Abstract
Cleavage and polyadenylation factor (CPF) is a multi-protein complex that functions in pre-mRNA 3′-end formation and in the RNA polymerase II (RNAP II) transcription cycle. Ydh1p/Cft2p is an essential component of CPF but its precise role in 3′-end processing remained unclear. We found that mutations in YDH1 inhibited both the cleavage and the polyadenylation steps of the 3′-end formation reaction in vitro. Recently, we demonstrated that an important function of CPF lies in the recognition of poly(A) site sequences and RNA binding analyses suggesting that Ydh1p/Cft2p interacts with the poly(A) site region. Here we show that mutant ydh1 strains are deficient in the recognition of the ACT1 cleavage site in vivo. The C-terminal domain (CTD) of RNAP II plays a major role in coupling 3′-end processing and transcription. We provide evidence that Ydh1p/Cft2p interacts with the CTD of RNAP II, several other subunits of CPF and with Pcf11p, a component of CF IA. We propose that Ydh1p/Cft2p contributes to the formation of important interaction surfaces that mediate the dynamic association of CPF with RNAP II, the recognition of poly(A) site sequences and the assembly of the polyadenylation machinery on the RNA substrate.
INTRODUCTION
All eukaryotic mRNA precursors (pre-mRNA) are extensively modified before they can serve as templates for protein synthesis. Pre-mRNA 3′-end processing is initiated by endonucleolytic cleavage at the poly(A) site. Subsequently, the upstream cleavage product is polyadenylated whereas the downstream fragment is rapidly degraded (for review see 1). The yeast 3′-end processing reaction can be reconstituted in vitro with the cleavage and polyadenylation factor IA (CF IA), cleavage and polyadenylation factor IB (CF IB), cleavage and polyadenylation factor (CPF) and the poly(A) binding protein (Pab1p) (2,3). Interactions between their subunits allow bridging of the different factors and ensure their coordinated action on the substrate. So far the CPF components Fip1p and Pfs2p have been shown to bridge CPF with CF IA by interacting with Rna14p, a subunit of CF IA (3,4). A table of the factors involved in 3′-end processing and their subunits is provided as supplementary material.
The polyadenylation signals that guide the processing machinery in yeast are redundant and more degenerate compared to the well-conserved sequences in higher eukaryotes. Still, conserved elements can be found, which are the efficiency element (EE), the positioning element (PE), the poly(A) site and U-rich sequences. The EE is located at variable distances upstream of the cleavage site (5). The PE is often found ∼20 nt upstream of the cleavage site and consists of an A-rich sequence (6). U-rich regions, located directly upstream and downstream of the cleavage site, and the poly(A) site itself act in concert to produce multiple recognition sites (5,7–9).
RNase H protection mapping experiments with a CYC1 pre-mRNA suggested that CPF is involved in the recognition of the poly(A) site by specific interactions of its subunits Yhh1p/Cft1p, Ydh1p/Cft2p and Yth1p with sequences surrounding the poly(A) site (9–11). Nab4p/Hrp1p (CF IB) binds to the EE (12,13) and, through interaction with Rna14p, possibly enables CF IA to bind to the PE (14). However, it was also reported that Nab4p/Hrp1p (15) as well as the PE and EE (9) are not essential for cleavage in vitro, underscoring the importance of the poly(A) site region itself.
The yeast and mammalian 3′-end processing factors are highly homologous (reviewed in 16). In mammals cleavage and polyadenylation specificity factor (CPSF) contributes to poly(A) site selection by binding to the well conserved AAUAAA element upstream of the cleavage site (17); CPSF-160 and possibly additional CPSF subunits are thought to mediate specific interactions to the RNA (18). The binding of purified CPSF is weak, but is strongly enhanced by a cooperative interaction with cleavage stimulation factor (CstF) bound to the downstream elements (19–21).
Transcription by RNA polymerase II (RNAP II) and pre-mRNA processing reactions are coupled events (reviewed in 22–25). The C-terminal domain (CTD) of RNAP II plays a central role in linking the processing reactions to transcription. Current models suggest that the CTD is hypo-phosphorylated during transcription initiation and that escape of RNAP II into the elongation phase is accompanied by phosphorylation of the CTD. It has been proposed that the change in charge upon phosphorylation enables proteins involved in pre-mRNA processing to bind to the CTD (26–28). This includes proteins involved in capping (29–32), splicing (33) and 3′-end formation (10,28,34–37). It is assumed that the assembled proteins subsequently travel with the elongating RNAP II during transcription and act on the nascent RNA transcript.
In mammals the CTD was suggested to play a direct role in 3′-end cleavage in vivo (34) and in vitro (38,39). Experiments in yeast provided evidence that transcription in the absence of the CTD was accompanied by a reduction of cleavage efficiency and the resulting mRNAs had shorter poly(A) tails (28,40). Correct transcription termination requires a functional poly(A) signal in all organisms (41,42, reviewed in 43), and yeast strains carrying mutations in CPF and CF IA were shown to be deficient in correct transcription termination, indicating the coupling between 3′-end processing and transcription termination (10,35,44–47). Furthermore, correct transcriptional termination was found to require the transcription factors Sub1p (48) and Res2p (49) and chromatin remodeling factors (50). The Nrd1p complex was shown to be required for correct termination at snoRNA genes (51).
Ydh1p/Cft2p (which will be referred to as Ydh1p in the remainder of this paper) is the 105 kDa subunit of CPF. It shares 24.4% identity and 43% similarity with the mammalian CPSF-100 protein. It is also significantly related to Ysh1p and to CPSF-73 (52). Ydh1p is essential for cell viability (53) and was shown to bind RNA (54). RNase H protection experiments suggested that the protein interacts with the poly(A) site region (9). Here, we show that Ydh1p is required for cleavage and polyadenylation in vitro and for poly(A) site recognition in vivo. Furthermore, we provide evidence that Ydh1p interacts with several subunits of CPF, with Pcf11p, a subunit of CF IA, and with the CTD of RNAP II. The results suggest that Ydh1p is part of an interaction surface that mediates important contacts with CF IA, the CTD of RNAP II and the pre-mRNA substrate.
