Abstract
Real-time PCR is being used increasingly as the method of choice for mRNA quantification, allowing rapid analysis of gene expression from low quantities of starting template. Despite a wide range of approaches, the same principles underlie all data analysis, with standard approaches broadly classified as either absolute or relative. In this study we use a variety of absolute and relative approaches of data analysis to investigate nocturnal c-fos expression in wild-type and retinally degenerate mice. In addition, we apply a simple algorithm to calculate the amplification efficiency of every sample from its amplification profile. We confirm that nocturnal c-fos expression in the rodent eye originates from the photoreceptor layer, with around a 5-fold reduction in nocturnal c-fos expression in mice lacking rods and cones. Furthermore, we illustrate that differences in the results obtained from absolute and relative approaches are underpinned by differences in the calculated PCR efficiency. By calculating the amplification efficiency from the samples under analysis, comparable results may be obtained without the need for standard curves. We have automated this method to provide a means of streamlining the real-time PCR process, enabling analysis of experimental samples based upon their own reaction kinetics rather than those of artificial standards.
INTRODUCTION
The study of gene function requires the ability to accurately quantify temporal and spatial patterns of gene expression and, given recent genomic advances, this requirement has become even more essential. Traditional approaches, such as northern blots and RNase protection assays are in many cases unsuitable, as their low sensitivity necessitates high concentrations of starting template (1,2). The advent of real-time PCR has enabled rapid and reproducible high throughput RT–PCR quantification, with an unparalleled dynamic range and extremely high sensitivity. As such, real-time PCR is fast becoming the method of choice for the quantification of gene expression, and is often recommended for the validation of microarray data (3–6). The speed with which this technique has been adopted has led to a range of potential instrumentation with a corresponding variety of fluorescent chemistries (2,4). However, the same fundamental concepts underlie all approaches to gene quantification by fluorescent real-time PCR. Firstly, the principle that accumulation of fluorescence is proportional to accumulation of amplification products underpins the whole concept of quantitative PCR (7). Secondly, the amplification efficiency must be comparable in all samples. A difference of 5% in amplification efficiency between two initially equal samples can result in one sample having twice as much product after 26 cycles of PCR (3). Finally, the threshold used for analysis must be within the linear phase of all the reactions, to ensure that the threshold cycle (Ct) is truly representative of initial template differences and not just a change in reaction kinetics.
Real-time PCR data analysis methods may be broadly classified as ‘absolute’ or ‘relative’ (8). Absolute quantification involves the construction of a standard curve based upon known copy numbers, whereas relative approaches involve determining the change in expression level relative to another set of experimental samples, typically the experimental control group (8). Whilst determining exact copy number is intuitively appealing, the generation of stable and reliable standards is both time-consuming and requires precise quantification (9). Moreover, the use of readily available nucleic acids such as plasmids introduces considerable risks of contamination. For most research applications a relative approach to quantification is more practical as it compares experimental samples against controls directly.
Original methods of relative quantification were based upon assuming an ideal amplification efficiency with a doubling of product every cycle, allowing the fold change to be calculated using the formula 2–ΔΔCt (8,10). This approach was refined to include the amplification efficiency of target and internal control genes as calculated from cDNA standard curves (9). An alternative method, requiring no standard curve, determines amplification efficiency from the actual slope of the amplification plot (11). The major problem facing this approach is that amplification efficiency changes throughout the PCR, with efficiency declining in later cycles as amplification products compete for DNA polymerase binding (12).
We have developed a simple algorithm for calculating the amplification efficiency from every sample within a real-time PCR assay and have furthermore automated this calculation to allow rapid data analysis and calculation of the sources of assay variability. To test the validity of this novel method against conventional absolute and relative approaches to quantitative real-time PCR we compared the ocular expression of the immediate early gene c-fos in wild-type and retinally degenerate rd/rd cl animals (13,14). In darkness, retinal photoreceptors are the major source of ocular c-fos expression (15,16) and the loss of photoreceptors in the rd/rd cl retina would therefore be expected to result in a major attenuation of nocturnal c-fos expression. Standard curves were included in the assay to allow a comparison of five different approaches to real-time PCR data analysis: (i) using plasmid DNA standard curves to determine copy numbers; (ii) cDNA standard curves; (iii) the 2–ΔΔCt method and a novel approach deriving amplification efficiency from amplification plots, applied as either (iv) mean amplification efficiency or (v) as individual corrections.
