Abstract
The degradation of mRNA in the yeast Saccharomyces cerevisiae takes place through several related pathways. In the most general mRNA-decay pathway, that of poly(A)-dependent decay, the normal shortening of the poly(A) tail on an mRNA molecule by deadenylation triggers mRNA decapping by the enzyme Dcp1p, followed by exonucleolytic digestion by Xrn1p. A specialized mRNA-decay pathway, termed nonsense-mediated decay, comes into play for mRNAs that contain an early nonsense codon. This pathway operates through the Upf proteins in addition to Dcp1p and Xrn1p. Previously, we identified a different specialized mRNA-decay pathway, the initiation-mediated decay pathway, and showed that it affects two Hsp70 heat-shock mRNAs under conditions of slowed translation initiation. Here we report that initiation-mediated mRNA decay also works through the Dcp1 and Xrn1 enzymes, and requires ongoing transcription by RNA polymerase II. We show that several other heat-shock mRNAs, including two from the Hsp90 gene family and three more from the Hsp70 gene family, are also subject to initiation-mediated decay, whereas a variety of non-heat-shock mRNAs are not affected.
INTRODUCTION
Molecules of mRNA generally have short half-lives, and in the budding yeast are degraded by several pathways (reviewed in 1,2). The major route for global mRNA degradation is termed the poly(A)-dependent pathway; this pathway is triggered when the 3′ poly(A) tail of an mRNA becomes shortened, by normal exonucleolytic activity, to a length of 10–12 nt. Deadenylation to this extent then allows removal of the highly modified m7G ‘cap’ nucleotide from the 5′ end of the mRNA by the decapping enzyme Dcp1p, followed by degradation of the remainder of the mRNA, in a 5′-to-3′ direction, by the exonuclease Xrn1p. A secondary pathway for general mRNA degradation is also dependent on deadenylation, but does not require decapping. Instead, degradation of an mRNA occurs 3′-to-5′ by a different exonuclease, termed the exosome (3). These two pathways are thought to be the primary pathways for general mRNA decay in yeast, because cells that contain mutations affecting both pathways are inviable (4,5).
Specialized pathways for yeast mRNA degradation have also been described. One of these is the nonsense-mediated mRNA-decay pathway, which primarily degrades aberrant mRNAs that contain a nonsense mutation (creating a stop codon) within what would otherwise be the open reading frame (ORF) of the mRNA. Recognition of this inappropriately positioned stop codon causes the translating ribosome to pause, leading to activation of the Upf proteins, decapping of the aberrant mRNA by Dcp1p and degradation by Xrn1p (reviewed in 6).
Another specialized pathway for mRNA degradation affects some yeast mRNAs under certain limiting conditions. This pathway, termed the initiation-mediated mRNA-decay pathway, is seen when the process of translation initiation is slowed down through the effects of mutations in general translation-initiation factors such as eIF3, eIF4E and eIF2B (leading to limiting amounts of eIF2) (7). We have shown that the mRNAs from two Hsp70 heat-shock genes, SSA1 and SSA2, are degraded by initiation-mediated decay, and that two proteins that facilitate nonsense-mediated mRNA decay, Upf1p and Upf2p, are also involved in initiation-mediated decay of these heat-shock mRNAs (7). The mechanistic relationship between initiation-mediated decay and nonsense-mediated decay is unclear, although the situations that trigger these two pathways are different.
One eukaryotic translation factor important in the process of translation initiation is eIF3. This protein operates at multiple steps in translation initiation, including the maintenance of 60S and 40S ribosomal subunits in dissociated form in preparation for initiation, association with the 40S subunit for interaction with the eIF2–GTP–Met-tRNAi ternary complex, thus forming the 43S pre-initiation complex, prevention of 43S complex disruption by the 60S subunit and assistance in the binding of 43S complex to mRNA (reviewed in 8,9). eIF3 is a multi-subunit protein; one of its essential subunits is Prt1p. Several temperature-sensitive mutations altering Prt1p have been found to affect translation initiation at a restrictive temperature, and do so with differing degrees of severity depending on the restrictive temperature (10). In previous work defining the initiation-mediated mRNA-decay pathway, we used two different prt1 mutant alleles, prt1-1 and prt1-63, which slow the initiation of translation to a similar degree but at different temperatures (10,11). The prt1-1 mutation, at a more elevated temperature, was also used to show that some translation initiation is a precondition for initiation-mediated mRNA decay (7). Here we again use the prt1-63 mutant allele to impose moderately decreased rates of translation initiation at the restrictive temperature of 37°C; under these conditions we characterize the initiation-mediated decay of several more mRNAs, all of the heat-shock variety, and show that initiation-mediated decay goes through the Dcp1p–Xrn1p mRNA-degradation pathway. We also report that several other mRNAs, from genes not of the heat-shock variety, are not subject to this specialized form of mRNA decay, raising the possibility that initiation-mediated mRNA decay may have a physiological basis related to heat shock or the products of heat-shock genes.
MATERIALS AND METHODS
Strains and plasmids
Strains are listed in Table 1. The HSP82-lacZ plasmid YCp83 was constructed by replacing the small HindIII–SacI fragment of YCp102 (7) with the 2.5-kb HindIII–SacI fragment from YIp144 (11) comprising base pairs –334 to +282 of HSP82 fused in frame to lacZ (12). The HSC82-lacZ plasmid YCp90C was constructed in two steps: first, replacing the small XbaI–SacI fragment of YCp102 with the 2.8-kb XbaI–SacI fragment from YIp232C (11) to create YCp90INT; then replacing the small PstI–XbaI fragment of YCp90INT with the 1.2-kb PstI–XbaI fragment of YIp232C. YCp90C contains HSC82 sequences from –600 (approximately) to +600 fused to lacZ (12). Both HSP82-lacZ and HSC82-lacZ fusion genes contain the same lacZ sequences and differ only in their 5′ ends. The SSA3-lacZ plasmid YCpSSA3 was constructed by replacing the small XhoI–SacI fragment of YCp102 with the 2.3-kb XhoI–SacI fragment from pWB204Δ468 (13). YCpSSA3 contains SSA3 sequences from –468 to codon 3 of the SSA3 gene fused in frame to lacZ. All three fusion genes were sequenced through the HSP-lac fusion junction and all fusion genes produce a functional β-galactosidase protein (data not shown).
Table 1. Strains used in this study.
