Abstract
The TspDTI restriction endonuclease, which shows a novel recognition specificity 5′-ATGAA(N11/9)-3′, was isolated from Thermus sp. DT. TspDTI appears to be a ‘twin’ of restriction endonuclease TspGWI from Thermus sp. GW, as we have previously reported. TspGWI was isolated from the same location as TspDTI, it recognizes a related sequence 5′-ACGGA(N11/9)-3′ and has conserved cleavage positions. Both enzymes resemble two other class-IIS endonucleases from Thermus sp.: TaqII and Tth111II. N-terminal amino acid sequences of TspGWI tryptic peptides exhibit 88.9–100% similarity to the TaqII sequence. All four enzymes were purified to homogeneity; their polypeptide sizes (114.5–122 kDa) make them the largest class-IIS restriction endonucleases known to date. The existence of a Thermus sp. sub-family of class-IIS restriction endonucleases of a common origin is herein proposed.
INTRODUCTION
Restriction endonucleases are traditionally divided into three major classes or types: I, II and III, with the vast majority of endonucleases included in class-II (1,2).
Class-I, exemplified by EcoK and EcoB, consists of multimeric enzymes, each composed of three distinct types of subunits: S, for recognition; R, for cleavage; and M, for methylation. The endonuclease complex contains all three subunits, while the methylation complex contains only S and M subunits. The endonuclease recognizes 6–8 bp asymmetric, interrupted cognate sequence; whereas for cleavage it requires Mg2+, S-adenosylmethionine (SAM) and ATP. SAM and ATP are utilized both as cofactors and allosteric effectors, which determine, based on methylation of substrate DNA, whether methylation or restriction activity will be turned on. During this complicated sequence of events, including ATP-driven translocation of DNA helix, scission takes place at random and far from the recognition site—up to several thousands base pairs (3,4).
Class-II was originally distinguished as featuring homodimeric or homomultimeric endonucleases, such as EcoRI, requiring Mg2+ as the only obligatory cofactor. They recognize 4–8 bp palindromic sequences and cleave within this sequence (1). Cognate methyltransferases are monomeric proteins, which require only SAM for activity.
In class-III, exemplified by EcoPI, an endonuclease contains two different subunits or a single subunit, and recognizes asymmetric sequences of 6 bp, cleaving outside at a distance of ∼25 bp. The enzymes require Mg2+ as a cofactor and ATP as an allosteric activator. In addition, they are stimulated by SAM (4).
Currently, with increasing numbers of new endonucleases found—over 250 specificities and several thousands of isoschizomers (2), traditional classification is no longer adequate to cover the diversity of restriction endonucleases known. Class-II turned out to be very heterogeneous, with numerous enzymes not fitting the original classification. Several new endonuclease categories are distinguishable, either temporarily described as sub-classes within class-II or as proposed separate types. In particular, (sub)class-IIS significantly differs from the class-II paradigm by recognizing 4–7 bp asymmetric sequences, cleavage at the defined distance of 0–20 bp downstream (5), monomeric architecture (6–8) and the utilization of different mechanism of recognition and scission (9,10). Some class-IIS endonucleases, in addition to their requirement for Mg2+, are stimulated by SAM, although this is not an obligatory cofactor. This unusual mode of interaction with DNA prompted detailed function–structure studies. In particular, FokI (11) has become a model protein for endonuclease–DNA interaction studies. FokI and other so far characterized class-IIS endonucleases are monomers in solution (6,12,13), although transient dimerization during cleavage has been observed (9,10). FokI and StsI both have two functional domains: one for binding to the cognate site and another for DNA cleavage (5,7–10,14–16). Such modular enzyme architecture allows for remarkable protein engineering experiments, such as the design and construction of artificial chimeric restriction endonucleases, composed of the FokI endonuclease cleavage domain fused with a site-specific DNA binding protein. Examples of this include Ubx homeodomain of Drosophila (17) and zinc-finger transcription factors fused to the C-terminal domain of FokI (18). Such hybrid proteins cleaved DNA at novel sites, with target recognition specificities imposed by the DNA-binding protein fusion partners. Separation of a recognition sequence from its cleavage site led to the development of a universal restriction endonuclease, capable of cleaving a single-stranded DNA target at a predetermined sequence (14,19).
Proposed class-IV is exemplified by Eco57I. The enzyme is composed of just a single polypeptide, which is a fusion of endonuclease and methyltransferase moieties. Monomeric enzyme recognizes an asymmetric sequence of 6 bp, but cleaves 16/14 bp downstream from its cognate site. It requires Mg2+ and is heavily dependent on SAM (20,21).
Type-BcgI-like contains unusual enzymes, which form an asymmetric protein complex of three subunits, of which two are identical. Such complexes recognize 5–7 bp continuous or interrupted asymmetric sites and can perform both cleavage and methylation. Methylation requires SAM and is stimulated by Mg2+. These endonucleases cleave both upstream and downstream of the cognate site (10/12nCGAnnnnnnTGCn12/10 in the case of BcgI), excising the recognition site along with flanking sequences from the target DNA. Cleavage requires Mg2+ and is stimulated by SAM (22,23).
Type-IIG partially overlaps (sub)class-IIS, class-IV and BcgI-like enzymes in that they bind to asymmetric sequences and cleave on one or both sides of their recognition sites. In addition, they are invariably stimulated by SAM, and both methyltransferase and restriction activity are located in the same polypeptide. This type is exemplified by HaeIV, which is specific to 7/13nGAYnnnnnRTCn14/9 sequence and forms a homodimer (24), as opposed to monomeric class-IIS enzymes (6,12,13).
