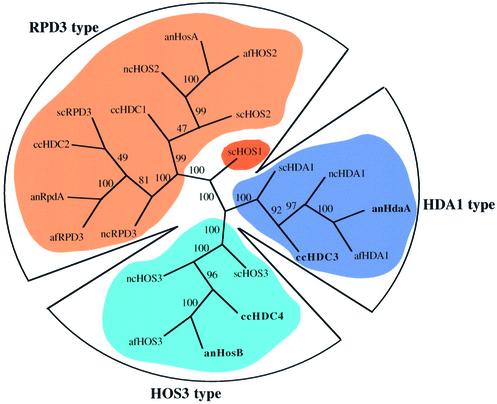
To determine the copy number of the genes, Southern blot analyses were performed with genomic DNA of the fungi. Blotting results showed that all genes are single copy sequences (data not shown). This finding was supported by the fact that sequence searches in the N.crassa genome database revealed just one single contig of high similarity to each of the Aspergillus/Cochliobolus genes (Table 1). HDAC sequences of A.nidulans and C.carbonum were aligned together with the corresponding sequences of S.cerevisiae and the putative orthologs of N.crassa and A.fumigatus. An unrooted phylogenetic tree was generated based on the alignment in CLUSTALW using PAUP* version 4.0b10 (Fig. 7). Sequence analysis suggests that the fungal HDAC enzymes have diverged into three distinct (sub)classes. The HOS3-type enzymes thereby show a clear distance to the RPD3 and the HDA1 group.
Figure 7.
Unrooted phylogenetic tree of fungal HDACs. anHosA, anRpdA, anHdaA (bold) and anHosB (bold) are from A.nidulans (32 and this paper). ccHDC1, ccHDC2, ccHDC3 (bold) and ccHDC4 (bold) are from C.carbonum (33,34 and this paper). Predicted HDACs of N.crassa are from contigs 1.1059 (ncHOS2), 1.91 (ncRPD3), 1.17 (ncHDA1) and 1.451 (ncHOS3) of the eponymous genomic DNA sequences of N.crassa (http://www.tigr.org). Predicted HDACs of A.fumigatus are from contigs 22 (afHOS2), 322 (afRPD3), 610 (afHDA1) and 143 (afHOS3) of the genomic DNA sequence of A.fumigatus (http://www-genome.wi.mit.edu). scHOS2, scRPD3, scHDA1, scHOS1 and scHOS3 are from S.cerevisiae (14,54). Sequences were aligned using ClustalW 1.8 (46) and trees were calculated using PAUP*4.0b10. Numbers indicate the bootstrap probability values of observing the branch topology shown (1000 re-sampling; a value of 100 indicates the maximum of probability that the split is certain). The given tree was calculated with the ‘neighbor joining’ (NJ) algorithm.
Transcription analysis of hdaA and hosB
To determine the transcription levels of the novel HDAC genes hdaA and hosB in A.nidulans, northern blot analyses were performed using poly A RNA of the fungus. The detected transcripts corresponded to the length of the coding sequences including the UTRs of the genes (data not shown). However, expression of HosB was low under used growth conditions of the fungus.
Recombinant expression of Aspergillus HDACs
Recombinant proteins were used to raise polyclonal antibodies for the identification of HDACs of A.nidulans by immunoblotting and IP assays. Recombinant HdaA and HosA, a recombinant fragment of the C-terminus of RpdA (32), and the conserved core domain of HosB (Fig. 2A) were expressed in E.coli M15 cells and purified via a 6× His affinity tag on the termini of the fragments (Fig. 2B) as described above. An extraordinary shift of ∼20 kDa of the recombinant RpdA fragment relative to its predicted molecular weight might be due to a specific property of the C-terminal part of fungal Rpd3-like proteins. A similar shift was observed for HDC2, the RpdA homolog of C.carbonum (34). Finally, the purified recombinant proteins were used for the production of polyclonal antibodies in rabbits. Since all recombinant His-tagged HDACs form inclusion bodies in the host cells, they were solubilized for the immunization procedure under denaturing conditions in 8 M urea. In order to attain active soluble HosB, the coding sequence of HosB was expressed in fusion with a GST tag (Fig. 2A). After induction with IPTG, significant enzyme activity was detected in the supernatant of the cell lysate but not in the lysate of an induced control strain expressing the GST protein only (Fig. 3A and B). To determine the sensitivity of this activity against HDAC inhibitors, we repeated the assay under various concentrations of TSA and HC toxin, respectively. Inhibition of recombinant HosB activity was compared with that of purified HdaA of the fungus. HdaA was highly sensitive against TSA treatment (Fig. 3C), whereas HosB turned out to be remarkably resistant (84 versus 4% reduction of activity at a concentration of 30 nM TSA and 98.5 versus 33% at 300nM TSA, respectively). Even at a TSA concentration of 1 mM HosB showed an activity of >40%. A similar resistance of the enzyme was obtained when HC toxin was used as inhibitory reagent (data not shown).
