Abstract
Prokaryotic transcriptional regulatory elements have been adopted for controlled expression of cloned genes in mammalian cells and animals, the cornerstone for gene-function correlations, drug discovery, biopharmaceutical manufacturing as well as advanced gene therapy and tissue engineering. Many prokaryotes have evolved specific molecular communication systems known as quorum-sensing to coordinate population-wide responses to physiological and/or physicochemical signals. A generic bacterial quorum-sensing system is based on a diffusible signal molecule that prevents binding of a repressor to corresponding operator sites thus resulting in derepression of a target regulon. In Streptomyces, a family of butyrolactones and their corresponding receptor proteins, serve as quorum-sensing systems that control morphological development and antibiotic biosynthesis. Fusion of the Streptomyces coelicolor quorum-sensing receptor (ScbR) to a eukaryotic transactivation domain (VP16) created a mammalian transactivator (SCA) which binds and adjusts transcription from chimeric promoters containing an SCA-specific operator module (PSPA). Expression of erythropoietin or the human secreted alkaline phosphatase (SEAP) by this quorum-sensor-regulated gene expression system (QuoRex) could be fine-tuned by non-toxic butyrolactones in a variety of mammalian cells including human primary and mouse embryonic stem cells. Following intraperitoneal implantation of microencapsulated Chinese hamster ovary cells transgenic for QuoRex-controlled SEAP expression into mice, the serum levels of this model glycoprotein could be adjusted to desired concentrations using different butyrolactone dosing regimes.
INTRODUCTION
Integrated molecular interventions in mammalian cells and animals which require precisely adjusted expression of desired transgenes (1) have fostered recent advances in prototype gene therapy scenarios (2), drug discovery (3), biopharmaceutical manufacturing (4,5), gene-function analysis (6–8) and the design of animal models mimicking prominent human disease phenotypes (7).
In recent years, a variety of heterologous transgene control systems have been designed for use in mammalian cells and transgenic animals (1,9). Most modulate target gene transcription in response to antibiotics (10–12), immunosuppressive agents (13) or steroid hormones and their analogs (14,15). Although well established in basic research, ongoing concerns about low or leaky expression, promotion of antibiotic resistance, difficulties in managing the levels of regulating chemicals and interference of inducers with host regulatory networks may jeopardize long-term use of current gene control arrangements in human therapy and substantiate the need for gene regulation systems responsive to a new class of molecules (16–18).
Quorum-sensing systems regulate prokaryotic gene expression in response to fluctuations in cell-population density. Quorum-sensing bacteria produce and release chemical signal molecules and communicate within and across species to regulate a diverse array of activities including symbiosis, virulence, competence, conjugation, motility, sporulation, biofilm formation and antibiotic production (19,20). In Streptomyces, a family of related butyrolactones and their corresponding receptor proteins serve as quorum-sensing systems to coordinate morphological development and antibiotic biosynthesis (20,21).
Here we report the use of Streptomyces coelicolor and Streptomyces pristinaespiralis butyrolactone-responsive quorum-sensing systems to adjust transgene expression in mammalian cells and mice. The use of quorum-sensing pheromones in vivo did not elicit prominent side effects suggesting that butyrolactones are safe inducers. Since most bacterial species are endowed with a specific quorum-sensing system (19,22), Streptomyces-derived QuoRex technology is a promising prototype of a new mammalian gene control system using an alternative class of inducing molecules.
MATERIALS AND METHODS
Synthesis and formulation of SCB1 and MP133
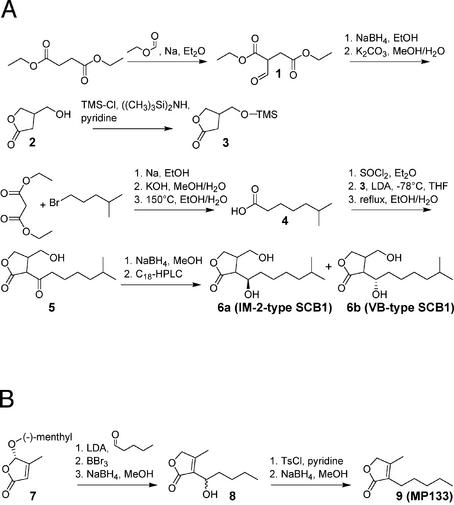
Racemic 2-(1′-hydroxy-6-methylheptyl)-3-(hydroxymethyl)-butanolide (SCB1) was synthesized as depicted in Figure 1A. The butanolide (2) was formed by the reduction of diethyl formylsuccinate (1) (23) with NaBH4, and the hydroxyl group of this butanolide was subsequently protected with a trimethylsilyl ether (24). The alkyl chain of SCB1 was synthesized starting from diethyl malonate and 1-bromo-4-methylpentane (25), which resulted in a 42% (w/w) yield of 6-methylheptanoic acid (4) over three steps. The acid was converted into its acid chloride with thionyl chloride and attached to 3-(trimetylsilyloxymethyl)-butanolide (3) using lithium diisopropylamine in dry tetrahydrofuran. After removal of the trimethylsilyl group, racemic 2-(6-methylheptanoyl)-3-(hydroxymethyl)-butanolide (5) or A-factor was obtained. The reduction of the ketone carbonyl group of 5 with NaBH4 and separation by preparative C18 reversed-phase HPLC resulted in racemic mixtures of the diastereoisomers of SCB1 (6). Diastereomers were over 99% pure as assessed by HPLC, and NMR-based analysis revealed no cross contamination by other diastereomers. A detailed protocol for the synthesis of SCB1 is provided at http://www.biotech.biol.ethz.ch/martinf/download/QuoRex.zip.
Figure 1.
Synthesis of SCB1 (A) and MP133 (B). More detailed information is provided at http://www.biotech.biol.ethz.ch/martinf/download/QuoRex.zip.
Synthesis of 4-methyl-3-pentyl-2(5H)-furanone (MP133) is shown in Figure 1B. Two synthetic pathways were designed for MP133. Whereas the first strategy follows Demnitz’s approach to furanones (26), the second protocol is analogous to the preparation of some furanones which naturally occur in Streptomyces antibioticus (27). According to the synthesis by Grossmann and Séquin (27), the readily available methylated furanone (7) was condensed with pentanal. Subsequent deprotection with BBr3 followed by NaBH4-reduction resulted in a racemate (8). This racemate was then tosylated at the side chain hydroxy group, and the tosylate obtained reduced with NaBH4 to give MP133 (9). Supplementary information on the synthesis of MP133 is available at http://www.biotech.biol.ethz.ch/martinf/download/QuoRex.zip.
