Abstract
Nonsense mutations are usually assumed to affect protein function by generating truncated protein products. Nonetheless, it is now clear that these mutations affect not just protein synthesis but also messenger RNA stability. The surveillance mechanism responsible for the detection and degradation of ‘nonsense’ RNA messages is termed nonsense-mediated RNA decay (NMD). Essential biochemical components of the NMD machinery have been defined in several species. Here we identify the Drosophila orthologue of one of these factors, Upf1, and document its expression during embryogenesis. To test whether NMD acts during Drosophila development, we make use of a mutation that introduces a stop codon into a variably spliced exon of the Hox gene Ultrabithorax (Ubx). Using real-time quantitative RT-PCR we demonstrate that Ubx transcripts containing the premature stop codon are expressed at lower levels than their wild type counterpart. Unexpectedly, we also find that the same mutation significantly increases the levels of a Ubx splicing isoform that lacks the exon containing the premature termination codon. These findings indicate that NMD is operational during Drosophila development and suggest that nonsense mutations may affect development by altering the spectrum of splicing products formed, as well as by reducing or eliminating protein synthesis.
INTRODUCTION
Eukaryotes as diverse as yeast and humans use a common biochemical pathway to detect and degrade RNA messages that contain premature termination codons (PTCs) (1). Although the mechanistic details underlying this type of RNA surveillance are not entirely understood, information from the animal systems analysed so far, indicates that the triggering of effective nonsense-mediated RNA decay (NMD) demands two signals to be present in the mRNA: a termination codon, and the presence of an intron downstream of this termination codon (1). Consistent with these requirements, mammalian intron-less genes are immune to NMD (2). Recent work performed in mammalian systems indicates that the second signal is probably not the intron itself but a ‘molecular label’ left after the intron is removed (3,4). This molecular label, the exon junction complex (EJC), has also been shown to be involved in the regulation of RNA export out of the nucleus (5,6). The binding of the EJC at a specific position on a given RNA message appears to be a key element for the distinction of PTCs from ordinary stop codons, which are mostly located in the final exon of the message (1,7,8). Additional support for this view is provided by experiments showing that artificial tethering of some members of the EJC, downstream of a regular termination codon, is sufficient to trigger NMD (9,10).
Since NMD also occurs in yeast, where only a small fraction of genes possesses introns, a somehow different mechanism must operate to identify PTC-bearing messages in this organism. Indeed, downstream sequence elements composed of degenerate AU-rich motifs have been shown to be essential cis-acting elements for the recognition of nonsense messages (11). In spite of these differences, in yeast as well as in humans, three interacting proteins, Upf1, Upf2 and Upf3, are necessary for effective NMD. Of these, Upf1 has been the one studied in most detail. The Upf1 protein is a member of the group I RNA helicases and has been shown to interact with the translation release factors eRF1 and eRF3, well in line with its role in NMD and translation termination (12). Tethering of Upf1 downstream of a termination codon was shown to be sufficient to trigger NMD (13).
In spite of the growing interest to explain the molecular mechanisms underlying NMD, fundamental aspects of this process remain mysterious. One unresolved issue is the question of where, within the cell, nonsense transcripts are destroyed. Mainstream opinion sustains the view that RNA degradation must occur somewhere in the cytoplasm (14). However, a growing number of examples suggests that nonsense messages can be recognised and degraded within the nucleus after detection by a mechanism of intra-nuclear scanning (15,16). Recent reports describing the existence of translation in the nucleus provide further support for these models (17,18).
Nonsense mutations have also been shown to affect pre-mRNA processing events, such as RNA splicing. This response, termed nonsense-associated altered splicing (NAS) consists of the increase of the levels of alternatively spliced transcripts that omit exons carrying nonsense codons (16,19). Two different types of NAS exist. One of them is triggered by nonsense mutations that disrupt splicing regulatory elements, which modulate the performance of alternative splicing reactions. The other type of NAS is based on an unidentified mechanism able to detect disruptions of the reading frame. Two recent articles provide strong evidence that, although both, NMD and frame-dependent NAS require the existence of a nonsense mutation and a translation-like process (20,21), they are functionally distinct processes that rely on different, but overlapping sets of proteins (22,23) [reviewed in (24)].
Another unresolved matter is the role of NMD (and NAS) in the normal biology of different organisms. In yeast for example, Upf1 null mutants are viable showing only minor metabolic problems (25). In nematodes, elimination of the NMD pathway results in viable animals with slight morphological defects (26). In contrast, targeted disruption of the mouse gene Rent1, the murine orthologue of Upf1, is embryonic lethal, indicating that some functions of Upf1 must be essential for some aspects of mouse development (27). Surprisingly, beyond this point there is no other information on the role played by the NMD pathway in animal development. This fact prompted us to investigate whether NMD was active during the embryonic development of the fruit fly Drosophila melanogaster.