Partial purification of the Aspergillus HDACs
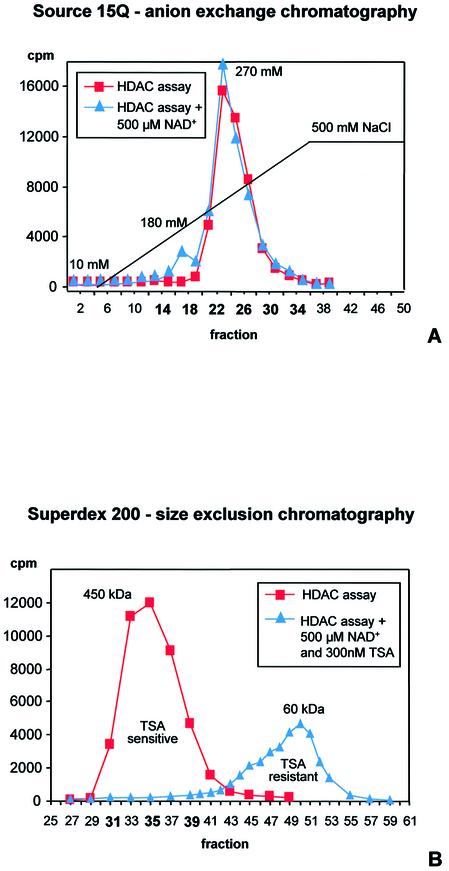
Protein extracts of A.nidulans shake culture mycelium were subjected to SourceQ-anion exchange chromatography. The main HDAC activity peak eluted at a salt concentration of 270 mM NaCl (Fig. 4A). However, an additional minor activity peak could be detected at ∼180 mM when the enzyme assay was performed in the presence of 0.5 mM NAD+. This NAD+-dependent HDAC activity is likely to represent a SIR2 homologous enzyme of Aspergillus. For further purification and improved separation of the detected activities, peak fractions 14–34 of the SourceQ-chromatography were pooled, concentrated and loaded onto an S200 gel filtration column. The major HDAC activity eluted at ∼450 kDa (Fig. 4B) indicating that the corresponding enzyme(s) obviously act(s) as high molecular mass complex(es). When the HDAC activity assay was performed in the presence of 0.5 mM NAD+, a second minor peak was detected at ∼60 kDa. To confirm the assumption that the main activity is due to the presence of a class 1 or class 2 HDAC and the small peak represents a SIR2 homolog of Aspergillus, enzyme activity assays were repeated in the presence of TSA; HOS3 or NAD+-dependent SIR2-related enzymes are known to be largely resistant to TSA. As expected, the HDAC activity of the major peak was sensitive to TSA. An inhibition of >80% was obtained at a concentration of 30 nM TSA; 300 nM TSA resulted in complete inhibition of enzymatic activity. However, no inhibition by TSA was observed for the low molecular weight NAD+-dependent enzyme activity (Fig. 4B).
Figure 4.
Purification of HDAC complexes of A.nidulans. (A) Elution profile of a Source 15Q anion exchange column of fungal protein extract. Elution was performed with 60 ml of a linear gradient from 10 to 500 mM NaCl in buffer B at a flow rate of 1 ml/min. Fractions of 1.5 ml were collected and assayed for HDAC activity with (triangles) and without (squares) 0.5 mM NAD+. Salt concentrations of eluting peak fractions are indicated. Fractions used for size exclusion chromatography are bold. (B) Size exclusion chromatography (Superdex 200). SourceQ chromatography peak fractions were concentrated and applied to an FPLC column equilibrated with 0.2 M NaCl in buffer B. The flow rate was 1 ml/min, fractions of 1.5 ml were collected and assayed for HDAC activity with (triangles) and without (squares) 0.5 mM NAD+ and 300 nM TSA, respectively. Estimated native size of complexes are indicated. Fractions used for IP experiments with specific antibodies against RpdA (fraction 31), HdaA (fraction 35) and HosA (fraction 39) are bold.