SCB1 and MP133 were dissolved in DMSO at 100 mg/ml and diluted to reach a final DMSO concentration of 0.1% (v/v) in all cell culture experiments. Both butyrolactones were used at a concentration of 10 µg/ml unless stated otherwise.
Plasmid construction
The S.coelicolor-derived butyrolactone-dependent transactivator SCA (ScbR-VP16) was constructed by PCR-mediated amplification of scbR from S.coelicolor A3(2) scbR using oligonucleotides OWW36 (5′-gtacgaattcccaccatggccaagcaggaccgggcg-3′) and OWW37 (5′-gcgcgcggctgtacgcggagtccttcccggtcggtgccagtt-3′) followed by cloning (EcoRI/BssHII) into pWW35 (10), thereby resulting in pWW122. Similarly, the S.pristinaespiralis-derived transactivator SPA (SpbR-VP16) was designed by cloning PCR-amplified [oligonucleotides OWW34 (5′-gtacgaattcccaccatggcgcggcaggagcgg-3′) and OWW35 (5′- gcgcgcggctgtacgcggagtggg cggcgggctggg-3′)] spbR from S.pristinaespiralis (strain NRRL2958) EcoRI/BssHII into pWW35 to give pWW123. SCA- and SPA-responsive promoters [PSCA (OscbR-PhCMVmin); PSPA (OpapRI-PhCMVmin)] were assembled by amplifying PhCMVmin-encoding fragments from pRevTre (Clontech, Palo Alto, CA) with OWW39 (5′-gatcgacgtcgccattgaaaaccgaccgtg ccgtttttttcctgcaggtcgagctcggtacccgggtc-3′; OpapRI site underlined) and OWW22 (10) as well as OWW49 (5′-gatcgacg tctaagatacagactgagcggtttttttcctgcaggtcgagctcggtacccgggtc-3′; OscbR site underlined) and OWW22 (10), respectively. The PSCA and PSPA-encoding fragments were cloned (AatII/EcoRI) into pWW36 (10) to result in plasmids pWW124 (PSCA-SEAP-pA) and pWW135 (PSPA-SEAP-pA), respectively. The EPO expression vector pWW156 (PSPA-EPO-pA) was constructed by EcoRI/XhoI-mediated swapping of SEAP (pWW135) with EPO encoded by pMF242 (10–12). The SCA-encoding retroviral vector pWW158 was constructed by excising SCA (EcoRI/BamHI) from pWW122 and cloning it (EcoRI/BglII) into pMSCVneo (Clontech). pWW122-derived retroviruses were produced following Clontech’s protocol and used for transduction of CHO-K1. The bacterial scbR expression vector pWW314 (Pn-scbR-his6-pA) was constructed by PCR-mediated amplification of scbR from pWW122 using primers OWW304 (5′- cggaattcccaccatgcatatggccaagcagg accgggc-3′) and OWW305 (5′-gctctagag caagcttttaatggtgatggtgatgatgggatccacgcggaaccagaccgtccttccc ggtcggtgcca-3′) followed by cloning (NdeI/HindIII) into pRSETmod (Weber,C.C., Link,N., Fux,C., Zisch,A.H., Weber,W. and Fussenegger,M., submitted for publication). Plasmid pWW56 has been described previously (10).
Gel retardation assays
PSCA- and PSPA-encoding probes were generated by PCR using plasmids pWW124 and pWW135 as templates, [32P]dATP and primers OWWF (5′-ggttccgccacatttcccc-3′) and OWWR (5′-aagctgactctagaggatcc-3′). Escherichia coli XL1-Blue (Stratagene, La Jolla, CA) were transformed with pWW314 and grown in LB to an OD600 of 0.6 prior to induction of scbR with 1 mM IPTG for 3 h. Transgenic E.coli were collected by centrifugation and disrupted in TA buffer [10 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1% (w/v) Triton X-100, 50 mM NaCl]. Crude protein extract (10 µg) was incubated with SCB1 for 10 min in the presence of 2 µg of competitor DNA [poly(dI–dC)·(dI–dC)] and then mixed with the probe. Protein–DNA complexes were resolved on 5% (w/v) polyacrylamide-TBE [90 mM Tris, 90 mM borate, 2 mM EDTA (pH 8)] gels, run at room temperature and constant voltage (7 V cm–1) for 2 h, dried and visualized by autoradiography.
Cell culture, transfection, stable cell lines and reporter gene assays
Chinese hamster ovary cells (CHO-K1, ATTC CCL-61) were cultured in FMX-8 complete medium (FMX-8; Cell Culture Technology, Zürich, Switzerland) supplemented with 10% (v/v) FCS and 1% (v/v) of a penicillin/streptomycin solution (cat. no. P4458; Sigma, St Louis, MO), African green monkey kidney cells (COS-7, ATCC CRL-1651), human embryonic kidney cells [HEK293-T (28)], baby hamster kidney cells (BHK-21, ATCC CCL-10) and human hepatocellular carcinoma cells (HepG2, ATCC HB-8065) were cultivated in DMEM-complete medium [DMEM supplemented with 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution]. Human umbilical vein endothelial cells (HUVEC; Promocell, Heidelberg, Germany) and mouse embryonic stem cells (StemCell Technologies, Meylan, France) were cultivated according to the suppliers’ protocols.
Twelve hours prior to transfection, 60 000 CHO-K1 cells were seeded per well of a 24-well plate. Plasmid DNA (1.2 µg per well; if a cotransfection was made, equal amounts of each plasmid were used) in 0.5 M CaCl2 (final volume: 12 µl) was mixed with 12 µl of phosphate solution (50 mM HEPES, pH 7.05, 280 mM NaCl, 1.5 mM Na2HPO4), vortexed for 5 s and incubated for 25 s prior to addition of 0.4 ml of FMX-8 medium containing 2% (v/v) FCS. The culture medium was replaced by the transfection mixture and cells were incubated for 5 h prior to performing a 30 s glycerol shock [0.5 ml of 5% (v/v) glycerol in FMX-8 medium containing 2% (v/v) FCS] and one wash step (1 ml of FMX-8 complete medium). The cells were cultivated in 0.5 ml of FMX-8-complete medium in the presence and absence of the regulating butyrolactone until reporter gene expression was quantified.
