Abstract
In an effort to understand the role of the linker histone in chromatin folding, its structure and location in the nucleosome has been studied by molecular modeling methods. The structure of the globular domain of the rat histone H1d, a highly conserved part of the linker histone, built by homology modeling methods, revealed a three-helical bundle fold that could be described as a helix–turn–helix variant with its characteristic properties of binding to DNA at the major groove. Using the information of its preferential binding to four-way Holliday junction (HJ) DNA, a model of the domain complexed to HJ was built, which was subsequently used to position the globular domain onto the nucleosome. The model revealed that the primary binding site of the domain interacts with the extra 20 bp of DNA of the entering duplex at the major groove while the secondary binding site interacts with the minor groove of the central gyre of the DNA superhelix of the nucleosomal core. The positioning of the globular domain served as an anchor to locate the C-terminal domain onto the nucleosome to obtain the structure of the chromatosome particle. The resulting structure had a stem-like appearance, resembling that observed by electron microscopic studies. The C-terminal domain which adopts a high mobility group (HMG)-box-like fold, has the ability to bend DNA, causing DNA condensation or compaction. It was observed that the three S/TPKK motifs in the C-terminal domain interact with the exiting duplex, thus defining the path of linker DNA in the chromatin fiber. This study has provided an insight into the probable individual roles of globular and the C-terminal domains of histone H1 in chromatin organization.
INTRODUCTION
In eukaryotic cells the genome is packaged into a 5–10 µm nucleus due to the existence of a highly ordered nucleoprotein architecture called chromatin. In the first order of organization of chromatin structure, 146 bp of DNA is packaged into a nucleosome core particle around a histone octamer (1,2). The linear array of the 10 nm polynucleosomal filament is folded into 30 nm irregular fibers facilitated and stabilized by the linker histone H1, whose architecture and mode of compaction have been matters of controversy over the last two decades (reviewed in 1–3). The controversy centers on the arrangement and interactions of nucleosome cores in three- dimensional space and the locations of linker DNA and the linker histone. Recent accumulating evidence suggests that chromatin structure above the level of the core particle plays a key role in determining the transcriptional status of genes and genetic loci (reviewed in 4–6) and therefore illustrates the critical importance of understanding the fundamental folding properties of nucleosomal arrays.
Although many models for the condensed fiber structure were proposed, the solenoid model of Finch and Klug (7) or variants thereof, gained initial acceptance by most researchers. A corollary of the solenoid model is that the linker DNA between adjacent nucleosomes must be bent or curled in some fashion, at least at physiological salt concentration to allow adjacent nucleosomes to contact each other. This salt-dependent bending or coiling was postulated to be facilitated by the interaction of linker DNA with the linker histone, which has gained experimental support from the studies of Butler and Thomas (8) and Thoma et al. (9). Other suggestions for the architecture of the 30 nm fiber have considered the zigzag conformation of nucleosomes and linker DNA that is observable at intermediate ionic strengths to constitute the basic structural motif (10–13). In these models, further compaction of the zigzag leads to helical structures with quite different properties from the solenoid. Here, the linker DNA is not coiled, but remains extended such that consecutive nucleosomes in the beaded chain are not consecutive in the 30 nm fiber. Bednar et al. (14) find that the center to center distance between nucleosomes in dimers does not decrease as the salt concentration is raised, suggesting the presence of straight linkers. The fiber architecture based on a three-dimensional zigzag implies quite different mechanisms for formation and stability than proposed for helical models.
Irrespective of the model, it has now been unambiguously established that the presence of the linker histone is essential for proper folding of the chromatin fiber (15). It is believed that the linker histone facilitates, in part, charge neutralization and helps guide the proper folding of chromatin fiber by contributing to the free energy of chromatin folding (16). Within chromatin, the basic unit consisting of the core particle and histone H1 with 166 bp of DNA wrapped around the complex is termed the ‘chromatosome’ or the ‘chromatin particle’. There are also two views on the location and interaction of histone H1 with the mononucleosome core. In one model, the globular domain of histone H1d from rat (gH1d) is shown to interact with both the entering and exiting DNA at the dyad axis (17). In another model, the histone H1 is shown to interact with one side of the DNA duplex within the core particle (18). Although the crystal structure of the nucleosome core particle has given us a detailed arrangement of the histone octamer and their interaction with the DNA helix (19), the structure of the chromatosome is not available yet. Elucidation of this structure will have a direct bearing on our understanding of the folding of chromatin fiber into higher order structures. Recently, we have identified by extensive mutagenesis analysis a 34 amino acid stretch within the C-terminus of histone H1d that is responsible for DNA condensation (20). By molecular modeling using fold recognition approaches, we have found that this 34 amino acid stretch is capable of adopting the high mobility group (HMG)-box fold (21). This has encouraged us to model the entire chromatosome particle using the presently available computational approaches. We are now reporting here the model for the chromatosome particle using the structural data of the nucleosome core particle, the structure of the histone globular domain (22) and the predicted structure of the C-terminus of histone H1 (21).
MATERIALS AND METHODS
The sequence of rat histone H1d, P15865 obtained from the SWISSPROT database, was used for all analyses. The SMART (23) and DART (24) tools were used to recognize domain boundaries. Regions 34–109 and 110–219 were identified as separate domains, which indeed correspond to the previous assignments of globular domain (gH1d) and the C-terminal domain (H1d_C), respectively (25). Database searches were carried out using the standard BLAST (26) and FASTA (27) algorithms. Pairwise sequence alignments were obtained using the Smith and Waterman algorithm (28) as implemented in the GCG package. Multiple alignments were carried out using CLUSTALW (29). The PRINTS (30) database was used to analyze sequence profiles. Secondary structure prediction was carried out using several well known methods working on different principles and a consensus obtained, using the Network Protein Sequence Analysis server available at http://npsa-pbil.ibcp.fr/ (31). Visualization, manipulations and analysis of various structures and structure superpositions were carried out using various modules available within Insight-II (Accelrys Inc.). The structural models of the different elements of the chromatosome particle were first obtained individually and then docked together into a complex as detailed in the Results.
The globular domain of rat H1d was built by standard homology modeling since a high confidence structural template [crystal structure of globular domain of chicken H5 (gH5); PDB: 1HST (22,32)] was available for it in the Protein Data Bank (PDB). The model was regularized by energy minimization (soaked in a 5 Å layer of water molecules and minimized using a steepest descent minimization followed by conjugate gradient minimization with a gradual introduction of cross-terms or Morse potential terms) using DISCOVER interfaced with Insight-II.