For this purpose, we first searched the Drosophila genome for conserved genes encoding components of the NMD biochemical machinery. Our search identified the Dm-Upf1 gene, which we show is dynamically expressed during embryogenesis. Secondly, we tested the activity of the NMD pathway by making use of a nonsense mutation that affects the Hox gene Ultrabithorax (Ubx) (28). Drosophila Ubx encodes a homeodomain transcription factor that regulates development in a particular region of the animal body (29). In normal development, the Ubx gene produces a family of protein isoforms through alternative splicing. Quantification of the abundance of two Ubx isoforms in wild type and Ubx nonsense-mutant embryos shows that: (i) messages containing the nonsense codon are expressed at lower levels than their wild type counterparts and (ii) in mutant embryos, mRNAs coding for a Ubx isoform that lacks the nonsense codon are present at higher levels than expected. These observations suggest that NMD is operational during Drosophila embryonic development and that nonsense mutations could potentially influence development by altering the ratio among splicing isoforms of the affected genes.
MATERIALS AND METHODS
Sequence alignment and phylogenetic analysis
A sequence similarity search of Drosphila data sets (Fly BLAST) using the human Upf1 gene (HUPF1) as probe, identified the Drosophila gene CG1559 and its protein product (SWISS PROT accession code Q9VYS3). Sequences were aligned using ESPript and loaded up in the TREE-puzzle program.
Fly stocks and embryo collections
Stocks carrying the Ubx195 allele were a kind gift from Javier Lopez. Ubx mutant chromosomes were balanced over TM6B balancer chromosomes carrying P{w+mW.hs:Ubi-GFP.S65T}PAD2 as provided from the Bloomington stock centre (http://fly.bio.indiana.edu). Populations of Oregon Red and Ubx195 adult flies (age 7–14 days) were transferred to cages where embryos were collected on apple juice plates for ∼1 h and then allowed to develop at 25°C for variable times.
RNA in situ hybridisations and other procedures with embryo and larvae
An exonic fragment of DmUpf1 was amplified from Oregon Red genomic DNA by PCR (primers were 5′-CAA TCTCCTCCTGTCCCTGTG-3′ and 5′-ATCTCATCGCGA TCCCTGCGG-3′) and subsequently subcloned in the pGEMT vector (Promega). Sense and antisense DIG-labelled riboprobes were synthesised by run-off transcription from SP6 and T7 promoters respectively (using a Boehringer kit). Whole mount RNA in situ hybridisations with DmUpf1 sense and antisense probes were carried out essentially as described by Tautz and Pfeifle (30). After hybridisation and washes, anti-DIG-AP antibody (Boehringer, dilution 1:2000) was added for 2 h at room temperature. Following a series of washes in 1× PBT (1× PBS, 0.01% Triton X-100), alkaline phosphatase (AP) enzymatic reactions were carried out in AP buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris–HCl pH 9.5, 0.01% Tween-20) with NBT/BCIP (Roche). Reactions were stopped by five rinses with 1× PBT. Embryos were dehydrated and rehydrated in a series of ethanol dilutions [30, 50, 70, 100% (v/v) and back] mounted in 80% glycerol and visualised under Nomarski optics in a Zeiss Axioplan microscope. Larval cuticles were mounted in Hoyer’s:lactic acid (1:1), left to dry on a hot plate for ∼24 h and observed using phase-contrast and dark-field microscopy. Digital photographs were taken with a Leica DFC 300 F camera and processed with Adobe Photoshop 5.0 software for Macintosh. Selection of Ubx195 mutant embryos was performed through the use of GFP balancer chromosomes (see details above). Embryos were manually sorted according to their fluorescence level under a Leica MZ FLIII UV dissecting microscope and transferred to Eppendorf tubes. Parallel samples of wild type and mutant embryos were swiftly frozen in liquid N2 and kept at –80°C until further processing.