Immunological identification of the enzymes
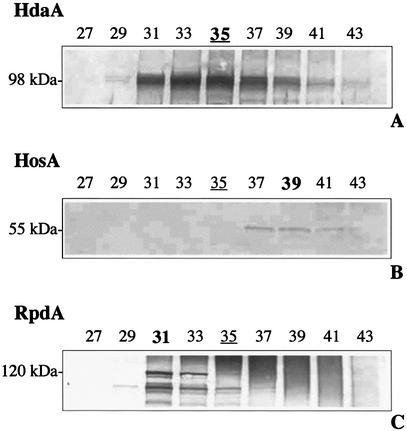
In order to further characterize the partially purified enzyme complex(es) of the major HDAC peak of Aspergillus, fractions of the S200 chromatography were subjected to immunoblotting with antibodies against RpdA, HosA, HdaA and HosB. Whereas anti-HosB antibodies did not reveal an immunosignal, anti-HdaA antibodies detected a protein band at an apparent molecular weight of ∼98 000, which is in accordance with the predicted molecular weight. The intensity of the immunosignal exactly corresponded with the pattern of HDAC activity in the S200 fractions (Fig. 5A). Incubation of the same blots with anti-RpdA antibodies yielded signals at molecular weights of ∼120 000 and 100 000. Although the predicted molecular weight of RpdA is 75 000 the enzyme migrates at a much higher apparent molecular weight in SDS–PAGE. This finding was substantiated by the unusual electrophoretic mobility of the C-terminal recombinant RpdA fragment (Fig. 2). Furthermore, we recently observed a similar shift for the recombinant HDC2, the RpdA ortholog of C.carbonum (34). However, the maximum intensity of the immunosignal in fraction 31 did not exactly correspond to the HDAC activity with its maximum in fraction 35 but was shifted to the left end (higher molecular weight) of the activity peak (Fig. 5C). When the blots were probed with antibodies against HosA, a faint band at a molecular weight of ∼55 000 could be detected, which corresponds to the predicted molecular weight of the HOS2 ortholog. In the case of HosA, the maximum of intensity was shifted to the right end (lower molecular weight) of the activity peak (Fig. 5B).
Figure 5.
Immunological identification of HDACs of A.nidulans. Aliquots of peak fractions 27–43 of the size exclusion chromatography were subjected to SDS–PAGE with subsequent blotting onto nitrocellulose membranes. Specific antibodies against HdaA, HosA and RpdA were used for immunodetection. Fraction of the activity peak is underlined, fractions with the maximum amount of each HDAC used for IP are bold. Molecular weights of the detected proteins are indicated.
These findings suggest that HDACs of A.nidulans elute as components of high molecular mass complexes of ∼400–500 kDa and that the major part of total enzyme activity is due to HdaA.
Imunoprecipitation experiments
To rule out the possibility that further (still unidentified) HDACs significantly contribute to HDAC activity, IP experiments were performed. Peak fraction 35 was incubated with purified HdaA antibodies and immunocomplexes were precipitated using protein A–Sepharose. Supernatant and pellet were tested for HDAC activity and subjected to immunoblotting. HDAC activity was quantitatively precipitated by HdaA antibodies with a concomitant depletion of the protein from the supernatant (Fig. 6A). In a control reaction using protein A–Sepharose without antibody, neither depletion of HDAC activity from the supernatant nor HdaA precipitation was observed. Obviously, HdaA contributes the vast majority of total HDAC peak activity.
Figure 6.