For transfection of HEK293-T, COS-7, BHK-21 and HepG2 cells, 120 000 cells were seed per well of a 12-well plate containing 1 ml of DMEM complete medium and grown for 12 h prior to transfection. Total plasmid DNA (0.6 µg) in 50 µl of 0.25 M CaCl2 was added dropwise to 50 µl of 2× HBS (100 mM HEPES, pH 7.1; 280 mM NaCl, 1.5 mM Na2HPO4) while vortexing, followed by addition to the cells. After a 4 h incubation time, the supernatant was removed and the cells were washed once with pre-warmed DMEM complete medium. Subsequently, the cells were cultured in 1 ml of DMEM complete medium per well in the presence or absence of regulating butyrolactone until assessment of reporter gene expression. Mouse ES cells (60 000 cells/well of a 12-well plate) were transfected using FuGENE 6 (Roche Molecular Biochemicals, Rotkreuz, Switzerland) according to the suppliers instructions. The regulating butyrolactone was added 4 h post-FuGENE 6-mediated transfection.
For HUVEC transfection, 300 000 cells were seeded per well of a 6-well plate 12 h prior to transfection. Total plasmid DNA (20 µg) in 150 µl of PBS (10 mM NaH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl) were mixed with 150 µl of DEAE Dextran solution (0.83 µg/µl in PBS; cat. no. D9885; Sigma). The cells were washed twice with PBS and incubated with the transfection mix for 30 min at 37°C in a humidified atmosphere containing 5% (v/v) CO2-containing atmosphere at 37°C. The plate was gently shaken every 10 min. After 30 min, 2 ml of medium (EC medium; Promocell, Heidelberg, Germany) supplemented with 100 µM chloroquine (cat. no. C6628; Sigma) was added to the cells followed by incubation for another 2.5 h. The medium was removed and the cells were treated with 1 ml of 9% (v/v) DMSO in EC medium for 90 s followed by a washing step and incubation in the presence and absence of the regulating butyrolactone for expression profiling of the reporter gene.
The stable SCA production cell line CHO-SCA was constructed by transducing CHO-K1 with pWW158-derived retroviruses. For construction of the double transgenic cell line CHO-SCA-SEAP, CHO-SCA1 was cotransfected with pWW124 and pZeoSV2 (Invitrogen, Carlsbad, CA). Stable cell lines were selected and maintained in culture medium supplemented with appropriate antibiotics (400 µg/ml G418, 100 µg/ml zeocin). Mixed stable cell populations were cloned using FACS-mediated single cell sorting (FACStarPlus; Beckton Dickinson, San Jose, CA). For all analyses CHO-SCA-SEAP was used at a density of 66 000 cells/ml. SEAP and EPO expression were quantified as described previously (12). All data represent average values of three independent transfections/cultures.
In vivo methods
Cells were encapsulated in alginate-poly-(l-lysine)-alginate beads (alginate-PLL-alginate; 200 cells/per capsule, 2 × 106 cells/animal) as described previously and injected intraperitoneally into female OF1 mice (oncins france souche 1; Iffa-Credo, Lyon, France) (10,29) obtained from Iffa-Credo. One hour after capsule implantation, mice were injected with different IM-2-SCB1 doses, diluted in 0.9% (w/v) NaCl from a stock solution of 100 mg/ml in ethanol. Serum SEAP levels were quantified from at least eight mice per dose as described (29, Fux,C., Weber,W. and Fussenegger,M., submitted for publication). All experiments involving animals were approved by the French Ministry of Agriculture and Fishery (Paris, France) and performed by M.D.E. at the Institut Universitaire de Technologie, IUTA, F-69622 Villeurbanne Cedex, France.
RESULTS
Design of a mammalian transcription control system based on Streptomyces quorum-sensing components
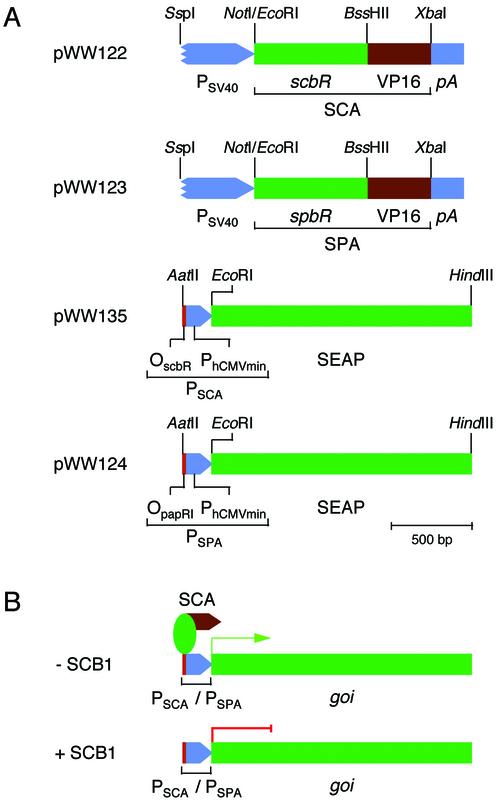
Exploiting molecular design concepts of the butyrolactone quorum-sensing system which evolved in Streptomyces to control antibiotic biosynthesis and/or coordinate morphological differentiation, we have constructed binary mammalian transcription control units called the quorum-sensor-regulated gene expression system (QuoRex). The receptors of the S.coelicolor [ScbR; S.coelicolor butyrolactone transcrip tional repressor; (30)] and S.pristinaespiralis [SpbR; S.pristinaespiralis butyrolactone transcriptional repressor (21)] butyrolactone quorum-sensing systems were each fused to the Herpes simplex virus-1 (HSV-1)-derived mammalian transactivation domain VP16 (31) thus creating the artificial mammalian transactivators SCA (ScbR-VP16) and SPA (SpbR-VP16) (Fig. 2). The SCA/SPA-specific mammalian promoters PSCA/PSPA were assembled by cloning the ScbR-/SpbR-specific operator modules OscbR/OpapRI 5′ of a minimal human cytomegalovirus immediate early promoter (PhCMVmin, Fig. 2). Cotransfection of the SCA/SPA expression vectors with plasmids encoding PSCA- and PSPA-driven human secreted alkaline phosphatase (SEAP) into CHO-K1 cells resulted in high SEAP expression levels (21.7 ± 2.7 U/l for SCA/PSCA-SEAP-pA and 29.6 ± 3.5 U/l for SPA/PSPA-SEAP-pA) which were comparable to those of a PSV40-driven SEAP control construct (pWW56; PSV40-SEAP-pA) (46.8 ± 6.7 U/l). Since the S.coelicolor and S.pristinaespiralis butyrolactone quorum-sensing systems are highly homologous, their derived mammalian transcription control units recognize the same operator sites in CHO-K1 cells: SCA binds and activates transcription from PSPA (85.8 ± 5.8 U/l for SCA/PSPA-SEAP-pA) and SPA promotes PSCA-driven expression (36.0 ± 2.2 U/l for SPA/PSCA-SEAP-pA). These findings demonstrate that the Streptomyces quorum-sensing components ScbR/OscbR and SpbR/OpapRI retain their specific (cross)interaction and form a potent heterologous transcription unit in mammalian cells.