MATERIALS AND METHODS
Tissue preparation, RNA extraction and reverse transcription
Wild-type and rd/rd cl mice (n = 4 each) were housed under a 12:12 h light:dark cycle for 3 weeks and killed at zeitgeber time 14 (2 h after lights off) according to Schedule 1 of the Animals (Scientific Research) Act. Eyes were enucleated in darkness using an infrared viewer and immediately placed in 0.5 ml of RNAlater™ (Ambion). Paired whole eyes were homogenised in 0.5 ml of TriReagent (Sigma Aldrich) using Fastprep tubes in a FastPrep FP 120 (Q-Biogene). Total RNA was then extracted in TriReagent according to the manufacturer’s instructions. RNA was resuspended at 60°C in 20 µl of RNA Secure (Ambion). RNA concentration was determined by spectrophotometry using an Eppendorf Biophotometer. An aliquot of 1 µg of total RNA was then treated with 2 U RNase-free DNase (Sigma Aldrich) for 30 min at 37°C to remove any traces of genomic DNA. DNase-treated RNA was reverse transcribed with random decamers using a RetroScript kit (Ambion), according to the manufacturer’s instructions. Once synthesised, cDNA fidelity was tested by PCR and samples were then stored at –20°C.
Real-time PCR assay
Primers for c-fos and β-actin were designed using MacVector software (Accelrys, UK) and tested to ensure amplification of single discrete bands with no primer-dimers. Primer sequences were as follows: c-fos forward, 5′-ATCGGCAGAAGGG GCAAAGTAG-3′; c-fos reverse, 5′-GCAACGCAGACTT CTCATCTTCAAG-3′ (174 bp product, spanning a 522 bp intron); β-actin forward, 5′-ACCAACTGGGACGATAT GGAGAAGA-3′; β-actin reverse, 5′-CGCACGATTTCCC TCTCAGC-3′ (403 bp product). All primers were synthesised by Sigma Genosys. PCR products for c-fos and β-actin were ligated into pGEM-T Easy vector (Promega) and transformed in DH5α competent cells (Invitrogen). Minipreps of isolated plasmid DNA were then prepared (Promega). Before use, plasmid concentration was determined by spectrophotometry using an Eppendorf BioPhotometer and serial dilutions were performed to give final concentrations between 102 and 105 (c-fos) or 103 and 106 (β-actin) copies. Standard curves of cDNA were composed of three 10-fold dilutions of wild-type ocular cDNA. Real-time PCR was conducted using Sybr Green I Mastermix (Applied Biosystems) using an ABI PRISM™ 7700 Sequence Detection System. Each reaction was run in triplicate and contained 1 µl of cDNA template along with 300 (c-fos) or 50 (β-actin) nM primers in a final reaction volume of 25 µl. Cycling parameters were 95°C for 10 min to activate DNA polymerase, then 40 cycles of 95°C for 15 s and 60°C for 1 min, with a final recording step of 78°C for 20 s to prevent any primer-dimer formation. Melting curves were performed using Dissociation Curves software (Applied Biosystems) to ensure only a single product was amplified, and samples were also run on a 3% agarose gel to confirm specificity.
Data analysis
Data were analysed initially using SDS 1.7 (Applied Biosystems). For absolute copy numbers, standard curves were plotted in SDS 1.7, and also from exported data with identical results obtained by performing linear regression of log concentration against threshold cycle (Ct).