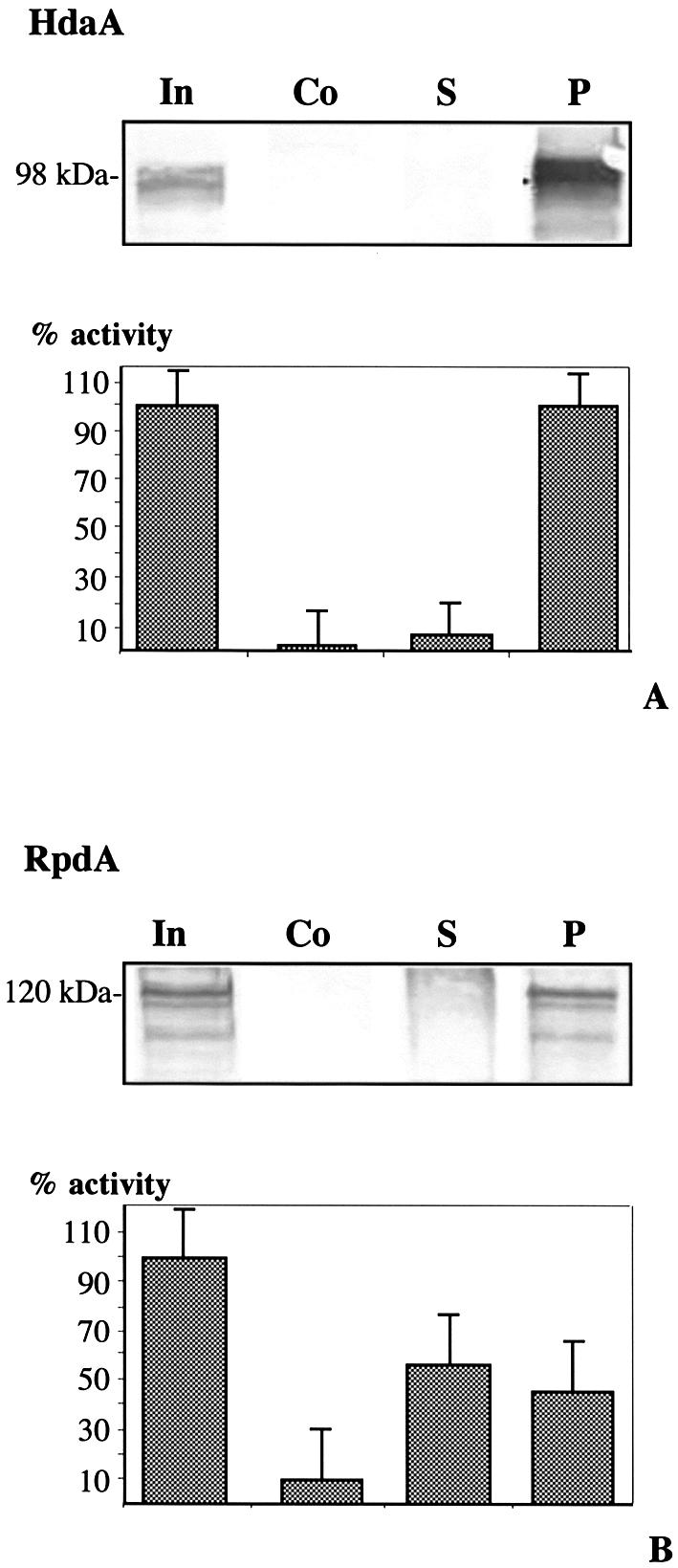
IP of HDACs of A.nidulans. Fifty microliters of fraction 35 or 31 of the S200 size exclusion chromatography was incubated with 2 µg of immunopurified antibodies against (A) HdaA or (B) RpdA together with 30 µl of protein A–Sepharose, equilibrated with buffer B. Incubation was performed under continuous shaking at 4°C for 2 h. After centrifugation, the supernatant (S) was used for the HDAC assay and immunoblotting. Pellets (P) were washed three times and finally resuspended in 20 µl of buffer B. The entire pellet and 50 µl of supernatant was used for the HDAC-assay. (Co), Control: incubation was performed without specific antibody but with protein A–Sepharose, (In) input. For immunoblotting, 20 µl of supernatant and the entire pellet were applied to SDS–PAGE with subsequent immunoblotting using anti-HdaA and anti-RpdA antibodies. Molecular weight of detected proteins is indicated. Data represent results of three independent experiments (A + B).
IP experiments were also performed with fraction 31 containing the major portion of RpdA and fraction 39 containing the major portion of HosA using the corresponding RpdA and HosA antibodies. In the case of fraction 31, 45% of total HDAC activity was precipitated (Fig. 6B). Immunoblots with anti-RpdA antibody revealed that RpdA was quantitatively precipitated and removed from the supernatant. Additionally, subsequent immunoblotting of the same IP with anti-HdaA antibody revealed a HdaA signal in the supernatant but not in the pellet (data not shown), indicating that the remaining enzyme activity in the supernatant derives from the HDA1 ortholog HdaA. This assumption is supported by an HdaA immunosignal in fraction 31 of the S200 Superdex chromatography (Fig. 5A).