Real-time quantitative PCR
Total RNA was extracted from freshly collected, liquid N2-frozen embryo populations of selected ages using the RNeasy kit from Qiagen. RNAs were examined by gel, quantified by spectrophotometry, re-examined by gel and finally retro-transcribed using a SuperScript RT–PCR kit (Invitrogen) following manufacturer’s instructions. Amplifications were performed with real-time TaqMan technology (PE Biosystems) and analysed using an ABI PRISM 7700 sequence detection system (Applied Biosystems). Thermocycler conditions were step 1: 50°C 2 min, step 2: 95°C 10 min, step 3: 95°C 15 s, step 4: 60°C 1 min, cycle back 45 times to step 3. The primers for Ubx Ia are as follows: forward 5′-CCGCCGTATTGTGTTAAATCAG-3′ (P2); reverse, 5′-GGCCAGCAATCACACATTCTAC-3′ (P1), and the TaqMan probe, 5′-CTTATC TTACCTGCGATAGCCATCCAGGG-3′ (FAM labelled). The primers for Ubx IVa are as follows: forward 5′-TGTCGGCCGCGTCTTC-3′ (P3); reverse, 5′-GGCCAGCAATCACACATTCTAC-3′ (P1), and the TaqMan probe, 5′-CAGACCATTTGTACCTGCGATAGCCATCC-3′ (VIC labelled). Expression of the ribosomal protein 49 gene (Rp49) was used as an additional control for RNA loading and integrity. Rp49 primers are forward 5′-GCGCACCAAGCACTTCATC-3′; reverse, 5′-GACGCACTCTGTTGTCGATACC-3′ and Rp49 TaqMan probe, 5′-ATATGCTAA GCTGTCGCACAAATGGC-3′ (VIC labelled). Primer and probes were designed using Primer Express™ software (Applied Biosystems). TAMRA was used as the 3′ quencher in all cases. After primer and probe optimisation experiments, all PCRs included 300 nM of each primer, 200 nM probe (Ubx Ia, Ubx IVa or Rp49) and 1× TaqMan® Universal PCR Master Mix (Applied Biosystems) containing the passive reference ROX. Experiments using different dilutions of plasmid templates for Ubx Ia, Ubx IVa and the mix (Ubx Ia + Ubx IVa) certified the specificity of the quantification procedure. The significance of differences in RNA abundance for different Ubx isoforms was assessed by evaluating thres hold cycle (CT) values, according to the ‘relative standard curve’ method (ABI Prism 7700 Manual, available from http://www.appliedbiosystems.com). For this, a sample obtained from a long collection of wild type embryos, showing high signals for both Ubx isoforms, was used as a calibrator. Reactions involving probes for Ubx Ia (FAM), Ubx IVa (VIC) and Rp49 (VIC) were carried out in separate optical tubes fed from a series of reaction pre-mixes containing all PCR reagents (including time and genotype-specific templates) with the exception of probes and primers (which were supplied from gene-specific detection pre-mixes). Ratios of Ubx Ia and Ubx IVa abundances to Rp49 were calculated to normalise signals. All real-time Taqman PCRs were done in triplicate. Whole experiments were repeated three times starting from new and independent embryo collections. Figures thus show each value as the mean ± SD of three independent experiments (n = 3), with each one of them represented by the mean ± SD of three separate quantitative PCRs.
RESULTS
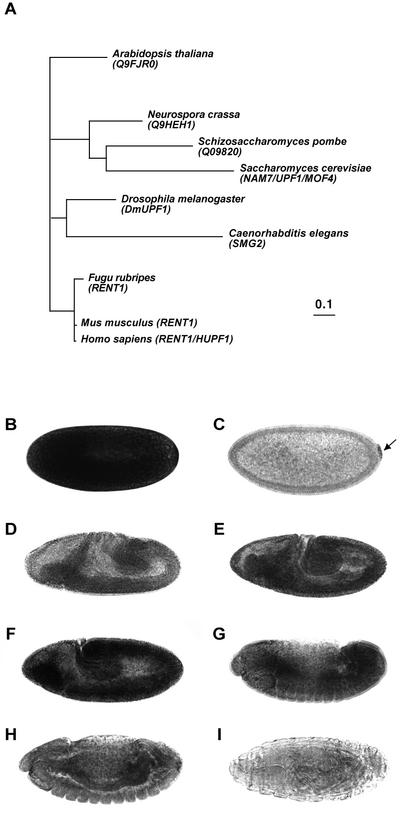
To explore the possibility that NMD affects developmental processes in Drosophila, we started by looking for the presence of conserved genetic components of the NMD machinery within the Drosophila genome. We focused our attention on the gene encoding Upf1, a protein that has been shown to be essential for an effective NMD response in all systems studied to date (25,26,31). Screening Drosophila genome databases (32) with the sequence of HUPF1 identifies one predicted gene, CG1559, encoding a protein with high amino acid similarity to HUPF1 (82%; Fig. 1). When CG1559 is used as a probe within trans-specific databases it returns signals coding for Upf1 products from yeast, plant, worm, mouse and human. Sequence analysis of CG1559, here re-named DmUpf1, revealed that all the characteristic biochemical functions of Upf1 are accommodated within the predicted protein including putative zinc fingers in its N-terminal portion and all seven RNA helicase motifs that are present in Upf1 and other group I RNA helicases (33). Alignment of DmUpf1 with other members of the Upf1 protein family reveals a high degree of amino acid conservation throughout the protein (Fig. 1). A phylogenetic analysis of the Upf1 protein family retrieves the major phylogenetic relationships among the groups included in the study, confirming the orthology of DmUpf1, and suggesting also that Upf1 genes may be useful for phylogenetic purposes (Fig. 2A).
Figure 1.