The simplest method of accurate relative quantification is by calculating the theoretical value R0. This is based upon the simple formula used to simulate a PCR up to the point of plateau:
Xn = X0 × (1 + E)n1
where Xn is the concentration of template at cycle n, X0 is the starting template concentration and E is the amplification efficiency, having a value of 1 when exact doubling of product occurs with every additional cycle and 0 when no increase in product occurs (8). As the basic concept underlying real-time PCR is that accumulation of fluorescence is proportional to accumulation of amplification product, the above equation may be reformulated to give the starting fluorescence (R0), which is proportional to the starting template quantity:
R0 = RCt × (1 + E)–Ct2
where Ct is the threshold cycle and RCt is the fluorescence at this cycle (the actual threshold used for analysis). As the desired blunt-ended fragments first appear in the third cycle of PCR (1), the efficiency component should more accurately be E–(Ct – 2), but when conducting relative quantification this discrepancy is cancelled out.
Amplification plot method
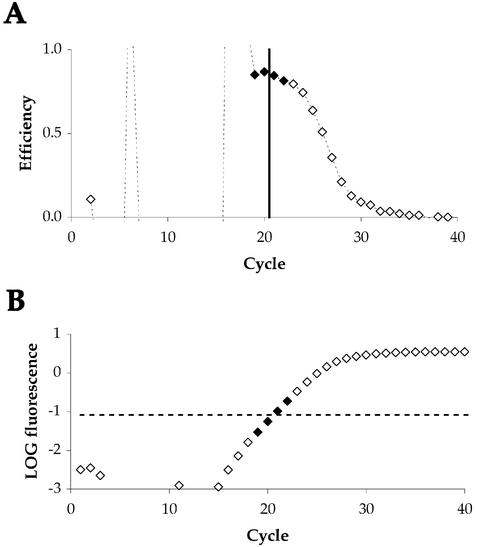
The amplification plot produced during real-time PCR can be used to determine the amplification efficiency by analysing the change of fluorescence throughout the linear phase. A novel model was applied based upon a linear regression to defined cycles of exponential amplification, without modelling saturation kinetics. The fluorescence maximum (Rmax) of each plot was determined and the background noise of the sample (Rnoise) was calculated from the standard deviation of cycles 1–10, to reflect the background level of fluorescence prior to detectable amplification. This provides the signal range, in which the amplification rate can be determined most accurately around the midpoint. The midpoint (M) of the transformed signal range can then be determined from:
M = Rnoise × √(Rmax/Rnoise)3
This is illustrated in Figure 1B. Using a 10-fold range around this midpoint, a minimum of three cycles within the linear phase will be utilised. This range may be extended, although an eventual decline in efficiency will occur due to the inclusion of cycles where the amplification rate is declining or influenced by background noise (see Fig. 1A). Using a linear regression to calculate the slope of log fluorescence around M yields a very good fit (R2 > 0.99) and inaccuracies introduced by low signals and reaction saturation are limited. As shown in Figure 1A, the amplification efficiency is highest around this midpoint. Amplification efficiency can then be calculated from the slope:
Figure 1.
Determination of amplification efficiency from the linear phase of real-time PCR data. (A) Illustration of decline in amplification efficiency throughout a sample PCR reaction. Efficiency is plotted as rate of change in fluorescence on a cycle-to-cycle basis [(RCt/RCt – 1) – 1]. The cycle at which fluorescence equals M is indicated by a solid black line. Filled symbols represent a 10-fold range around this midpoint. (B) Illustration of the same sample data plotted as fluorescence on a logarithmic scale against cycle number. The midpoint of the detectable linear phase is shown as a broken black line. Filled symbols again represent the range around the midpoint used to calculate amplification efficiency.
Efficiency = 10(1/slope) – 14
Figure 1B illustrates an amplification plot for β-actin analysed using this method, with the midpoint and selected cycles indicated.
Data handling
Data handling was simplified by automating all calculations in an Excel workbook (Microsoft), entitled Data Analysis for Real-Time PCR (DART-PCR). This workbook enables the rapid calculation of threshold cycles, amplification efficiency and resulting R0 values (along with the associated error) from raw data exported from SDS 1.7. Differences in amplification efficiency are assessed using one-way analysis of variance (ANOVA), based upon the null hypotheses: (i) that amplification efficiency is comparable within sample groups (outlier detection); (ii) that amplification efficiency is comparable between sample groups (amplification equivalence).
Differences between normalised wild-type and rd/rd cl c-fos expression were assessed using a two-tailed Student’s t-test assuming unequal variance.