A crystal structure of the ‘open’ form of the Holliday junction (HJ), available in the PDB as part of a RuvA nucleic acid complex [PDB: 1C7Y; NDB: PD0136 (33,34)], was used to model the gH1d–HJ complex, as described below. The minimized globular domain was manually docked onto the crystal structure of the HJ, using key pointers from the available experimental data as described later, thus resulting in a model of the gH1d–HJ complex. Modeling gH1d–HJ as well as the next few intermediates in obtaining the chromatin particle, primarily involves positioning one molecule with respect to the other. To achieve this, an integrated approach has been adopted here, wherein knowledge from sequence and structural bioinformatics analysis of the corresponding molecules/domains has been combined with the extensive biochemical data available in the literature.
The crystal structure of the nucleosomal core particle determined by Luger et al. (19) reveals that it consists of four core histones assembled into an octamer and 146 bp of nucleosomal DNA arranged into two duplexes around the octamer. The gH1d–HJ complex was superposed on the nucleosomal core particle so as to obtain a best fit between the phosphate backbone atoms of the two arms (PD0136: D and F) of the HJ DNA with those of the entering duplex (1AOI: I1–I12, J271–J292) and the central gyre of the nucleosomal core particle (1AOI: I70–I81, J213–J224) to obtain the positioning of gH1d. The position of the additional 13 bp of DNA was also obtained from this superposition. The model indicates an interaction of gH1d with the first to the sixth bases of the HJ DNA, accounting for interactions with an additional 6 bp as compared with the nucleosome structure. However, the model has 13 bp as it was directly derived from the HJ DNA. Similarly, the model of H1d_C has 15 bp of DNA associated with it, again derived directly from the template (2LEF) used in its building. It must be noted that the exact number of base pairs placed have no special significance in this study. Modeling of H1d_C by a combination of fold recognition methods and integration with biochemical data has been described previously (21).
The model of the chromatosome particle was obtained by docking together gH1d and H1d_C along with the pieces of DNA associated with them onto the nucleosome core particle. The interactions between the protein domains and the DNA were estimated in terms of hydrogen bonds. Distances of 2.5 to 3.6 Å between the donor and acceptor atoms were considered as hydrogen bonds. The individual elements in the complexes represent low energy models by themselves, since they are either crystal structures or have been obtained by standard modeling protocols and have been energy minimized. The final model has also been subjected to limited energy minimization, primarily to normalize the geometry at the splice junctions. However, the model presented here has been built using models of individual domains and disjointed pieces of DNA at the entry and exit sites on the duplexes. There is no clear evidence pointing to the extent of bend in DNA at these sites. Therefore, the final model reflects one of the possible local conformations at each of the splice junctions. Nonetheless, the methodology used here for modeling the complexes tacitly ensures not only structural correctness, but also the correct conformations and the relative positioning of their constituents, to a major extent.
RESULTS AND DISCUSSION
Elements of chromatin structure
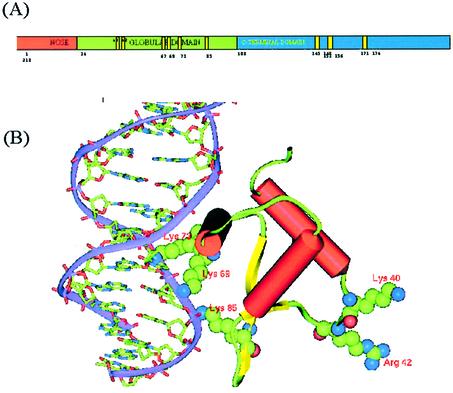
A typical eukaryotic nucleus contains ∼5 × 109 bp of DNA associated non-covalently with five types of basic proteins called histones, resulting in its compact packaging into chromatin, exhibiting a periodicity of ∼200 bp (1). The four core histones H2A, H2B, H3 and H4, together with 146 bp of DNA, form a nucleosome, the basic structural unit of chromatin. The folding of the nucleosomes into higher order chromatin structure is facilitated by the fifth histone H1, also termed as the linker histone due to its association with the linker DNA. Micrococcal nuclease digestion of chromatin results in a 166 bp complex called the ‘chromatosome particle’, consisting of the nucleosome core, histone H1 and 20 bp of linker DNA (35). Further digestion of this particle leads to 146 bp of DNA containing the nucleosomal core. The crystal structure of the nucleosomal core has been determined to 2.8 Å resolution (19). Two of each type of the core histone form an octamer around which DNA is wrapped, organized into two gyres of DNA superhelix. While the interactions of the core histones with the DNA wrapped around it are clearly understood, very little is known from the literature about the location and role of histone H1 in chromatin structure, the knowledge of which is essential to understand the accessibility of gene promoters and gene-specific transcription. The histone H1 protein consists of three distinct domains, a small 34 residue N-terminal fragment (nose), the central 74 residue globular domain (head) and a slightly larger 110 residue C-terminal domain (tail), as depicted schematically in Figure 1A. In aqueous media, at physiological pH and ionic strength, the N-terminal nose is believed to have no regular structure, while the C-terminal domain, that is otherwise unfolded, is believed to fold into an ordered structure in the presence of nucleic acids.
Figure 1.
(A) Domain architecture in rat histone H1d. The binding site residues in the globular domain as well as the three S/TPKK motifs in the C-terminal domain are highlighted. Residue numbers for the binding site residues of gH1d (40, 42, 67, 69, 73, 85) correspond to the numbering scheme used in the literature and in PDB: 1HST and will correspond to residues 50, 52, 78, 80 and 95 in the sequence P15865, respectively. (B) Model of the gH1d (cartoon representation) complexed with DNA (ribbon). Residues Lys69, Arg73 and Lys85 in the primary site and Lys40 and Arg42 in the secondary binding site are shown as space-filled objects.