MATERIALS AND METHODS
Yeast strains
For random mutagenesis of YDH1, mutagenic PCR was carried out with a low concentration of ATP (3). The primers used for PCR were: Ydh1-N (5′-CCCTTACGGATTGAAGTCATT-3′) and Ydh1-C (5′-TTGAACCTTTTATTTGTGCTG-3′). Plasmid pBD63 (YDH1–LEU2–CEN) was obtained by subcloning of the YDH1 gene from pIA115 (YDH1–URA3–CEN) (53) into the BamHI and SacI restriction sites of pRS415. The plasmid was digested with the restriction enzymes NsiI and SpeI. The fragment containing plasmid sequences and sequences flanking the YDH1 gene was co-transformed together with the mutagenized PCR product into the yeast strain YPP106 (53). Transformants were selected on minimal medium lacking leucine and replica-plated onto 5-FOA plates in order to shuffle out the pIA115 plasmid. The colonies were then replica-plated onto YPD-plates and incubated at 25, 33 or 37°C, respectively. Plasmids of candidate colonies that showed a ts phenotype were isolated by standard procedures. The conditional growth phenotype was then verified by retransformation of the isolated plasmids into YPP106, 5-FOA treatment and growth tests at elevated temperatures.
Genotypes of yeast strains used in this study were: YPP106: MATα; ura3-1; ade2-1; leu2-3,112; his3-11,15; trp1Δ; ydh1::TRP1 [pIA115; CEN4–URA3–YDH1] (53); YAK1: MATα; ura3-1; ade2-1; leu2-3,112; his3-11,15; trp1Δ; ydh1::TRP1 [pAK21 CEN–LEU2–ydh1-1]; YAK2: MATα; ura3-1; ade2-1; leu2-3,112; his3-11,15; trp1Δ; ydh1::TRP1 [pAK22 CEN–LEU2–ydh1-2]; YAK3: MATα; ura3-1; ade2-1; leu2-3,112; his3-11,15; trp1Δ; ydh1::TRP1 [pAK23 CEN–LEU2–ydh1-3]; Y190: ura3-52; trp1-901; ade2-101; leu2-3; 112 his3-200r; gal4D; gal 80D; URA3:GAL1-lacZ; LYS2::GAL1-HIS3; cyhr; Clontech.
Plasmids and primers
The plasmids encoding the C-terminally truncated Ydh1p fragments were obtained by digestion of plasmid pBD71 (9) with restriction enzymes AccI (pBD91; encoding the recombinant protein ΔC338), BamHI/XbaI (pBD92; encoding ΔC182), BamHI/SpeI (pAK5; encoding ΔC137), BamHI/AflII (pAK6; encoding ΔC555), BamHI/AgeI (pAK7; encoding ΔC613). GST-Ysh1p was encoded by pBD38, which was obtained by subcloning of Ysh1p into p20 (GST expression vector) using the restriction enzymes NdeI and BamHI. GST-Tev-Pta1p was encoded by pBD51, which was obtained by PCR amplification of Pta1p followed by digestion with NdeI and BamHI and ligation into p26 (GST-Tev expression vector). GST-Yhh1p-H6 was encoded by pBD75 (10).
The plasmid encoding the protein used to produce antibody directed against Ydh1p was constructed by digestion of pQE-9 (His6 expression vector Qiagen) with BamHI/HindIII. The insert was constructed by PCR, amplifying the sequence between primer Ydh5′ (5′-ATCGCGGATCCATGACTTATAAATACAATTG-3′) and Ydh3′ (5′-AGCCCAAGCTTATTTACTCAATTCGTTTGGT-3′) of YDH1. For details about pBD-CTD, pACT2-YHH1, pACT2-YSH1 and pACT2-PCF11 see (10).
Expression of recombinant proteins in Escherichia coli
BL21 E.coli cells carrying the respective plasmid were grown at 25°C in 2× YT until they reached an OD600 of ∼1. Following induction by 0.5 mM IPTG, incubation was continued for 6 h. The proteins were purified at 4°C on glutathione–Sepharose 4B as recommended (Pharmacia) and the protein was eluted with GST-elution buffer [75 mM KCl, 50 mM Tris–HCl pH 7.9, 10% glycerol, 10 mM glutathione (reduced), 0.01% NP-40, 1 mM DTT].
Protein–protein interactions
In vitro translations were performed with the TNT-coupled transcription–translation system (Promega). GST fusion protein (100 ng) was incubated with in vitro translated [35S]methionine-labeled proteins for 1 h. The mixture was bound in a total volume of 860 µl to 20 µl glutathione sepharose (Pharmacia), which was previously equilibrated in 1 ml PBS, 0.01% NP-40 and 100 µg BSA. The matrix was washed three times with IPP150 (150 mM KCl, 20 mM Tris–HCl pH 8.0, 0.01% NP-40) and the proteins were eluted by addition of protein loading buffer and incubation at 95°C. Bound proteins were separated by SDS–PAGE and visualized by autoradiography. Phosphorylation of GST–CTD was performed as described previously (38) and the assay of the CTD interaction was carried out as described (10). The two-hybrid tests were carried out as described (55).
Extract preparation and in vitro cleavage and polyadenylation assays
Extracts competent for in vitro processing were prepared following the procedure previously described (3). The cleavage and polyadenylation assays were carried out as described (56). To restrict the assay to cleavage only, EDTA replaced MgAc and CTP replaced ATP. For each reaction 30–40 µg total protein was used and, in the case of the reactions carried out at 34°C, pre-incubated at this temperature for 5 min. The RNA substrates were prepared by run-off transcription following the previously described procedure (9).
RNA analyses
Northern analyses and RNase H experiments were carried out as described (10). In addition we employed the oligonucleotides anti-U24 (5′-TCAGAGATCTTGGTGATAAT-3′) and anti–snR13 (5′-GGCAAAAGCCAAACAGCAACTCGAGCCAAATGCACTCATATTCATCATAT-3′), which were labeled with [γ-32P]ATP by T4 polynucleotide kinase.
The reverse transcription analysis was performed as described (45) with primers downstream of snoRNA genes as previously described (51).