RESULTS
Absolute quantification
Absolute quantification was conducted using copy numbers derived from plasmid standards. The term ‘absolute’ is perhaps misleading as the copy numbers derived are relative to the standard used.
β-Actin levels in wild-type and rd/rd cl eyes were determined to be highly comparable, with around 409 186 copies in wild-type and 421 917 copies in rd/rd cl. The comparable levels of β-actin expression given equivalent RNA loading demonstrate that this gene is a suitable internal control for normalising RNA loading in this experimental context (17,18).
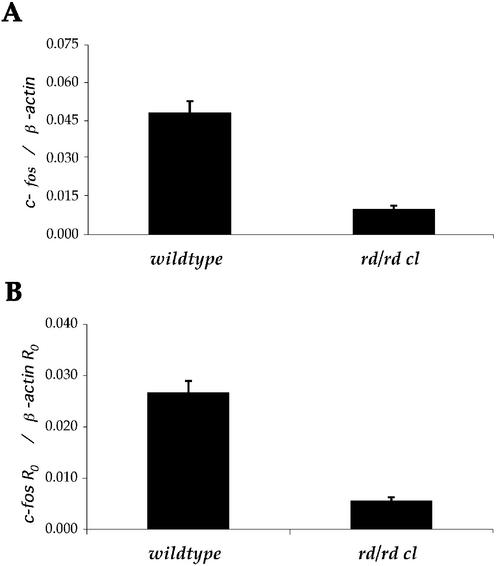
As expected, in comparison to wild-type mice, rd/rd cl animals demonstrate a much lower expression of c-fos in ocular tissues. The expression of ocular c-fos was determined as 19 646 copies in wild-type, compared with 4256 copies in rd/rd cl. When c-fos expression was normalised to β-actin, this resulted in a significant decrease of 4.78-fold (P < 0.001), as illustrated in Figure 2A.
Figure 2.
Comparison of absolute and relative approaches to quantifying expression of c-fos in the eyes of wild-type and retinally degenerate (rd/rd cl) mice. (A) Absolute quantification, plotted as copies of c-fos normalised to copies of β-actin as determined from plasmid standard curves. (B) Relative quantitation expressed as c-fos R0 normalised to β-actin R0. Mean efficiency was derived from amplification plots without the use of a standard curve.
The intra-assay variability was calculated from reaction triplicates as 4.2 and 17.5% for c-fos (wild-type and rd/rd cl) and 4.3 and 2.3% for β-actin (wild-type and rd/rd cl). This compares favourably with previous calculations of real-time variability, of 14.2% for Sybr Green and 24.0% for TaqMan™ (19), and furthermore suggests that intra-assay variability may be both primer and template dependent. Higher intra-assay variability for c-fos in rd/rd cl may be an effect of lower starting template concentration. Both plasmid and cDNA standard curves demonstrated higher variability at lower concentrations (data not shown).
Relative quantification
To compare the accuracy of efficiency-corrected relative quantification, amplification efficiency was derived from cDNA standard curves (see Table 2). Using equation 2, R0 was calculated for c-fos expression normalised to β-actin. The result showed a significant decrease (P < 0.001), with a 4.60-fold lower expression of c-fos in rd/rd cl compared to wild-type eyes.
Table 2. PCR efficiency derived from various approaches to data analysis.
| Method | c-fos efficiency | β-actin efficiency |
|---|---|---|
| Absolute | 0.853 | 0.819 |
| cDNA standard curve | 0.824 | 0.845 |
| 2–ΔΔCt | 1.000 | 1.000 |
| Amplification plot | 0.846 (± 0.036) | 0.874 (± 0.023) |
| Individual correction | 0.781–0.885 | 0.839–0.905 |
A value of 0 corresponds to no amplification, whereas 1 indicates a perfect doubling of product with every cycle. Efficiency was calculated from plasmid and cDNA standard curves using a modified version of equation 4, where E = 10(–1/slope) – 1 and from amplification plots using equations 3 and 4 (see text for details). The standard deviation associated with this efficiency is noted where calculated. Discrepancies between the efficiency values for c-fos and β-actin may be due to errors in efficiency calculation or standard curve construction or may represent differences in efficiency between the templates used.