In the case of fraction 39, no significant HDAC activity could be precipitated with the anti-HosA antibody (data not shown). Immunoblotting with anti-HdaA antibodies revealed HdaA in the supernatant; this portion of HdaA together with the corresponding enzyme activity could be precipitated with anti-HdaA antibody (data not shown). Obviously, the bulk of HDAC activity of fractions 37–41 is due to the activity of HdaA and, under the employed experimental conditions, HosA plays a minor role as an active enzyme in the fungus.
DISCUSSION
The enzymes
Since the first HDAC was identified in 1996 (13) the number of newly identified HDAC enzymes in a wide spectrum of organisms has rapidly increased. Within the past 2 years the number of human class 1 and class 2 HDACs rose from 5 to 11 (47). In addition to these classical enzymes, two classes of structurally unrelated HDACs have been identified: the NAD+-dependent sirtuins (48) and the plant specific HD2 class (16). HOS3 of S.cerevisiae was the founding member of another group of classical enzymes that have been characterized only in yeast systems as yet (22,49). Here we report the first identification of HOS3 members from multicellular eukaryotes, from the filamentous fungi A.nidulans and C.carbonum. Furthermore, we found putative HOS3 orthologs in the genomic sequence of the fungi A.fumigatus and N.crassa but not in other eukaryotic organisms. These findings led to the conclusion that HOS3-type enzymes are not unique for yeast (as assumed) but may be a peculiarity of fungi, similar to the HD2 class in plants. Understanding the function of HOS3 orthologs in fungi will require further studies. However, a HOS3-deficient yeast mutant displayed elevated acetylation at lysines 5, 8 and 12 of histone H4 (49). Furthermore, yeast HOS3 has intrinsic enzymatic activity and is largely insensitive towards known HDAC inhibitors. In particular, the latter represents a remarkable feature that is very unusual for class 1 and class 2 HDACs. Here we could demonstrate intrinsic activity and resistance against inhibitors for HosB, which confirms its affiliation to the HOS3 group. Despite conspicuous sequence similarities and the analogical biochemical properties between HOS3 and the orthologs presented in this paper some differences exist as well. HDC4 and HosB are among the largest HDACs identified so far. With molecular weights of 128 000 and 121 000 HDC4 and HosB are considerably larger than HOS3 (79 000). The extraordinary size of these enzymes was substantiated by the search for putative HOS3 homologs in A.fumigatus and N.crassa (Fig. 7) that also revealed predicted molecular weights between 120 000 and 130 000. In contrast to its yeast ortholog, HosB apparently did not contribute significantly to the total HDAC activity of A.nidulans. From our point of view there are several possible explanations for the apparent lack of in vivo activity of the enzyme. (i) HosB may, when associated with specific protein complexes in vivo, only be active with intact chromatin but may be enzymatically inactive in an in vitro assay using isolated core histones as substrate. (ii) A very low expression rate of the enzyme may prevent the detection of the enzyme and its activity. This assumption is supported by the fact that northern analysis revealed a rather weak hosB transcript under the used growth conditions. HOS3-type proteins of filamentous fungi may represent a kind of resistant safeguard enzymes that are upregulated and/or become catalytical active in cases when other HDAC complexes are disturbed or inhibited. Very recently, this was suggested for a putative inhibitor-insensitive HDAC activity of C.carbonum (34,50). (iii) HosB may be more susceptible to degradation processes than other HDACs and thereby might be lost under the used chromatographically purification procedures.
In contrast to HosB and HDC4, HdaA and HDC3 have molecular weights that correspond well to the yeast counterpart HDA1 (80 000–95 000). HDA1 proteins are characterized by a large proportion of identical regions in the middle part of the sequence with an absolute conservation of amino acids that are essential for catalytic activity (see Fig. 1). The exon/intron structure of the HDC3 and hdaA gene is remarkable. HDC3 has 7 introns, hdaA has 14, which is, to our knowledge, the highest number of introns that ever has been reported for a fungal gene. Interestingly, the shortest exon in the hdaA gene encodes only four amino acids.