Figure 2.
Design and function of the quorum-sensor-derived mammalian gene regulation system (QuoRex). (A) Diagram of QuoRex’ genetic elements: the butyrolactone-dependent transactivators SCA and SPA were assembled by fusing the S.coelicolor/S.pristinaespiralis quorum-sensing receptors ScbR/SpbR 3′ to the Herpes simplex virus 1 (HSV-1)-derived mammalian transactivation domain. SCA and SPA were cloned into a constitutive mammalian expression configuration (pWW122/pWW123) driven by the simian virus 40 promoter (PSV40) and terminated by an SV40-derived polyadenylation site (pA). SCA/SPA-specific promoters were constructed by cloning the ScbR-/SpbR-specific operator modules (OscbR/OpapRI) adjacent to a minimal version of the human cytomegalovirus immediate early promoter (PhCMVmin). (B) In the absence of regulating butyrolactones (–SCB1), the transactivator (SCA) binds to its cognate PSCA/PSPA promoters and initiates transcription of the cloned gene of interest (goi). However, addition of SCB1 (+SCB1) results in disruption of the SCA-PSCA/PSPA interactions and repression of goi.
Butyrolactone responsiveness of QuoRex-controlled transgene expression in mammalian cells
Expecting that SCA-PSCA, SCA-PSPA, SPA-PSCA and SPA-PSPA interactions could be engineered for adjustable transgene expression in mammalian cells by specific butyrolactone derivatives, we have designed novel/refined protocols for the synthesis of racemic SCB1 which elicits antibiotic production in S.coelicolor (25,30) and MP133, a butenolide first isolated from S.antibioticus (32) (Fig. 1). The enantiomers of SCB1 were obtained as two racemic mixtures, IM-2 type SCB1 and VB type SCB1 (IM-2-SCB1 and VB-SCB1).
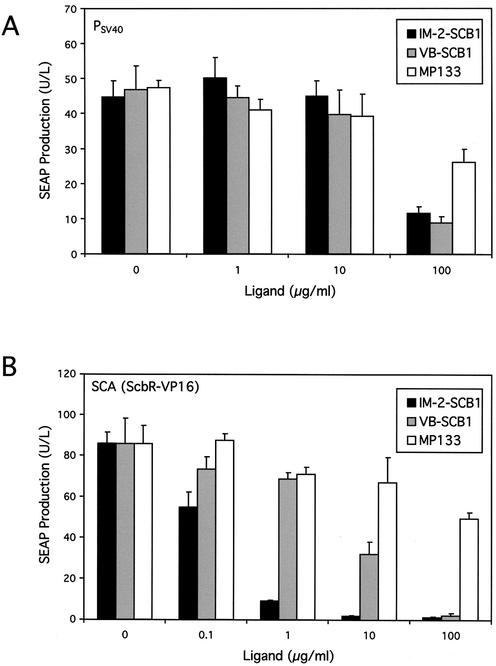
In order to qualify as molecules for controlling transgene expression in mammalian cells, SCB1 and MP133 should not have any undesirable physiological impact on host cells. An indicator for possible toxicity of SCB1 and MP133 is their influence on constitutive reporter gene expression. We have therefore added increasing SCB1 and MP133 concentrations to a CHO-K1 derivative engineered for constitutive SEAP expression. Both butyrolactones showed no effect on SEAP expression at concentrations up to 10 µg/ml. At a concentration of 100 µg/ml, both SCB1 and MP133 were toxic/growth-retarding for mammalian cells as indicated by a depression in SEAP expression levels which was more prominent for both stereoisomers of SCB1 than for MP133 (Fig. 3A).
Figure 3.
Impact of butyrolactone ligands on constitutive and QuoRex- controlled gene expression in CHO-K1. (A) Effect of the SCB1 diastereomers IM-2-SCB1 and VB-SCB1 as well as MP133 on constitutive SEAP expression in CHO-K1. Triplicate CHO-K1 cultures were transfected with a PSV40-driven SEAP expression vector (pWW56) and grown for 48 h at the indicated ligand concentrations prior to quantification of SEAP in the culture medium. (B) Modulation of PSPA-driven SEAP expression in CHO-K1 cells transiently transfected with pWW122 (PSV40-SCA-pA) and pWW124 (PSPA-SEAP-pA) following addition of different IM-2-SCB1, VB-SCB1 and MP133 concentrations. SEAP assays were performed 48 h post-transfection (all assays in triplicate).
When used to adjust transient PSPA-driven SEAP expression in CHO-K1 cells transfected with an SCA expression vector, addition of increasing concentrations of SCB1 correlated with decreasing SEAP production profiles indicating that this butyrolactone releases SCA from PSPA which terminated transcriptional activation of SEAP expression (Fig. 3B). However, the structurally related MP133 was unable to modulate SEAP production significantly within its non-toxic concentration range. Interestingly, IM-2-SCB1 was more efficient than VB-SCB1 in changing SCA’s allostery to prevent PSPA binding. This was illustrated by a progressive IM-2-SCB1-mediated decrease of SEAP activity reaching basal expression at 10 µg/ml corresponding to 1.9% of fully induced expression. In contrast, addition of VB-SCB1 reduced expression of this human glycoprotein only to half-maximum levels at this concentration (Fig. 3B). The remarkable stereoselective discrimination of SCA between the two SCB1 diastereomers emphasizes the specificity of quorum-sensing phenomena. IM-2-SCB1 retained its excellent regulation performance in an isogenic QuoRex-controlled erythropoietin expression setting (PSPA-EPO-pA; –IM-2-SCB1: 690 ± 28 mIU/ml; +IM-2-SCB1: 23.7 ± 5.2 mIU/ml).
In contrast to SCA-dependent transactivation of target promoters, neither SCB1 nor MP133 were able to disrupt SPA-PSPA or SPA-PSCA interactions suggesting that SCB1 is specific for SCA (in vitro and in vivo; data not shown). Thus, SPA awaits isolation and characterization of SpbR-specific butyrolactones before implementation of the S.pristinaespiralis-derived mammalian gene regulation system can proceed (20).