Protein extraction for western blotting
The cells were grown at 25°C and shifted to 37°C; during incubation their growth was kept in the log phase. At each time point, 40 ml of the culture (OD600 = 0.4) was harvested. The following procedure was carried out on ice. The cells were resuspended in IPP150 and an equal volume of glass beads was added. The cells were opened by rigorous vortexing. Five millilitres of IPP150 and protease inhibitors were added and the mixture was centrifuged for 1 h at 8000 g r.p.m. Four millilitres of the supernatant was thereafter centrifuged for 2 h at 200 000 g and the protein was concentrated by centrifugation in a centricon YM10 (Millipore). The protein concentration was determined by Bradford analysis and equal amounts of total proteins were separated by SDS–PAGE.
RESULTS
Ydh1p is required for cleavage and polyadenylation in vitro
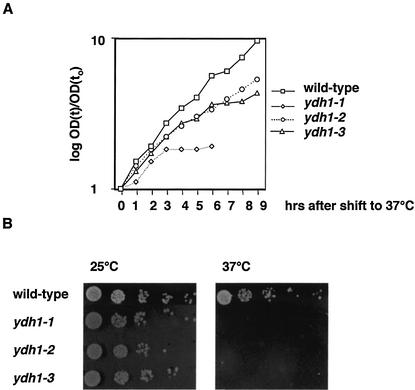
In order to investigate the role of Ydh1p in 3′-end processing we generated temperature sensitive ydh1 alleles (see Materials and Methods). Figure 1A shows growth curves of ydh1 mutant and isogenic wild-type cells following shift from 25 to 37°C. The ydh1-1 strain displayed growth arrest at 37°C after 3 h, whereas the strains ydh1-2 and ydh1-3 ceased growth after ∼5 h. A drop-test revealed that the mutant strains did not form colonies at the restrictive temperature (Fig. 1B).
Figure 1.
YDH1 mutant strains display a temperature sensitive phenotype. (A) Growth curves of wild-type and mutant ydh1 strains at 37°C. (B) 10-fold serial dilution of cultures spotted on YPD plates followed by incubation at the indicated temperature for 2 days.
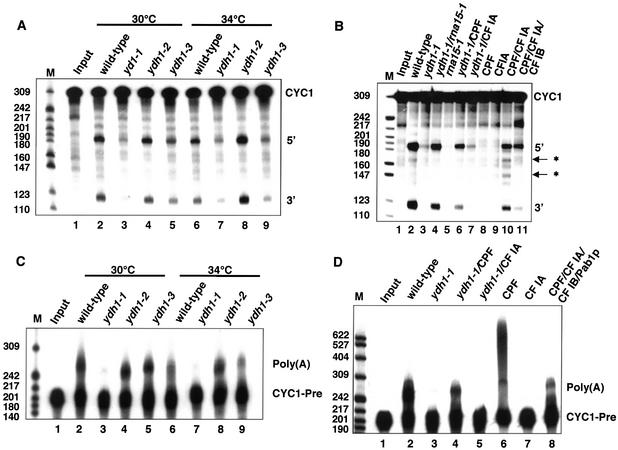
Next, we analysed whether mutations in YDH1 affect cleavage and polyadenylation in vitro and tested extracts from wild-type, ydh1-1, ydh1-2 and ydh1-3 cells for their ability to cleave a synthetic CYC1 pre-mRNA (Fig. 2A). Extracts from wild-type (lanes 2 and 6), ydh1-2 (lanes 4 and 8) and ydh1-3 cells (lanes 5 and 9) accurately cleaved the substrate RNA at 30 and 34°C. Notably, ydh1-3 extract (lane 9) showed a reduced efficiency of cleavage at 34°C compared to wild-type (lane 6). In contrast, extract of ydh1-1 cells was deficient in cleavage at both temperatures (lanes 3 and 7).
Figure 2.
Ydh1p is required for cleavage and polyadenylation in vitro. In vitro analysis of cleavage (A and B) and polyadenylation activities (C and D) of ydh1 mutant extracts. Input lanes (1) represent mock-treated reactions. The migration positions of the CYC1 and CYC1-precleaved (CYC1-Pre) RNA substrate, specific (5′ and 3′) and cryptic (asterisks) cleavage products and the polyadenylation products [Poly(A)] are indicated on the right of each panel. The position and size (in number of nucleotides) of the marker bands are indicated on the left. (A) Extracts prepared from yeast strains (as indicated above the lanes) were monitored for their ability to cleave internally 32P-labelled CYC1 RNA substrate at 30 and 34°C. (B) Reconstitution of cleavage activity in ydh1-1 extract at 30°C. As indicated above the lanes, ydh1-1 extract was combined with equal amounts of rna15-1 extract (lane 4), purified CPF (lane 6) or CF IA (lane 7). (C) Specific polyadenylation was analysed at 30 and 34°C with internally 32P-labelled precleaved CYC1 RNA substrate that ends at the natural poly(A) site. (D) Reconstitution of specific polyadenylation activity in ydh1-1 extract at 30°C. ydh1-1 extract was combined with purified CPF (lane 4) or CF IA (lane 5).
To show that the deficiency in cleaving the RNA substrate is due to inactive Ydh1p or CPF, respectively, we carried out reconstitution assays either with extract which is mutant in a CF IA subunit (rna15-1) or with purified factors at 30°C (Fig. 2B). The cleavage activity of ydh1-1 extract was restored upon addition of rna15-1 extract (lane 4) or purified CPF (lane 6), but not by addition of purified CF IA (lane 7). As expected, the rna15-1 extract on its own lacked cleavage activity (lane 5) (56) and purified CPF and CF IA alone were not able to cleave the substrate (lanes 8 and 9); cleavage occurred at specific and at cryptic sites when both factors were added to the reaction (lane 10); site-specific cleavage was obtained by including CF IB in the reaction (lane 11) (15).