| Strain | Genotype | Sourcea |
|---|---|---|
| 21R | MATa ura3-52 leu2-3,112 ade1 | 54 |
| TC3-212-3 | MATα ura3-52 leu2-3,112 prt1-63 | 55 |
| TC3-21-1 | MATa ura3-52 leu2-3,112 ade1 his prt1-63 | A |
| PLH6C | MATa ura3 leu2-3,112 ade his3-11,15 prt1-63 | NMD2-1D × TC3-212-3, then × 21R |
| NMD2-1D | MATa ura3-1 leu2-3,112 ade2-1 his3-11,15 trp1-1 can1-100 nmd2::HIS3 | B; 7 |
| ZPU70-H | MATa ura3-52::YIp100 leu2-3,112 ade1 prt1-1 | 7 |
| PLY103 | MATα ura3-52 leu2-3,112 trp1-7 upf1::URA3 | E |
| CBPU9BM | MATα ura3-52::YIp100 leu2-3,112 ade trp1-7 prt1-1 upf1::URA3 | ZPU70-H × PLY103 |
| P6U9A | MATα ura3-52 leu2-3,112 ade1 his trp1-7 prt1-63 upf1::URA3 | 7 |
| P26U3A | ura3-52 leu2-3,112 ade1 trp1-7 his6 prt1-63 | 21R × P6U9A |
| PLH3C | ura3 leu2-3,112 ade his3-11,15 trp1-1 prt1-63 | 7 |
| MW326 | MATα ura3-52 leu2-3,112 his3-11,15 trp1-Δ1 lys2 ssa1::HIS3 ssa2::LEU2 [CEN GAL1p-SSA1] | C |
| W303-1A | MATa ura3-1 leu2-3,112 ade2-1 his3-11,15 trp1-1 | A |
| PS2D5C | ura3 leu2-3,112 ade his3-11,15 trp1 lys2 prt1-63 ssa2::LEU2 | MW326 × W303-1A, then × PLH3C |
| PS2D5D | ura3 leu2-3,112 ade his3-11,15 trp1 ssa2::LEU2 | MW326 × W303-1A, then × PLH3C |
| CB54A | MATa ura3-52::YIp102 leu2-3,112 ade1 prt1-63 | 7 |
| CB61A | MATa ura3-52::YIp100 leu2-3,112 ade prt1-63 | 7 |
| CLD82α | MATα ura ade his trp hsc82::LEU2 | D; 56 |
| PLD82α | MATα ura ade his trp hsp82::LEU2 | D; 56 |
| C4X9D | MATα ura3-52 leu2-3,112 ade his3 prt1-63 hsc82::LEU2 | CLD82α × PLH6C, then 3 × to 21R |
| 3H15A | ura3-52 leu2-3,112 ade his3 prt1-63 hsp82::LEU2 | PLD82α × PLH6C, then 3 × to 21R |
| PLR62B | MATα ura3-52 leu2-3,112 ade1 rpb1-1 | 7 |
| PLR62C | MATa ura3-52 leu2-3,112 ade trp1 prt1-63 | Sister segregant to PLR62B |
| PLR66C | ura3-52 leu2-3,112 rpb1-1 prt1-63 | Sister tetrad to PLR62B |
| NMD2-1C | MATα ura3-1 leu2-3,112 ade2-1 his3-11,15 trp1-1 can1-100 nmd2::HIS3 | B; 7 |
| NM4A3D | ura3-52 leu2-3,112 nmd2::HIS3 prt1-63 | NMD2-1C × CB54A |
| SL4D | ura3-52 leu2-3,112 his4 xrn1::URA3 prt1-63 | TC3-212-3 × yRP1331 (44), then 2 × to TC3-212-3 |
| SL2B | ura3-52 leu2-3,112 his4 trp1 prt1-63 | TC3-212-3 × yRP1331 (44), then 2 × to TC3-212-3 |
| DCP8A2 | ura3 dcp1::URA3 prt1-63 | TC3-212-3 × yRP1328 (44), then to TC3-212-3 |
aA, R.A.Singer (Dalhousie University); B, A.Jacobson (University of Massachusetts); C, M.Werner-Washburne (University of New Mexico); D, S.Lindquist (University of Chicago); E, M.Culbertson (University of Wisconsin).
Culture conditions and RNA analysis
Yeast cultures were grown in glucose-supplemented YNB minimal medium at 23°C overnight to a density of 2–4 × 106 cells/ml, and then transferred to 32 or 37°C for further incubation. For northern analysis, equal amounts of total RNA (determined spectrophotometrically) were resolved by electrophoresis through 1.2% agarose plus 2 M formaldehyde and transferred to Hybond N membrane (Amersham) for hybridization. Equal loading was confirmed by in-gel ethidium staining of rRNA (data not shown). ACT1 mRNA or U3 RNA was used as an internal control.
Probes used were a 3.1-kb BamHI–DraI fragment of the SSA2-lacZ gene that contains only lac sequences, a 0.5-kb EcoRI–HindIII CYH2 fragment that hybridizes to both the pre-mRNA and the mRNA (from pGEM4ZCYH2, kindly supplied by A. Jacobson), a 2.9-kb HindIII PGK1 fragment (from pB1, kindly supplied by M. Dobson), a 0.9-kb BamHI–PstI URA3 fragment, a 1.7-kb EcoRI–XhoI CLN3 fragment, a 1.2-kb BamHI HIS3 fragment, an XhoI–HindIII ACT1 fragment, a 1.2-kb EcoRI fragment of pUTX232 that contains the first 600 bp of the HSC82 ORF (14) and cross-hybridizes with HSP82 transcripts due to 97% coding-sequence identity (12,14), a NcoI–HindIII fragment of SSA2 that cross-hybridizes to the virtually identical (15,16) SSA1 sequences, a 0.6-kb fragment of the SSA3 coding region that is derived from the 0.7-kb RsaI fragment that does not cross-hybridize with the other Hsp70 genes that are similar in sequence (17) (Fig. 1), an internal 1.4-kb Cla1 fragment of the SSC1 gene, and a 0.4-kb fragment of KAR2 that encompasses the 3′ coding region. The U3 (SNR17A) probe was a PCR product derived using oligonucleotides: 5′-gtcgacgtacttcagtatgtaatataccccaaac and 5′-gtcagactgccatttgtacccacccatagagc. Northern analyses were performed at least in duplicate and blots were exposed for several different times to obtain signals in the linear range. Densitometry was performed using the NIH Image program (developed at the US National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). Relative abundance is expressed with respect to values for U3 RNA or 18S rRNA in the same lanes.
Figure 1.
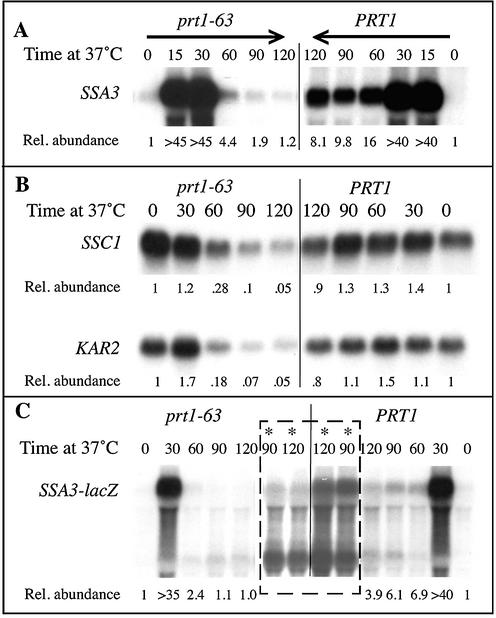
SSA3, SSC1 and KAR2 mRNAs are rapidly decreased in abundance when translation initiation is slowed. Cultures of cells proliferating at 23°C were transferred to 37°C for further incubation and at the indicated times (in min) RNA was extracted, then resolved electrophoretically, blotted and probed for the indicated mRNAs, and also for ACT1 mRNA as an internal loading control (data not shown). Strains used were (A and B) TC3-212-3 (prt1-63) and 21R (PRT1); (C) TC3-212-3 containing YCpSSA3, and 21R containing YCpSSA3. Note that for the SSC1 data a 2-fold increased amount of RNA was loaded from the prt1-63 strain at time 0. (C) The starred lanes enclosed by the dotted lines are a longer exposure of the 90- and 120-min samples. The relative abundance of each mRNA was calculated with respect to that of 18S rRNA.