DNA-binding specificity of SCA
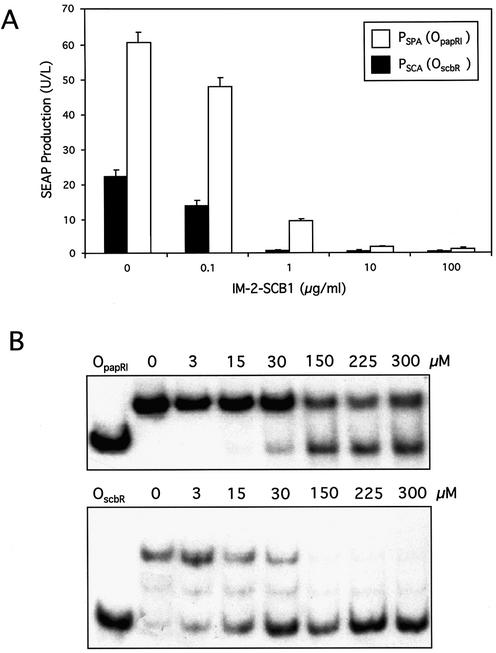
The regulatory activity of SCA for promoters PSCA and PSPA was analyzed following cotransfection of pWW122 (PSV40-SCA-pA) with either pWW135 (PSCA-SEAP-pA) or pWW124 (PSPA-SEAP-pA) into CHO-K1. Surprisingly, SCA activated the promoter containing the heterologous operator sequence (OpapRI) more efficiently than the same promoter with its cognate operator (OscbR). At all IM-2-SCB1 concentrations tested (0–100 µg/ml), the SCA-PSCA configuration only reached approximately one-third of the SEAP expression levels compared to the heterologous SCA-PSPA combination (Fig. 4A).
Figure 4.
Specificity and affinity of SCA for S.coelicolor/S.pristinaespiralis OscbR/OpapRI operator modules. (A) SCA-dependent SEAP expression levels driven by PSPA/PSCA promoters in CHO-K1 cells in the presence of different IM-2-SCB1 concentrations. CHO-K1 cells were cotransfected with the SCA expression plasmid pWW122 (PSV40-SCA-pA) and the SCA-dependent SEAP expression vector pWW124/pWW135 (PSPA-SEAP-pA/PSCA-SEAP-pA). Transfected cell lines were grown for 48 h in medium containing the indicated IM-2-SCB1 concentrations prior to quantification of SEAP expression levels. (B) Gel retardation assays showing interactions of ScbR with OscbR and ScbR with OpapRI. The ScbR and radioactively labeled OscbR and OpapRI protein–DNA complexes were incubated with various IM-2-SCB1 concentrations and resolved on a polyacrylamide gel. ScbR is released from OscbR at lower IM-2-SCB1 concentrations compared to OpapRI indicating higher affinity of this S.coelicolor quorum-sensing receptor for S.pristinaespiralis OpapRI than for its autologous OscbR.
In order to investigate whether variations in expression levels were associated with different affinities of SCA for PSCA and PSPA, in vitro-formed SCA-PSCA and SCA-PSPA complexes incubated at increasing IM-2-SCB1 concentrations were resolved on polyacrylamide gels. These gel retardation assays provided biochemical evidence that SCA did indeed have higher affinity for the heterologous PSPA than for PSCA which apparently renders the SCA-PSPA configuration more effective for controlling transgene expression in mammalian cells (Fig. 4B).
Detailed expression analysis of the SCA-PSPA-based QuoRex system
The performance of the SCA-PSPA-based QuoRex system was evaluated in a variety of mammalian cell types including human primary cells and mouse embryonic stem cells. To this end, we cotransfected the SCA expression vector pWW122 (PSV40-SCA-pA) and the SCA-responsive SEAP expression vector pWW124 (PSPA-SEAP-pA) into different cell lines and assayed the SEAP levels in their culture supernatants following cultivation for 48 h in the presence and absence of IM-2-SCB1. In all cell lines tested, high SEAP expression could be monitored in the absence of IM-2-SCB1 whereas complete repression was achieved following addition of this butyrolactone (Table 1).
Table 1. Expression characteristics of SCA/PSPA-driven secreted alkaline phosphatase in different cell lines in the presence and absence of IM-2-SCB1.
| Cell line | –IM-2-SCB1 | +IM-2-SCB1 |
|---|---|---|
| CHO-K1 | 60.4 ± 8.3 | 1.81 ± 0.17 |
| BHK-21 | 5.0 ± 0.4 | 0.23 ± 0.04 |
| COS-7 | 7.5 ± 0.5 | 0.14 ± 0.04 |
| HEK 293-T | 228.0 ± 7.6 | 33.5 ± 0.11 |
| Hep G2 | 2.1 ± 0.1 | 0.096 ± 0.035 |
| HUVEC | 0.5 ± 0.08 | 0.0001 ± 0.0001 |
| Mouse ES | 1.0 ± 0.02 | 0.027 ± 0.007 |
Cells were transfected with plasmids pWW122 (PSV40-SCA-pA) and pWW124 (PSPA-SEAP-pA) and grown for 48 h in the presence (10 µg/ml) and absence of IM-2-SCB1 before scoring of SEAP production (in U/l).
We also evaluated QuoRex’s key regulation characteristics (dose-dependence, expression kinetics and reversibility) in a stable expression configuration. QuoRex was engineered into CHO-K1 cells following a two step procedure. The SCA expression unit was transduced using a transgenic retrovirus (5′LTR-ψ+-SCA-PPGK-neoR-3′LTR) and individual CHO-K1-derived cell clones were assessed for QuoRex performance after transient transfection with the PSPA-driven SEAP expression construct pWW124. The regulation performance of four independent CHO-SCA clones in the presence and absence of IM-2-SCB1 is shown in Table 2. Because of its balanced expression profiles combining high maximum SEAP expression levels as well as tight repression, we chose CHO-SCA35 for further studies. The PSPA-driven SEAP expression unit was stably integrated into CHO-SCA35 and individual clones were scored for their regulation performance in the presence and absence of IM-2-SCB1. Table 2 shows four different double transgenic clones of which CHO-SCA35-SEAP16 was chosen for further analysis.