Sequence alignment of Upf1 proteins. Amino acid sequences corresponding to Upf1 proteins from different species. Drosophila melanogaster Upf1 (DmUpf1) shares extensive sequence homology with other members of the Upf1 protein family. Red letters indicate residues shared by many members of the family; red background indicates strict conservation of residues in all the species included in the study. Species names and SWISS PROT accession codes/ references for each sequence are as follows: D.melanogaster (SWP:Q9VYS3), Caenorabditis elegans (SWP:O76512), Fugu rubripes (Q98TR3), Mus musculus (Q9EPU0), Homo sapiens (Q92900), Saccharomyces cerevisiae (NAM7/UPF1/MOF4), Schizosaccharomyces pombe (Q09820), Neurospora crassa (Q9HEH1) and Arabidopsis thaliana (Q9FJR0).
Figure 2.
Phylogenetic and expression data on DmUpf1. (A) Phylogeny of the Upf1 protein family. Diagram showing a phylogenetic tree displaying the major relationships among sequences of the Upf1 protein family. Protein sequences are indicated by species name followed by gene name or accession code. (B–I) DmUpf1 RNA expression at different developmental stages during Drosophila embryogenesis: (B) stage 2, (C) stage 5, (D) stage 7, (E) stage 8, (F) stage 9, (G) stage 12, (H) stage 14 and (I) stage 17. Note the high level of DmUpf1 signal observed by stage 2, the localised signal detected at stage 5 (arrow) and the decrease of DmUpf1 expression observed in late stages (G–I). Stages are according to Campos-Ortega and Hartenstein (41). In all panels, anterior is to the left and dorsal is up.
To study DmUpf1 expression during Drosophila development we cloned a fragment of the DmUpf1 gene by PCR and hybridised RNA probes prepared from this sequence to Drosophila embryos (see Materials and Methods). Con sistent with data from other systems, we observe that DmUpf1 is expressed in most tissues throughout development (Fig. 2B–I). Nonetheless, different expression phases can be distinguished. DmUpf1 RNA is present at very high levels throughout early cleavage stage embryos, indicating a maternal contribution (Fig. 2B). The presence of this maternal product may reflect the necessity to have a fully operational RNA surveillance system while critical early embryonic genes are being expressed. At late blastoderm stages, levels of transcript decline, but localised DmUpf1 RNA accumulation now becomes evident in the pole cells (Fig. 2C). After stage 5, DmUpf1 RNA is expressed at varying levels in most or all tissues (Fig. 2D–F). Expression persists throughout much of embryogenesis, though in very late stages we note a decrease in the levels of DmUpf1 RNA (Fig. 2G–I). Altogether, these sequence and expression data suggest that essential components of the NMD machinery are present during Drosophila embryonic development.
To test if NMD is actually operational during Drosophila development, we employed a nonsense mutation that affects the Hox gene Ultrabithorax (Ubx) (28). Ubx is expressed in a specific region along the antero–posterior axis of the animal body where it determines many aspects of segment morphology. This region, defined as parasegments 5 and 6, extends from the posterior compartment of the second thoracic segment to the anterior compartment of the first abdominal segment (29).
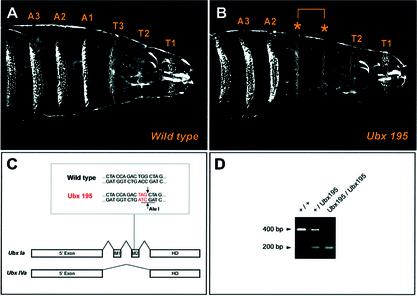
As a result of alternative splicing, the Ubx gene produces a family of closely related proteins or isoforms. Ubx isoforms differ from one another by the presence or absence of optional exons that separate the 5′ exon from the 3′ exon, which encodes the Ubx homeodomain (34,35). The Ubx nonsense allele that we used, Ubx195, carries a point mutation in the differentially spliced exon M2 (Fig. 3C) (36). This exon is only present in the longer versions of the protein, isoforms I and II (types a and b), and is spliced out from the short isoform IV (34,35) (Fig. 3C). Ubx195 homozygotes die as larvae with defects in the CNS and tracheal system characteristic of a homeotic transformation of parasegments 5 and 6 to parasegment 4. Externally this transformation resembles that of a Ubx null mutant (Fig. 3A and B), but in the CNS Ubx195 clearly retains some function, presumably mediated by Ubx isoform IV (36).
Figure 3.