The same data was also analysed using the 2–ΔΔCt method, again resulting in a significant decrease in ocular c-fos in rd/rd cl animals (P < 0.001). In this case, the fold decrease was determined as 5.80. The overestimation of the difference in expression is produced by the assumption of an ideal amplification efficiency of 1.
Amplification plot method
Using the slope of real-time PCR amplification plots as described, the mean amplification efficiency was determined as 0.846 for c-fos and 0.874 for β-actin. When the mean amplification efficiency was used with equation 2 to calculate relative expression, the reduction in c-fos expression in the rd/rd cl eye was determined as 4.79 (P < 0.001). The results of data analysis are shown in Figure 2B and summarised in Table 1.
Table 1. Relative quantification of c-fos expression in wild-type and rd/rd cl mice using five alternative approaches to data analysis.
| Method | Fold change | SD | |
|---|---|---|---|
| Plasmid SC | Wild-type | 1.000 | 0.092 |
| rd/rd cl | 0.209 | 0.028 | |
| cDNA SC | Wild-type | 1.000 | 0.093 |
| rd/rd cl | 0.217 | 0.028 | |
| 2–ΔΔCt | Wild-type | 1.000 | 0.105 |
| rd/rd cl | 0.173 | 0.025 | |
| Amplification plot | Wild-type | 1.000 | 0.085 |
| rd/rd cl | 0.209 | 0.026 | |
| Individual correction | Wild-type | 1.000 | 0.507 |
| rd/rd cl | 0.282 | 0.274 |
Most methods give comparable results, but differences do occur in the exact magnitude of the change and the associated variance. For copy numbers, fold change is determined from the expression ratio (copies of c-fos/copies of β-actin) relative to the wild-type. For other approaches, fold changes are determined from the mean normalised expression (R0 c-fos/R0 β-actin) relative to the mean normalised wild-type expression. Fold decrease equals the reciprocal of the fold change. Relative standard deviation is obtained similarly by expressing the standard deviation of the normalised expression relative to the mean normalised wild-type expression.
To test the effects of applying individual reaction corrections, relative quantification using the amplification efficiency of every individual sample was also conducted. Following these corrections a significant decrease in expression was still apparent, although this difference was determined as 3.55-fold (P < 0.05).
There was no significant difference between the reaction efficiencies of wild-type (c-fos 0.839, β-actin 0.872) and rd/rd cl (c-fos 0.852, β-actin 0.876) samples as tested by ANOVA and no individual sample within any of the groups demonstrated a significantly different efficiency. Applying individual corrections appears unjustified based upon these findings and rather than improving accuracy, introduces systematic errors which exaggerate the difference in expression and increase the assay noise.
Validation of the amplification plot method on known concentrations
To test the accuracy of the amplification plot method on samples of known concentration, data from standard curves were also analysed. Table 3 shows data from β-actin standard curves analysed using R0 values and shows a very close approximation to the actual dilutions used in the standard curves (r2 > 0.998). These calculated dilutions are not exact, even when using the efficiency derived from the standard curve, indicating that errors must be apparent in the standard curve construction.
Table 3. Relative quantitation of known concentrations of β-actin plasmid and cDNA.
| Sample | Dilution | R0 | Calculated dilution |
|---|---|---|---|
| β-Actin plasmid | 1 000 000 copies | 4.063 × 10–7 | 1.000 |
| 100 000 copies | 4.386 × 10–8 | 0.108 | |
| 10 000 copies | 3.299 × 10–9 | 0.008 | |
| 1000 copies | 2.620 × 10–10 | 0.001 | |
| β-Actin cDNA | 1 | 8.805 × 10–8 | 1.000 |
| 0.1 | 9.200 × 10–9 | 0.104 | |
| 0.01 | 8.618 × 10–10 | 0.010 |
Dilutions are calculated from the R0 of each dilution as a fraction of the R0 of the undiluted sample.
Given extremely precise pipetting a 1% relative error may be expected (Gilson), and following a 10-fold dilution this translates to an error of up to 1% in efficiency. Pipetting errors directly affect the calculated amplification efficiency, and cumulative error, particularly given imprecise pipetting or poorly calibrated pipettes, can therefore result in considerable effects on the accuracy of the amplification efficiency calculated from standard curves.