Table 2. Expression characteristics of stable CHO-SCA and CHO-SCA-SEAP clones in the presence and absence of IM-2-SCB1.
| CHO-SCA | ||||
| Clone no. | 15 | 16 | 35 | 38 |
| −IM-2-SCB1 | 8.8 ± 2.9 | 6.1 ± 0.6 | 6.8 ± 0.3 | 6.7 ± 1.0 |
| +IM-2-SCB1 | 0.13 ± 0.02 | 0.10 ± 0.12 | 0.15 ± 0.08 | 0.15 ± 0.03 |
| CHO-SCA-SEAP | ||||
| Clone no. | 10 | 16 | 63 | 109 |
| –IM-2-SCB1 | 8.4 ± 0.4 | 5.2 ± 0.2 | 3.0 ± 0.1 | 13.7 ± 0.3 |
| +IM-2-SCB1 | 0.67 ± 0.06 | 0.09 ± 0.02 | 0.17 ± 0.15 | 0.17 ± 0.17 |
CHO-SCA cell clones were transfected with pWW124 (PSPA-SEAP-pA) and grown for 48 h in the presence and absence of IM-2-SCB1 before measuring SEAP activity (in U/l). CHO-SCA-SEAP cell clones were grown for 48 h with (10 µg/ml) and without IM-2-SCB1 before SEAP assays.
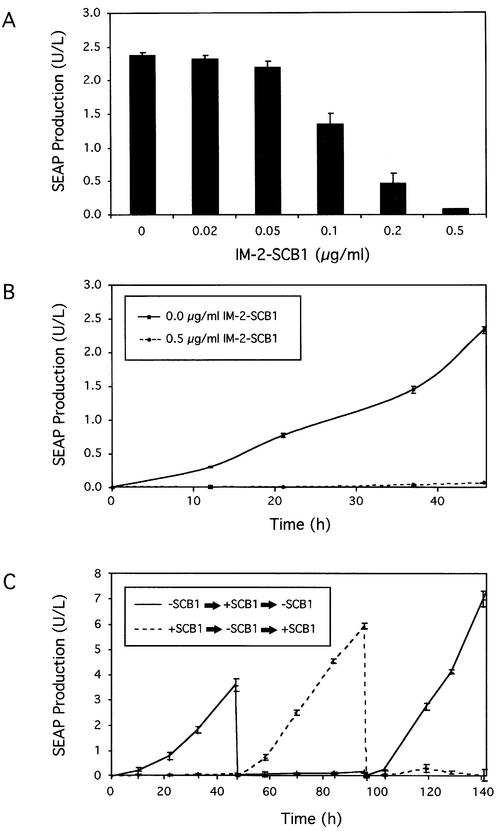
Upon exposure of CHO-SCA35-SEAP16 to increasing IM-2-SCB1 concentrations, SEAP expression could be adjusted to intermediate levels until it was completely repressed to detection limits when the butyrolactone concentration reached 0.5 µg/ml (Fig. 5A). Interestingly, the sensitivity of CHO-SCA35-SEAP16 to IM-2-SCB1 was one order of magnitude higher compared to transient transfections, a phenomenon which has previously been reported for double-transgenic cell lines harboring the tetracycline-response gene regulation system (33).
Figure 5.
Regulation characteristics of CHO-SCA35-SEAP16 transgenic for QuoRex-controlled SEAP expression. (A) IM-2-SCB1-adjustable SEAP production. CHO-SCA35-SEAP16 cells (66 000 cells/ml) were cultivated for 48 h at indicated IM-2-SCB1 concentrations prior to quantification of SEAP production. (B) Expression kinetics of CHO-SCA35-SEAP16 cells cultivated in the presence and absence of IM-2-SCB1 for 48 h. (C) Reversibility of QuoRex-controlled transgene expression. Two cultures (66 × 106 CHO-SCA35-SEAP16 cells/ml) were grown initially in the presence and absence of IM-2-SCB1. At 48 h intervals the IM-2-SCB1 status was reversed (–IM-2-SCB1 → +IM-2-SCB1 → –IM-2-SCB1; +IM-2-SCB1 → –IM-2-SCB1 → +IM-2-SCB1). SEAP production was monitored during the entire cultivation.
SEAP expression kinetics of CHO-SCA35-SEAP16 grown in the presence of 0.5 µg/ml IM-2-SCB1 or in the absence of this regulating butyrolactone were monitored for 48 h (Fig. 5B). Despite its excellent regulation profiles, high expression levels in the absence of IM-2-SCB1 and tight repression in its presence, the QuoRex system proved to be completely reversible following repeated cycles of addition and withdrawal of IM-2-SCB1 from the culture for a period of over 140 h (Fig. 5C).
In vivo quorum-sensing-responsive gene expression
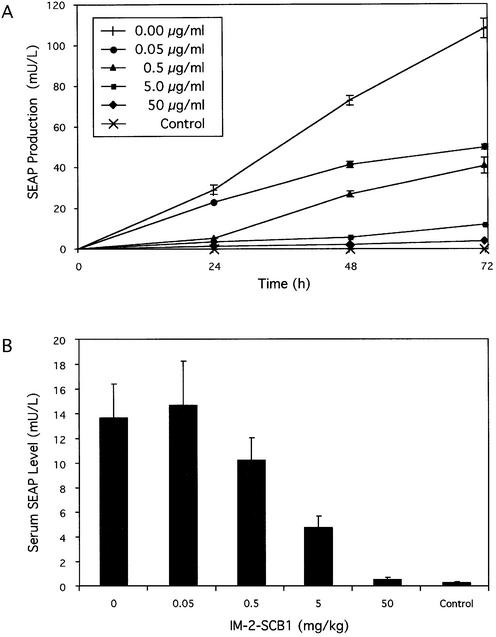
Following successful validation of QuoRex-controlled gene expression in mammalian cells, we assessed its potential for adjustable transgene expression in vivo. Mimicking current gene therapy scenarios, we implanted CHO-SCA35-SEAP16 cells, encapsulated in biocompatible alginate-PLL-alginate beads, intraperitoneally into immunocompetent mice. Microencapsulation protects implanted cells from the mouse immune system and prevents any inflammatory response as well as the elimination of transgenic cells. Microencapsulated cells remained responsive to IM-2-SCB1 as this butyrolactone can diffuse through the alginate-PLL-alginate layers and SEAP, a model for therapeutic proteins, was secreted across the capsule (Fig. 6A). Following intraperitoneal implantation of the same in vitro-validated cell batch, the mice received different IM-2-SCB1 doses by injection (0–50 mg/kg). Serum SEAP levels of treated mice were quantified 72 h post-implantation. IM-2-SCB1-adjustable SEAP production including full repression to undetectable serum levels was reached in these animals at butyrolactone concentrations which showed no undesired effects on mouse physiology or behavior suggesting that quorum-sensing molecules were safe for in vivo applications. Control mice harboring non-engineered CHO-K1 cells did not contain elevated serum SEAP levels in the presence and absence of IM-2-SCB1. IM-2-SCB1-responsive SEAP expression profiles in mice resembled those observed in vitro with the same batch of microencapsulated cells, thereby confirming the in vivo compatibility of the QuoRex system (Fig. 6).