Half-life determinations
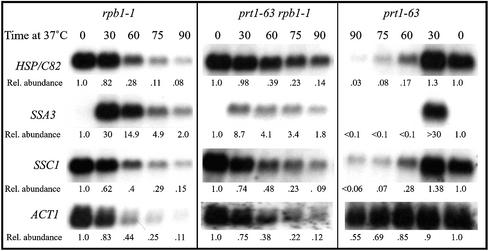
Abundances were determined for mRNAs in one rpb1-1 strain (PLR62B), one prt1-63 strain (PLR62C) and two prt1-63 rpb1-1 double-mutant strains (PLR66C and PLR68B). Each of these strains is 81% identical to wild-type strain 21R and was derived from three backcrosses with this strain. Cells were grown in the same supplemented minimal medium and transferred to the same 37°C water bath for further incubation. By this procedure, cultures reach 37°C within 3 min. At least two independent analyses were done for each set of samples, and several exposures of each northern blot were quantified to ensure that the linear range of the film was used. Normalization was with respect to U3 RNA and 18S rRNA. The results for these determinations differed by <10% for each mRNA, and were averaged to yield the values in Table 2.
Table 2. mRNA half-livesa.
| mRNA | Genotype of mRNA source | ||
|---|---|---|---|
| rpb1-1 | rpb1-1 prt1-63 | prt1-63b | |
| HSP/C82 | 17 | 20.2 | 10.5 |
| SSA3 | 15.3 | 17.6 | <5 |
| SSC1 | 20 | 20 | 11.2 |
| ACT1 | 20.1 | 19.8 | 32 |
aHalf-life values were determined as described in Materials and Methods.
bAbundance half-lives, due to ongoing mRNA synthesis in these RPB1 cells.
To ensure reproducibility, HSP/C82 and ACT1 mRNA half-lives were also measured for rpb1-1 prt1-63 double-mutant cells (PLR68B) that contained either a plasmid-borne wild-type PRT1 gene or a vector control. These cells were grown in synthetic complete medium lacking uracil (for plasmid maintenance). The half-life values for HSP/C82 and ACT1 mRNAs from these strains were within 15% of the values in Table 2 (data not shown).
RESULTS
SSA3 mRNA is rapidly degraded when translation initiation is slowed
In yeast cells that carry the prt1-63 mutation, the mRNAs from two Hsp70 genes, SSA1 and SSA2, and that from a cognate SSA2-lacZ reporter gene are all subject to more rapid degradation compared with what is seen in wild-type cells (7). This increased mRNA degradation is observed when the rate of mRNA translation initiation is slowed down by the effects of the prt1-63 mutation, and is termed initiation-mediated mRNA decay. To investigate whether this phenomenon is more widespread, the mRNA stabilities for three other members of the Hsp70 gene family, SSA3, SSC1 and KAR2, were assessed. The prt1-63 mutation was again used to decrease the rate of translation initiation.
Immediately after cells are transferred to a high temperature, transcription of the yeast heat-shock genes undergoes a rapid increase (the induction phase) followed by a slow decline (the recovery phase) that results in a level of heat-shock mRNA, at heat-shock temperatures, somewhat higher than at non-heat-shock temperatures. The SSA3 member of the Hsp70 gene family displays this classic heat-shock transcriptional response (18), as is evident in Figure 1A, which shows SSA3 mRNA abundance in heat-shocked wild-type cells. The same heat shock for prt1-63 mutant cells resulted in the induction of SSA3 mRNA to the same degree and with the same kinetics (>40-fold by 15 min at 37°C) as in wild-type cells. However, this transfer to 37°C also causes the prt1-63 mutation to slow the rate of translation initiation; a decrease in translation to ∼30% the level in wild-type cells similarly treated (at 60 min) (10) was paralleled by a decrease in SSA3 mRNA abundance to levels far lower than those in wild-type cells. By 90 min after temperature shift, SSA3 mRNA was nearly undetectable in prt1-63 mutant cells but easily detected in wild-type cells. Therefore, slowing down translation initiation has an effect on SSA3 mRNA analogous to that seen for SSA1 and SSA2 mRNAs (7).
The Hsp70 mRNAs that up to this point have been shown to be affected by initiation-mediated decay are all encoded by the same SSA subgroup of Hsp70 genes. These genes are highly homologous to each other: SSA1 and SSA2 are >90% identical at the DNA level, and SSA3 is nearly 85% identical to SSA1 and SSA2 (19). These Ssa-type Hsp70 proteins also function in the same intracellular compartment, the cytoplasm and individually each protein is non-essential (17,20,21). To assess if diverse Hsp70-family mRNAs are also affected by initiation-mediated decay we measured mRNA abundance for the SSC1 and KAR2 genes under conditions of limited translation initiation. SSC1 and KAR2 are only 50–60% identical to each other and to the SSA group of Hsp70 genes (19,22). Unlike SSA3, these Hsp70-family genes are significantly expressed at non-heat-shock temperatures and show only minimal induction upon heat shock (18,22) (Fig. 1B). Moreover, each is essential for growth, and their encoded proteins function in different, non-cytoplasmic compartments, with Ssc1p in the mitochondrion and Kar2p in the endoplasmic reticulum (22,23). Nonetheless, Figure 1B shows that SSC1 and KAR2 mRNAs are also markedly decreased in abundance (at least 10-fold lower by 2 h at 37°C compared with PRT1 wild-type cells) under the same conditions that trigger initiation-mediated decay of the SSA mRNAs (Fig. 1A). Thus, a variety of Hsp70-family mRNAs are subject to initiation-mediated decay.
For SSA2 mRNA, experiments using SSA2-lac reporter genes indicated that targeting sequences for initiation- mediated mRNA decay are within SSA2 mRNA 5′ sequences (7). To assess if the 5′ end of SSA3 mRNA also contains sequences responsible for rapid mRNA degradation when translation initiation is slowed down, we used a chimeric SSA3-lacZ fusion gene and monitored the abundance of its mRNA in wild-type and prt1-63 mutant cells. The fusion gene comprises the SSA3 sequences –468 to +9 (+1 is the A of the AUG) fused in frame to the Escherichia coli lacZ gene, and produces functional β-galactosidase enzyme (13; data not shown). As seen in Figure 1C, SSA3-lacZ mRNA levels, like SSA3 mRNA levels, were decreased in abundance under conditions of decreased translation initiation in prt1-63 mutant cells. The starred lanes are a longer exposure of the later time points that clearly indicate this 4-fold difference in abundance. The SSA3 sequences present in the SSA3-lacZ mRNA therefore confer instability. However, this decrease was less than the 7-fold decrease seen for endogenous SSA3 mRNA (Fig. 1A), suggesting that additional sequences may be involved in SSA3 mRNA instability. To confirm that lacZ sequences do not contribute to this instability we measured the abundance of a fusion mRNA that comprises the 5′ sequences from HIS4 mRNA fused in-frame to lacZ mRNA. This HIS4-lacZ fusion mRNA was as abundant in prt1-63 mutant cells as in wild-type cells even after 2 h at 37°C (Fig. 4). This observation, plus our similar finding for a CYC1-lacZ reporter mRNA (7), confirms that lacZ sequences do not contribute to the decreased SSA3-lacZ mRNA levels seen under conditions of slowed translation initiation, and strengthens our earlier conclusion that the actions of initiation-mediated mRNA decay are limited to a subset of mRNA species.
Figure 4.