The activity
One central claim of this paper is that HdaA and RpdA are major contributors to total HDAC activity of A.nidulans. The class 2 enzyme HdaA accounts for the main part of the HDAC activity and exists as a part of a multiprotein complex of ∼450 kDa (Fig. 5A). However, IP experiments showed that the class 1 enzyme RpdA also contributes to the HDAC activity and is part of an enzyme complex that is even larger in size. These results are in line with findings in C.carbonum where an anti-HDC2 (RpdA homolog) antibody could immunoprecipitate only a minor part of the HDAC activity (34). Furthermore, our results also correspond with biochemical evidence of yeast HDAC complexes (14) where the 350 kDa HDA complex (containing HDA1) was shown to contribute significantly more to the overall HDAC activity than the HDB complex (containing RPD3). Although HosA, another class 1 enzyme of Aspergillus, could be detected on immunoblots of enzymatically active fractions, IP yielded only negligible activity; we therefore conclude that HosA plays only a minor role if any for total Aspergillus HDAC activity. This assumption is confirmed by the lack of enzymatic activity of the corresponding yeast HOS2. Although several lines of genetic evidence indicate that HOS2 posesses HDAC activity (51–53), it never has been characterized as a functional enzyme. However, discrepant results exist with respect to HOS2-deficient mutants as well. Whereas in yeast HOS2 deletion has no apparent effect on growth or phenotype, the corresponding HDC1 deletion strain of C.carbonum displays significantly altered capabilities of utilizing different carbon sources and also exhibits a change in the pathogenicity against its host maize (33).
Apart from the well known class 1 and class 2 HDACs, it is evident that, as in higher eukaryotes, an NAD+-dependent SIR2-like HDAC also accounts for a part of the total HDAC activity in filamentous fungi. This was confirmed just recently by the identification and cloning of a SIR2 homologous gene in Aspergillus and Cochliobolus (unpublished data of our lab). However, it is quite clear from our chromatographic analysis that SIR2-type HDACs contribute a minor part of the total HDAC activity and are well separated from the other HDACs in different types of chromatography (Fig. 4).
The classification
Currently we refer to four different classes of HDACs: RPD3 homologs, HDA1 homologs, sirtuins and the plant HD2-specific enzymes. Unfortunately, nomenclature of (putative) HDACs is confusing (15) and classification is unsatisfactory for a number of reasons. On one hand RPD3-related class 1 enzymes and HDA1-related class 2 HDACs are characterized by considerable sequence homologies/similarities (21), on the other hand two additional, totally different classes of HDACs exist (plant HD2; sirtuins) that have neither any relation to each other nor to the class 1 and class 2 enzymes. Furthermore, a growing number of HOS3-type enzymes like HosB and HDC4 have conserved domains with both, class 1 and class 2 enzymes (Fig. 1) but do not really fit in either class (Fig. 7). In addition, these proteins share several unique sequence motives that may be important domains responsible for the insensitivity towards HDAC inhibitors and other specific biochemical properties of this enzyme group. Consequently, they are members of an additional HDAC class (class 5). Moreover it should be mentioned that the RPD3 group is divided into two branches (namely the RPD3/HOS2 enzymes and yeast HOS1). However, there are no further HOS1 orthologs identified so far, which at present would justify a separate HOS1 group.
To make HDACs with pronounced sequence similarities clearly distinguishable from enzymes of other groups, we suggest a reconsideration of the current nomenclature of HDAC enzymes. One suggestion is to distinguish three main classes of HDACs: (i) a class 1, the ‘classical’ HDACs that contains subclasses with strong sequence similarities to RPD3, HDA1 and HOS3; (ii) a class 2 that contains the NAD+-dependent sirtuines with the subclasses SIR2 and HST1-4 and (iii) a class 3 that contains plant specific HD2-related enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Jonathan D. Walton (East Lansing) for critical comments and discussion. This work was supported by grants from the Austrian Science Foundation to S.G. (P15439) and G.B. (P13209), and a grant from the Austrian National Bank to P.L. (P-9000). Sequencing of A.fumigatus was accomplished with support from National Institute of Allergy and Infectious Diseases (NIAID) to Dr David Denning, University of Manchester, UK, and Dr William Nierman at TIGR. The genomic sequence of N.crassa was obtained from the Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research.