Figure 6.
Butyrolactone-responsive SEAP expression from microencapsulated CHO-K1 cell derivatives in culture and in mice. (A) IM-2-SCB1-responsive SEAP expression from encapsulated cells transgenic for QuoRex-controlled SEAP expression. CHO-SCA35-SEAP16 or CHO-K1 control cells (2 × 106 cells per culture) were microencapsulated in alginate-poly-l-lysine-alginate microcapsules (400 µm in diameter, 200 cells/ capsule) and cultivated in medium supplemented with different IM-2-SCB1 concentrations and analyzed for SEAP production after 72 h. (B) SCB1-adjustable SEAP expression in mice. The same microencapsulated batch of CHO-SCA35-SEAP16 and CHO-K1 control cells as used for in vitro cultivation [2 × 106 cells, 200 cells/capsule; see details in (A)] was implanted intraperitoneally into immunocompetent mice. At 1 h post-implantation the mice received IM-2-SCB1 injections at indicated doses. Blood samples were collected after 72 h and quantified for SEAP levels.
DISCUSSION
Pharmacological control of transgene expression is a current priority in therapeutic engineering of mammalian cells to titrate levels of desired protein pharmaceuticals in gene therapy scenarios, or to reprogram preferred cell phenotypes for tissue engineering (6,10,34–38).
Quorum-sensing systems enabling coordination of gene expression within and between prokaryotic populations represent a vast reservoir of small-molecule response regulators, which could be adopted for use as heterologous mammalian gene regulation systems (19,22). As proof of principle we converted a butyrolactone quorum-sensing regulator, which controls antibiotics biosynthesis in S.coelicolor and S.pristinaespiralis, for use as gene control systems in mammalian cells and mice (21,25). ScbR and SpbR, as well as their cognate operators OscbR and OpapRI, are highly homologous and recognize similar DNA sequences (20). Interestingly, ScbR/SCA had stronger affinities for heterologous OpapRI/PSPA than for their cognate OscbR/PSCA modules which is reflected in higher maximum expression levels of the SCA-PSPA configuration compared to the sister SCA-PSCA set-up. In contrast to the flexibility in sequence recognition, the interaction of these quorum-sensing receptors with regulating butyrolactones was highly specific. SCB1 exclusively interacts with ScbR and ScbR-derived mammalian transactivators but not with SpbR derivatives. Furthermore, SCA discriminates between the SCB1 diastereomers IM-2-SCB1 and VB-SCB1 since IM-2-SCB1 enables fine-tuning of transgene expression in mammalian cells with a lower concentration window than VB-SCB1.
An important characteristic of the QuoRex system is its in vivo compatibility showing negligible basal transcriptional activity and IM-2-SCB1-adjustable target gene expression in mice. Pioneering in vivo use of SCB1 suggested that this type of butyrolactone is well tolerated in animals when administered at regulating dosing regimes.
The prototype QuoRex system provides the first evidence that molecular interactions enabling bacteria to communicate as multicellular systems can be successfully transferred to a multicellular organism to fine-tune heterologous transgene expression to desired levels. There are many bacterial quorum-sensing systems, all consisting of protein receptors, their cognate butyrolactones and operator sequences. Thus, QuoRex represents a vast potential for development of a new generation of small molecule-adjustable gene control systems for clinical application (39).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Zhi Li and Dongliang Chang for HPLC-MS analysis, Eva Niederer (Institute for Biomedical Engineering, ETH Zurich) for FACS sorting and Beat Kramer for critical comments on the manuscript. This work was supported by the Swiss National Science Foundation (grant 631-065946) and Cistronics Cell Technology GmbH, Einsteinstrasse 1-5, POB 145, CH-8093 Zurich, Switzerland.
REFERENCES
- 1.Fussenegger M. (2001) The impact of mammalian gene regulation concepts on functional genomic research, metabolic engineering and advanced gene therapies. Biotechnol. Prog., 17, 1–51. [DOI] [PubMed] [Google Scholar]
- 2.Bohl D., Naffakh,N. and Heard,J.M. (1997) Long-term control of erythropoietin secretion by doxycycline in mice transplanted with engineered primary myoblasts. Nature Med., 3, 299–305. [DOI] [PubMed] [Google Scholar]
- 3.Aubel D., Morris,R., Lennon,B., Rimann,M., Kaufmann,H., Folcher,M., Bailey,J.E., Thompson,C.J. and Fussenegger,M. (2001) Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot. (Tokyo), 54, 44–55. [DOI] [PubMed] [Google Scholar]
- 4.Fussenegger M., Schlatter,S., Datwyler,D., Mazur,X. and Bailey,J.E. (1998) Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol., 16, 468–472. [DOI] [PubMed] [Google Scholar]
- 5.Umana P., Jean-Mairet,J., Moudry,R., Amstutz,H. and Bailey,J.E. (1999) Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat. Biotechnol., 17, 176–180. [DOI] [PubMed] [Google Scholar]
- 6.Malleret G., Haditsch,U., Genoux,D., Jones,M.W., Bliss,T.V., Vanhoose,A.M., Weitlauf,C., Kandel,E.R., Winder,D.G. and Mansuy,I.M. (2001) Inducible and reversible enhancement of learning, memory and long-term potentiation by genetic inhibition of calcineurin. Cell, 104, 675–686. [DOI] [PubMed] [Google Scholar]
- 7.Ewald D., Li,M., Efrat,S., Auer,G., Wall,R.J., Furth,P.A. and Hennighausen,L. (1996) Time-sensitive reversal of hyperplasia in transgenic mice expressing SV40 T antigen. Science, 273, 1384–1386. [DOI] [PubMed] [Google Scholar]
- 8.Chin L., Tam,A., Pomerantz,J., Wong,M., Holash,J., Bardeesy,N., Shen,Q., O’Hagan,R., Pantginis,J., Zhou,H. et al. (1999) Essential role for oncogenic Ras in tumour maintenance. Nature, 400, 468–472. [DOI] [PubMed] [Google Scholar]
- 9.Weber W. and Fussenegger,M. (2002) Artificial mammalian gene regulation networks-novel approaches for gene therapy and bioengineering. J. Biotechnol., 98, 161–187. [DOI] [PubMed] [Google Scholar]