Type-CviJI-like contains endonucleases of eukaryotic origin, found only in viruses infecting unicellular Chlorella algae. The enzymes resemble class-II in their homodimeric structure and Mg2+ requirement as the only cofactor. However, they have features not found in any other class of restriction endonuclease: recognizing more frequent sequences than their prokaryotic counterparts—degenerated 4 bp (statistically equivalent to 3 bp), and their specificity changes in the presence of an adenine nucleotide derivative to cleave even more frequently—essentially a 2 bp sequence (25–27).
Type-BfiI-like shares characteristics of class-IIS enzymes, except it does not require Mg2+ for cleavage, which is a radical exception amongst restriction endonucleases of all types, and involves a different mechanism of DNA scission (28).
There are more variations within these groups or putative classes, and even evolutionary traces of other functions preceding restriction of DNA can be found, such as a ligase moiety present in NaeI. It has been shown that just a single mutation converted NaeI to topoisomerase/recombinase (29).
In this paper we present evidence for the existence of a Thermus sp. sub-family of related class-IIS restriction endonucleases, which have a combination of exclusive features in addition to those found in class-IIS and class-IV.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and reagents
Thermus sp. DT was from EURx Ltd bacterial strains collection, isolated during the company research program. The bacterium is an obligatory thermophile, which grows between 56 and 75°C. The optimum cultivation conditions were at 60°C in a modified Luria broth (0.5% tryptose, 0.3% yeast extract, 0.2% NaCl, pH 7.2, Nitsch’s trace elements) (30). Escherichia coli DH11S {mcrA [mrr-hsdRMS(rK–, mK+)-mcrBC] Λ(lac-proAB) Λ(recA1398) deoR, rpsL, srl, thi/F′ proAB+ lacIQZΛM15} (Life Technologies, Gaithersburg, MD) was used for transformation of ligation mixtures and DNA propagation.
Difco media components were from Becton-Dickinson (Franklin Lakes, NJ). Agarose GTG was from FMC (Rockland, MA). Phosphocellulose P11 resin was from Whatman (Springfield Mill, UK). Hydroxyapatite HTP was from Bio-Rad (Hercules, CA). Other chromatographic resins were from Pharmacia Biotech AB (Uppsala, Sweden). Immobilized TPCK-trypsin was from Pierce (Rockford, IL). All other reagents were from Amresco (Solon, OH) or Sigma-Aldrich (St Louis, MO), of the highest available purity.
Cloning vector pTZ18U (ApR, MCS, f1 ori, T7 promoter) (31), was obtained from Dr David Mead, Molecular Biology Inc., WI. Plasmids pBR322, pUC19 and pACYC184, miniprep DNA purification kit, SmaI endonuclease, T4 DNA ligase, T4 DNA polymerase and lambda DNA were from EURx Ltd (Gdańsk, Poland).
TspDTI purification
The TspDTI restriction endonuclease was isolated using the following stages, performed at 4°C.
Polyethyleneimine (PEI) removal of nucleic acids. Thermus sp. DT cells were resuspended in buffer A [50 mM Tris–HCl pH 7.0, 100 mM NaCl, 5 mM EDTA, 10% glycerol, 5 mM β-mercaptoethanol (βME), 0.1% Triton X-100, 1 mM PMSF and 20 µg/ml benzamidine] and lysozyme was added to a concentration of 1 mg/ml. The suspension was incubated for 1 h and sonicated. Bacterial debris was spun down, the NaCl concentration was increased to 200 mM and PEI (pH 7.0) was added to 0.4%. The suspension was stirred for 1 h at 4°C and the nucleic acid–PEI complex was removed by centrifugation.
Ammonium sulphate (AmS) fractionation. The PEI supernatant was adjusted to 30% AmS saturation and stirred for 2 h. Precipitated contaminating proteins were removed by centrifugation and AmS was added to the supernatant to increase its concentration to 60% saturation. The protein fraction from the 30–60% AmS precipitation was spun down and the supernatant was discarded.
Phosphocellulose chromatography. Peletted TspDTI was dissolved in buffer B (20 mM KPO4 pH 7.0, 30 mM NaCl, 0.5 mM EDTA, 5% glycerol, 5 mM βME, 1 mM PMSF, 20 µg/ml benzamidine), dialysed against buffer B and adsorbed into phosphocellulose P11. The column was washed with buffer B and eluted with a gradient of 30 mM to 1 M NaCl in buffer B.
Heparin–agarose chromatography. Fractions containing restriction activity were dialysed against buffer C (20 mM Tris–HCl pH 7.5, 30 mM NaCl, 0.5 mM EDTA, 5% glycerol, 5 mM βME) and applied to a Heparin–agarose column. The column was washed with buffer C and TspDTI was eluted with a 30 mM to 1 M NaCl gradient in buffer C.
DEAE–Sephadex chromatography. TspDTI was dialysed against buffer D (20 mM Tris–HCl pH 7.5, 70 mM NaCl, 0.5 mM EDTA, 5% glycerol, 5 mM βME), applied to a DEAE–Sephadex column, which was eluted with buffer D.
Molecular sieving on Sephadex G-120. Active fractions from DEAE–Sephadex were concentrated to 3 ml and subjected to molecular sieving on a Sephadex G-120 column, equilibrated in buffer E [20 mM Tris–HCl pH 8.3, 3 mM MgCl2, 25 mM (NH4)2SO4, 25 mM KCl, 0.5 mM DTT, 5% glycerol].