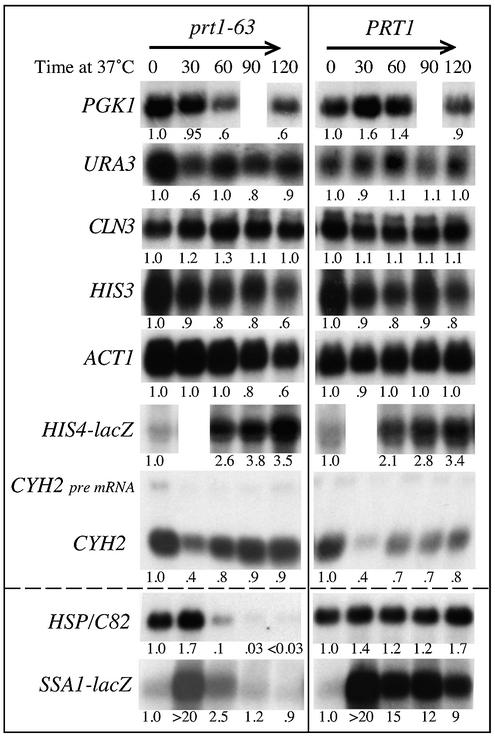
Not all mRNAs are rapidly degraded when translation initiation is slowed. Cell growth, sampling and RNA blotting were as described in Figure 1. The probes are described in the Materials and Methods. Strains were 21R (lanes 6–10) and the prt1-63 strains P26U3A for URA3, CB61A for CLN3 and TC3-212-3 for the remainder (lanes 1–5). Note that there is no 30-min sample for the HIS4-lacZ probe and no 90-min sample for the PGK1 probe. The relative abundance of each mRNA was calculated with respect to that of 18S rRNA.
HSP82 and HSC82 mRNAs are also rapidly degraded when translation initiation is slowed
The finding that several mRNAs from the Hsp70 gene family are markedly decreased in abundance in prt1-63 mutant cells raised the possibility that other heat-shock mRNAs might also be subject to initiation-mediated decay. The mRNAs encoded by the Hsp90 gene family were considered to be good candidates. We have shown that Hsp90-based fusion proteins are underrepresented in prt1-1 mutant cells when translation was slowed down at 32°C (the temperature for this prt1 mutant allele that slows down the rate of translation initiation without blocking protein synthesis), and that Hsp90-gene mRNAs are virtually absent (11) (Fig. 5A). Similarly, Hsp90-lacZ fusion proteins were also underrepresented in prt1-63 mutant cells at the restrictive temperature (data not shown). To test if decreased Hsp90 mRNA levels account for this decreased synthesis of Hsp90 proteins when translation initiation is slowed down, we monitored the abundance of Hsp90-gene mRNAs and Hsp90-lacZ fusion-gene mRNAs in prt1-63 mutant cells.
Figure 5.
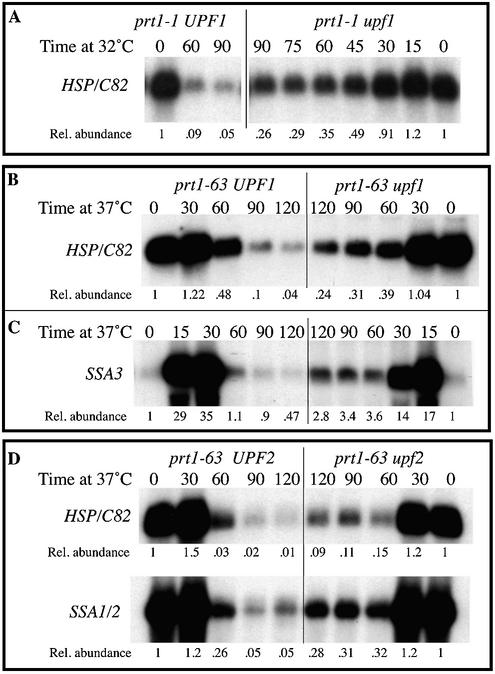
upf1 and upf2 mutations minimize HSP/C82 and SSA3 mRNA degradation in prt1-1 and prt1-63 mutants. Cell growth, sampling and RNA blotting for (B), (C) and (D) were as described in Figure 1. (A) Cells proliferating at 23°C were transferred to 32°C for further incubation prior to RNA extraction. Blots were probed with either an internal fragment of the HSC82 gene that cross-hybridizes with HSP82, with the SSA3 probe, or with an internal fragment of SSA2 that cross-hybridizes with SSA1, then stripped and re-probed for ACT1 mRNA as an internal loading control. Strains used were (A) ZPU70H (prt1-1) and CBPU9BM (prt1-1 upf1); (B) and (C) TC3-212-3 (prt1-63) and P6U9A (prt1-63 upf1); (D) TC3-212-3 (prt1-63) and NM4A3D (prt1-63 upf2). The relative abundance of each mRNA was calculated with respect to that of 18S rRNA.
The yeast Hsp90 gene family comprises two nearly identical genes, the heat-inducible HSP82 gene and the cognate HSC82 gene that is more constitutively transcribed (12,14). In hybridization experiments it is difficult to distinguish between HSP82 and HSC82 mRNA, because the two protein products are 97% identical (14) and probes cross-hybridize to both mRNAs. Therefore, to assess the abundance of individual Hsp90 mRNAs we constructed strains that contained either an hsp82::LEU2 disruption mutation or an hsc82::LEU2 disruption mutation, thus allowing the detection of each Hsp90-gene mRNA in the absence of the other.
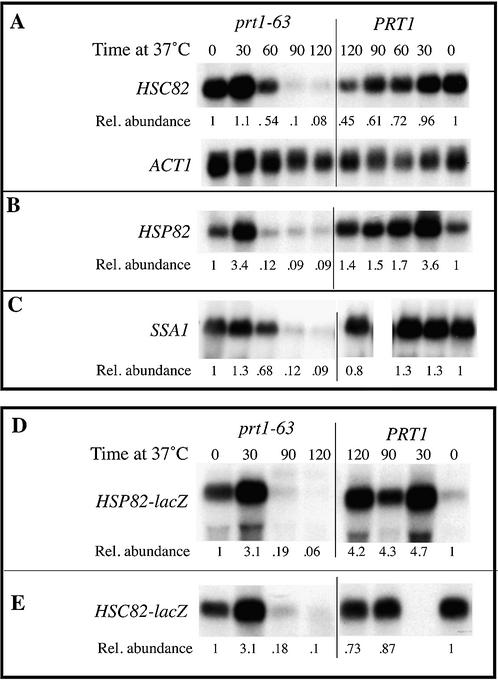
As expected from a heat-shock cognate gene, HSC82 mRNA levels were easily detectable prior to heat shock, and were increased only mildly by transfer to a heat-shock temperature (14), followed by a decline during the recovery phase (Fig. 2A). In prt1-63 hsp82::LEU2 mutant cells there was also a normal induction of HSC82 mRNA, but during the recovery phase the levels of HSC82 mRNA were decreased to levels 5-fold lower than in hsp82::LEU2 cells with normal translation initiation.
Figure 2.
Rapid degradation of HSP82 and HSC82 mRNAs when translation initiation is slowed. Cell growth, sampling, RNA blotting and quantitation were as described in Figure 1. Blots were probed: (A and B) with an internal fragment of the HSC82 gene that cross-hybridizes to the virtually identical HSP82 gene; (C) with an internal fragment of SSA2 that cross- hybridizes under the conditions used here with SSA1; (D and E) with an internal lacZ fragment. All blots were then stripped and re-probed for ACT1 mRNA as an internal loading control. Strains used were (A) 3H15A containing control vector pRS313 (prt1-63), and 3H15A containing wild-type PRT1 plasmid pDEHISPRT (PRT1); each strain also has an hsp82::LEU2 disruption. (B) C4X9D carrying control vector pRS316 (prt1-63), and C4X9D containing wild-type PRT1 plasmid pR16P (PRT1); each strain also has an hsc82::LEU2 disruption. (C) PS2D5C (prt1-63) and PS2D5D (PRT1); each strain also has an ssa2::HIS3 disruption. (D) TC3-212-3 containing YCp82 (prt1-63), and 21R containing YCp82 (PRT1). (E) TC3-212-3 containing YCp90C, and 21R containing YCp90C. Note that the 90-min sample for the PRT1 strain in (D) is under loaded, and that there is no 30-min sample for the PRT1 strain for (E). The relative abundance of each mRNA was calculated with respect to that of 18S rRNA.