The following sections detail the structure of both the domains of H1 and how they assemble together to form the chromatin particle. The model has been built in various stages: (i) the globular domain, built by homology modeling techniques; (ii) a complex of the globular domain with a segment of DNA at the primary site, built based on biochemical data and structural analysis of several helix–turn–helix (HTH) variants; (iii) a complex of globular domain with the HJ, obtained by docking using biochemical data; (iv) a complex of the globular domain with the nucleosomal core, using models derived in the previous two steps; and (v) a complex of the H1d C-terminal domain with the globular domain and the nucleosomal core, based on the intermediate at stage (iv) and a previously built model of the C-terminal domain along with the DNA associated with it. The five steps together give a comprehensive picture of the chromatosome particle, consisting of the head and tail regions of H1, the nucleosomal core as well as the entry and exit strands of nucleosomal DNA. We believe this to be the first attempt to model the whole chromatosome particle and that it will serve as a framework in understanding the biology of chromatin formation.
The globular domain of rat H1d: its structure and interactions with DNA
The structure of gH5 has been well characterized both by NMR (36) and X-ray crystallography (22), and primarily consists of a three-helical bundle fold, belonging to the ‘winged-helix DNA binding domain’ structural super-family. The globular domain is fairly well conserved in linker histones across diverse species, thus enabling homology modeling of the domain in rat H1d (Fig. 1B). The globular domain is also necessary for generating the kinetic intermediate ‘chromatosome particle’ observed during micrococcal nuclease digestion of the nucleosome containing 166 bp of DNA and histone H1. The location of the globular domain, however, within the nucleosome has been controversial (17,18,37).
A molecular model of the 96 residues constituting the gH1d was built using homology modeling techniques. The crystal structure of gH5 was used as the template with which gH1d shared 78% sequence similarity over the entire region, without any major insertions and deletions. The model reveals a three-helical bundle with a β-hairpin at the C-terminus that is anchored to the base of the helical core. The crystal structure of gH5 has been shown to bear significant similarity to those of catabolite gene activator protein and the hepatocyte nuclear factor (HNF-3γ), both known to be DNA binding proteins, thus leading to the identification of the DNA binding site in gH5 (38), which has been correlated well with site-directed mutagenesis studies (39). The structure can be described as belonging to a family of HTH variants since the characteristic four residue turn between the helices has been lengthened to a loop (38). The model built for rat H1d also contained all the features of gH5 identified as important for nucleic acid binding. Based on the structures of the CAP and HNF-3γ, the protein–DNA complex was modeled, in which a cluster of positively charged residues consisting of Arg67, Lys69 and Lys73 all located on the third helix, with side chains pointing to the exterior, formed several hydrogen bonds with the DNA molecule. Similar to other HTH-like proteins, this model revealed binding of the third helix at the major groove of the DNA molecule. Lys85, another residue implicated to be part of the basic cluster (40), was found to interact with the phosphates about half a turn away from the major groove. Site-directed mutagenesis and crystal structure studies have indicated the existence of a second DNA binding site formed by Lys40 and Arg42 (41,42). The structure of the domain indicates that these residues are located on the opposite face, separated from the primary recognition cluster by ∼30 Å (Fig. 1B). These residues are highly conserved in the globular domains of all somatic H1s, consistent with their involvement in the formation of a second binding site.
Positioning of the globular domain in the nucleosome structure
The best characterized component of the nucleosomes is the crystal structure of the nucleosome core particle, consisting of 145 bp of DNA wound around an octamer of ‘core’ histones. Linker DNA interconnects core particles and is clearly known to be associated with the linker histone. Reports in the literature on the location of the globular domain in the nucleosome are conflicting. The model by Allan et al. proposed that the globular domain of the linker histone was centered on the ‘pseudo dyad’ of the nucleosome, probably contacting the entering and the exiting duplexes thereby protecting them symmetrically ∼10 bp at each end from extended nucleosomal digestion (43). Neutron diffraction studies by Lambert et al. (44) suggested that the binding of the globular domain to the nucleosomes may be asymmetrical. Yet another model suggested by studies on the nucleosome reconstituted onto the unique sea urchin 5S rDNA sequence, indicated that H1 was located internal to a DNA gyre and contacted only one duplex at a position distal from the dyad axis (45). However, the validity of such interpretations have been questioned (17,18).
Despite the controversies, the facts that remain clearly established are that: (i) histone H1 has a crucial role in stabilizing nucleosome structure (37); (ii) mononuclesomes released upon micrococcal nuclease digestion of chromatin are trimmed in two stages, the first stage leading to a 166 bp particle, where H1 is retained and the second, a 146 bp particle containing the core histone octamer but not the linker histone (1); (iii) the globular domain alone is sufficient to confer protection to the extra 20 bp upon nuclease digestion (17,18); (iv) compelling evidence by mobility shift assays has established the stoichiometry of linker histone H5 to the core histone octamer (mononucleosome) to be 1 (46); (v) the globular domain has the ability to bind to two DNA duplexes, as demonstrated by clear dual contacts in a site-specific cross-linking DNA cleavage study, in which a photo-activatable group was attached to SH groups of unique cysteines engineered into the surface of the globular domain of H5 (47); (vi) site-directed mutagenesis studies as well as the crystal structure of the globular domain clearly identify two DNA binding sites, one on each face of the domain (41,42); and (vii) both the sites are shown to be required for the formation of the chromatosome. These data along with the crystal structure of the nucleosomal core particle as well as the structural level understanding of the interactions of the globular domain provide a basis to model the structure of the entire chromatosome particle.
The globular domain has been shown to bind preferentially, over naked duplex B-DNA, to distorted DNA structures such as the HJ and cis-platinated DNA (48,49). The crystal structure of the HJ DNA is available from the PDB (34), thus providing the framework to model its complex with the globular domain. Crystal structures of the HJ DNA determined in the past few years (50) indicate that they adopt a classical X-stacked conformation in their free forms. However, it has been well recognized (51) that the conformation of the junction is quite plastic and the global shape is readily manipulated by the proteins that specifically recognize the junction. The crystal structures of the HJs recognized by the branch migration machinery as illustrated by the RuvA–HJ complex as well as by site-specific recombination enzymes such as the Flp recombinase (52) are now available in the structure databases (32,33). Both these proteins, which exhibit striking selectivity and very high affinity to the junction DNA, bind HJ in a completely open, near-planar, near-square conformation in which all co-axial helical stacking is absent. Such structures have also been shown to be present in the crystal packing of the dodecameric DNA crystals and have been said to reflect the biologically relevant HJ structures (53,54). The free form of HJ is also believed to be in the extended ‘open’ conformation in the absence of cations (50). Upon preliminary docking and structural analysis, it became apparent that the globular domain could bind easily to the open form of the HJ, as in the other protein–HJ complexes. The HJ structure from the RuvA complex has therefore been used in this study. Similar modeling could have been carried out using either the HJ structure complexed to the FLP recombinase or that obtained from the crystal packing of the DNA oligomers by Timsit et al. (53) and Timsit and Moras (54).