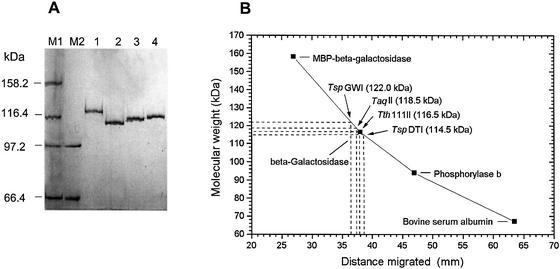
Hydroxyapatite chromatography. The enzyme was further dialysed against buffer F (20 mM KPO4 pH 7.0, 30 mM NaCl, 0.1 mM EDTA, 5% glycerol, 5 mM βME), adsorbed to a Hydroxyapatite HTP column, washed with buffer F and eluted with a 20–900 mM KPO4 pH 7.0 gradient in buffer F. Purified, homogeneous TspDTI (Fig. 2) was dialysed against buffer G [20 mM Tris–HCl pH 7.5, 3 mM MgCl2, 25 mM (NH4)2SO4, 2 mM DTT, 50% glycerol] and stored at –20°C.
Figure 2.
Determination of polypeptide molecular sizes for Thermus class-IIS endonucleases: TspDTI, TspGWI, TaqII and Tth111II. (A) SDS/PAGE of purified, homogeneous TspDTI, TspGWI, TaqII and Tth111II endonucleases. Lane M1, protein marker broad range (New England Biolabs); lane M2, low molecular weight marker (Amersham-Pharmacia). Bands marked in lanes M1 and M2 are as follows: 158.2 kDa, MBP-β-galactosidase; 116.4 kDa, β-galactosidase; 97.2 kDa, phosphorylase b; 66.4 kDa, bovine serum albumin. Lane 1, TspGWI endonuclease; lane 2, TspDTI endonuclease; lane 3, Tth111II endonuclease; lane 4, TaqII endonuclease. (B) Graph showing estimation of polypeptide sizes for TspDTI, TspGWI, TaqII and Tth111II.
Determination of TspDTI recognition and cleavage sites
The TspDTI recognition site and cleavage positions were established by shotgun cloning and sequencing of the partial digestion products of bacteriophage lambda DNA. The TspDTI-generated restriction fragment ends were blunted with T4 DNA polymerase in the presence of dNTPs (30), cloned into the SmaI site of a modified pTZ18u vector (31), transformed into E.coli DH11S and plated onto X-Gal/IPTG plates (30). Miniprep plasmid DNA was isolated from white colonies and the fragment–vector junctions were sequenced using the ABI Prism 310 automated sequencer with ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer Applied Biosystems, Foster City, CA). The obtained sequence data were then analysed using ABI Chromas 1.45 software (Perkin Elmer Applied Biosystems) and DNASIS 2.5 software (Hitachi Software, San Bruno, CA).
Proteolysis of TspGWI and amino acid sequence determination
Purified TspGWI was subjected to limited TPCK-trypsin digestion to obtain internal polypeptides for N-terminal amino acid sequencing. Proteolysis of 30 µg TspGWI was conducted in 110 µl of buffer T (20 mM Tris–HCl pH 8.3, 25 mM KCl, 3 mM MgCl2, 5% glycerol, 0.05% Tween 20, 0.5 mM DTT) with 30 µl gel-immobilized TPCK-trypsin and gentle shaking at 24°C for 3 h. The immobilized TPCK-trypsin was removed by centrifugation. The supernatant, containing TspGWI fragments was run on a 6% SDS/PAGE denaturing gel and electroblotted onto PVDF membrane in 100 mM CAPS-NaOH buffer pH 11.0. The N-terminal amino acid sequence analysis of blotted polypeptides was performed on a gas-phase sequencer (Model 491, Perkin Elmer-Applied Biosystems). The phenylthiohydantoin derivatives were analysed by online gradient high performance liquid chromatography on Microgradient Delivery System Model 140C equipped with Programmable Absorbance Detector Model 785A and Procise software (Perkin Elmer-Applied Biosystems).
RESULTS AND DISCUSSION
Purification and properties of Thermus class-IIS endonucleases
TspDTI activity is present in very small quantities in its natural host Thermus DT strain, thus the development of an extensive isolation procedure was essential. Seven purification stages were needed to obtain a homogeneous protein: PEI and AmS fractionations, followed by five chromatographic steps.
The optimum reaction conditions for TspDTI are in 10 mM Tris–HCl pH 8.0 at 25°C, 10 mM MgCl2, 10 mM DTT. The temperature activity range extends from 42 to 85°C, with maximum activity observed at 65–75°C. Under all digestion conditions tested, a stable partial cleavage pattern was observed (Fig. 1). Spermidine does not affect TspDTI activity, while SAM stimulates the enzyme several-fold. In the presence of SAM and without Mg2+ the enzyme methylates its recognition site, which becomes resistant to TspDTI cleavage upon the subsequent addition of Mg2+ (data not shown). TspDTI can be inactivated by 20 min incubation at 89°C.
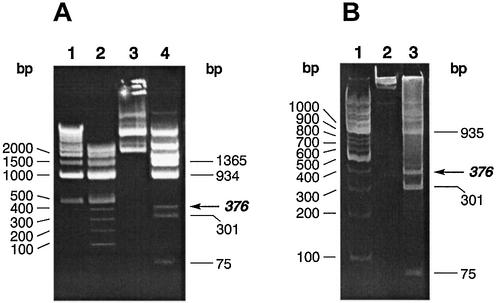
Figure 1.
Partial digestion of pUC19 plasmid DNA with TspDTI restriction endonuclease. (A) TspDTI cleavage of pUC19 DNA, 1.5% agarose/TAE. Lane 1, 1 kb ladder; lane 2, 100 bp ladder; lane 3, untreated pUC19 DNA; lane 4, TspDTI-cut pUC19 DNA. (B) TspDTI cleavage of pUC19 DNA, 6% polyacrylamide/TBE. Lane 1, 100 bp ladder; lane 2, untreated pUC19 DNA; lane 3, TspDTI-cut pUC19 DNA. The smallest partial digestion band of 376 bp is indicated in bold italics with horizontal arrow.