Next we analysed whether mutations in YDH1 affect the polyadenylation reaction of pre-cleaved CYC1 substrate (CYC1-Pre) in vitro (Fig. 2C). No polyadenylation activity could be observed in ydh1-1 extract at 30 and 34°C (lanes 3 and 7). In contrast, the ydh1-2 and ydh1-3 extracts specifically and efficiently polyadenylated the substrate at both temperatures (lanes 4, 5, 8 and 9), comparable to wild-type (lanes 2 and 6). Specific polyadenylation activity was restored in ydh1-1 extract by addition of purified CPF (Fig. 2D, lane 4) but not by addition of purified CF IA (lane 5). As shown before, CPF by itself unspecifically polyadenylated the substrate (lane 6) (3), whereas CF IA alone displayed no polyadenylation activity (lane 7). Specific polyadenylation activity was restored upon combination of CPF, CF IA, CF IB and Pab1p (lane 8).
Notably, addition of 100–250 ng recombinantly expressed Ydh1p failed to reconstitute the cleavage and polyadenylation activities in ydh1-1 extract, possibly because the recombinant protein was not able to replace its mutant counterpart in the CPF complex (results not shown).
The above results showed that the ydh1-1 extract was strongly reduced in cleavage and polyadenylation activities in vitro and that both activities could be reconstituted by addition of purified CPF. Furthermore, the cleavage activity of the ydh1-3 mutant was lower at 34°C compared to 30°C. The results indicate that Ydh1p is involved in both steps of the 3′-end processing reaction.
mRNAs are unstable in ydh1 mutant strains at restrictive temperature
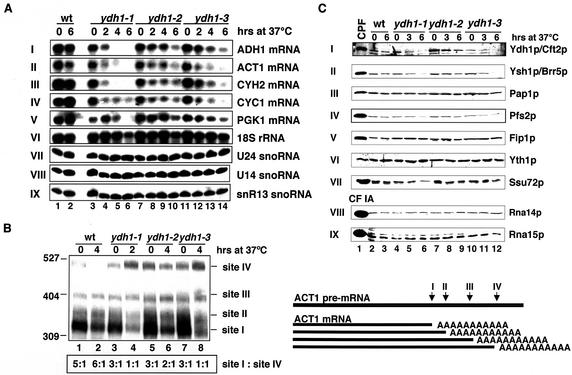
mRNAs without poly(A) tails are rapidly degraded in living cells. Yeast strains with a 3′-end processing deficiency are therefore expected to under-accumulate mRNAs after shift to the restrictive temperature. For this reason, we performed northern blot analyses on total RNA extracted from strains grown at 25°C and after shift to 37°C (Fig. 3A). The amount of 18S rRNA served as control for the loading of the RNA (panel VI). ADH1, ACT1, CYH2 and CYC1 mRNA levels diminished in the ydh1-1 mutant cells after 2 h shift to 37°C (panels I–IV, lanes 3–6). The level of the stable PGK1 mRNA (t1/2 = 45 min) dropped significantly only after the cells were shifted to 37°C for 6 h (panel V, lanes 3–6). In comparison the ADH1, ACT1, CYH2 and CYC1 mRNA levels of the ydh1-2 mutant strain only dropped slightly after 4 h, and the PGK1 mRNA level remained stable even after shift to 37°C for 6 h (panels I–V, lanes 7–10). The mutant strain ydh1-3 under-accumulated ACT1 mRNA after only 2 h at 37°C; ADH1, CYH2 and CYC1 mRNA levels were reduced after 4 h (panels I–IV, lanes 11–14). The PGK1 mRNA level showed a slight decrease after 4 h and more so after 6 h (panel V, lanes 11–14). The RNA polymerase II transcribed U24, U14 and snR13 snoRNAs were stable in all mutants tested, even after shift to 37°C for 6 h (panels VII–IX), indicating that snoRNA processing was not affected in ydh1 mutant strains. These results showed that ydh1 mutant strains under-accumulated mRNAs at restrictive temperature. The ydh1 mutant phenotypes are possibly due to a deficiency of the strains in poly(A) site recognition (see below) which might impair the efficiency in 3′-end processing.
Figure 3.
mRNAs are unstable in ydh1 mutant strains at restrictive temperature and the mutant strains are deficient in the recognition of ACT1 poly(A) site in vivo. (A) Northern analysis of total RNA extracted from wild-type and mutant ydh1 cells grown at 23°C, or after shift to 37°C for 2, 4 and 6 h as indicated above each lane. The RNAs were separated on formaldehyde/1.2% agarose gels (panels I–VI) or 8.3 M urea/8% polyacrylamide gels (panels VII–IX). The filters were developed with random-primed labeled probe (panels I–V) or end-labeled oligonucleotides (panels VI–IX) directed against the RNA species indicated at the right of each panel. (B) Analysis of ACT1 poly(A) site usage in wild-type and mutant ydh1 cells. The schematic on the right shows the relative positions of the different poly(A) sites. Total RNA extracted from wild-type and mutant ydh1 cells after growth at 23°C, or following shift to 37°C as indicated above each lane, was treated with an oligonucleotide complementary to the 3′ region of the ACT1–mRNA (ACT1–RnaseH) and RNaseH. The RNAs were separated on an 8 M urea/6% polyacrylamide gel and the filters were incubated with random primed labeled probe directed against the 3′-end of the ACT1 mRNA. RNA levels were quantified by PhosphorImager scanning (Molecular Dynamics); the ratios of poly(A) site I:site IV usage for each lane are indicated at the bottom. (C) Western analysis of wild-type and ydh1 mutant extracts prepared from cells grown at 23°C, or after shift to 37°C for 3 and 6 h. Equal amounts of total protein were loaded in each lane. Lane 1 shows purified CPF (panels I–VII) and CF IA (panels VIII and IX). The filters were treated with antibodies directed against the proteins indicated at the right.
Ydh1p is required for poly(A) site selection of ACT1 pre-mRNA
The previous observation that Ydh1p binds RNA around the poly(A) site (9), raised the possibility that the protein is required for poly(A) site recognition. To test this, we analysed poly(A) site usage of ACT1 pre-mRNA in wild-type and mutant cells (57). The ACT1 3′ untranslated region (UTR) contains at least four polyadenylation sites, of which the most proximal one (site I) is used with highest frequency in wild-type cells (Fig. 3B; lanes 1 and 2). All tested ydh1 mutants used site I three times more often than site IV at 25°C (lanes 3, 5 and 7). At restrictive temperature, however, processing shifted from site I towards site IV in a ratio of 1:1 in ydh1-1 and ydh1-3 cells (lanes 4 and 8); the ydh1-2 mutant showed a ratio of 2:1 (lane 6). These results indicated that mutations in ydh1-1, ydh1-2 and ydh1-3 impair poly(A) site recognition in vivo. To verify that the ACT1 mRNAs were polyadenylated we incubated the reactions with RNase H and the oligos ACT1-RnaseH and dT to digest the poly(A) tails. This resulted in a slight downward shift and sharpening of the RNA bands. This showed that the ACT1 mRNAs were polyadenylated (results not shown).