Based upon this data, an approximate fluorescence per copy can be calculated as ∼4.06 × 10–13 for β-actin (based on 1 × 106 copies to minimise dilution errors), which corrected for amplicon size results in ∼1.01 × 10–13 per 100 bp of product. Fluorescence per copy may be expected to be proportional to product size with a double-stranded DNA-binding dye such as Sybr Green I. In this study, we calculated the fluorescence per copy for c-fos as 2.03 × 10–13, which corresponds to 1.17 × 10–13 per 100 bp.
DISCUSSION
The nocturnal expression of the immediate early gene c-fos within the murine eye was found to be significantly lower in rd/rd cl animals in comparison to wild-type. Given the loss of rod and cone photoreceptors in this model, this supports previous reports that the source of nocturnal c-fos expression in the rodent eye is the photoreceptor layer (15,16).
These data demonstrate that absolute (copy number) and relative (fold change) approaches to real-time PCR produce very similar results, as summarised in Table 1. All of the five methods applied to data analysis demonstrate that c-fos expression is significantly reduced in the rd/rd cl eye in comparison to the wild-type. The magnitude of this change and associated variance are extremely comparable between plasmid DNA standard curves, cDNA standard curves and the mean efficiency amplification plot methods. The 2–ΔΔCt method provides a good approximation, although the assumption of an amplification efficiency of 1 exaggerates the difference. The least comparable method of data analysis is applying individual sample corrections, as this introduces considerable systematic errors in data analysis, as discussed below. As can be seen from Table 2, the only practical difference between these approaches is the way in which amplification efficiency is calculated and how this correction is subsequently applied.
Standard curve methods have become widely used for the purpose of calibrating real-time PCR reactions against known concentrations of nucleic acids. Whilst standard preparations such as amplicon, plasmid, oligonucleotide or synthesised RNA provide a readily available source for standard curve construction, use of these purified standards is dependent upon the assumption that the amplification efficiency of standard and cDNA samples is identical. Whilst this may be true, it is rarely tested and samples of cDNA may in fact possess secondary structure or contain PCR inhibitors remaining from the RNA extraction, DNase treatment or reverse transcription steps, all of which may subtly affect PCR efficiency (1,20). By using cDNA to construct standard curves, differences between the amplification efficiency of standard and template may be circumvented (9). However, the range of a cDNA standard curve is limited by the expression level in the sample used (particularly for rare transcripts) and only represents that sample. The dilution steps involved in constructing standard curves raise additional problems. Lower starting template concentrations result in greater assay variability, making the higher dilutions of a standard curve less reliable. Furthermore, the concentration of nucleic acids present in the reaction may adversely affect amplification efficiency (12), necessitating the use of a carrier nucleic acid.
Copy numbers derived from standards are also prone to error, and in many cases may be meaningless. Cumulative errors introduced by spectrophotometry, calculations of molecular weight and pipetting errors result in copy numbers being an approximation rather than an absolute unit. Furthermore, the copy number in experimental samples may not be comparable due to differences in RNA concentration, RNA quality and reverse transcription rates, all of which may vary considerably and are difficult to control for (3). For example, a sample containing partially degraded RNA will return a lower copy number, whereas a sample undergoing efficient reverse transcription will contain a higher copy number.
In most biological applications, gene expression data are normalised to one or more internal controls to account for these differences (17,18). This results in an expression ratio of target gene to internal control. This process of normalisation is not what is meant by ‘relative’ quantification; it is when the normalised experimental samples are then calibrated to the normalised control samples that a relative expression value is derived (10). The relative expression is simply the ratio between normalised samples, and the result is the same whether the measurements used are copy numbers or theoretical values such as R0. This is illustrated by calculating the R0 using the amplification efficiency derived from the plasmid DNA standard curves in this study. When calculated as relative expression, the fold change and data variance observed are mathematically identical to those obtained with copy numbers.
