Abstract
Proteolytic activity of the 20S proteasome is regulated by activators that govern substrate movement into and out of the catalytic chamber. However, the physiological relationship between activators, and hence the relative role of different proteasome species, remains poorly understood. To address this problem, we characterized the total pool of cytosolic proteasomes in intact and functional form using a single-step method that bypasses the need for antibodies, proteasome modification, or column purification. Two-dimensional Blue Native(BN)/SDS-PAGE and tandem mass spectrometry simultaneously identified six native proteasome populations in untreated cytosol: 20S, singly and doubly PA28-capped, singly 19S-capped, hybrid, and doubly 19S-capped proteasomes. All proteasome species were highly dynamic as evidenced by recruitment and exchange of regulatory caps. In particular, proteasome inhibition with MG132 markedly stimulated PA28 binding to exposed 20S α-subunits and generated doubly PA28-capped and hybrid proteasomes. PA28 recruitment virtually eliminated free 20S particles and was blocked by ATP depletion. Moreover, inhibited proteasomes remained stably associated with distinct cohorts of partially degraded fragments derived from cytosolic and ER substrates. These data establish a versatile platform for analyzing substrate-specific proteasome function and indicate that PA28 and 19S activators cooperatively regulate global protein turnover while functioning at different stages of the degradation cycle.
INTRODUCTION
The 20S proteasome is a cylindrical multicatalytic protease comprised of two stacked inner rings of proteolytically active β-subunits flanked by two outer rings of α-subunits (Lowe et al., 1995; Groll et al., 1997; Voges et al., 1999; Glickman and Ciechanover, 2002; Pickart and Cohen, 2004). In mammalian cells, proteasome activity is controlled by regulatory complexes (caps) that bind to the exposed ends of α-subunits and open the gate into and out of the catalytic chamber (Hoffman et al., 1992; DeMartino and Slaughter, 1999; Voges et al., 1999; Groll et al., 2000; Kloetzel and Ossendorp, 2004; Rechsteiner and Hill, 2005). One such cap, the 19S regulatory complex (PA700/RC), contains a hexameric ring of AAA-ATPases (base) and at least 12 additional subunits (lid) that recognize, unfold, and translocate polyubiquitinated proteins into the axial opening of the 20S core (Tanaka, 1998; Glickman et al., 1999; Strickland et al., 2000; Leggett et al., 2005; Liu et al., 2005). Two 19S RCs bind the 20S particle to form the doubly capped (30.3S) proteasomes (Yoshimura et al., 1993) that are generally believed to be the major species responsible for degrading polyubiquitinated proteins in the cell (Tanaka and Tsurumi, 1997; Voges et al., 1999; Wolf and Hilt, 2004). 19S RC binding to the 20S core particle requires ATP (Orino et al., 1991), and ATP hydrolysis transiently dissociates 20S and 19S particles, possibly to allow release of degradation products (Babbitt et al., 2005). Thus it has been proposed that the degradation cycle involves dynamic interactions between proteasome subcomplexes.
An alternate proteasome cap consists of ring-shaped heptamers formed by the small 28-kDa protein PA28α, β, or γ (REG α, β, or γ). PA28 was identified nearly a decade ago by the groups of DeMartino and Rechsteiner as an ATP-independent activator that stimulates the degradation of small peptides but not intact or polyubiquitinated proteins (Dubiel et al., 1992; Ma et al., 1992). High-resolution crystal structures have demonstrated that the PA28 homolog, PA26, induces a conformational change in the N-termini of 20S α-subunits that opens the entrance of the catalytic chamber (Whitby et al., 2000; Forster et al., 2005). PA28 (PA26) also alters the proteolytic properties of 20S and/or 19S-20S proteasomes by modifying the pattern, but not the overall size of cleaved products (Harris et al., 2001; Li et al., 2001; Cascio et al., 2002; Wang et al., 2003). PA28αβ subunits are particularly abundant in immune tissues and are coordinately regulated by IFNγ together with β1i, β2i, and β5i subunits of the immunoproteasome, ER peptide transporters (TAP1, TAP2), and MHC class I molecules (Früh and Yang, 1999; Rechsteiner et al., 2000; Kloetzel and Ossendorp, 2004). PA28 expression also stimulates formation of “hybrid” proteasomes containing both 19S and PA28 caps (Hendil et al., 1998; Tanahashi et al., 2000). Collectively, these and related findings have suggested that PA28 plays a primary role in antigen presentation by generating a distinct set of peptide fragments that preferentially bind MHC class I molecules (Kloetzel, 2004; Rechsteiner and Hill, 2005).
An important and unresolved issue in proteasome biology is how different regulatory caps contribute to proteolytic function in cells. This is in part due to limitations of conventional purification methodologies, e.g., multistep column chromatography, which are prone to loss of activators in high ionic strength buffers (Tanaka et al., 1988; Driscoll and Goldberg, 1990; Udvardy, 1993; Shibatani and Ward, 1995). Information on the function of PA28 and hybrid proteasomes has therefore been largely derived from in vitro reconstitution experiments using purified PA28, 20S and singly capped 19S-20S proteasomes (Kopp et al., 2001; Cascio et al., 2002; Rechsteiner and Hill, 2005). Although, 20S, 26S, and hybrid proteasomes have been isolated by immunoaffinity purification (Hendil et al., 1998; Saeki et al., 2000; Verma et al., 2000) and column chromatograpy under low ionic strength (Tanahashi et al., 2000), such approaches have had limited success in analyzing the in vivo relationship between different proteasome pools.