The pattern of transcription of HSP82 differs from that of HSC82 in that HSP82 mRNA is present at only low levels prior to heat shock and increases to much higher levels immediately after heat shock; during the recovery phase mRNA levels decline to values that are still higher than those pre-heat shock (14) (Fig. 2B). In prt1-63 hsc82::LEU2 mutant cells the levels of HSP82 mRNA were increased to the same degree by heat shock as in PRT1 hsc82::LEU2 cells, but during the recovery phase HSP82 mRNA abundance decreased markedly, to levels 15-fold lower than in cells with normal translation initiation. This decrease began earlier for HSP82 mRNA than for HSC82 mRNA, as indicated by the 60-min time point. We compared the kinetics of this decrease in HSP82 mRNA abundance with those for another heat-inducible mRNA, from SSA1, to determine if this earlier decrease was a feature common to highly inducible heat-shock mRNAs. As shown in Figure 2C, SSA1 mRNA (the SSA2 gene was disrupted so that our probe hybridized only with SSA1 mRNA) still persisted in prt1-63 mutant cells 60 min after transfer to 37°C. We conclude from this and the above Hsp70 gene family observations that the rapidity with which initiation-mediated decay affects mRNA levels is a characteristic of the individual mRNA rather than a hallmark of a pattern of transcriptional regulation.
As we did above for SSA3 mRNA, we constructed Hsp90-lacZ fusion genes and measured mRNA levels to identify RNA sequences that are determinants of this decreased mRNA abundance. The HSC82-lac and HSP82-lac fusion genes described earlier (11) were modified to eliminate lac operon sequences downstream of the lacZ gene, thus making the mRNAs from these reporter genes identical (in lac sequences) with that from the SSA2-lacZ reporter gene described earlier (7), and virtually identical to the lac sequences of SSA3-lacZ described above (Fig. 1C). Although these downstream lac sequences have no effect on initiation-mediated mRNA decay (7), this structural modification simplifies the analysis in that only a single, 3.4-kb fusion mRNA is expressed from these modified HSC82-lacZ and HSP82-lacZ genes. As seen in Figure 2D and E, HSP82-lacZ and HSC82-lacZ mRNA levels, like those from the endogenous HSP82 and HSC82 genes, were decreased in abundance in prt1-63 mutant cells (compared with wild-type cells) under conditions of decreased translation initiation. These results indicate that the 5′ ends of the two Hsp90 mRNAs are sufficient to sensitize these mRNAs to initiation-mediated mRNA decay.
Initiation-mediated decay is faster than predicted by mRNA half-lives
The parameters of initiation-mediated decay were investigated further. For the SSA1 and SSA2 mRNAs, the decreased abundance noted in prt1-63 mutant cells at an elevated temperature has been shown to be due to an increased degradation rate (7). To see if the decreased levels of SSA3 mRNA in prt1-63 mutant cells are also a consequence of more active mRNA degradation we measured the stability of several mRNAs. Direct mRNA-stability experiments in yeast often use cells that contain a temperature-sensitive mutation in the gene encoding the largest subunit of RNA polymerase II, Rpb1p. Cells with this rpb1-1 mutation cease transcription within 5–15 min after transfer to 37°C (24). Sampling at various times after this transcriptional shutoff allows the abundance of pre-existing mRNA molecules to be measured, with decreased abundance indicating the rate of mRNA degradation (reviewed in 25).
As shown in Figure 3, SSA3 mRNA was induced 30-fold by heat shock even in rpb1-1 mutant cells. This newly made SSA3 mRNA is most likely transcribed in the first few minutes at 37°C, before the rpb1-1 mutation inhibits further transcription. SSA3 mRNA was reasonably stable in rpb1-1 mutant cells, with a half-life of ∼15 min (Table 2). Even by 2 h at 37°C the SSA3 mRNA was easily detectable. In prt1-63 cells, however, although SSA3 was also induced 30-fold after heat shock, by 60 min at 37°C the mRNA was undetectable. Similar rapid decreases in abundance were seen in prt1 mutant cells for SSC1 and for the combined HSP82 and HSC82 mRNAs, whereas without the influence of the prt1 mutation the SSC1 mRNA half-life (as measured in the rpb1-1 mutant) was ∼20 min, and for HSP/C82 the half-life of the combined mRNAs was 17 min (Table 2). These results indicate that SSA3, SSC1 and HSP/C82 mRNAs are degraded more rapidly under conditions of decreased translation (prt1-63), suggesting that initiation-mediated decay has a significant effect on SSA3, SSC1 and HSP/C82 mRNA abundance.
Figure 3.
Ongoing transcription is required for the rapid decrease in the abundance of HSP/C82, SSA3 and SSC1 mRNAs when translation is impaired. Cell growth, sampling and RNA blotting were as described in Figure 1. Blots were probed first with an internal fragment of the HSC82 gene that cross-hybridizes to the virtually identical HSP82 gene (HSP/C82), then stripped and re-probed for SSA3, SSC1 and ACT1 mRNAs and then for U3 RNA as an internal loading control. Strains used were PLR62B (rpb1-1), PLR66C (rpb1-1 prt1-63) and PLR62C (prt1-63). The relative abundance of each mRNA was calculated with respect to that of U3 snoRNA (SNR17A) and used to calculate the half-lives as shown in Table 2.
In the same rpb1-1 mutant cells the ACT1 mRNAs disappeared with a half-life of 20 min (Fig. 3 and Table 2) and the URA3 transcript, with a very short half-life, was undetectable at the 30-min time point (data not shown), whereas in prt1-63 cells these mRNAs were easily detected even at 2 h at 37°C (Fig. 4).
Initiation-mediated decay is limited to a subset of yeast mRNAs
We assessed the mRNA levels for a diverse group of yeast genes to determine if mRNA degradation is a common feature when translation initiation is slowed down. For this, mRNAs with widely differing half-lives were chosen. We monitored two long half-life mRNAs, from PGK1 and ACT1, the moderate half-life HIS3 mRNA and the short half-life URA3 mRNA (26–28). None of these mRNAs were markedly different in abundance in prt1-63 mutant cells compared with wild-type cells, even under conditions in which heat-shock mRNAs are markedly decreased in abundance (Fig. 4). In general, therefore, the half-life of an mRNA is not a significant determinant of susceptibility to initiation-mediated decay.
We looked next at an mRNA that contains an upstream (u)ORF, because uORFs have been implicated in the degradation of the uORF-containing mRNAs (29,30). Furthermore, the uORF-containing mRNA in question here, from the CLN3 gene, is reported to be poorly translated in prt1-63 mutant cells at 37°C (31), and poorly translated mRNAs have been shown to be susceptible to increased degradation (32). Nonetheless, despite the presence of an uORF and even though CLN3 mRNA is translated as inefficiently as SSA2 mRNA (11), an mRNA that is subject to initiation-mediated decay (7), CLN3 mRNA abundance was not decreased in prt1-63 mutant cells (Fig. 4). These results indicate that neither the presence of an uORF nor an inherently low translation efficiency is sufficient to target that mRNA for degradation via the initiation-mediated decay pathway.