Phenotype of Ubx195 mutants and structure of Ubx splicing isoforms investigated in this study. (A and B) Ventral cuticle phenotypes of wild type (A) and Ubx195 (B) larvae. In Ubx195 mutants, denticle belts of the third thoracic segment (T3, in parasegment 5) and the first abdominal segment (A1, in parasegment 6) are transformed to resemble that of T2 (parasegment 4) (asterisks). (C) Scheme of Ubx protein isoforms including (Ubx Ia) or excluding exon M2 (Ubx IVa). In Ubx195 mutants, a G to A base substitution generates a PTC (red); this change also creates an AluI restriction site absent from the wild type sequence (underlined sequence). For simplicity, other Ubx isoforms that are not part of this study, such as Ubx II and all isoforms b, have been excluded from the diagram. (D) Presence of a restriction site for AluI within a 400 bp genomic PCR product including the exon M2, results in two small products of 200 bp that indicate the purity of the mutant embryo populations used in our study.
The location of the stop codon in Ubx195 mutants establishes an experimental system that provides the exceptional opportunity of generating two qualitatively different types of mature mRNAs from the same promoter: PTC-bearing and PTC-exempt (Fig. 3C). If NMD is operational during Drosophila development, the abundance of messages for Ubx isoform Ia, including the premature stop codon, should be lower in Ubx195 mutant embryos than in wild type embryos. On the other hand, the abundance of messages encoding Ubx isoform IVa, without the premature stop codon, should in principle be similar in both Ubx195 and wild type embryos (see below). To test these predictions, we developed a real-time quantitative RT-PCR assay that allowed us to determine relative RNA levels coding for two Ubx isoforms: Ubx Ia and Ubx IVa. TaqMan fluorogenic probes were designed to match isoform-specific exon–exon junctions, thus avoiding amplification from genomic DNA (Fig. 4A) (see also Materials and Methods). Ubx195 mutant chromosomes were maintained over a GFP-balancer, allowing the isolation of a homogeneous Ubx195/Ubx195 embryo population (Fig. 3D) (see also Materials and Methods). Wild type and Ubx195 embryos were collected for ∼1 h and allowed to develop at 25°C. Total RNA was prepared from embryos of selected ages, quantified, retro-transcribed and used as template for real-time PCRs. An additional TaqMan probe was designed to detect the abundance of mRNA encoding Rp49, which served as an external control for RNA loading and integrity.
Figure 4.
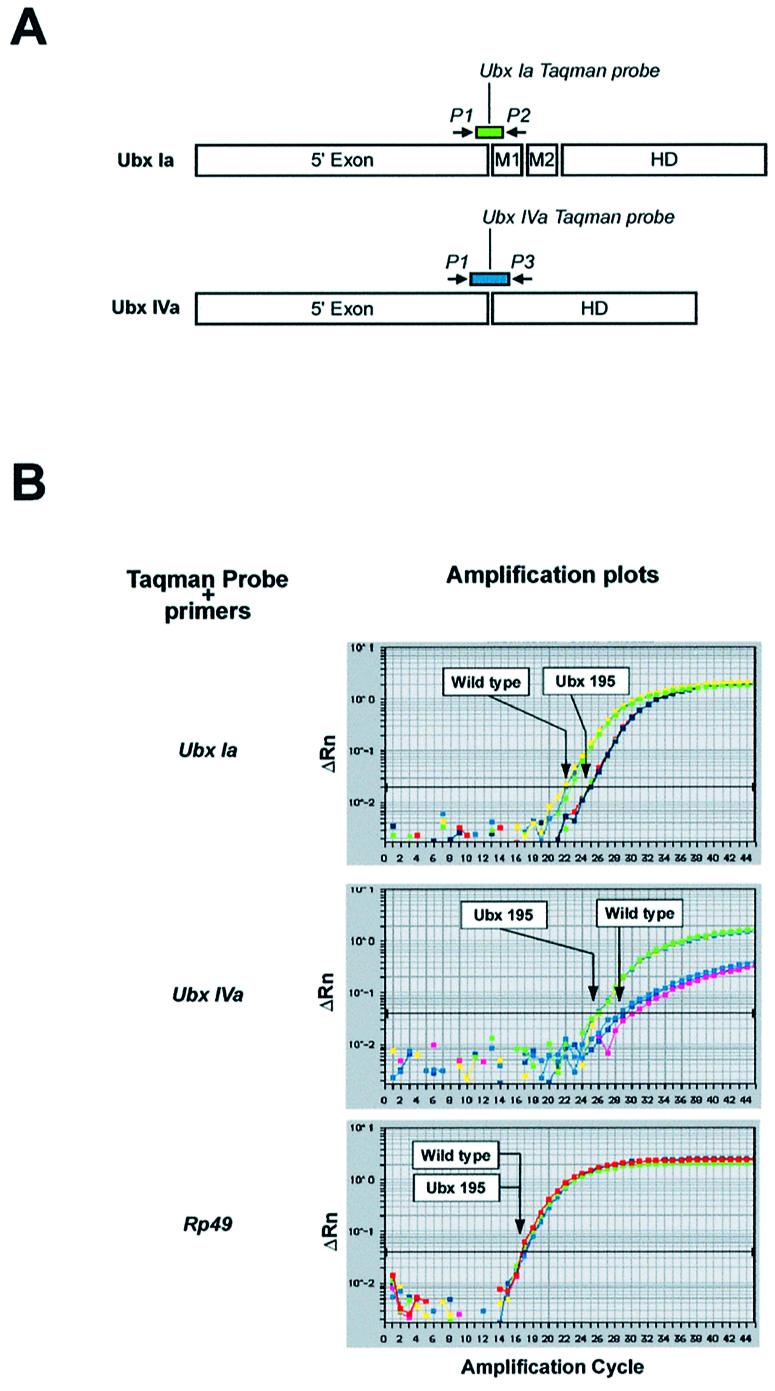