The available experimental data pertaining to the interactions of gH1d with HJ have been interpreted and used in model building in a hierarchical fashion, which has resulted in deciphering the unique mode of interaction between gH1d and HJ. At the gross level, data suggesting the preferential binding of histone H1d to HJ presents a vast number of possibilities for the manner in which the two can interact. Next, incorporating the nature of gH1d and its interactions with DNA clearly identified the anchor points of interaction on gH1d, thus reducing the number of possibilities to only a handful. Essentially, the data that defined the interaction-anchors originate from site-directed mutagenesis studies of the residues in the primary and secondary binding sites, sequence and structural analysis of gH1d as well as inferences from a structural similarity search and analysis of the PDB. Next, utilizing the knowledge that both binding sites of gH1d were required for binding either to HJ or to the chromatin particle, combined with the inferences from the analysis of the features of HJ, indicating that any face of HJ was made of two arms of DNA segments and formed two potential sites for interacting with the protein, further reduced the number of modes of protein–DNA interaction to only a few. Docking of gH1d onto HJ indicated that gH1d could only be positioned on any one face of HJ based on structural considerations, in which gH1d interacted through the basic cluster on helix 3 with one arm of HJ and the basic patch made of Lys40 and Arg42 interacted with the other arm of the same face of HJ, thus leading to a choice between two models. Finally, incorporating the experimental data suggesting that the primary site on helix 3 of gH1d interacts with the major groove while the secondary binding site interacts with the minor groove of a DNA duplex led to the understanding of the precise mode of interaction of gH1d with HJ, thus leading to a unique possibility for positioning gH1d with respect to HJ. Thus, the gH1d was positioned onto the four-arm HJ structure, such that all the features of the globular domain binding to DNA, such as (i) primary DNA binding through residues Arg67, Lys69 and Lys73 at the major groove, (ii) a second DNA binding site formed by residues Lys40 and Arg42 at the minor groove and (iii) a strong interaction of Lys85 with the DNA phosphate, are maintained (Fig. 2A). Therefore, the model becomes intrinsically validated both in terms of the structural considerations as well as the vast amount of biochemical data available. An architectural motif of DNA, formed by the combination of self-fitted segments in the oligomer crystal packing resembles the open planar HJ structures closely. This ‘self-fitted cross’ has been suggested to be a common recognition motif for various proteins, including recombinases, topoisiomerases and architectural proteins involved in chromatin assembly (55). It is indeed remarkable that the conformation of the two duplexes in one face of the HJ as seen in the crystal structures is just appropriate for accommodating the globular domain. Any other conformation of HJ would not satisfy all the experimental and analytical data described above. It is quite likely that the HJ structure mimics the sub-structure of the two DNA duplexes of the nucleosome, as required for binding to gH1d. This again is consistent with the suggestion by Timsit and Moras that the ‘self-fitting cross’ could occur in many biological processes where DNA–DNA interactions are involved, such as packing of genomic DNA and assembly of the nucleosomes (54).
Figure 2.
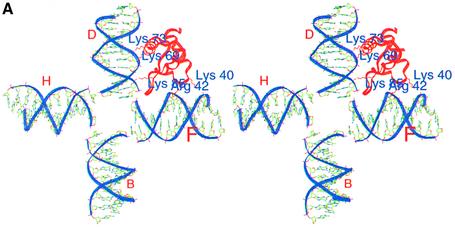
(A) A stereo view of the globular domain (red)–HJ (blue) complex. Interactions of the globular domain at both the primary and secondary sites with two arms of the HJ are clearly seen. Residues in the binding site are labeled. (B) Stereo view of the globular domain docked onto the nucleosome by superposing two arms of the gH1d–HJ (red–blue) complex onto the nucleosomal core (pink). The fourth arm of the HJ is not shown for clarity.
The resulting gH1d–HJ complex was superposed onto the nucleosome structure such that one arm (arm F where D–F arms form the face where gH1d will bind) of the HJ structure matched exactly with the inner duplex of the nucleosome while the other arm (arm B, opposite to the face where the globular domain was placed) could be approximately positioned onto the entering duplex (Fig. 2B). For convenience, the duplex interacting with the globular domain is referred to as the entering duplex while the other one will be referred to as the exiting duplex in this article, but it is entirely possible for the nomenclature to be swapped. The second arm (arm D) forming the face (D–F) of the gH1d-binding motif would now indicate the position and conformation of the extra 15–20 bp at the entry point. The interactions in the complex modeled here, indeed suggest that the conformation of the two duplexes of the nucleosome, upon interacting with the globular domain, are similar to that seen in the HJ structure.
It must be emphasized that only two arms of the HJ structure have been used here for modeling the gH1d complex and these two arms mimic the sub-structure of the nucleosomal duplexes, whereas the other two arms of the planar HJ structure cannot possibly represent the path of the duplexes around the nucleosomes. Consistent with this, the third (B) arm did not match in conformation with that of the entering duplex. We expect a gradual decrease in the bend of the DNA of the nucleosome at the entry point prior to the 145 bp where the extra 15–20 bp will be located so as to have a smooth transition between the bend angles at this point to that observed in the nucleosome core particle. Positioning of the globular domain within the nucleosome structure as modeled in Figure 2B, is also substantiated by the observations of Zhou et al. (18) based on site-specific protein–DNA cross-linking experiments with gH5. They have also proposed that helix III of gH5 binds within the first helical turn of the chromatosomal DNA, while the secondary DNA binding site makes contact with the nucleosomal DNA in the dyad axis.