We also monitored the levels of an intron-containing RNA. The primary transcript from the CYH2 gene contains a single intron (33). In wild-type cells the ratio of CYH2 intron-containing pre-mRNA to mature mRNA is usually 1:10 (34) (Fig. 4). However, in cells that are defective in some aspect of mRNA decay, such as those that contain upf1, upf2, dcp1 or xrn1 mutations, CYH2 pre-mRNA is less rapidly degraded and the ratio of pre-mRNA to mature mRNA is nearly 1:1 (34–37).
We monitored the levels of CYH2 pre-mRNA and mature mRNA in prt1-63 mutant cells after shift to conditions that impaired translation. As shown in Figure 4, at 23°C the CYH2 pre-mRNA and mature mRNA were present at the expected 1:10 ratio in both prt1-63 and wild-type cells, and that ratio remained the same for mutant and wild-type cells even 2 h after the temperature shift. Thus, slowing translation does not mimic the effect of mutations affecting mRNA decay. Except for the expected transient decrease in abundance shortly after transfer to 37°C (38,39), the levels of CYH2 pre-mRNA and mature mRNA were unaffected after transfer to 37°C in both prt1 and wild-type cells. Thus, neither the intron-containing CYH2 pre-mRNA nor the spliced CYH2 mRNA is affected by the initiation-mediated decay pathway.
Upf1p, Upf2p, Dcp1p and Xrn1p mediate the decreased Hsp90 (HSP82 and HSC82) mRNA abundance when translation initiation is slowed
In yeast there are well-described mRNA-decay pathways that share several common components (reviewed in 1). Most mRNAs targeted for degradation are first decapped, mainly by the Dcp1 decapping enzyme, and are then degraded in a 5′-to-3′ direction by the exonuclease Xrn1p. mRNAs comprising a specialized subset are degraded via the nonsense-mediated-decay pathway (NMD). One function of this mRNA-decay pathway is thought to be mRNA surveillance, because an mRNA with an aberrant Stop codon within the normal ORF is degraded rapidly by NMD. The NMD pathway also includes decapping and 5′-to-3′ exonuclease digestion of the mRNA, and requires the presence of Upf proteins for proper function.
The initiation-mediated mRNA-decay pathway, which also affects a small subset of mRNAs but is triggered by a different mechanism (slowed translation initiation), also draws on the functions of at least two Upf proteins, Upf1p and Upf2p. We therefore assessed the involvement of these ‘NMD’ proteins and the mRNA-degradation proteins Dcp1p and Xrn1p in the initiation-mediated decay of the combined Hsp90 (HSP/C82) and SSA1/2 mRNAs.
We had shown previously that both Upf1 and Upf2 proteins are involved in the initiation-mediated decay of SSA1 and SSA2 mRNAs (7). That analysis was extended to the HSP/C82 and SSA3 mRNAs, monitoring first the effects of a upf1 mutation. After transfer of prt1-63 cells to 37°C, HSP/C82 mRNA levels (detected by a probe that cross-hybridizes to both HSP82 and HSC82 mRNAs) were decreased 10-fold by 90 min (Fig. 5B). However, in prt1-63 upf1 double-mutant cells this decrease was much less, only 3-fold. This smaller decrease in HSP/C82 mRNA levels due to a upf1 mutation was also seen for cells carrying another prt1 mutation, prt1-1. In prt1-1 cells, transfer to 32°C elicits the same slowdown in translation as does the transfer of prt1-63 cells to 37°C, to global translation rates ∼30% of those of wild-type cells by 60 min (10). Figure 5A shows that in these prt1-1 mutant cells the HSP/C82 mRNA abundance was decreased 10-fold by 60 min and 20-fold by 90 min at 32°C. However, in prt1-1 upf1 double-mutant cells the HSP/C82 mRNA levels decreased to a much lesser extent, only 3-fold by 60 min and 4-fold by 90 min. This upf1 effect was reproducible. Four independent but related prt1-63 upf1 double-mutant strains were assessed in several replicate experiments for each, and all gave the same findings, a 3–4-fold difference in HSP/C82 levels each time. All of these double mutants are the products of related prt1-63 strains (sharing 75% identity) crossed to a upf1 strain. Finally, a prt1-1 upf1 strain was similarly constructed and tested twice, each time giving results equivalent to those from the four related prt1-63 upf1 strains described above. Thus, the upf1 mutation results in reproducibly attenuated initiation-mediated mRNA decay.
The absence of a functional Upf1 protein caused a similar 3–4-fold increase in SSA3 mRNA levels in prt1-63 mutant cells (Fig. 5C).
We also monitored the effects of the Upf2 protein on HSP/C82 mRNA levels under conditions of limited translation. Again we saw higher HSP/C82 mRNA levels in prt1-63 upf2 mutants, a ∼5-fold difference that is similar to that seen for SSA1/2 mRNA (7) (Fig. 5D). Two independent prt1-63 upf2 double mutants were tested; both gave the same result. (In cells with wild-type PRT1 function, the upf mutations have no effect on the abundance of these mRNAs at 23, 32 or 37°C; data not shown.) We therefore conclude that both Upf1p and Upf2p are involved in the initiation-mediated decay of several heat-shock mRNAs.
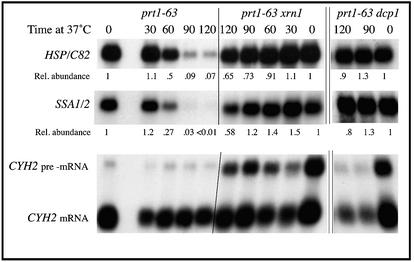
As shown in Figure 6, HSP/C82 mRNA abundance was decreased to a significantly lesser degree in prt1-63 dcp1 and prt1-63 xrn1 double-mutant cells compared with that in prt1-63 DCP1 XRN1 single mutant cells, differing by 8–10-fold after a 2-h incubation at 37°C, to the extent that HSP/C82 mRNA abundance in these double-mutant cells resembled that in wild-type cells (data not shown). Similarly, the abundance of the combined Hsp70 mRNAs from SSA1 and SSA2 (cross-hybridizing probe) in prt1-63 dcp1 and prt1-63 xrn1 mutant cells resembled that in wild-type cells (5-fold greater after a 2-h incubation at 37°C). Neither the dcp1 nor the xrn1 mutation had any effect on the abundance of HSP/C82 or SSA1/2 mRNAs in cells with normal Prt1p function and normal translation initiation (data not shown). Cells carrying the dcp1 mutation accumulate suppressor mutations at a high frequency (D.Schwartz, personal communication), so to ensure that our dcp1 cells exhibited the expected phenotype we re-probed the RNA blots to detect RNA encoded by the CYH2 gene. One diagnostic phenotype of dcp1 and xrn1 mutations is the accumulation of CYH2 pre-mRNA (36,37). All of the dcp1 and xrn1 strains used here accumulated CYH2 pre-mRNA, showing a 1:1 ratio of CYH2 pre-mRNA to CYH2 mature mRNA, whereas in our wild-type DCP1 (and XRN1) strains the ratio of CYH2 pre-mRNA to mature mRNA was the expected 1:10 (Fig. 6) (36,37). As further confirmation that our dcp1 strains were expressing the dcp1 phenotype without genetic interference, we repeated this diagnostic CYH2 RNA hybridization on three other independent prt1-63 dcp1 strains that showed the dcp1 effect on HSP/C82 and SSA1/2 mRNA levels; all of these strains displayed the dcp1 phenotype (data not shown). Similarly, three other independent prt1-63 xrn1 strains showed the CYH2 RNA effect (data not shown). These results therefore indicate that Dcp1p and Xrn1p, and therefore in all likelihood decapping and 5′-to-3′ degradation, participate in initiation-mediated mRNA decay of the heat-shock mRNAs investigated here.