Quantification of Ubx isoforms in wild type and Ubx195 embryos. (A) The location of primers (arrows) and Taqman probes (rectangles) is indicated. Please note that Taqman probes only hybridise to isoform-specific exon–exon junctions. This experimental design also prevents the detection of non-specific signals from traces of genomic DNA present in RNA preparations. (B) Amplification plots of real-time PCRs using Ubx Ia, Ubx IVa and Rp49 probes on wild type and Ubx195 templates (late embryos). The plots relate PCR cycle number to changes (Δ) in detected fluorescence with background removed (Rn) on a logarithmic scale. Arrows highlight threshold cycle values (Ct) obtained for each genotype in the different reactions. The lower the Ct values, the higher the abundance of the RNA species detected by the system.
Results of Ubx and Rp49 RNA quantification for wild type and Ubx195 late embryos are shown in Figure 4B. All panels display amplification plots showing values of normalised fluorescence intensity (ΔRn) versus amplification cycle, for both, wild type and Ubx195 embryos (all assays in triplicate – see Materials and Methods). All samples show the same Ct for the control probe Rp49, thus certifying equal RNA loading in all cases. For Ubx Ia, Ct values for wild type samples are lower than those for Ubx195 samples, indicating that levels for Ubx Ia are higher in wild type than in Ubx195 embryos of similar age. This finding is consistent with a Drosophila NMD pathway operating on RNA messages containing PTCs. To test whether PTC-free RNA isoforms derived from the Ubx transcription unit were or were not affected by NMD, we looked at the levels of Ubx IVa in the same RNA samples. Contrary to our expectations, we detected an increase in the levels of Ubx IVa message when comparing Ubx195 embryos with wild types. This result suggests that the presence of a nonsense mutation could alter the balance between splicing isoforms of Ubx.
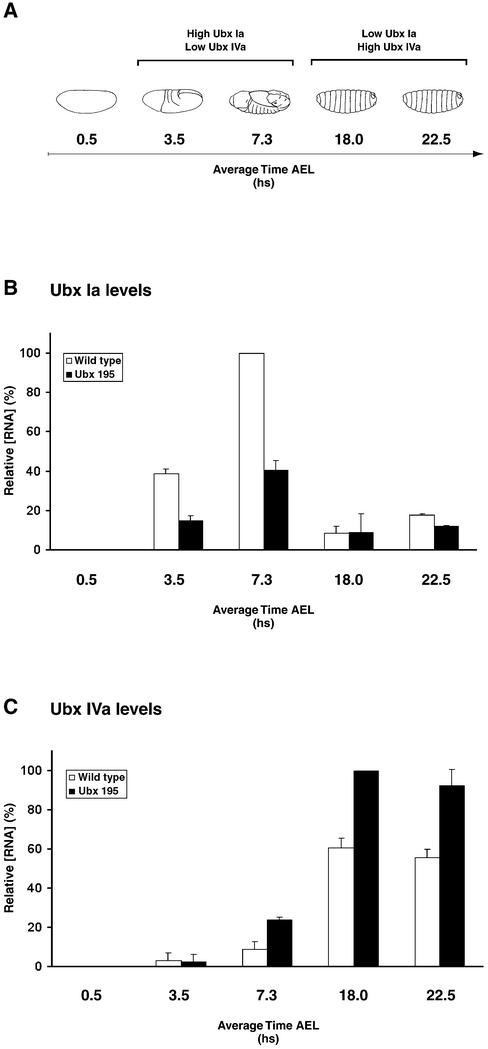
RNA levels coding for different Ubx isoforms fluctuate during embryonic development (34,35) so any discrepancy in the age distributions of embryos in wild type and Ubx195 samples might cause an alteration in the ratio of isoforms. To control for this, we extracted RNA from a series of closely timed samples, choosing points in development when Ubx isoforms display major differences in tissue distribution (Fig. 5A). The outcome of these experiments is summarised in Figure 5 (B and C). Ubx Ia levels appear to be reduced in Ubx195 embryos during most of embryogenesis (Fig. 5B). At the same time, levels of Ubx IVa RNA appear to be increased in Ubx195 embryos (Fig. 5C). There is also some suggestion from our data that the magnitude of both these effects may vary during development.