The C-terminal domain: its structure and interactions with DNA
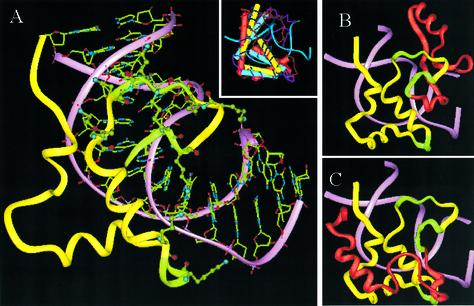
It is rather surprising that there has been virtually no information about the structure of the C-terminal domain. Recently, however, we have predicted the structure of this domain using a combination of fold recognition and other bioinformatics approaches (Fig. 3A). The sequence of the C-terminal domain contains the three octapeptide repeat units housing the S/TPKK motifs, highlighted in Figure 1. The first two of these repeating units are in tandem and span amino acids 144–159, while the third octapeptide unit is present 10 amino acids away from the first two and spans amino acids 170–177. We have also shown by extensive site-directed and deletion mutagenesis studies that the 34 amino acid segment encompassing the three repeat units to be responsible for DNA condensing properties of H1 and therefore can be termed as the DNA condensing segment (20). The C-terminus of histone H1 is mostly random coil in solution, which however, attains significant α-helicity in 60% trifluoroethanol. It is believed that the C-terminus of histone H1 attains its optimum secondary and hence tertiary structure upon interaction with DNA. The structure prediction studies indicate that H1d_C can adopt a structure similar to that of the HMG-box domain present within proteins like rHMG1, sex-determining region within chromosome Y (hSRY) and lymphoid enhancing binding factor-1 (mLEF-1). The model illustrates the expected role of the DNA-condensing segment, thus corroborating the mutational data very well. This observation not only provides an understanding of the structural basis of DNA condensation by histone H1, but also explains the role of H1d_C in competing with the other major chromatin architectural proteins, the HMG proteins, for binding to various target sites within chromatin in vivo.
Figure 3.
(A) The HMG-box fold in the structure of the C-terminal domain of rat H1d (yellow) and its interactions with DNA (pink). Lysine residues in the three S/TPKK motifs (green) are also shown. The inset shows a superposition of different HMG-box proteins that were used as templates to model the H1d_C. (B and C) Two alternate models proposed for the whole H1d_C. The two models differ from each other only in their first 26 residues (shown in red). The first figure shows the model where this segment was modeled based on the sub-structure in malate dehydrogenase while the second picture indicates a model where the segment is based on cytochrome C4. The latter model was found to fit better in the context of the whole chromatin particle (see text).
Positioning the C-terminal domain
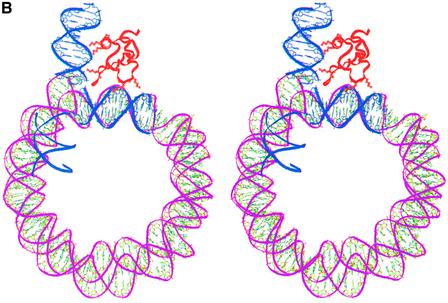
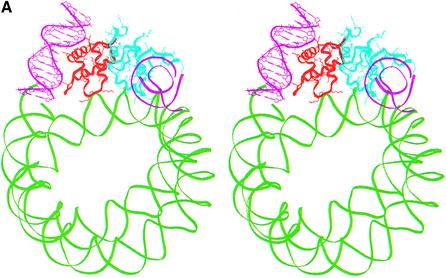
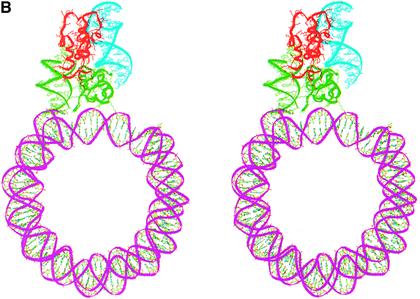
Placing the globular domain on the nucleosome provided an anchor point at residue 108 (the last residue of the globular domain) at which point H1d_C will have to be positioned. Several orientations of this domain with respect to the globular domain are, however, possible and were considered systematically. During structure prediction, we arrived at two models through fold recognition methods, each one differing from the other only in the relative positions of their N-terminal 26 amino acid residues (Fig. 3B and C) (21). A rigid body rotation search with both these models of the H1d_C with respect to the gH1d indicated that the first model of H1d_C, where the N-terminal segment was built based on the sub-structure of malate dehydrogenase, resulted in orientations that were either not interacting with the second duplex of DNA or had severe steric clashes with the globular domain or the region around the dyad axis of the nucleosome core. This model was therefore not considered any further. On the other hand, a systematic search with the second model for H1d_C proposed by us previously, which contained a HMG-box-like fold and in which the N-terminus was modeled based on the sub-structure in cytochrome C4 (Fig. 3C), yielded encouraging results both in terms of the possibility of positioning it juxtaposed to the globular domain as well as in terms of its interactions with the nucleosome. The search identified two plausible positions for this model of H1d_C and therefore two models of H1d (including both globular and C-terminal domains) were constructed in the context of the nucleosome. The first one places the C-terminal domain side by side with the globular domain such that H1d_C interacts with the region around the dyad axis and the exiting duplex (Fig. 4A), whereas the second model places H1d_C somewhat on top of the gH1d making it interact with both the entering and the exiting duplexes but not with the region around the dyad axis (Fig. 4B), thus forming a stem-like structure. Further analysis and correlation with electron microscopy (EM) data described below, indicated that the latter model where the C-terminal domain bridges both the duplexes to be more plausible.
Figure 4.
(A) Stereo view of a model of the chromatosome particle containing the nucleosomal core shown as a green ribbon [core histones not shown for clarity in this and in (B)], globular domain (red) and the C-terminal domain (cyan) placed side-by-side with the globular domain. The DNA segments interacting with the globular and the C-terminal domains are shown as pink ribbons. (B) Stereo view of an alternate model of the chromatosome particle which differs from the first in the position of the C-terminal domain. Here, H1d_C (protein in red and DNA in cyan) is positioned on top of the globular domain (green).
The stem structure and the path of DNA
Although there is no structural data available regarding the chromatosome particle to date, there have been some experiments that suggest the probable organization within the chromatosome which is influenced by the path of the linker DNA. There is unequivocal evidence that the entering and the exiting DNAs remain uncrossed in linear mononucleosomes. Conventional EM of histone H1-depleted chromatin fibers have shown that DNA arms do not cross each other at nucleosome entry and exit sites even at the physiological ionic strength (46). This has also been substantiated by cryo-EM of both mononucleosomes (47) and soluble native chromatin (13). This conclusion is further supported by the experiments of Toth et al. (56), who studied the DNA end to end distance using a fluorescent resonance energy transfer (FRET) technique. The strong bends of the DNA arms in the entry–exit region may reflect steric obstacles on the histone surface or electrostatic repulsion between the two DNA arms. This uncrossing persists upon binding of the gH5 despite further increase of wrapping from the 1.6–1.7 turns to 1.8–1.9 turns (46). These conclusions are also supported by the linking number measurements on nucleosomes reconstituted on DNA mini circles (57,58). EM experiments have further shown the formation of a stem-like structure in histone H1 containing nucleosomes, wherein the C-terminal tail is expected to interact with both the entering and the exiting duplexes resulting in a 3-fold compaction of the linker DNA (46), which is again supported by the FRET experiments of Toth et al. (56).