Figure 6.
HSP/C82 and SSA3 mRNA degradation in prt1-63 mutants occurs through the Xrn1 and Dcp1 mRNA-decay pathway. Cell growth, sampling, RNA blotting and quantitation were as described in Figure 1. Blots were probed with either an internal fragment of the HSC82 gene that cross-hybridizes with HSP82, with an internal fragment of SSA2 that cross-hybridizes with SSA1, or with an internal fragment of CYH2 that hybridizes to both CYH2 pre-mRNA and CYH2 mature mRNA, then stripped and re-probed for ACT1 mRNA as an internal loading control. Strains used were TC3-212-3 (prt1-63), SL4D (prt1-63 xrn1) and DCP8A-2 (prt1-63 dcp1).
Initiation-mediated decay depends on ongoing transcription
As described above, mRNA half-lives were estimated through the use of the temperature-sensitive rpb1-1 mutation to block further transcription and thus halt the production and export of new mRNAs into the cytoplasmic pool. This procedure for half-life measurement was also applied to cells bearing the prt1-63 mutation. This approach was possible because both the rpb1-1 and prt1-63 mutations have the same 37°C restrictive temperature, allowing the simultaneous imposition of blocking conditions for both mutations. As shown in Figure 3 and summarized in Table 2, this experiment showed that in cells harboring the rpb1-1 mutation the prt1-63 effect on mRNA abundance is abolished. The mRNA half-lives in rpb1-1 prt1-63 double-mutant cells were the same as in rpb1-1 single-mutant cells, and longer than the values for mRNA-abundance half-life in prt1-63 single-mutant cells. The same results were obtained using isogenic rpb1-1 prt1-63 double-mutant cells carrying either a plasmid-borne wild-type PRT1 gene or empty vector (data not shown), thus verifying the lack of prt1-63 effects on mRNA decay under conditions of transcription shutdown. Initiation-mediated mRNA decay, manifested at the translation level, is apparently possible only under conditions of ongoing gene transcription in the nucleus. Implications of this novel finding are discussed below.
DISCUSSION
We show here that several yeast heat-shock mRNAs are subject to initiation-mediated decay, a mode of mRNA degradation that is seen when translation initiation is slow. To decrease the rate of translation initiation, the experiments here used a temperature-sensitive mutation affecting the Prt1p component of the initiation factor eIF3, but analogous effects are also seen when translation initiation is slowed by use of an mutant version of eIF4E, the mRNA cap-binding protein, or a mutant version of eIF2B, the exchange factor that activates eIF2 (7,40). Here and elsewhere (7,40) we report initiation-mediated decay under these conditions for a variety of heat-shock mRNAs and for chimeric heat-shock mRNAs in which lacZ sequences replace coding sequence.
Initiation-mediated mRNA decay and the degradation of several heat-shock mRNAs has so far been found when translation initiation, although slowed to ∼30% of the wild-type rate, is still taking place, thus allowing significant ongoing protein synthesis (10). In contrast, when translation initiation is virtually abolished the rapid degradation of the SSA1/SSA2 Hsp70 mRNAs and the HSP82/HSC82 Hsp90 mRNAs is also abolished (7; unpublished observations). These experiments used two different prt1 mutations: the prt1-63 mutation studied here but at the higher restrictive temperature of 40°C, which decreases translation to <5% (10), and the prt1-1 mutation, which also rapidly decreases translation to <5% at 37°C (7,41). Thus, slowing translation initiation and actually blocking translation initiation have different consequences for heat-shock mRNA degradation. These effects are mRNA-specific: analogous experiments show that blocking translation initiation by use of the prt1-1 mutation causes more rapid degradation of SDH2 mRNA, which encodes the iron protein subunit of succinate dehydrogenase (42), but has no effect on half-life for several non-heat-shock mRNAs (43). Similarly, severe inhibition of translation initiation by use of the prt1-63 mutation at an elevated restrictive temperature causes increased degradation of mRNAs for the PGK1 and MFA2 genes (44).
The initiation-mediated decay of heat-shock mRNAs was shown here to rely on Upf1p and Upf2p, as suggested previously (7), and also on the mRNA-decapping enzyme Dcp1p and the 5′-to-3′ exoribonuclease Xrn1p. Initiation-mediated mRNA decay of heat-shock mRNAs is virtually abolished when either of these latter two enzymes is absent, suggesting that the trigger for initiation-mediated decay shunts an mRNA molecule into the pathway for degradation that is common to all known mRNA-decay schemes (reviewed in 1). On the other hand, initiation-mediated decay is only partially prevented when the Upf proteins are absent, suggesting the existence of Upf-dependent and -independent sub-pathways for initiation-mediated decay.
The importance of ongoing transcription by RNA polymerase II for initiation-mediated mRNA decay was an unexpected finding. This dependence on transcription could be a manifestation of several different interactions. For example, initiation-mediated decay may be physically associated with the transcription process. A round of ‘pioneer translation’ in the nucleus has been hypothesized, weeding out newly made but mutant or improperly spliced mRNA molecules (45). Initiation-mediated decay may act at this intranuclear stage. Halting new RNA synthesis would therefore halt the rapid, initiation-mediated decay of newly synthesized RNA molecules, while leaving those mRNAs already in the cytoplasm to be degraded at the usual rate. However, initiation-mediated decay causes the abundance of affected mRNAs to decline at faster rates than can be accounted for by half-life measurements (Table 2). Therefore, initiation-mediated decay must also affect cytoplasmic mRNAs, regardless of any effects during nuclear translation. In another model, ongoing transcription may be necessary for the continued production of a labile mRNA-decay factor. Alternatively, ongoing transcription may be necessary for the continued export from the nucleus of an mRNA-associated decay factor. Several proteins are exported from the nucleus in association with mRNA molecules, and are then imported back into the nucleus for another round of mRNA export. For several of these mRNA-associated proteins, including Npl3p and Nab2p, blocking new transcription using the rpb1-1 mutation shuts off the supply of new mRNA molecules for export and results in the concentration of each of these proteins in the nucleus (46,47). One candidate for an initiation-mediated decay component that is exported from the nucleus as a consequence of new transcription is Upf3p. This protein recruits Upf2 (48), is required for nonsense-mediated mRNA decay and shuttles between the nucleus and the cytoplasm (49), while the human Upf3 has been shown to bind to spliced mRNAs prior to export (50).