DDBJ/EMBL/GenBank accession nos+ AF306859, AF307341, AF537125 and AF537126
REFERENCES
- 1.Felsenfeld G., Boyes,J., Chung,J., Clark,D. and Studitsky,V. (1996) Chromatin structure and gene expression. Proc. Natl Acad. Sci. USA, 93, 9384–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loidl P. (1988) Towards an understanding of the biological function of histone acetylation. FEBS Lett., 227, 91–95. [DOI] [PubMed] [Google Scholar]
- 3.Loidl P. (1994) Histone acetylation: facts and questions. Chromosoma, 103, 441–449. [DOI] [PubMed] [Google Scholar]
- 4.Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone deacetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- 5.Davie J.R. (1998) Covalent modifications of histones: expression from chromatin templates. Curr. Opin. Genet. Dev., 8, 173–178. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- 8.Lee D.Y., Hayes,J.J., Pruss,D. and Wolffe,A.P. (1993) A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell, 72, 73–84. [DOI] [PubMed] [Google Scholar]
- 9.Ura K., Kurumizaka,H., Dimitrov,S., Almouzni,G. and Wolffe,A.P. (1997) Histone acetylation: influence on transcription, nucleosome mobility and positioning and linker histone-dependent transcriptional repression. EMBO J., 16, 2096–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Rodas G., Brosch,G., Georgieva,E.I., Sendra,R., Franco,L. and Loidl,P. (1993) Histone deacetylase. A key enzyme for the binding of regulatory proteins to chromatin. FEBS Lett., 317, 175–180. [DOI] [PubMed] [Google Scholar]
- 11.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 12.Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- 13.Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 14.Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leipe D.D. and Landsman,D. (1997) Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res., 25, 3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lusser A., Kölle,D. and Loidl,P. (2001) Histone acetylation: lessons from the plant kingdom. Trends Plant Sci., 6, 59–65. [DOI] [PubMed] [Google Scholar]
- 17.Graessle S., Loidl,P. and Brosch,G. (2001) Histone acetylation: plants and fungi as model systems for the investigation of histone deacetylases. Cell. Mol. Life Sci., 58, 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khochbin S. and Wolffe,A.P. (1997) The origin and utility of histone deacetylases. FEBS Lett., 419, 157–160. [DOI] [PubMed] [Google Scholar]
- 19.Robyr D., Suka,Y., Xenarios,I., Kurdistani,S.K., Wang,A., Suka,N. and Grunstein,M. (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell, 109, 437–446. [DOI] [PubMed] [Google Scholar]
- 20.Leipe D.D., Aravind,L. and Koonin,E.V. (1999) Survey and summary: did DNA replication evolve twice independently? Nucleic Acids Res., 27, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grozinger C.M., Hassig,C.A. and Schreiber,S.L. (1999) Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl Acad. Sci. USA, 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srikantha T., Tsai,L., Daniels,K., Klar,A.J. and Soll,D.R. (2001) The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol., 183, 4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- 24.Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- 25.Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- 26.Landry J., Sutton,A., Tafrov,S.T., Heller,R.C., Stebbins,J., Pillus,L. and Sternglanz,R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA, 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarente L. (2000) Sir2 links chromatin silencing, metabolism and aging. Genes Dev., 14, 1021–1026. [PubMed] [Google Scholar]
- 28.Frye R.A. (2000) Phylogenetic classification of prokaryotic and eukaryotic SIR2-like proteins. Biochem. Biophys. Res. Commun., 273, 793–798. [DOI] [PubMed] [Google Scholar]
- 29.Lusser A., Brosch,G., Loidl,A., Haas,H. and Loidl,P. (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science, 277, 88–91. [DOI] [PubMed] [Google Scholar]
- 30.Dangl M., Lusser,A., Brosch,G., Loidl,A., Haas,H. and Loidl,P. (1998) Second family of histone deacetylases: response. Science, 280, 1167a. [Google Scholar]
- 31.Wu K., Tian,L., Malik,K., Brown,D. and Miki,B. (2000) Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J., 22, 19–27. [DOI] [PubMed] [Google Scholar]