Western blot analyses were carried out to test whether the observed phenotypes were the result of an under-accumulation of Ydh1p or other subunits of the 3′-end processing machinery (Fig. 3C). As control, purified CPF or CF IA was analysed in parallel (lane 1). In wild-type and ydh1-2 cells Ydh1p was stable even after 6 h at 37°C (panel I, lanes 2, 3 and 7–9), whereas it was reduced in ydh1-1 and ydh1-3 cells after 3 h at 37°C (lanes 5, 6, 11 and 12). Ysh1p/Brr5p, Pfs2p and Ssu72p (panels II, IV and VII) under-accumulated in ydh1-1 and ydh1-3 cells after 6 h at 37°C (lanes 6 and 12). The levels of Pap1p, Fip1p and Yth1p were constant even after 6 h at 37°C (panels III, V and VI). In addition, mutations in YDH1 did not affect the levels of the CF IA subunits Rna14p and Rna15p (panels VIII and IX).
These results indicate that the defects observed in the ydh1 mutant strains might be caused by a destabilization of the Ydh1p protein. Since the destabilization at restrictive temperature of Ysh1p/Brr5p, Pfs2p and Ssu72p was detectable at later time-points compared to the appearance of phenotypes [see northern blot and ACT1 poly(A) site selection], we consider it unlikely that lower levels of factors other than Ydh1p are responsible for the deficiencies of the ydh1 mutant cells.
Ydh1p interacts with the C-terminal domain of RNA polymerase II
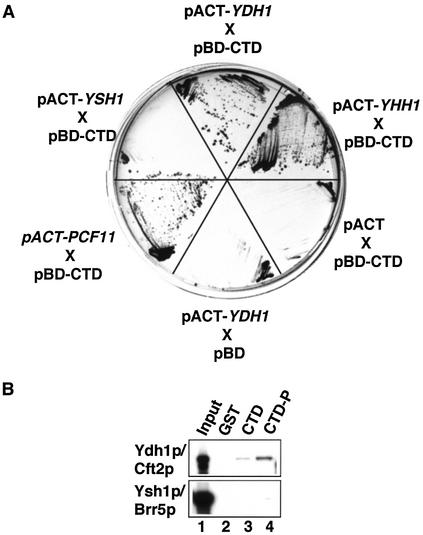
The CTD of RNAP II plays an important role in coupling transcription and 3′-end formation (reviewed in 22,23). We were interested to know whether Ydh1p contributes to this process by physically interacting with the CTD. To test this, we carried out GAL4-based yeast two-hybrid tests and assayed for the activation of expression of the HIS3 and lacZ reporter genes. Strain Y190 was co-transformed with plasmids carrying the GAL4 DNA binding domain fused to the CTD (pBD-CTD) and the GAL4 activation domain fused to test genes. As positive controls we analysed the known CTD interactors YHH1/BRR5 (pACT2-YHH1) and PCF11 (pACT2-PCF11) (10,28,47) and pACT2-YSH1 as negative control. As shown in Figure 4A the plasmids carrying YDH1, YHH1 and PCF11 in combination with the plasmid pBD-CTD enabled cells to grow on medium lacking histidine under stringent conditions (in the presence of 35 mM 3′-aminotriazole). In contrast, no growth was observed with pACT2-YSH1 or when empty pBD and pACT2 plasmids were tested. These results were confirmed by an X-Gal filter lift assay, which monitors β-galactosidase expression (results not shown). Thus, YDH1 showed a two-hybrid interaction with the CTD of RNAP II.
Figure 4.
Ydh1p interacts with the CTD of RNAP II. (A) GAL4-based two-hybrid analysis. Y190 cells were co-transformed with a plasmid encoding the GAL4 DNA binding domain fused to the CTD (pBD-CTD) and plasmids (pACT2) encoding the GAL4 activation domain fused to the genes indicated. Transformants were tested for expression of the HIS3 reporter gene on medium lacking histidine, and containing 35 mM 3-amino-1,2,4-triazole. As control, pBD CTD was co-transformed with the empty pACT2 plasmid and pACT2-YDH1 with the empty pBD plasmid. (B) Pull-down experiments with 1 µg GST (lane 2), GST–CTD (lane 3), phosphorylated GST–CTD (lane 4) and in vitro translated, [35S]methionine-labeled proteins (indicated on the left). Lane 1 shows 10% of the in vitro translated reactions used in the assay. Bound proteins were separated by SDS–PAGE and visualized by autoradiography.
To confirm that Ydh1p interacts with the CTD, and to test whether this interaction is influenced by the phosphorylation state of the CTD we performed GST pull-down experiments with in vitro translated radioactively labeled proteins and recombinant GST–CTD fusion protein. Ydh1p interacted with the CTD (Fig. 4B, lane 3) and this interaction was enhanced upon phosphorylation of the CTD (lane 4); no signal was observed with GST alone (lane 2). Ysh1p/Cft1p interacted with neither form of the GST–CTD protein. These results suggested that Ydh1p binds the CTD and that the interaction is enhanced upon phosphorylation of the CTD.
Correct transcription termination by RNAP II requires a functional poly(A) site on the nascent RNA and the CTD plays an important role in coupling transcription and pre-mRNA processing (reviewed in 43). The observations that Ydh1p is involved in poly(A) site recognition and that it interacted with the CTD raised the possibility that Ydh1p might also be involved in transcriptional termination. Therefore, we carried out transcriptional run-on analysis with the well-characterized GAL1/10 controlled CYC1 gene in vivo (44). We did not observe a significantly increased RNAP II density downstream of the CYC1 terminator in ydh1-1 and ydh1-3 cells grown at 25°C or following shift to 37°C compared to wild-type (results not shown). Similarly, we did not detect the accumulation of read-through products in ydh1 strains in reverse transcription experiments with primers annealing downstream of the cleavage site of different snoRNA species (snR39b, snR45, snR3, snR50, snR71) (51). This suggested that Ydh1p is not generally required for termination of RNAP II during transcription of pre-mRNAs and snoRNAs.