Fluorescence must be proportional to DNA content for real-time PCR techniques to be valid (3). This is confirmed by the calculation of R0 per molecule (when normalised to amplicon size), which suggests a value for both c-fos and β-actin of around 1 × 10–13 per 100 bp of amplicon. This also suggests that increasing amplicon size may be a simple means of increasing assay sensitivity when using double-stranded DNA binding dyes such as Sybr green 1.
As the determination of amplification efficiency critically underpins accurate real-time PCR, a means of monitoring amplification efficiency of all samples is desirable. Whilst the exact magnitude of a difference in expression may not be essential, ensuring that all samples exhibit comparable amplification efficiency certainly is (3). The use of raw data to determine amplification efficiency provides a powerful means of determining PCR efficiency in every single reaction.
The novel method we present here allows an automated calculation of amplification efficiency for every sample in a real-time PCR assay, which we have found to be remarkably robust. We have found that using the mean efficiency provides a good representation of the amplification kinetics, and the results derived from this approach compare favourably with those obtained via conventional standard curve approaches. A comparison of plasmid DNA standard curve and this novel approach is shown in Figure 2, illustrating that the final results are indistinguishable.
Individual sample efficiency corrections can be applied using this method and one may assume that would provide improved precision. However, applying individual corrections leads to the introduction of additional error (see Table 1). This apparently counter-intuitive result reflects the impact of amplification efficiency on final results. The number of cycles over which the amplification efficiency is near maximum (but free of background noise) is surprisingly limited and there may only be a small number of available data points above the detection threshold before the amplification rate starts to decline (see Fig. 1A). As such, determination of amplification efficiency is limited by errors in measurement.
Several previous methods have suggested the use of individual corrections (11,21), and whilst this would seem the obvious approach, it is in fact impractical due to the likelihood of introducing considerable systematic errors, as demonstrated herein. These errors are due to the relationship between amplification efficiency (E) and R0, as described in equation 2. Any correction based upon E is capable of having dramatic effects on the resulting R0 value due to the exponential magnification of this value. Therefore, if different E values are used for each sample, any error in the measured E will be exponentially magnified, and even very small errors in E will result in a considerable effect on R0.
This is simply illustrated by comparing three hypothetical samples, A–C, each with a Ct of 26.15 at a threshold (RCt) of 0.06, but with differing amplification efficiencies. If E = 0.85 for Sample A, 0.84 for Sample B and 0.80 for Sample C, the R0 values would therefore be 6.19 × 10–9, 7.13 × 10–9 and 1.27 × 10–8, respectively. Relative to Sample A, expression in Sample B is therefore 15% higher, whereas Sample C is 105% higher. As can be seen from this example, even a difference in amplification efficiency of 0.01 can have a significant influence on the R0 value, and a difference of just 0.05 yields a wholly different interpretation of the data.
All measurements of amplification rate are estimates, and the most reliable estimates are based upon repeated measurements. As such, when using amplification efficiencies derived from individual sample kinetics the mean amplification efficiency provides a more accurate measure of efficiency than individual corrections, which if even slightly inaccurate may distort results rather than providing increased precision. We have found this to be the case in all data sets examined, with significantly increased data variance following application of individual corrections.
As such, one must assume that amplification efficiency is comparable unless there is sufficient evidence to suggest otherwise. By using the mean efficiency, and incorporating tests for outlier detection within groups and for differences between groups, we can be certain that samples exhibit comparable kinetics and corrections are only applied when sufficient statistical evidence exists to justify them.
Finally, measures of intra-assay variability are easily calculated (for efficiency and R0) using replicate reactions, and this provides a measure of the inherent experimental error, to which the operator may contribute significantly (4). The calculation of intra-assay variability should be based upon copy numbers or R0 and not upon Ct values, as the latter are logarithmic units and as such result in a misleading representation of reproducibility (19).
CONCLUSIONS
This study supports the hypothesis that nocturnal expression of ocular c-fos originates from the photoreceptor layer. In addition, we illustrate that absolute and relative approaches to real-time PCR data analysis provide very comparable end results and that the key difference between these approaches are produced by the amplification efficiency applied. Whilst standard curves are widely used for calculating amplification efficiency, they are based upon the assumption that the amplification efficiencies of the diluted standard and unknown samples are identical. Furthermore, the copy numbers often produced may actually give a misleading interpretation.