Determination of TspDTI recognition and cleavage sites
The TspDTI cleavage pattern of pUC19, pBR322, pACYC184 and lambda DNA indicated a high frequency of cleavage. The digested plasmid DNAs were run on an agarose gel (Fig. 1) and compared with the digestion patterns of known restriction endonucleases. The comparison suggested that TspDTI is an enzyme with a novel specificity. However, even repeated cleavage with concentrated TspDTI preparations failed to yield a complete reaction, resulting in a stable partial cleavage pattern, with ∼50% of the substrate DNA converted to complete digestion bands (Fig. 1). Thus, the inability to completely cut substrate DNA is either an intrinsic feature of the enzyme or a key cofactor was missing in the reaction.
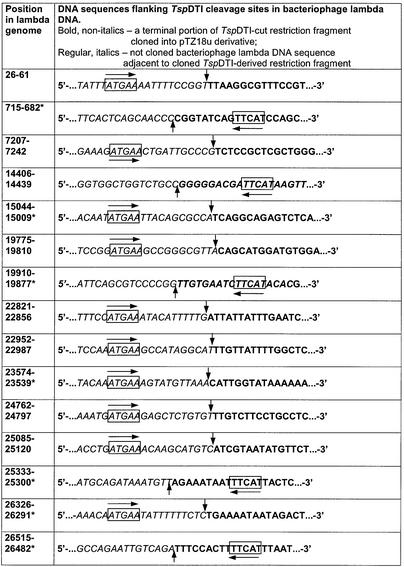
Comparison of the 35 junction sequences (29 are shown in Table 1) indicated that TspDTI belongs to the class-IIS restriction endonucleases, since a putative non-palindromic recognition site was found in the cloned inserts at a constant distance from the vector junction. TspDTI is a novel prototype of restriction specificity. The enzyme recognizes a 5 bp asymmetric cognate site (boxed) and cleaves DNA downstream, after nt 11 and 9 (vertical arrows) in the top and the bottom strand, respectively, yielding 2 nt 3′ single-stranded termini:
Table 1. Determination of the TspDTI recognition sequence and cleavage positions by shotgun cloning and sequencing of TspDTI restriction fragments.
*Sequence obtained from TspDTI restriction fragments cloned in reverse complement orientation. Base numbering refer to the conventional orientation of lambda genome; bold text, a terminal portion of TspDTI-cut restriction fragment, T4 DNA polymerase repaired and cloned into pTZ18u derivative; italic text, not cloned bacteriophage lambda DNA sequence adjacent to cloned TspDTI-derived restriction fragment; box + horizontal arrow, TspDTI recognition sequence; vertical arrows, TspDTI cleavage positions.
A computer prediction of the cleavage frequency shows that there are four TspDTI sites within pUC19, 10 within pBR322, 11 within pACYC184 and 176 within lambda DNA. However, the actual TspDTI digestion pattern exhibits more bands than expected, due to partial cleavage (Fig. 1).
Evolutionary implications
Both the recognition sequence of TspDTI and its cleavage positions appear to be related to those reported by us previously for the Thermus sp. restriction endonuclease TspGWI, 5′- ACGGA(N11/9)-3′ (32). The TspDTI cognate site has only two differences as compared to the 5 bp recognition site of TspGWI: a transition from C to T and from G to A in the second and fourth bases, respectively. Moreover, the cleavage positions at nt 11 and 9 are exactly the same for both enzymes. Since both TspDTI and TspGWI originate from two Thermus sp. isolates found in the same hot spring sample, it is possible that both enzymes occurred as a result of a recent divergent evolution event. In addition, TspDTI and TspGWI might be closely related to two previously reported class-IIS endonucleases found in different Thermus species: TaqII from Thermus aquaticus (33) and Tth111II from Thermus thermophilus (34), and more distantly related to mesophilic endonucleases: EciI from E.coli (REBASE: rebase.neb.com) and BceAI and BcefI, both from Bacillus cereus (35) (REBASE: rebase.neb.com) (Table 2). Both TaqII and Tth111II recognize asymmetric 6 bp redundant sites and cleave 11 and 9 nt downstream: 5′-GACCGA(N11/9)-3′ or 5′-CACCCA(N11/9)-3′ (33) and 5′-CAARCA(N11/9)-3′ (34), respectively. Due to a redundancy of TaqII and Tth111II 6 bp recognition sites, their overall cleavage frequency is only slightly lower than that of TspDTI and TspGWI 5 bp non- redundant sites. One of several possible evolutionary scenarios would be that TspGWI and TspDTI have eliminated the first C or G from an ancestral TaqII/Tth111II-like recognition sequence, thus evolving toward more frequent restriction. As illustrated in Table 2, TspGWI and TspDTI 5 bp cognate sites show 2–3 differences, when compared to those of TaqII and Tth111II. All these changes are located within a variable 3 bp 5′-TGA-3′ ‘core’ region (bases 2–4) of the TspDTI recognition site 5′ATGAA-3′. The first and the last A residues (bases 1 and 5) of the TspGWI and TspDTI recognition sites remain conserved amongst all four enzymes. In addition, one of the two possible variants of Tth111II cognate sites, 5′-CAAGCA-3′, also shares an internal G residue (base 4) with the TspDTI site (Table 2). Recognition sequence similarities are further validated by strict conservation of cleavage positions at nt 11 and 9 for TspGWI, TspDTI, TaqII and Tth111II. The proposal for the existence of a class-IIS sub-family, within Thermus sp., is further reinforced by the following findings.