The C-terminal tail of the linker histone, in contrast to the globular domain, does not affect the wrapping much. Instead, by efficiently countering repulsion of the DNA arms, the highly positively charged C-terminal tail bridges them together into a stem over a distance of ∼30 bp. The two duplexes in the stem may not stay parallel but rather wind negatively around each other, up to approximately half a turn with the full-length H5. Our model is totally consistent with the above-mentioned observations and in fact provides a molecular basis for the understanding of each event. This stem structure also has implications for models of nucleosome folding into the 30 nm filament.
The model of the C-terminal domain, consistent with the DNA condensation data, has shown that the 34 amino acid segment identified by us as the ‘DNA condensing domain’ corresponds structurally to the DNA binding and bending region of the HMG-box domains of Lef-1 and hSRY. The structural data show that the concave surface of the L-shaped molecular architecture of these proteins indeed interacts with the DNA. Furthermore, the bent DNA is stabilized due to its position in the vicinity of the helix such that the lysine residues which project from within the helix interact with the DNA. We have suggested previously, in view of the extensive structural similarity with Lef-1, SRY and HMG-D proteins, that the DNA may be bent upon interaction with the C-terminus of histone H1d wherein the 34 amino acid stretch encompassing the octapeptide repeats plays a key role in defining the angle of bending with the octapeptide repeats containing the S/TPKK motifs functioning as the anchor points. Several biochemical studies (8,45,59) have in fact indicated that the compaction of linker DNA in nucleosomal templates is accompanied by bending or kinking of linker DNA. In the case of Lef-1, the DNA duplex binds to the concave surface of the Lef domain and is bent severely towards the major groove but retains the Watson–Crick base pairing (60). The domain makes extensive and continuous contacts in the minor groove which encompasses the entire region implicated in binding as evidenced by foot-printing and mutagenesis studies. Bending and opening of the minor groove is accompanied by substantial narrowing and deepening of the major groove.
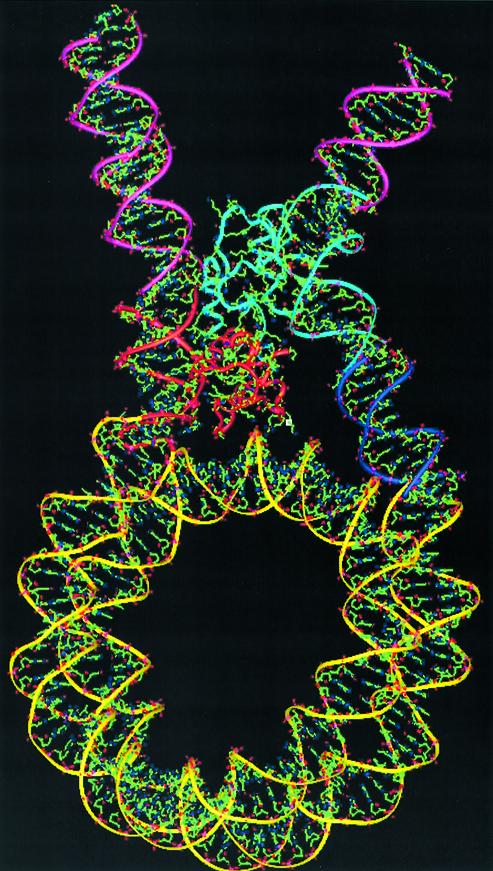
From the above discussion it is evident that the C-terminal tail is essential for the formation of a stem-like structure. The EM data has enabled us to choose the most suitable one of the two positions for the C-terminus. In this model (Fig. 5), the C-terminal domain interacts with both the entry and the exit duplexes, through Lys116 and Lys182 with the entry strand and the DNA condensation unit encompassing the three SPKK motifs (Lys147, Lys148, Lys155, Lys156, Lys173 and Lys174) with the exit strand. We have discussed the role of this motif in bending DNA (21) and have proposed that the bending may be similar to that seen in Lef-1 (60). This model explains the role of the C-terminal domain and its interactions with the DNA both at the single molecular level in isolation and also in the larger context of a mononucleosome. The model with H1d_C in this position also indicates that the DNA condensing unit interacts with DNA approximately 24 bases distant from the exiting end of the 145 bp nucleosomal DNA seen in the crystal structure (1AOI). Similarly, the domain interacts with the DNA segment at approximately 17–18 bases distant from the entering end of the same nucleosomal DNA (Fig. 5). The repeat length of DNA per nucleosome is widely believed to be ∼200 bp which implies the length of the linker DNA to be ∼54 bp. The model of the chromatosome particle accounts for the location and orientation of ∼40 bp on both the entering and the exiting duplex together. Given the lack of understanding of the distribution of the remaining part of the linker DNA between the entering and the exiting duplexes as well as the conformational flexibility in them, it is difficult to model the exact location of the adjacent nucleosome. Although this study presents distinct clues such as bending of the DNA by H1d_C as well as the stem formation upon H1d_C binding, there is still no compelling evidence to favor either the solenoid model or the zigag model over one another. However, the model, along with a clearer understanding of the individual roles of both the domains of the linker histone presented here, is expected to trigger further work towards a more complete understanding of the path of DNA in chromatin.
Figure 5.
Final model of the chromatosome particle based on the position of H1d_C as shown in Figure 4B. In this model, H1d_C is seen to bridge both the entering (left) and the exiting (right) duplexes while significantly bending the exiting duplex. The nucleosome core (yellow), the globular domain (red) and the C-terminal domain (cyan) of H1d, along with the segments of DNA directly associated with them, comprise the final model. The blue stretch indicates the additional segment that would be required to accommodate the H1d_C domain in this orientation (see text). When extended using standard B-DNA, the additional segments will be placed on both the entry and exit duplexes, as represented by the pink segments.