Given that every amplification plot contains information regarding the amplification kinetics of the sample, it is instead possible to use this data to determine the amplification efficiency. By calculating the amplification efficiency for every sample, it is then a simple matter to test for outliers and to ensure that sample and control populations exhibit similar kinetics. Due to the potential for introducing considerable systematic errors, amplification efficiency must be assumed to be comparable unless evidence exists to suggest otherwise. We have automated all the necessary calculations and statistical tests to allow a simple and effective means of analysing real-time PCR data, enabling the assay to be conducted rapidly based upon the kinetics of experimental samples, rather than additional artificial standards.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was partly funded by research grants from the UK Biotechnology and Biological Sciences Research Council and private funding. Thanks to Dr Robert Lucas for useful discussions and to all the staff in the Advanced Biotechnology Centre at Charing Cross Hospital.
REFERENCES
- 1.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 2.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 3.Freeman W.M, Walker,S.J. and Vrana,K.E. (1999) Quantitative RT-PCR: pitfalls and potential. Biotechniques, 26, 112–125. [DOI] [PubMed] [Google Scholar]
- 4.Bustin S.A. (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol., 29, 23–39. [DOI] [PubMed] [Google Scholar]
- 5.Ginzinger D.G. (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Haematol., 30, 503–512. [DOI] [PubMed] [Google Scholar]
- 6.Klein D. (2002) Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med., 8, 257–260. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi R., Fockler,C. Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 8.Livak K.J. (1997) ABI Prism 7700 Sequence Detection System. User Bulletin no. 2. PE Applied Biosystems, AB website, bulletin reference: 4303859B 777802-002. http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf [Google Scholar]
- 9.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res., 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak K.J. and Schmittgen,T.W. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 11.Liu W. and Saint,D.A. (2002) A new quantitative method of real-time RT-PCR assay based on simulation of PCR kinetics. Anal. Biochem., 302, 52–59. [DOI] [PubMed] [Google Scholar]
- 12.Kainz P. (2000) The PCR plateau phase: towards an understanding of its limitations. Biochim. Biophys. Acta, 1494, 23–27. [DOI] [PubMed] [Google Scholar]
- 13.Freedman M.S, Lucas,R.J., Soni,B., Von Schantz,M., Munoz,M., David-Gray,Z. and Foster,R.G. (1999) Regulation of mammalian circadian behaviour by non-rod, non-cone photoreceptors. Science, 284, 502–504. [DOI] [PubMed] [Google Scholar]
- 14.Lucas R.J., Freedman,M.S., Munoz,M., Garcia-Fernandez,J.M. and Foster,R.G. (1999) Regulation of the mammalian pineal by non-rod, non-cone photoreceptors. Science, 284, 505–507. [DOI] [PubMed] [Google Scholar]
- 15.Nir I. and Agarwal,N. (1993) Diurnal expression of c-fos in the mouse retina. Mol. Brain Res., 19, 47–54. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida K., Kawamura,K. and Imaki,J. (1993) Differential expression of c-fos mRNA in rat retinal cells: regulation by light/dark cycle. Neuron, 10, 1049–1054. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Higgins,P.J. and Crawford,D.R. (2000) Control selection for RNA quantitation. Biotechniques, 29, 332–337. [DOI] [PubMed] [Google Scholar]
- 18.Sturzenbaum S.R. and Kille,P. (2001) Control genes in quantitative molecular biological techniques: the variability of invariance. Comp. Biochem. Physiol., 130B, 281–289. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen T.D, Zakrajsek,B.A., Mills,A.G., Gorn,V., Singer,M.J. and Reed,M.W. (2000) Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem., 285, 194–204. [DOI] [PubMed] [Google Scholar]
- 20.McPherson M.J. and Moller,S.G. (2000) PCR. BIOS Scientific Publishers, Oxford, UK. [Google Scholar]
- 21.Liu W. and Saint,D. (2002) Validation of a quantitative method for real time PCR kinetics. Biochem. Biophys. Res. Commun., 294, 347–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.