Table 2. Comparison between TspDTI/TspGWI endonuclease ‘twins’ and class-IIS restriction endonucleases with related recognition and cleavage sites.
| Restriction endonucleasea | Bacterial hosta | Recognition sitea | Cleavage positionsa | Reaction temperaturea | Polypeptide sizeb | Native molecular sizec | Specific DNA methylation | Reference |
|---|---|---|---|---|---|---|---|---|
| TspDTI | Thermus sp. | ATGAA | N11/9 | 70°C | 114.5 kDa | 110–130 kDa | +++ | This work |
| TspGWI | Thermus sp. | ACGGA | N11/9 | 70°C | 122.0 kDa | 110–130 kDa | +/– | (32), This work |
| TaqII | Thermus aquaticus | GACCGA | N11/9 | 70°C | 118.5 kDa | 110–130 kDa | ++++ | (33), (36), This work |
| CACCCA | N11/9 | (125.6 kDad) | ||||||
| Tth111II | Thermus thermophilus | CAARCA | N11/9 | 70°C | 116.5 kDa | 110–130 kDa | ND | (34), This work |
| EciI | Escherichia coli | GGCGGA | N11/9 | 37°C | ND | ND | ND | REBASE |
| BcefI | Bacillus cereus | ACGGC | N12/13 | 30°C | ND | ND | ND | (35) |
| BceAI | Bacillus cereus | ACGGC | N12/14 | 30°C | ND | ND | ND | REBASE |
aThermus sp.-derived and TspDTI/TspGWI-related endonucleases, bases in a recognition sequence, cleavage positions and reaction temperatures are marked in bold.
bAs estimated by SDS/PAGE of homogeneous proteins.
cAs estimated by molecular sieving under native buffer conditions.
dAs calculated from sequencing/genetic analysis data obtained for the taqIIR gene.
ND, not determined.
(i) All the four putative related endonucleases have been purified to homogeneity from their native Thermus strains (A.Żylicz-Stachula, I.Sobolewski and P.M.Skowron, manuscript in preparation; S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation). Strikingly, their respective polypeptides migrate to the same position on SDS/PAGE gels. Only prolonged electrophoresis allows for distinguishing subtle variations in their molecular sizes (kDa): TspDTI, 114.5 ± 7; TspGWI, 122.0 ± 7; TaqII, 118.5 ± 7; and Tth111II, 116.5 ± 7 (Fig. 2). Such large polypeptides are rare amongst prokaryotes, and to our knowledge they are the largest class-IIS restriction endonucleases known to date.
(ii) TspDTI-containing fractions eluted from the Sephadex G-120 column, used for the enzyme purification, show a homogeneous protein band, with a relative molecular weight of ∼120 ± 10 kDa on SDS/PAGE denaturing gels. Relative band intensities in consecutive column fractions correlate perfectly with the restriction activity peak of TspDTI (data not shown). Subsequent comparison of the elution profiles of TspDTI, TspGWI, TaqII and Tth111II from a molecular sieving column shows that all the peaks of activity appear at nearly identical positions, characteristic of large proteins of native molecular size of 110–130 kDa (Table 2) (S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation). This indicates the same, monomeric structure for TspDTI, TspGWI, TaqII and Tth111II.
(iii) TaqII restriction endonuclease has been cloned, expressed and purified in our laboratory (36). The recombinant TaqII exhibits the same molecular size as the native TaqII, which matches sequencing/genetic analysis data obtained for the taqIIR gene: 3315 bp/1105 aa/125.6 kDa (36; S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation).
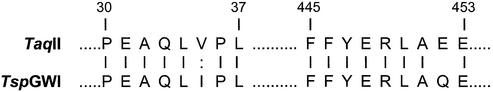
(iv) TspGWI has been subjected to partial trypsin digestion and the N-terminal amino acid sequences for two internal peptides have been determined. Comparison between two TspGWI tryptic fragments and the complete TaqII endonuclease amino acid sequence revealed near perfect homology: in peptide 1, an 8 aa continuous region contains seven identical amino acids and a single conservative substitution (100% similarity), while in peptide 2, a 9 aa region contains eight identical amino acids (88.9% similarity) (Fig. 3) (36; S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation).
Figure 3.
Amino acid sequence comparison between two tryptic fragments of TspGWI endonuclease and complete TaqII endonuclease sequence. Identical amino acid residues are indicated by straight lines, and similar residues by dots. The partial amino acid sequence of TspGWI restriction endonuclease was obtained by limited trypsin digestion, followed by N-terminal protein sequencing of two internal peptides. The TaqII amino acid sequence was translated from the cloned taqIIR coding gene (36; S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation). The first homologous region extends from amino acid 30 to 37 and the second region from amino acid 445 to 453 of TaqII restriction endonuclease.
(v) Both TspDTI and the TaqII are capable of specific methylation of their recognition sites in the presence of SAM (Table 2) (A.Żylicz-Stachula, I.Sobolewski and P.M.Skowron, manuscript in preparation). Some data suggest that TspGWI has residual methylation activity (data not shown). Analysis of the cloned taqIIR gene (36; S.M.Rutkowska, I.Jaworowska, I.Sobolewski and P.M.Skowron, manuscript in preparation) reveals that it is a fusion containing both an endonuclease moiety and a methyltransferase module with FGG and DPPY motifs (37).