CONCLUSIONS
Distinct roles for the globular domain and the C-terminal domain of histone H1 at the structural level are clearly discernible from the results presented in this study. The globular domain primarily serves as an anchor in binding to the nucleosomal DNA through its dual binding sites. It is also clear that the only way in which the globular domain can bind to DNA and explain the biochemical observation of protection of the extra 20 bp to nuclease digestion of chromatin, is with its primary binding site binding to the entering duplex at the major groove and its secondary binding site binding to the inner gyre of the DNA superhelix of the nucleosome, close to the dyad axis at a minor groove. The presence of two spatially separated DNA binding sites on the globular domain accounts for its affinity to the four-way HJ structure. From structural considerations, it is obvious that it would also be impossible for the globular domain to bind either at a site interior to the nucleosome or at a site outside the nucleosomal core but bridging both the entering and the exiting duplexes. The position suggested in the model also supports asymmetric protection of the 15–20 bp upon nuclease digestion.
The modeling studies indicate that the C-terminal domain, on the other hand, has a role of directing the path of DNA in chromatin fibers by its ability to bend DNA. The model clearly indicates that the three S/TPKK motifs present in the C-terminus are responsible for this property of H1. The positioning of the C-terminal domain in the chromatin particle clearly supports the formation of a stem-like structure resembling that observed by EM. The differences observed in EM data in the presence and absence of the C-terminal domain are also easily explained by the model. The model indicates that the C-terminal domain bridges the entering and the exiting duplexes by countering electrostatic repulsion between the duplexes.
Acknowledgments
ACKNOWLEDGEMENTS
The use of facilities at the Interactive Graphics Based Molecular Modeling Facility and the Distributed Information Center (both supported by the Department of Biotechnology), and the facilities at the Super Computer Education and Research Center are gratefully acknowledged. This work was financially supported by grants from the Council of Scientific and Industrial Research and the Department of Biotechnology, New Delhi.
REFERENCES
- 1.van Holde K.E. (1988) Chromatin. Spring Verlag, New York. [Google Scholar]
- 2.Wolffe A.P. (1998) Chromatin Structure and Function. Academic Press, London. [Google Scholar]
- 3.van Holde K.E. and Zlatanova,J. (1996) What determines the folding of chromatin fiber? Proc. Natl Acad. Sci. USA, 93, 10548–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenfeld G. (1992) Chromatin as an essential part of the transcriptional mechanism. Nature, 355, 219–223. [DOI] [PubMed] [Google Scholar]
- 5.Fedor M.J. (1992) Chromatin structure and gene expression. Curr. Opin. Cell Biol., 4, 436–443. [DOI] [PubMed] [Google Scholar]
- 6.Mizzen C.A. and Allis,C.D. (2000) New insights into an old modification. Science, 289, 2290–2291. [DOI] [PubMed] [Google Scholar]
- 7.Finch J.T. and Klug,A. (1976) Solenoid model for superstructure in chromatin. Proc. Natl Acad. Sci. USA, 73, 1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler P.J.G. and Thomas,J.O. (1980) Changes in chromatin folding in solution. J. Mol. Biol., 140, 505–529. [DOI] [PubMed] [Google Scholar]
- 9.Thoma F., Koller,T. and Klug,A. (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt dependent superstructure of chromatin. J. Cell Biol., 83, 403–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodcock C.L., Grigoryev,S.A. and Horowitz,R.A. (1993) A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl Acad. Sci. USA, 90, 9021–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leuba S.H., Yang,G., Robert,C., Samori,K., van Holde,K., Zlatanova,J. and Bustamente,C. (1994) Three dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl Acad. Sci. USA, 91, 11621–11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Holde K.E. and Zlatanova,J. (1995) Chromatin higher order structure: chasing a mirage. J. Biol. Chem., 270, 8373–8376. [DOI] [PubMed] [Google Scholar]
- 13.Bednar J., Horovitz,R.A., Grigoryev,S.A., Carruthers,L.M., Hansen,J.C., Koster,A.J. and Woodcock,C.L. (1998) Nucleosomes, linker DNA and linker histone form a unique structural motif that directs the higher order folding and compaction of chromatin. Proc. Natl Acad. Sci. USA, 95, 14173–14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bednar J., Horowitz,R.A., Dubochet,J. and Woodcock,C.L. (1995) Chromatin conformation and salt induced compaction: three dimensional structural information from cryo-electron micrsoscopy. J. Cell Biol., 131, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allan J., Cowling,G.J., Harborne,N., Cattani,P., Craigie,R. and Gould,H. (1981) Regulation of higher-order structure of chromatin by histones H1 and H5. J. Cell Biol., 90, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark D.J. and Kimura,T. (1990) Electrostatic mechanism of chromatin folding. J. Mol. Biol., 211, 883–896. [DOI] [PubMed] [Google Scholar]
- 17.An W., Leuba,S.H., van Holde,K.E. and Zlatanova,J. (1998) Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc. Natl Acad. Sci. USA, 95, 3396–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y.B., Gerchman,S.E., Ramakrishnan,V., Travers,A. and Muyldermans,S. (1998) Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature, 395, 402–405. [DOI] [PubMed] [Google Scholar]
- 19.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosomal core particle at 2.8 Å resolution. Nature, 389, 351–360. [DOI] [PubMed] [Google Scholar]
- 20.Bharath M.M.S., Ramesh,S., Chandra,N. and Rao,M.R.S. (2002) Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA condensing domain by site directed mutagenesis. Biochemistry, 41, 7617–7627. [DOI] [PubMed] [Google Scholar]