- 32.Graessle S., Dangl,M., Haas,H., Mair,K., Trojer,P., Brandtner,E., Walton,J.D., Loidl,P. and Brosch,G. (2000) Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta, 1492, 120–126. [DOI] [PubMed] [Google Scholar]
- 33.Baidyaroy D., Brosch,G., Ahn,J., Graessle,S., Wegener,S., Tonukari,N.J., Caballero,O., Loidl,P. and Walton,J.D. (2001) A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell, 13, 1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brosch G., Dangl,M., Graessle,S., Loidl,A., Trojer,P., Brandtner,E.M., Mair,K., Walton,J.D., Baidyaroy,D. and Loidl,P. (2001) An inhibitor-resistant histone deacetylase in the plant pathogenic fungus Cochliobolus carbonum. Biochemistry, 40, 12855–12863. [DOI] [PubMed] [Google Scholar]
- 35.Reichmann M., Jamnischek,A., Weinzierl,G., Ladendorf,O., Huber,S., Kahmann,R. and Kämper,J. (2002) The histone deacetylase Hda1 from Ustilago maydis is essential for teliospore development. Mol. Microbiol., 46, 1169–1182. [DOI] [PubMed] [Google Scholar]
- 36.Pontecorvo G., Roper,J.A., Hemmons,L.M., MacDonald,K.D. and Bufton,A.W.J. (1953) The genetics of Aspergillus nidulans. Adv. Genet., 141–238. [DOI] [PubMed] [Google Scholar]
- 37.van Hoof A. Leykam,J., Scheffer,H.J. and Walton,J.D. (1991) A single β-1,3-glucanase secreted by the maize pathogen Cochliobolus carbonum acts by an exolytic mechanism. Physiol. Mol. Plant Pathol., 39, 259–267. [Google Scholar]
- 38.Evans G.A. and Wahl,G.M. (1987) Cosmid vectors for genomic walking and rapid restriction mapping. Methods Enzymol., 152, 604–610. [DOI] [PubMed] [Google Scholar]
- 39.Gibson T.J., Coulson,A.R., Sulston,J.E. and Little,P.F. (1987) Lorist2, a cosmid with transcriptional terminators insulating vector genes from interference by promoters within the insert: effect on DNA yield and cloned insert frequency. Gene, 53, 275–281. [DOI] [PubMed] [Google Scholar]
- 40.Scott-Craig J.S., Panaccione,D.G., Pocard,J.A. and Walton,J.D. (1992) The cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7-kilobase open reading frame. J. Biol. Chem., 267, 26044–26049. [PubMed] [Google Scholar]
- 41.Brody H., Griffith,J., Cuticchia,A.J., Arnold,J. and Timberlake,W.E. (1991) Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res., 19, 3105–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 43.Fidel S., Doonan,J.H. and Morris,N.R. (1988) Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene, 70, 283–293. [DOI] [PubMed] [Google Scholar]
- 44.Smith D.B. and Johnson,K.S. (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene, 67, 31–40. [DOI] [PubMed] [Google Scholar]
- 45.Sendra R., Rodrigo,I., Salvador,M.L. and Franco,L. (1988) Characterization of pea histone deacetylases. Plant Mol. Biol., 11, 857–866. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J.D., Higgins,D.G. and Gibson,T.J.. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao L., Cueto,M.A., Asselbergs,F. and Atadja,P. (2002) Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem., 277, 25748–25755. [DOI] [PubMed] [Google Scholar]
- 48.Gray S.G. and Ekstrom,T.J. (2001) The human histone deacetylase family. Exp. Cell Res., 262, 75–83. [DOI] [PubMed] [Google Scholar]
- 49.Carmen A.A., Griffin,P.R., Calaycay,J.R., Rundlett,S.E., Suka,Y. and Grunstein,M. (1999) Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl Acad. Sci. USA, 96, 12356–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baidyaroy D., Brosch,G., Graessle,S., Trojer,P. and Walton,J.D. (2002) Characterization of inhibitor-resistant histone deacetylase activity in plant-pathogenic fungi. Eukaryotic Cell, 1, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edmondson D.G., Zhang,W., Watson,A., Xu,W., Bone,J.R., Yu,Y., Stillman,D. and Roth,S.Y. (1998) In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gcn5p activation. Cold Spring Harb. Symp. Quant. Biol., 63, 459–468. [DOI] [PubMed] [Google Scholar]
- 52.Wittschieben B.O., Fellows,J., Du,W., Stillman,D.J. and Svejstrup,J.Q. (2000) Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J., 19, 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson A.D., Edmondson,D.G., Bone,J.R., Mukai,Y., Yu,Y., Du,W., Stillman,D.J. and Roth,S.Y. (2000) Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev., 14, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidal M. and Gaber,R.F. (1991) RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]