Taken together, the near identity of very atypical molecular sizes, substantial similarities in recognition sequences, identity in cleavage sites, strong partial amino acid sequence homology between TspGWI and TaqII, cleavage stimulation by SAM, the presence of all four endonucleases in the same bacterial genus as well as biochemical similarities point to the common evolutionary origin of TspDTI, TspGWI, TaqII and Tth111II, thus defining a sub-family of Thermus class-IIS enzymes. These endonucleases are characterized by a unique combination of features found only in the Thermus class-IIS sub-family (such as extremely large polypeptides), as well as those present in class-IIS (asymmetric cognate sequence, cleavage outside recognition site) and sub-class IV (SAM stimulation, endonuclease–methyltransferase genes fusion). Moreover, considering SAM dependence, the Thermus class-IIS sub-family enzymes show continuity between class-IIS and class-IV features from barely detectable (TspGWI) to strong stimulation (TaqII) (data not shown).
Three more class-IIS endonucleases from unrelated bacterial species exhibit marked similarities to the putative Thermus sp. class-IIS endonuclease family. The mesophilic EciI endonuclease, from E.coli, recognizes 5′-GGCGGA(N11/9)-3′ (REBASE: rebase.neb.com). The enzyme shares the last 4 bp out of the 5 bp of the TspGWI recognition site (bases 2–5) and has the same cleavage positions of N11/9. Moreover, BcefI and BceAI, mesophilic endonucleases isolated from B.cereus, both recognize the 5′-ACGGC-3′ cognate site. Their recognition sequence differs in only one bp (last base, 5) from the TspGWI recognition sequence. However, BcefI and BceAI have cleavage positions shifted further downstream: 1 nt in the top strand and 4 or 5 nt in the bottom strand to N12/13 or N12/14, respectively (35; REBASE: rebase.neb.com).
In general, homologies between restriction endonucleases are very rare. They are usually limited to the epitopes, such as the highly variable catalytic motif PDX10–30(D/E)XK (38,39). Amongst class-IIS endonucleases, both primary sequence and structural homology were reported and studied in detail for imperfect isoschizomers FokI and StsI (15,16). Both enzymes bind to the 5 bp asymmetric site 5′-GGATG-3′ and cleave 9/13 (FokI) or 10/14 (StsI) nt downstream. Remarkably, even though the enzymes exhibit relatively high amino acid sequence homology (30%), they are very distinct biochemically: StsI is an acidic protein (pI 6.3), while FokI is very basic (pI 9.4), they do not cross-react immunologically and they have different reaction optima. Nevertheless, they share a common domain organization, suggesting very close similarities in their mechanism of action (5,7–10,15–18). In contrast, two other neoschizomers belonging to class-II, SmaI and XmaI, show no homology between their amino acid sequences. Both recognize 5′-CCCGGG-3′ sites, however, they leave blunt or 4 nt sticky ends, respectively. Apparently, the mechanism of recognition is different, as they bend the DNA helix in opposite orientations (40). Very few other cases of known homologous endonucleases in class-II are limited to isoschizomers, they are listed below.
(i) EcoRI/RsrI/MunI share 18–50% homologous amino acids and common active site architecture with XcyI and Cfr9I (39,41); (ii) XmaI/XcyI/Cfr9I are highly homologous, exhibiting 80% homology between Cfr9I and XmaI/XcyI (41); (iii) AvaI/BsoBI pair, the AvaI from cyanobacteria Anabaena variabilis has a thermophilic counterpart from a distant species, Bacillus stearothermophilus. Nevertheless, the enzymes show 55% homology and possess common amino acid residues critical for catalytic activities (42,43); (iv) BsuRI from prokaryotic Bacillus sphaericus and CviJI from IL-3A virus-infected eukaryotic Chlorella, in spite of the fact that they originate from two separate kingdoms, still exhibit 11.6% homology. Moreover, they show a substantial epitopic similarity (35%) over the 132 amino acids region (25–27,44); (v) TaqI isochizomers group contains eight related isoschizomers, with homology ranging from 54 to 100%, which was correlated with the geographical location of their Thermus host strains (45). Moreover, differences in their amino acid sequences allowed for an insight into observed varied thermostability (45,46). The corresponding methyltransferases are remarkably similar, indicating that both the endonuclease and the methyltransferase components of TaqI-related restriction–modification systems (RMs) were evolving as linked genes, in contrast to EcoRI and RsrI systems (47); (vi) BsuFI and MspI share 45% overall amino acid sequence similarity, including three smaller regions with 60% identity. Interestingly, in spite of their close enzyme relatedness, the mspIRM genes have a divergent arrangement, while bsuFIRM genes have convergent organization (48); (vii) EcoHK31I and EaeI share 92% identity; their corresponding methyltranferases are both composed of two homologous subunits, α and β, regulated by the same alternative open reading frame mechanism. Moreover, some evidence shows that EcoHK31I and EaeI RMs were subjected to intergenic tranfer (49); (viii) BsuBI and PstI share 46% amino acid identity. The RMs have different genetic organization: pstIRM genes are transcribed divergently, while bsuBIRM are arranged in head-to-tail orientation, with bsuBIM preceding bsuBIR (50).
According to the above examples, both primary sequences of endonuclease and methyltransferase coding genes, within related RMs, as well as the gene organization, are subjected to intense evolutionary pressure. This results in a high evolutionary rate and variability, even amongst related RMs, where methyltransferase can evolve either separately or together with an endonuclease component, or even be horizontally transferred amongst different bacterial species (49).
Whether a homology of recognition and cleavage sites of TspDTI, TspGWI, TaqII and Tth111II (possibly including EciI, BcefI and BceAI as well) is mirrored by the similarity of biochemical properties, homology and organization of their coding genes and amino acid sequences, remains to be further evaluated.