While we see no evidence of initiation-mediated decay for the mRNAs from several non-heat-shock genes, the mRNAs from five members of the yeast Hsp70 gene family, and both members of the Hsp90 gene family, are all subject to initiation-mediated decay. These findings, although derived from a limited sample of mRNAs, suggest that initiation-mediated mRNA decay may have an important physiological role under stress conditions (reviewed in 51,52). Susceptibility of an mRNA to initiation-mediated decay may be correlated with gene-product function: each of the Hsp70 and Hsp90 proteins encoded by these mRNAs participates in protein folding/refolding through chaperone activity (reviewed in 53). These observations raise the possibility that fine-tuning the expression of chaperone proteins through initiation-mediated decay of the encoding mRNAs is important during times of slow translation initiation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elizabeth Craig, Michael Culbertson, Melanie Dobson, Alan Jacobson, Arlen Johnson, Susan Lindquist, Roy Parker, Mark Rose, David Schwartz and Maggie Werner-Washburne for strains and plasmids, Rick Singer for helpful discussions and Kendra Gillis and Yahua Song for technical assistance. H.L.H. was supported by a Studentship from Cancer Research and Education Nova Scotia, and S.A.L. was supported by a Studentship from the Nova Scotia Health Research Foundation. This work was funded by a grant to C.A.B. from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Tucker M. and Parker,R. (2000) Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem., 69, 571–595. [DOI] [PubMed] [Google Scholar]
- 2.Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 4.Brown J.T., Bai,X. and Johnson,A.W. (2000) The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA, 6, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs J.S., Anderson,A.R. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culbertson M.R. (1999) RNA surveillance unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- 7.Barnes C.A. (1998) Upf1 and Upf2 proteins mediate normal yeast mRNA degradation when translation initiation is limited. Nucleic Acids Res., 26, 2433–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershey J.W.B. and Merrick,W.C. (2000) The pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88. [Google Scholar]
- 9.Hinnebusch A.G. (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Sonenberg,N., Hershey,J.W.B. and Matthews,M.B. eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 185–243. [Google Scholar]
- 10.Hanic-Joyce P.J., Johnston,G.C. and Singer,R.A. (1987) Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp. Cell Res., 172, 134–145. [DOI] [PubMed] [Google Scholar]
- 11.Barnes C.A., Singer,R.A. and Johnston,G.C. (1993) Yeast prt1 mutations alter heat-shock gene expression through transcript fragmentation. EMBO J., 12, 3323–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrelly F.W. and Finkelstein,D.B. (1984) Complete sequences of the heat shock-inducible HSP90 gene of Saccharomyces cerevisiae. J. Biol. Chem., 259, 5745–5751. [PubMed] [Google Scholar]
- 13.Boorstein W.R. and Craig,E.A. (1990) Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borkovich K.A., Farrelly,F.W., Finkelstein,D.B., Taulien,J. and Lindquist,S. (1989) hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol., 9, 3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellwood M.S. and Craig,E.A. (1984) Differential regulation of the 70K heat shock gene and related genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slater M.R. and Craig,E.A. (1989) The SSA1 and SSA2 genes of the yeast Saccharomyces cerevisiae. Nucleic Acids Res., 17, 805–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner-Washburne M., Stone,D.E. and Craig,E.A. (1987) Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 2568–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner-Washburne M., Becker,M., Kosic-Smithers,J. and Craig,E.A. (1989) Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol., 171, 2680–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig E.A., Kramer,J. and Kosic-Smithers,J. (1987) SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc. Natl Acad. Sci. USA, 84, 4156–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshaies R.J., Koch,B.D., Werner-Washburne,M., Craig,E.A. and Schekman,R.A. (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature, 332, 800–805. [DOI] [PubMed] [Google Scholar]
- 21.Craig E.A. and Jacobsen,K. (1984) Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell, 38, 841–849. [DOI] [PubMed] [Google Scholar]
- 22.Rose M.D., Misra,L.M. and Vogel,J.P. (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell, 57, 1211–1221. [DOI] [PubMed] [Google Scholar]
- 23.Craig E.A., Kramer,J., Shilling,J., Werner-Washburne,M., Holmes,S., Kosic-Smithers,J. and Nicolet,C.M. (1989) SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol. Cell. Biol., 9, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonet M., Scafe,C., Sexton,J. and Young,R. (1987) Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrick D., Parker,R. and Jacobson,A. (1990) Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer V. and Struhl,K. (1996) Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 28, 5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira C.C. and McCarthy,J.E.G. (1995) The relationship between translation and mRNA stability. J. Biol. Chem., 270, 8936–8943. [DOI] [PubMed] [Google Scholar]
- 30.Vilela C., Linz,G., Rodrigues-Pousada,C. and McCarthy,J.E.G. (1998) The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res., 26, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polymenis M. and Schmidt,E.V. (1997) Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev., 11, 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fried H.M. and Warner,J.R. (1982) Molecular cloning and analysis of yeast gene for cycloheximide resistance and ribosomal protein L29. Nucleic Acids Res., 25, 3133–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He F., Peltz,S.W., Donahue,J.L., Rosbash,M. and Jacobson,A. (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1 mutant. Proc. Natl Acad. Sci. USA, 90, 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He F. and Jacobson,A. (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev., 9, 437–454. [DOI] [PubMed] [Google Scholar]
- 36.Tharun S. and Parker,R. (1999) Analysis of mutations in the yeast mRNA decapping enzyme. Genetics, 151, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F. and Jacobson,A. (2001) Upf1, Nmd2, and Upf3 regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol., 21, 1515–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim C.H. and Warner,J.R. (1983) Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol. Cell. Biol., 3, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herruer M.H., Mager,W.H., Raue,H.A., Vreken,P., Wilm,E. and Planta,R.J. (1988) Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res., 16, 7917–7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes C.A., MacKenzie,M.M., Johnston,.G.C. and Singer,R.A. (1995) Efficient translation of an SSA1-derived heat-shock mRNA in yeast cells limited for cap-binding protein and eIF-4F. Mol. Gen. Genet., 246, 619–627. [DOI] [PubMed] [Google Scholar]
- 41.Hartwell L.H. and McLaughlin,C.S. (1969) A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl Acad. Sci. USA, 62, 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cereghino G.P., Atencio,D.P., Saghbini,M., Beiner,J. and Scheffler,I.E. (1995) Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol. Biol. Cell, 6, 1125–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch E.M. and Jacobson,A. (1999) An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J., 18, 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz D.C. and Parker,R. (1999) Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishigaki Y., Li,X., Serin,G. and Maquat,L.E. (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell, 106, 607–617. [DOI] [PubMed] [Google Scholar]
- 46.Lee M.S., Henry,M. and Silver,P.A. (1996) A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev., 10, 1233–1246. [DOI] [PubMed] [Google Scholar]
- 47.Green D.M., Marfatia,K.A., Crafton,E.B., Zhang,X., Cheng,X. and Corbett,A.H. (2002) Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem., 277, 7752–7760. [DOI] [PubMed] [Google Scholar]
- 48.Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. [DOI] [PubMed] [Google Scholar]
- 49.Shirley R.L., Ford,A.S., Richards,M.R., Albertini,M. and Culbertson,M.R. (2002) Nuclear import of Upf3p is mediated by importin-α/-β and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics, 161, 146514–146582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim V.N., Kataoka,N. and Dreyfuss,G. (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science, 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- 51.Estruch F. (2000) Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev., 24, 469–486. [DOI] [PubMed] [Google Scholar]
- 52.Kaufman R.J. (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev., 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- 53.Morano K.A., Liu,P.C.C. and Thiele,D.J. (1998) Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr. Opin. Microbiol., 1, 197–203. [DOI] [PubMed] [Google Scholar]
- 54.Johnston S.A. and Hopper,J.E. (1982) Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl Acad. Sci. USA, 79, 6971–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans D.R.H., Rasmussen,C., Hanic-Joyce,.P.J, Johnston,G.C., Singer,R.A. and Barnes,C.A. (1995) Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol., 15, 4525–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowley A., Singer,R.A. and Johnston,G.C. (1991) CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol., 11, 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]