Figure 5.
Expression profile of Ubx isoforms in wild type and Ubx195 mutant embryos. (A) Diagram of embryos at various points in development (hours) as those analysed in RNA quantification experiments (AEL: after egg laying). Note that in wild type embryos, the highest expression levels for isoforms Ubx Ia and Ubx IVa are detected at different stages of embryogenesis. See also references (34,35). (B and C) Real-time quantitative PCR estimates of relative RNA levels for Ubx Ia (B) and Ubx IVa (C) in embryos at developmental times as shown in (A). White and black columns represent wild type and Ubx195 samples, respectively. Sample groups displaying the highest RNA levels of their experimental series were arbitrarily assigned the value of 100%. For further details on real-time quantitative PCR, please refer to Materials and Methods.
DISCUSSION
The widespread expression of DmUpf1 throughout embryogenesis, and the effects of the Ubx195 mutation on Ubx Ia transcript levels, suggest that NMD operates during Drosophila development. Our identification of DmUpf1 together with a recent report identifying the Drosophila orthologues of the nematode Smg5 and Smg7 genes (37) provide the first information on the insect NMD genetic machinery.
It would be desirable to test the developmental requirement for DmUpf1 by eliminating its function. Unfortunately no candidate point mutations exist, and the available deficiencies covering the gene eliminate multiple gene functions, and severely disrupt embryonic development (data not shown). RNA interference (RNAi), in principle, provides an alternative way to knockout gene function, but in Caenorhabditis elegans, efficient operation of RNAi requires expression of Upf1 (38), making this approach somewhat problematical. These considerations become even more complex in the context of a maternal contribution for DmUpf1 as suggested by our expression data. Therefore, at present we do not know the null phenotype of DmUpf1.
The observed over-production of Ubx IVa in Ubx195 embryos implies that the presence of a nonsense mutation in the Ubx Ia transcript, is able to affect the probability that it will be re-spliced to generate other isoforms. Effects of nonsense codons on alternative splicing have been documented in other systems (1,16) but, to our knowledge, never shown to affect developmental genes or processes. As mentioned in the introduction, NAS has initially been explained as the consequence of the physical disruption of exonic splicing enhancers (ESEs) or silencers (ESSs) by nonsense mutations [reviewed in (16)]. However, very recent work shows that NAS could also result from reading frame disruptions (21,23,39). In spite of these stimulating advances, the molecular mechanisms behind NAS are still largely unclear.
It has been shown that Ubx isoform IVa is not produced by an exon-skipping mechanism, but instead through a complex re-splicing cycle from a Ubx Ia precursor transcript (40). In our system, Ubx195 embryos produce less isoform Ia and more isoform IVa than their wild type counterparts, suggesting that NMD is not reducing the splicing precursor pool of Ubx Ia available for re-splicing, and may in fact be increasing the probability of such a re-splicing event.
As judged by the effects on the Ubx system, the efficiency of NMD does not seem to be constant throughout development (Fig. 5). Maximum NMD efficiency (as fold difference in Ubx Ia transcript levels between wild type and nonsense mutants) occurs between ∼3 and 8 h after egg laying (Fig. 5 and data not shown). The magnitude of these effects is in line with previous reports on NMD activity in other systems. Interestingly, this also coincides with the times at which DmUpf1 is expressed at high levels in most embryonic tissues. The efficiency of NMD seems to decrease at a time when DmUpf1 signal falls (Figs 2 and 5). Perhaps the availability of different amounts of DmUpf1 accounts for the variable efficiency of NMD. However, here we must be cautious in assuming a causal relationship, since DmUpf1 is only one component of the NMD machinery and we only observed its RNA levels. Another possibility is that developmental oscillations in the transcriptional levels of nonsense-RNA templates susceptible to NMD could lead to variability in the nonsense-RNA fraction escaping NMD control.
Perhaps a limitation of our Ubx system in live embryos is that it does not allow us to estimate the individual contributions of NMD and NAS activities to the net Ubx RNA levels. Future studies involving nonsense and wild type Ubx splicing mini-genes, expressed in cell culture and transgenic animals, will help us to determine the extent of the developmental effects of NMD and NAS on isoform steady levels.
Biological processes as fundamental as sex determination have been shown to involve the generation of alternative transcripts containing PTCs, the usual assumption being that these messages would lead to the production of non-functional truncated proteins. Our findings, demonstrating that NMD is operational during Drosophila development, imply that other mechanisms may need to be considered.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Philippa Smith and Andrew McKenzie for their assistance with real-time PCR, Javier Lopez for kindly providing fly stocks, Chuck Cook for help with phylogenetic analysis and Juliane Mossinger, Cassandra Extavour, Elio Sucena, Henrique Teotonio and Vitor Barbosa for general advice and encouragement. We also thank two anonymous referees for their constructive criticism. This work was supported by The Wellcome Trust.