- 21.Bharath M.M.S., Chandra,N. and Rao,M.R.S. (2002) Prediction of an HMG-box fold in the C-terminal domain of histone H1: insights into its role in DNA condensation. Proteins, 49, 71–81. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.M. (1993) Crystal structure of the globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- 23.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,R.C. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartman P.G., Chapman,G.E., Moss,T. and Bradbury,E.M. (1977) Studies on the role and mode of operation of the very lysine rich histone H1 in eukaryotic chromatin: the three structural regions of the histone H1 molecule. Eur. J. Biochem., 77, 45–51. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 27.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence analysis. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith T.F and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attwood T.K. and Beck,M.E. (1994) PRINTS—a protein motif fingerprint database. Protein Eng., 7, 841–848. [DOI] [PubMed] [Google Scholar]
- 31.Combet C., Blanchet,C., Geourjon,C. and Deléage,G. (2000) NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci., 25, 147–150. [DOI] [PubMed] [Google Scholar]
- 32.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.N. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman H.M., Olson,W.K., Beveridge,D.L.,Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.H., Srinivasan,A.R. and Schneider,B. (1992) The Nucleic Acid Database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J., 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariyoshi M., Nishino,T., Iwasaki,H., Shinagawa,H. and Morikawa,K. (2000) Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA, 97, 8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widom J. (1998) Structure, dynamics and function of chromatin in vitro. Annu. Rev. Biophys Biomol. Struct., 27, 285–327. [DOI] [PubMed] [Google Scholar]
- 36.Cerf C., Lippens,G., Muyldermans,S., Segers,A., Ramakrishnan,V., Wodak,S., Halenga,K. and Wyns,L. (1993) Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: sequential assignment and secondary structure. Biochemistry, 32, 11345–11351. [DOI] [PubMed] [Google Scholar]
- 37.Thomas J.O. (1999) Histone H1: location and role. Curr. Opin. Cell Biol., 11, 312–317. [DOI] [PubMed] [Google Scholar]
- 38.Ramakrishnan V. (1994) Histone structure. Curr. Opin. Struct. Biol., 4, 44–50. [Google Scholar]
- 39.Buckle R.S., Maman,J.D. and Allan,J.(1992) Site directed mutagenesis studies on the binding of the globular domain of linker histone H5 to the nucleosome. J. Mol. Biol., 223, 651–659. [DOI] [PubMed] [Google Scholar]
- 40.Thomas J.O. and Wilson,C.M. (1986) Selective radiolabeling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J., 20, 3531–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goytisolo F.A., Gerchman,S.E., Yu,X., Rees,C., Graziano,V., Ramakrishnan,V. and Thomas,J.O. (1996) Identification of two DNA binding sites on the globular domain of histone H5. EMBO J., 15, 3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 42.Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- 43.Allan J., Mitchel,T., Harborne,N., Bohm,L. and Crane-Robinson,C. (1986) Roles of H1 domains in determining higher order structure and H1 location. J. Mol. Biol., 187, 591–601. [DOI] [PubMed] [Google Scholar]
- 44.Lambert S., Muyldermanas,S., Baldwin,J., Kilner,J., Ibel,K. and Wijns,L. (1991) Neutron scattering studies of chromatosomes. Biochem. Biophys. Res. Commun., 179, 810–816. [DOI] [PubMed] [Google Scholar]
- 45.Pruss D., Bartholomow,B., Persinger,J., Hayes,J.J., Arentes,G., Moudrianakis,E.N. and Wolffe,A.P. (1996) An assymetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science, 274, 614–617. [DOI] [PubMed] [Google Scholar]
- 46.Hamiche A., Scultz,P., Ramakrishnan,V., Oudet,P. and Prunell,A. (1996) Linker histone dependent DNA structure in linear mononucleosomes. J. Mol. Biol., 257, 30–42. [DOI] [PubMed] [Google Scholar]
- 47.Furrer P., Bednar,J., Dubochet,J., Hamiche,A. and Prunell,A. (1995) DNA at the entry-exit of the nucleosome observed by cryo-electron microscopy. J. Struct. Biol., 114, 177–183. [DOI] [PubMed] [Google Scholar]
- 48.Varga-Weisz P., Zlatanoava,J., Leuba,S.H., Schroth,G.P. and van Holde,K.E. (1994) Binding of histones H1 and H5 and their globular domains to four-way junction DNA. Proc. Natl Acad. Sci. USA, 91, 3525–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paneva E.G., Spassovska,P., Ramakrishnan,V., Oudet,P. and Prunell,A. (1998) Interaction of histone H1 with cis-platinated DNA. Z. Naturforsch., 53, 135–138. [DOI] [PubMed] [Google Scholar]
- 50.Ortiz-Lombardia M., Gonzalez,A., Eritja,R., Aymami,J., Azorin,F. and Coll,M.(1999). Crystal structure of a DNA Holliday junction. Nature Struct. Biol., 6, 913–917. [DOI] [PubMed] [Google Scholar]
- 51.Lilley D.M. and Norman,D.G. (1999) The Holliday junction is finally seen with crystal clarity. Nature Struct. Biol., 6, 897–899. [DOI] [PubMed] [Google Scholar]
- 52.Conway A.B., Chen,Y. and Rice,P.A. (2003) Structural plasticity of the flp-Holliday junction complex. J. Mol. Biol., 326, 425–434. [DOI] [PubMed] [Google Scholar]
- 53.Timsit Y., Westhof,E., Fuchs,R.P.P. and Moras,D. (1989) Unusual helical packing in crystals of DNA bearing a mutation hot spot. Nature, 341, 459–462. [DOI] [PubMed] [Google Scholar]
- 54.Timsit Y. and Moras,D. (1991) Groove-backbone interaction in B-DNA. Implication for DNA condensation and recombination. J. Mol. Biol., 221, 919–940. [DOI] [PubMed] [Google Scholar]
- 55.Timsit Y. and Moras,D. (1994) DNA self-fitting: the double helix directs the geometry of its supramolecular assembly. EMBO J., 13, 2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toth K., Brun,N. and Langowski,J. (2001) Trajectory of nucleosomal linker DNA studies by fluorescence resonance energy transfer. Biochemistry, 40, 6921–6928. [DOI] [PubMed] [Google Scholar]
- 57.Zivanovic Y., Duband-Goulet,I., Schultz,P., Stofer,E., Oudet,P. and Prunell,A. (1990) Chromatin reconstitution on small DNA rings III: histone H5 dependence of DNA supercoiling in the nuclsosome. J. Mol. Biol., 214, 479–495. [DOI] [PubMed] [Google Scholar]
- 58.Prunell A. (1998) A topological approach to nucleosome structure and dynamics: the linking number paradox and other issues. Biophys. J., 74, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao J., Lowary,P.T. and Widom,J. (1990) Direct detection of linker DNA bending in defined length oligomers of chromatin. Proc. Natl Acad. Sci. USA, 87, 7603–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love J.J., Li,X., Case,D.A., Giese,K., Grosschedl,R. and Wright,P.E. (1995) Structural basis for DNA bending by the architectural transcriptional factor LEF 1. Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]