Ydh1p interacts with other subunits of CPF and with Pcf11p, a subunit of CF IA
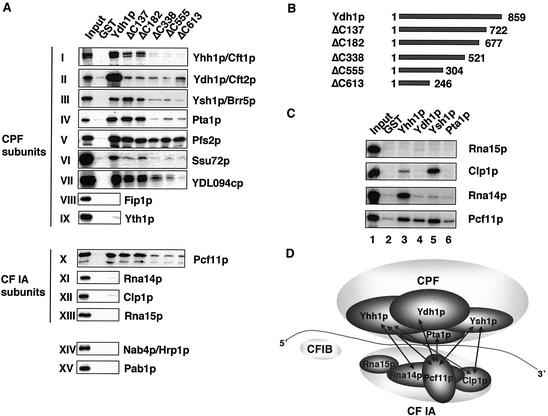
As Ydh1p is part of a multi-protein complex we were interested to examine with which of the other CPF subunits the protein interacts. Therefore, we expressed a N-terminally GST-tagged and C-terminally His-tagged version of the Ydh1 protein (G-Ydh1-H) in E.coli and carried out GST pull-down experiments with radioactively labeled in vitro translated proteins. As shown in Figure 5A, G-Ydh1-H interacted with itself and with the CPF subunits Yhh1p/Cft1, Ysh1p/Brr5p, Pta1p, Pfs2p, Ssu72p, YDL094cp (panels I–VII) and the CF IA subunit Pcf11p (panel X). G-Ydh1-H did not pull-down the CPF subunits Fip1p and Yth1p (panels VII and IX), nor the CF IA subunits Rna14p, Clp1p and Rna15p (panels XI–XIII), Nab4p/Hrp1 (CF IB) or Pab1p (panels XIV and XV). Since we identified a large number of potential Ydh1p interaction partners, we decided to investigate whether these interactions were specific and therefore assignable to distinct regions of Ydh1p. For this purpose the pull-downs were repeated with C-terminally truncated Ydh1p proteins (Fig. 5B). Figure 5A shows that the very C-terminus was required for interaction of Ydh1p with itself (panel II) as most of the interaction was lost upon deletion of the C-terminal 137 amino acids. The N-terminal 246 amino acids were sufficient for interaction with Pfs2p (panel V) and central protein sequences most likely mediated interactions with Yhh1p/Cft1p, Ysh1p/Brr5p, Pta1p, Ssu72, pYDL094c and Pcf11p (panels I, III, IV, VI, VII and X). These results suggested that the Ydh1p subunit might contribute to the stability and structural order of CPF and bridge the two factors CPF and CF IA.
Figure 5.
Ydh1p interacts with other subunits of CPF and with Pcf11p, a subunit of CF IA. (A) GST pull-down experiments with 0.5 µg GST, 100 ng G-Ydh1-H (Ydh1p) or 100 ng of the different C-terminally truncated, GST-tagged Ydh1 fragments (as indicated above each lane) and in vitro translated [35S]methionine-labeled proteins (indicated on the right). The first lane shows 10% of the in vitro translated protein used in the binding reactions. (B) Schematic drawing of the full length and C-terminal truncations of Ydh1p. The numbers indicate the length of the constructs in amino acids. (C) GST pull-down experiment with GST-tagged recombinant proteins (indicated above each lane) and in vitro translated, [35S]methionine-labeled proteins (indicated on the right). (D) Model of protein–protein interaction surface (not all CPF subunits and protein interactions are shown).
To obtain a better understanding of how CPF interacts with CF IA we decided to assay for further interactions between the two factors. Figure 5C shows a GST pull-down with in vitro translated protein and GST-tagged recombinant proteins. Of the analysed CPF subunits we found that only Yhh1p/Cft1p interacted strongly with Rna14p. Yhh1p/Cft1p also bound to Pcf11p, and more weakly to Clp1p (lane 3). Ydh1p pulled-down only Pcf11p (lane 4). Ysh1p/Brr5p interacted strongly with Clp1p and Pcf11p (lane 5). A weak signal was observed for the interaction between Pta1p and Pcf11p (lane 6). These results suggested that Ydh1p, Yhh1p/Cft1p, Ysh1p/Brr5p and possibly also Pta1p are involved in forming a protein–protein interaction surface between CPF and CF IA. Our combined results indicate that Ydh1p plays an important role in determining the RNA binding specificity of CPF and in the assembly of the 3′-end formation machinery and its tethering to RNAP II.
DISCUSSION
The yeast and mammalian 3′-end formation machineries display a surprisingly complex subunit composition. At present, up to 20 polypeptides are known that constitute the factors that catalyse the yeast 3′-end formation reaction in vitro (45,58). The ongoing characterization of the components suggests distinct and specialized tasks of the individual polypeptides in both steps of the 3′-end processing reaction as well as in the coupling of 3′-end formation to transcription. In this work we characterized the role of the yeast CPF subunit Ydh1p in these processes.
To analyse if Ydh1p is necessary for cleavage and polyadenylation we produced conditional ydh1 mutant strains and carried out in vitro assays. The experiments revealed a deficiency of the mutant extracts in both steps of the 3′-end processing reaction. The activity could be rescued upon addition of purified CPF. mRNAs without a poly(A) tail are prone to rapid degradation in vivo. Northern analysis revealed that the levels of a number of different mRNAs were reduced at restrictive temperature in Ydh1p mutants. The results support a requirement for Ydh1p in both steps of 3′-end processing. However, we do not expect Ydh1p to be directly involved in the catalysis of the 3′-end processing reaction, because it took several hours at the restrictive temperature before the mRNA levels were reduced. Furthermore, the ACT1 poly(A) site selection assay revealed that a substantial amount of pre-mRNA appeared to be cleaved and polyadenylated in vivo at the restrictive temperature in all mutants.