Acknowledgments
ACKNOWLEDGEMENTS
DNA sequencing by Ireneusz Sobolewski is greatly acknowledged. The helpful suggestions regarding the manuscript by Jakub Zaleski, the figure preparation by Barbara Skowron-Zaleska and bacterial isolation by Robert Atras are appreciated. The N-terminal amino acid sequence analysis was performed at BioCenter (Jagiellonian University, Krakow, Poland). This project was supported by EURx Ltd funds available to P.M.S.
REFERENCES
- 1.Roberts R.J. and Halford,S.E. (1993) Type II restriction endonucleases. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 35–88. [Google Scholar]
- 2.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE—restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endlish B. and Linn,S. (1985) The DNA restriction endonuclease of Escherichia coli B. I, II. J. Biol. Chem., 260, 5720–5738. [PubMed] [Google Scholar]
- 4.Bickle T.A. (1993) The ATP-dependent restriction enzymes. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 89–109. [Google Scholar]
- 5.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes—a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 6.Kaczorowski T., Skowron,P. and Podhajska,A.J. (1989) Purification and characterization of FokI restriction endonuclease. Gene, 80, 209–216. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.C., Skowron,P.M. and Szybalski,W. (1996) Structural requirements for the FokI–DNA interaction and oligodeoxynucleotide-instructed cleavage. J. Mol. Biol., 258, 638–649. [DOI] [PubMed] [Google Scholar]
- 8.Skowron P.M., Kaczorowski,T., Tucholski,J. and Podhajska,A.J. (1993) Atypical DNA-binding properties of class-IIS restriction endonucleases: evidence for recognition of the cognate sequence by a FokI monomer. Gene, 125, 1–10. [DOI] [PubMed] [Google Scholar]
- 9.Wah D.A., Bitinaite,J., Schildkraut,I. and Aggarwal,A.K. (1998) Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitinaite J., Wah,D.A., Aggarwal,A.K. and Schildkraut,I. (1998) FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugisaki H. and Kanazawa,S. (1981) New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI). Gene, 16, 73–78. [DOI] [PubMed] [Google Scholar]
- 12.Sêktas M.S., Kaczorowski,T. and Podhajska,A.J. (1992) Purification and properties of the MboII, a class-IIS endonuclease. Nucleic Acids Res., 20, 4333–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucholski J., Skowron,P.M. and Podhajska,A. (1995) MmeI, a class-IIS restriction endonuclease: purification and characterization. Gene, 157, 87–92. [DOI] [PubMed] [Google Scholar]
- 14.Kim S.C., Podhajska,A.J. and Szybalski,W. (1988) Cleaving DNA at any predetermined site with adapter-primers and class-IIS restriction enzyme. Science, 240, 504–506. [DOI] [PubMed] [Google Scholar]
- 15.Kita K., Kotani,H., Sugisaki,H. and Takanami,M. (1989) The FokI restriction-modification system. I. Organization and nucleotide sequences of the restriction and modification genes. J. Biol. Chem., 264, 5751–5756. [PubMed] [Google Scholar]
- 16.Kita K., Suisha,M., Kotani,H., Yanase,H. and Kato,N. (1992) Cloning and sequence analysis of the StsI restriction-modification gene: presence of homology to FokI restriction-modification enzymes. Nucleic Acids Res., 20, 4167–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.-G. and Chandrasegaran,S. (1994) Chimeric restriction endonuclease. Proc. Natl Acad. Sci. USA, 91, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang B.H., Schaeffer,C.J., Li,Q.H. and Tsai,M.D. (1996) SPlase—a new class IIS zinc-finger restriction endonuclease with specificity for SP1 binding sites. J. Prot. Chem., 15, 481–489. [DOI] [PubMed] [Google Scholar]
- 19.Szybalski W. (1985) Universal restriction endonucleases: designing novel cleavage specificities by combining adapter oligonucleotide and enzyme moieties. Gene, 40, 169–173. [DOI] [PubMed] [Google Scholar]
- 20.Janulaitis A.M., Petrusyte,Z., Maneliene,S., Klimasauskas,S. and Butkus,V. (1992) Purification and properties of the Eco57I restriction endonuclease and methylase—prototypes of a new class (type IV). Nucleic Acids Res., 20, 6043–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrusyte M.P., Bitinaite,J.B., Menkevicius,S., Klimasauskas,S., Butkus,V. and Janulaitis,A. (1988) Endonuclease of a new type. Gene, 74, 89–91. [DOI] [PubMed] [Google Scholar]
- 22.Kong H., Roemer,S.E., Waite-Rees,P.A., Benner,J.S., Wilson,G.G. and Nwankwo,D.O. (1994) Characterization of BcgI, a new kind of restriction-modification system. J. Biol. Chem., 269, 683–690. [PubMed] [Google Scholar]
- 23.Sears L.E., Zhou,B., Aliotta,J.M., Morgan,R.D. and Kong,H. (1996) BaeI, another unusual BcgI-like restriction endonuclease. Nucleic Acids Res., 24, 3590–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piekarowicz A., Gołaszewska,M., Sunday,A.O., Siwińska,M. and Stein,D.C. (1999) The HaeIV restriction modification system of Haemophilus aegypticus is encoded by a single polypeptide. J. Mol. Biol., 293, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 25.Xia Y., Burbank,D.E., Uher,L., Rabussay,D. and Van Etten,J.L. (1987) IL-3A virus infection of a Chlorella-like green alga induces a DNA restriction endonuclease with novel sequence specificity. Nucleic Acids Res., 15, 6075–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skowron P.M, Swaminathan,N., McMaster,K., George,D., Van Etten,J. and Mead,D. (1995) Cloning and application of the two/three-base restriction endonuclease R.CviJI from IL-3A virus-infected Chlorella. Gene, 157, 37–41. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan N., Mead,D., McMaster,K., George,D., Van Etten,J. and Skowron,P.M. (1996) Molecular cloning of the three base restriction endonuclease R.CviJI from eukaryotic Chlorella virus IL-3A. Nucleic Acids Res., 24, 2463–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapranauskas R., Sasnauskas,G., Lagunavicius,A., Vilkaitis,G., Lubys,A. and Siksnys,V. (2000) Novel subtype of type IIS restriction enzymes—BfiI endonuclease exhibits similarities to the EDTA-resistant Nuc of Salmonella typhimurium. J. Biol. Chem., 275, 30878–30885. [DOI] [PubMed] [Google Scholar]