REFERENCES
- 1.Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Maquat L.E. and Li,X. (2001) Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA, 7, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeHir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeHir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]
- 6.Dreyfuss G., Kim,V.N. and Kataoka,N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J., Belgrader,P., Zhou,X. and Maquat,L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- 9.Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson M.F. and Shyu,A.B. (2002) RNA surveillance by nuclear scanning? Nature Cell Biol., 4, E144–147. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez C.I., Bhattacharya,A., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene, 274, 15–25. [DOI] [PubMed] [Google Scholar]
- 12.Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J. (2001) mRNA quality control: marking the message for life or death. Curr. Biol., 11, R88–91. [DOI] [PubMed] [Google Scholar]
- 14.Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buhler M., Wilkinson,M.F. and Muhlemann,O. (2002) Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep., 3, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maquat L.E. (2002) NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev., 16, 1743–1753. [DOI] [PubMed] [Google Scholar]
- 17.Brogna S., Sato,T.A. and Rosbash,M. (2002) Ribosome components are associated with sites of transcription. Mol. Cell, 10, 93–104. [PubMed] [Google Scholar]
- 18.Iborra F.J., Jackson,D.A. and Cook,P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- 19.Valentine C.R. (1998) The association of nonsense codons with exon skipping. Mutat. Res., 411, 87–117. [DOI] [PubMed] [Google Scholar]
- 20.Li S., Leonard,D. and Wilkinson,M.F. (1997) T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med., 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Hamilton,J.I., Carter,M.S., Li,S. and Wilkinson,M.F. (2002) Alternatively spliced TCR mRNA induced by disruption of reading frame. Science, 297, 108–110. [DOI] [PubMed] [Google Scholar]
- 22.Mendell J.T., ap Rhys,C.M. and Dietz,H.C. (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science, 298, 419–422. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Chang,Y.F., Hamilton,J.I. and Wilkinson,M.F. (2002) Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell, 10, 951–957. [DOI] [PubMed] [Google Scholar]
- 24.Moore M.J. (2002) RNA events. No end to nonsense. Science, 298, 370–371. [DOI] [PubMed] [Google Scholar]
- 25.Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- 26.Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- 27.Medghalchi S.M., Frischmeyer,P.A., Mendell,J.T., Kelly,A.G., Lawler,A.M. and Dietz,H.C. (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet., 10, 99–105. [DOI] [PubMed] [Google Scholar]
- 28.Lewis E.B. (1978) A gene complex controlling segmentation in Drosophila. Nature, 276, 565–570. [DOI] [PubMed] [Google Scholar]
- 29.Morata G. and Kerridge,S. (1981) Sequential functions of the bithorax complex of Drosophila. Nature, 290, 778–781. [DOI] [PubMed] [Google Scholar]
- 30.Tautz D. and Pfeifle,C. (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 31.Sun X., Perlick,H.A., Dietz,H.C. and Maquat,L.E. (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 33.Koonin E.V. (1992) A new group of putative RNA helicases. Trends Biochem. Sci., 17, 495–497. [DOI] [PubMed] [Google Scholar]
- 34.Kornfeld K., Saint,R.B., Beachy,P.A., Harte,P.J., Peattie,D.A. and Hogness,D.S. (1989) Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev., 3, 243–258. [DOI] [PubMed] [Google Scholar]
- 35.O‘Connor M.B., Binari,R., Perkins,L.A. and Bender,W. (1988) Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J., 7, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinzierl R., Axton,J.M., Ghysen,A. and Akam,M. (1987) Ultrabithorax mutations in constant and variable regions of the protein coding sequence. Genes Dev., 1, 386–397. [Google Scholar]
- 37.Chiu S.Y., Serin,G., Ohara,O. and Maquat,L.E. (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA, 9, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domeier M.E., Morse,D.P., Knight,S.W., Portereiko,M., Bass,B.L. and Mango,S.E. (2000) A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science, 289, 1928–1931. [DOI] [PubMed] [Google Scholar]
- 39.Li B., Wachtel,C., Miriami,E., Yahalom,G., Friedlander,G., Sharon,G., Sperling,R. and Sperling,J. (2002) Stop codons affect 5′ splice site selection by surveillance of splicing. Proc. Natl Acad. Sci. USA, 99, 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatton A.R., Subramaniam,V. and Lopez,A.J. (1998) Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon–exon junctions. Mol. Cell, 2, 787–796. [DOI] [PubMed] [Google Scholar]
- 41.Campos-Ortega J.A. and Hartenstein,V. (1997) The embryonic development of Drosophila melanogaster. 2nd Edn. Springer Verlag, Berlin, Germany. [Google Scholar]