We reported previously that Ydh1p binds to a CYC1 pre-mRNA around the poly(A) site; this suggested a role for the protein in poly(A) site selection (9). Here we show that Ydh1p is involved in poly(A) site selection of ACT1 pre-mRNA in vivo, as all analysed ydh1 mutant strains displayed more frequent use of alternative poly(A) sites compared to the wild-type strain. These results underscore our previously postulated model that Ydh1p contributes to poly(A) site selection (9). In contrast to mammalian polyadenylation signals, the cis-acting sequences in yeast are highly degenerate (reviewed in 1,8). We proposed that the RNA binding subunits of CPF act in concert to achieve specific recognition of the correct poly(A) site (9). The preferential use of cleavage sites located downstream of the major site in Ydh1 mutants may be caused by the coupling of transcription elongation and 3′-end processing. Reduced RNA binding efficiency of CPF containing mutant Ydh1p could lead to skipping of the first cleavage site of the nascent pre-mRNA emerging from the elongating RNAP II. Ydh1p (9), Yhh1p/Cft1p (10) and Yth1p (11) were shown to bind preferentially to U-rich elements localized directly upsteam and downstream of the poly(A) site, thus colocalizing with the region to which the complete CPF factor binds (9). These interactions with the RNA substrate are thought to be crucial for poly(A) site selection and for cleavage activity. Thus, the observed in vitro cleavage deficiency of the ydh1 mutant cells might result from insufficient poly(A) site selection. It remains to be determined how Ydh1p interacts with RNA. The primary sequence of the protein does not display any clear similarities to known RNA binding domains. Preliminary RNA binding experiments with portions of Ydh1p indicated that the RNA binding activity is distributed throughout the entire length of the protein (results not shown). However, detailed analyses have not been done yet. Interestingly, RNA binding activity has not been demonstrated for the mammalian homolog of Ydh1p, CPSF 100 kDa. Considering the highly conserved cis-acting elements in mammalian pre-mRNAs (reviewed in 1), it seems possible that a smaller set of proteins is sufficient for specific poly(A) site recognition, whereas in yeast a cooperative interplay of more RNA binding proteins is required to recognize the more degenerate sequence elements in a specific fashion.
We have shown that Ydh1p interacted with the CTD of RNAP II both in a two-hybrid test and in a GST pull-down assay. This interaction was enhanced upon phosphorylation of the CTD. The only other CPF subunit found to interact with the phosphorylated CTD is Yhh1p/Cft1p (10). We suggest that Ydh1p might be involved in tethering CPF to transcribing RNAP II and thereby contributes to the coupling of 3′-end processing and transcription. However, the ydh1 mutants did not reveal a defect in transcriptional termination at the CYC1 terminator. As this was the only pre-mRNA that was tested by transcriptional run-on analysis we cannot exclude that Ydh1p might be involved in transcriptional termination of other genes. Moreover, reverse transcription analysis on snoRNAs in ydh1 mutant strains did not reveal 3′-extended transcripts, indicating that Ydh1p was not required for snoRNA termination either. We propose that Ydh1p helps to tether CPF to elongating RNAP II but that it is not needed for general transcription termination. Ydh1p and Yhh1p/Cft1p show a phenotypic similarity in that both proteins bind RNA, are involved in poly(A) site recognition and bind to the CTD (10). However, the observation that mutants in yhh1 are severely impaired in transcription termination at the CYC1 gene (10) whereas ydh1 mutants have no general termination defect, indicates that the functional roles of the individual 3′-end formation factor subunits can be substantially different.
It is not well understood how the subunits of the CPF and CF IA factors assemble to form functional 3′-end processing complexes on the RNA substrate. CPF consists of up to 15 polypeptides, which interact with each other and form a stable factor. We found that Ydh1p interacted with the CPF subunits Yhh1p/Cft1p, Ysh1p/Brr5p, Pta1p, Pfs2p, Ssu72p and pYDL094c. These many interactions indicate that the protein contributes substantially to the assembly and structural order of the CPF factor. Strikingly, Ydh1p also bound strongly to itself in the GST pull-down assays. This may indicate that there is more than one Ydh1p molecule present per 3′-end processing unit. However, silver stain analysis of purified CPF factor suggested an apparent stoichiometric presence of Ydh1p compared to other CPF subunits (45). Interestingly, we found that the CF IA subunit Pcf11p interacted with the CPF components Ydh1p, Yhh1p/Cft1p, Ysh1p/Brr5p, and weakly with Pta1p. In addition, we showed that the CPF subunits Yhh1p/Cft1p and Ysh1p/Brr5p interacted with the CF IA subunits Rna14p and Clp1p, respectively. The latter interactions were also observed between the homologous subunits of the mammalian 3′-end formation machinery. CPSF 160 kDa interacts with the CstF 77 kDa protein (18) and CPSF 73 kDa interacts with hClp1p (59). So far, only Pfs2p and Fip1p were shown to bridge CPF and CF IA (3,4). We propose that Ydh1p, Yhh1p/Cft1p, Ysh1p/Brr5p, Pfs2p, Fip1p, and possibly Pta1p, contribute to a protein–protein interaction surface which acts in the assembly of the 3′-end formation machinery and which appears to be conserved in evolution (Fig. 5D).
Our analysis of Ydh1p suggests that the protein is an important constituent of protein interaction surfaces which act in the association of 3′-end processing factors with RNAP II, the recognition of poly(A) signal sequences and the assembly of an active 3′-end formation complex on the RNA substrate.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Isabelle Kaufmann, Myriam Schaub and Jeannette Wolf for comments on the manuscript. This work was supported by the University of Basel, the Swiss National Science Fund, the European Community (www.euronomics.org) via the Bundesamt für Bildung und Wissenschaft, Bern (grant 01.0123) and the Louis-Jeantet-Foundation for Medicine.