- 10.Weber W., Fux,C., Daoud-el Baba,M., Keller,B., Weber,C.C., Kramer,B.P., Heinzen,C., Aubel,D., Bailey,J.E. and Fussenegger,M. (2002) Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol., 20, 901–907. [DOI] [PubMed] [Google Scholar]
- 11.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussenegger M., Morris,R.P., Fux,C., Rimann,M., von Stockar,B., Thompson,C.J. and Bailey,J.E. (2000) Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol., 18, 1203–1208. [DOI] [PubMed] [Google Scholar]
- 13.Rivera V.M., Clackson,T., Natesan,S., Pollock,R., Amara,J.F., Keenan,T., Magari,S.R., Phillips,T., Courage,N.L., Cerasoli,F.,Jr et al. (1996) A humanized system for pharmacologic control of gene expression. Nature Med., 2, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 14.Wang X.J., Liefer,K.M., Tsai,S., O’Malley,B.W. and Roop,D.R. (1999) Development of gene-switch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc. Natl Acad. Sci. USA, 96, 8483–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., O’Malley,B.W.,Jr, Tsai,S.Y. and O’Malley,B.W. (1994) A regulatory system for use in gene transfer. Proc. Natl Acad. Sci. USA, 91, 8180–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapunisk-Uner J.E., Sande,M.A. and Chambers,H.F.S. (1996) Tetracycline, Chloramphenicol, Erythromycin and Miscellaneous Antibacterial Agents, 9th Edn. McGraw-Hill, New York. [Google Scholar]
- 17.Cohlan S.Q. (1977) Tetracycline staining of teeth. Teratology, 15, 127–129. [DOI] [PubMed] [Google Scholar]
- 18.Iuliucci J.D., Oliver,S.D., Morley,S., Ward,C., Ward,J., Dalgarno,D., Clackson,T. and Berger,H.J. (2001) Intravenous safety and pharmacokinetics of a novel dimerizer drug, AP1903, in healthy volunteers. J. Clin. Pharmacol., 41, 870–879. [DOI] [PubMed] [Google Scholar]
- 19.Miller M.B. and Bassler,B.L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol., 55, 165–199. [DOI] [PubMed] [Google Scholar]
- 20.Folcher M., Gaillard,H., Nguyen,L.T., Nguyen,K.T., Lacroix,P., Bamas-Jacques,N., Rinkel,M. and Thompson,C.J. (2001) Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis and expression of a superoxide dismutase. J. Biol. Chem., 276, 44297–44306. [DOI] [PubMed] [Google Scholar]
- 21.Folcher M., Morris,R.P., Dale,G., Salah-Bey-Hocini,K., Viollier,P.H. and Thompson,C.J. (2001) A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem., 276, 1479–1485. [DOI] [PubMed] [Google Scholar]
- 22.Fuqua C., Parsek,M.R. and Greenberg,E.P. (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet., 35, 439–468. [DOI] [PubMed] [Google Scholar]
- 23.Payet P. and Grob,C.A. (1954) N-Methyl-3-piperidon-5-carbonsäure. Helv. Chim. Acta, 37, 1266–1272. [Google Scholar]
- 24.Yamada Y., Sugamura,K., Kondo,K., Yanagimoto,M. and Okada,H. (1987) The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. (Tokyo), 40, 496–504. [DOI] [PubMed] [Google Scholar]
- 25.Takano E., Nihira,T., Hara,Y., Jones,J.J., Gershater,C.J., Yamada,Y. and Bibb,M. (2000) Purification and structural determination of SCB1, a gamma-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem., 275, 11010–11016. [DOI] [PubMed] [Google Scholar]
- 26.Demnitz F.W.J. (1989) The Mukaiyama reaction of ketene bis(trimethylsilyl) acetals with -halo acetals: a convenient butenolide synthesis. Tetrahedron Lett., 30, 6109–6112. [Google Scholar]
- 27.Grossmann G. and Séquin,U. (2001) Synthetic access to biologically active butenolides from Streptomyces antibioticus. Synlett, 2, 278–280. [Google Scholar]
- 28.Aiello L., Guilfoyle,R., Huebner,K. and Weinmann,R. (1979) Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology, 94, 460–469. [DOI] [PubMed] [Google Scholar]
- 29.Weber W. and Fussenegger,M. (2002) In Balbas,P. and Laurence,A. (eds), Recombinant Protein Protocols, 2nd Edn. Humana Press, New Jersey, in press. [Google Scholar]
- 30.Takano E., Chakraburtty,R., Nihira,T., Yamada,Y. and Bibb,M.J. (2001) A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol., 41, 1015–1028. [DOI] [PubMed] [Google Scholar]
- 31.Triezenberg S.J., Kingsbury,R.C. and McKnight,S.L. (1988) Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev., 2, 718–729. [DOI] [PubMed] [Google Scholar]
- 32.Braun D., Pauli,N., Sequin,U. and Zahner,H. (1995) New butenolides from the photoconductivity screening of Streptomyces antibioticus (Waksman and Woodruff) Waksman and Henrici 1948. FEMS Microbiol. Lett., 126, 37–42. [DOI] [PubMed] [Google Scholar]
- 33.Baron U., Gossen,M. and Bujard,H. (1997) Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res., 25, 2723–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auricchio A., Rivera,V.M., Clackson,T., O’Connor,E.E., Maguire,A.M., Tolentino,M.J., Bennett,J. and Wilson,J.M. (2002) Pharmacological regulation of protein expression from adeno-associated viral vectors in the eye. Mol. Ther., 6, 238–242. [DOI] [PubMed] [Google Scholar]
- 35.Favre D., Blouin,V., Provost,N., Spisek,R., Porrot,F., Bohl,D., Marme,F., Cherel,Y., Salvetti,A., Hurtrel,B. et al. (2002) Lack of an immune response against the tetracycline-dependent transactivator correlates with long-term doxycycline-regulated transgene expression in nonhuman primates after intramuscular injection of recombinant adeno-associated virus. J. Virol., 76, 11605–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa H., Miyazaki,J. and Smith,A.G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genet., 24, 372–376. [DOI] [PubMed] [Google Scholar]
- 37.Pollock R., Giel,M., Linher,K. and Clackson,T. (2002) Regulation of endogenous gene expression with a small-molecule dimerizer. Nat. Biotechnol., 20, 729–733. [DOI] [PubMed] [Google Scholar]
- 38.Weber W., Marty,R.R., Keller,B., Rimann,M., Kramer,B.P. and Fussenegger,M. (2002) Versatile macrolide-responsive mammalian expression vectors for multiregulated multigene metabolic engineering. Biotechnol. Bioeng., 80, 691–705. [DOI] [PubMed] [Google Scholar]
- 39.Allison A.C. (2000) Immunosuppressive drugs: the first 50 years and a glance forward. Immunopharmacology, 47, 63–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.