- 29.Jo K. and Topal,M.D. (1995) DNA topoisomerase and recombinase activities in NaeI restriction endonuclease. Science, 267, 1817–1820. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Mead D.A., Szczęsna-Skorupa,E. and Kemper,B. (1986) Single-stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng., 1, 67–74. [DOI] [PubMed] [Google Scholar]
- 32.Żylicz-Stachula A., Harasimowicz-Słowińska,R.I., Sobolewski,I. and Skowron,P.M. (2002) TspGWI, a thermophilic class-IIS restriction endonuclease from Thermus sp., recognizes novel asymmetric sequence 5′-ACGGAN11/N9-3′. Nucleic Acids Res., 30, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker D., Hoff,M., Oliphant,A. and White,R. (1984) A second type II restriction endonuclease from Thermus aquaticus with unusual sequence specificity. Nucleic Acids Res., 12, 5567–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinomiya T., Kobayashi,M. and Sato,S. (1980) A second site-specific endonuclease from Thermus thermophilus 111, Tth1111II. Nucleic Acids Res., 8, 3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venetianer P. and Orosz,A. (1988) BcefI, a new type IIS restriction endonuclease. Nucleic Acids Res., 16, 3053–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutkowska S.M., Jaworowska,I. and Skowron,P.M. (2002) GenBank accession no. AY057443.
- 37.Klimasauskas S., Timinskas,A., Menkevicius,S., Butkiene,D., Butkus,V. and Janulaitis,A.A. (1989) Sequence motifs characteristic of DNA [cytosine-N4] methylases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res., 17, 9823–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson J.E. (1993) Restriction endonucleases and modification methylases. Curr. Opin. Struct. Biol., 3, 24–30. [Google Scholar]
- 39.Siksnys V., Timinskas,A., Klimasauskas,S., Butkus,V. and Janulaitis,A. (1995) Sequence similarity among type-II restriction endonucleases, related by their recognized 6-bp target and tetranucleotide-overhang cleavage. Gene, 157, 311–314. [DOI] [PubMed] [Google Scholar]
- 40.Withers B.E. and Dunbar,J.C. (1993) The endonuclease isoschizomers, SmaI and XmaI, bend DNA in opposite orientations. Nucleic Acids Res., 21, 2571–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubys A., Menkevicius,S., Timinskas,A., Butkus,V. and Janulaitis,A. (1994) Cloning and analysis of translational control for genes encoding the Cfr9I restriction-modification system. Gene, 141, 85–89. [DOI] [PubMed] [Google Scholar]
- 42.Ruan H., Lunnen,K.D., Scott,M.E., Moran,L.S., Slatko,B.E., Pelletier,J.J., Hess,E.J., Benner,J.,II, Wilson,G.G. and Xu,S. (1996) Cloning and sequence comparison of AvaI and BsoBI restriction-modification systems. Mol. Gen. Genet., 252, 695–699. [PubMed] [Google Scholar]
- 43.Ruan H., Lunnen,K.D., Pelletier,J.J. and Xu,S. (1997) Overexpression of BsoBI restriction endonuclease in E. coli, purification of the recombinant BsoBI, and identification of catalytic residues of BsoBI by random mutagenesis. Gene, 188, 35–39. [DOI] [PubMed] [Google Scholar]
- 44.Kiss A., Sain,B., Csordas-Toth,E. and Venetianer,P. (1977) A new sequence-specific endonuclease (Bsp) from Bacillus sphaericus. Gene, 1, 323–329. [DOI] [PubMed] [Google Scholar]
- 45.Cao W., Lu,J. and Barany,F. (1997) Nucleotide sequences and gene organization of TaqI endonuclease isoschizomers from Thermus sp. SM32 and Thermus filiformis Tok6A1. Gene, 197, 205–214. [DOI] [PubMed] [Google Scholar]
- 46.Cao W., Lu,J., Welch,S.G., Williams,R.A. and Barany,F. (1998) Cloning and thermostability of TaqI endonuclease isoschizomers from Thermus species SM32 and Thermus filiformis Tok6A1. Biochem. J., 333, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barany F., Danzitz,M., Zebala,J. and Mayer,A. (1992) Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes: comparison with the isoschizomeric TaqI enzymes. Gene, 112, 3–12. [DOI] [PubMed] [Google Scholar]
- 48.Kapfer W., Walter,J. and Trautner,T.A. (1991) Cloning, characterization and evolution of the BsuFI restriction endonuclease gene of Bacillus subtilis and purification of the enzyme. Nucleic Acids Res., 19, 6457–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K.F., Shaw,P.C., Picone,S.J., Wilson,G.G. and Lunnen,K.D. (1998) Sequence comparison of the EcoHK31I and EaeI restriction-modification systems suggests an intergenic transfer of genetic material. Biol. Chem., 379, 437–441. [DOI] [PubMed] [Google Scholar]
- 50.Xu G., Kapfer,W., Walter,J. and Trautner,T.A. (1992) BsuBI—an isospecific restriction and modification system of PstI: characterization of the BsuBI genes and enzymes. Nucleic Acids Res., 20, 6517–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]