REFERENCES
- 1.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J. and Moore,C. (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol., 12, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohnacker M., Barabino,S.M., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preker P.J., Lingner,J., Minvielle-Sebastia,L. and Keller,W. (1995) The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell, 81, 379–389. [DOI] [PubMed] [Google Scholar]
- 5.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo P., Li,W.Z., Hampsey,D.M., Zaret,K.S. and Sherman,F. (1991) Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J., 10, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Helden J., del Olmo,M. and Perez-Ortin,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Z. and Sherman,F. (1996) 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–481. [DOI] [PubMed] [Google Scholar]
- 9.Dichtl B. and Keller,W. (2001) Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J., 20, 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dichtl B., Blank,D., Sadowski,M., Hübner,W., Weiser,S. and Keller,W. (2002) Yhh1p/Cft1p directly links poly(A) site selection and RNA polymerase II transcription termination. EMBO J., 21, 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barabino S.L.M., Ohnacker,M. and Keller,W. (2000) Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S. and Hyman,L.E. (1998) A specific RNA–protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res., 26, 4965–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross S. and Moore,C.L. (2001) Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol., 21, 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′-end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatkin A.J. and Manley,J.L. (2000) The ends of the affair: capping and polyadenylation. Nature Struct. Biol., 7, 838–842. [DOI] [PubMed] [Google Scholar]
- 17.Keller W., Bienroth,S., Lang,K.M. and Christofori,G. (1991) Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J., 10, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murthy K.G.K. and Manley,J.L. (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald C.C., Wilusz,J. and Shenk,T. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz J., Shenk,T., Takagaki,Y. and Manley,J.L. (1990) A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol., 10, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmartin G.M. and Nevins,J.R. (1991) Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol. Cell. Biol., 11, 2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol., 14, 336–342. [DOI] [PubMed] [Google Scholar]
- 23.Proudfoot N.J., Furger,A. and Dye,M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 502–512. [DOI] [PubMed] [Google Scholar]
- 24.Howe K. (2002) RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta, 1577, 308. [DOI] [PubMed] [Google Scholar]
- 25.Proudfoot N. and O’Sullivan,J. (2002) Polyadenylation: a tail of two complexes. Curr. Biol., 12, R855–R857. [DOI] [PubMed] [Google Scholar]
- 26.Cho E.J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komarnitsky P., Cho,E.J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licatalosi D.D., Geiger,G., Minet,M., Schroeder,S., Cilli,K., McNeil,J.B. and Bentley,D.L. (2002) Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell, 9, 1101–1111. [DOI] [PubMed] [Google Scholar]
- 29.McCracken S., Fong,N., Rosonina,E., Yankulov,K., Brothers,G., Siderovski,D., Hessel,A., Foster,S., Shuman,S. and Bentley,D.L. (1997) 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev., 11, 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho E.J., Takagi,T., Moore,C.R. and Buratowski,S. (1997) mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev., 11, 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho C.K., Sriskanda,V., McCracken,S., Bentley,D., Schwer,B. and Shuman,S. (1998) The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem., 273, 9577–9585. [DOI] [PubMed] [Google Scholar]
- 32.Cho E.J., Rodriguez,C.R., Takagi,T. and Buratowski,S. (1998) Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev., 12, 3482–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuryev A., Patturajan,M., Litingtung,Y., Joshi,R.V., Gentile,C., Gebara,M. and Corden,J.L. (1996) The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl Acad. Sci. USA, 93, 6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G., Greenblatt,J., Patterson,S.D., Wickens,M. and Bentley,D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- 35.Barilla D., Lee,B.A. and Proudfoot,N.J. (2001) Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez C.R., Cho,E.J., Keogh,M.C., Moore,C.L., Greenleaf,A.L. and Buratowski,S. (2000) Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol., 20, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong N. and Bentley,D.L. (2001) Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev., 15, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirose Y. and Manley,J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- 39.Ryan K., Murthy,K.G., Kaneko,S. and Manley,J.L. (2002) Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol., 22, 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNeil J.B., Agah,H. and Bentley,D. (1998) Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev., 12, 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orozco I.J., Kim,S.J. and Martinson,H.G. (2002) The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem., 277, 42899–42911. [DOI] [PubMed] [Google Scholar]
- 42.Tran D.P., Kim,S.J., Park,N.J., Jew,T.M. and Martinson,H.G. (2001) Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol. Cell. Biol., 21, 7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proudfoot N.J. (1989) How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci., 14, 105–110. [DOI] [PubMed] [Google Scholar]
- 44.Birse C.E., Minvielle-Sebastia,L., Lee,B.A., Keller,W. and Proudfoot,N.J. (1998) Cleavage of the primary transcript couples 3′-end RNA-processing with termination of pol II transcription. Science, 280, 298–301. [DOI] [PubMed] [Google Scholar]
- 45.Dichtl B., Blank,D., Ohnacker,M., Friedlein,A., Roeder,D., Langen,H. and Keller,W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell, 10, 1139–1150. [DOI] [PubMed] [Google Scholar]
- 46.Hammell C.M., Gross,S., Zenklusen,D., Heath,C.V., Stutz,F., Moore,C. and Cole,C.N. (2002) Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol., 22, 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadowski M., Dichtl,B., Hübner,W. and Keller,W. (2003) Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J., 22, 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvo O. and Manley,J.L. (2001) Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell, 7, 1013–1023. [DOI] [PubMed] [Google Scholar]
- 49.Aranda A. and Proudfoot,N. (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell, 7, 1003–1011. [DOI] [PubMed] [Google Scholar]
- 50.Alen C., Kent,N.A., Jones,H.S., O’Sullivan,J., Aranda,A. and Proudfoot,N.J. (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell, 10, 1441–1452. [DOI] [PubMed] [Google Scholar]
- 51.Steinmetz E.J., Conrad,N.K., Brow,D.A. and Corden,J.L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature, 413, 327–331. [DOI] [PubMed] [Google Scholar]
- 52.Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- 53.Preker P.J., Ohnacker,M., Minvielle-Sebastia,L. and Keller,W. (1997) A multisubunit 3′-end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J., 16, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Kessler,M.M. and Moore,C.L. (1997) Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem., 272, 10831–10838. [DOI] [PubMed] [Google Scholar]
- 55.Fromont-Racine M., Rain,J.C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- 56.Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- 57.Mandart E. and Parker,R. (1995) Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol., 15, 6979–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 59.deVries H., Rüegsegger,U., Hübner,W., Friedlein,A., Langen,H. and Keller,W. (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J., 19, 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.