We now report a single-step purification scheme using Blue Native(BN)-PAGE that enables us for the first time, to characterize dynamic changes in the global complement of native cytosolic proteasomes. BN-PAGE was initially developed to purify and analyze multimeric membrane protein complexes from mitochondria (Schägger and von Jagow, 1991; Schägger et al., 1994) and has subsequently been extended to the endoplasmic reticulum, chloroplast and peroxisome (Caliebe et al., 1997; Wang and Dobberstein, 1999; Reguenga et al., 2001; Shibatani et al., 2005) as well as cytosolic proteins including proteasomes (Camacho-Carvajal et al., 2004). This technique allows intact multimeric protein complexes to be separated by size with a high degree of precision in the first dimension using Coomassie G250 dye as the negative charge carrier. Components from each complex can then be identified by SDS-PAGE in the second dimension. In combination with liquid chromatography/tandem mass spectrometry, (LC-MS/MS), we simultaneously isolated six major proteasome populations from resting cytosol and identified these species as 20S, singly and doubly PA28-capped (PA28-20S and PA28-20S-PA28), singly 19S-capped (26S), hybrid (PA28-20S-19S), and doubly 19S-capped (30.3S) proteasomes. All proteasome species were highly dynamic, as evidenced by rapid exchange of regulatory complexes in response to cytosolic manipulation. Moreover, proteasome inhibition further stimulated PA28 recruitment and rapidly generated doubly PA28-capped and hybrid proteasomes that remained stably associated with specific subsets of degradation intermediates. These data indicate that PA28 and 19S activators cooperate in general protein turnover and function at different stages of the degradation cycle.
MATERIALS AND METHODS
Preparation and Analysis of Proteasome Fractions
Rabbit reticulocytes lysate (RRL) was prepared by hypotonic lysis (1:1 volume with ddH2O) of reticulocytes and stored at −80°C as described (Carlson et al., 2005). Ribosomes were removed from 25-μl aliquots of RRL by centrifugation (350,000 × g for 1 h at 4°C). Ribosome-free RRL was then diluted fivefold in 20 mM Tris-HCl, pH 7.6, 1 mM DTT, 1 mM ATP, and 1 mM MgCl2 and centrifuged at 350,000 × g for 2 h at 4°C. The resulting proteasome pellets were resuspended in 30 μl of buffer A (50 mM HEPES-NaOH, pH 7.8, 75 mM NaCl, 7.5 mM MgOAc2, 2.5 mM DTT, and 7.5% [wt/vol] glycerol). Where indicated, ribosome-free RRL was incubated with the following: 1) 1.0 mM ATP, 12 mM creatine phosphate, and 80 μg/ml creatine kinase (Roche, Indianapolis, IN); or 2) 20 U/ml hexokinase (Sigma, St. Louis, MO) and 20 mM 2-deoxyglucose; or 3) 100 μM Z-Leu-Leu-Leu-CHO (MG132; Sigma) in buffer containing 18 mM sucrose, 18 mM KCl, 3 mM DTT, 5 mM MgCl2, 10 mM Tris-HCl, pH7.5, for 0–60 min at 37°C. Proteasomes were then isolated by ultracentrifugation as described above.
BN-PAGE, Native-PAGE, and Two-dimensional BN/SDS-PAGE
BN-PAGE was based on the protocol described by Schägger et al. (1994). Crude proteasome pellets were resuspended in buffer A, loaded directly onto BN-PAGE gels (cast as a 5–13% acrylamide gradient [acrylamide:bis-acrylamide = 32:1] in 50 mM BisTris/HCl, pH 7.0, 500 mM ε-aminocaproic acid [EACA], and overlaid by 4% stacking gel in the same buffer). Electrophoresis was carried out at 4°C at 100 V for 17 h (cathode buffer = 50 mM Tricine, 15 mM BisTris-HCl [pH 7.0] and 0.02% Coomassie G250 [Serva Blue G; Serva Electrophoresis GmbH, Heidelberg, Germany], anode buffer = 50 mM BisTris-HCl, pH 7.0) and then at 500 V for 24 h after reducing Coomassie G250 dye concentration to 0.002%. Thyroglobulin (670 kDa) ferritin (440 kDa) and bovine serum albumen (dimer,133 kDa) were used as molecular weight markers. Clear Native-PAGE (Schägger et al., 1994; Wittig and Schägger, 2005) was carried out in the same manner as BN-PAGE except that the EACA concentration in gel was reduced to 50 mM and Coomassie G-250 dye was omitted in cathode buffer. For Coomassie staining, the gel was fixed in 35% (vol/vol) methanol, 10% (vol/vol) acetic acid, washed three times with H2O, and incubated in 8% ammonium sulfate, 0.816% phosphoric acid, 0.08% Coomassie G-250, and 20% methanol. Two-dimensional (2D) SDS-PAGE was performed by treating BN-PAGE strips with 1% DTT in 62.5 mM Tris-HCl, pH 6.8, 10% (wt/vol) glycerol, 2% SDS for 20 min at 24°C and alkylating with 2.5% iodoacetamide in 62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS for 20 min at 24°C. Gel strips were then placed over an SDS-PAGE (12–17% gradient, 4% stacking gel) and run at room temperature. Gels were scanned using a Umax Power Look III transmission scanner (Lexmark, Dallas, TX). Coomassie-stained bands were quantitated using a Li-Cor Odyssey infrared imaging system (700-nm channel) V.1.2 (Lincoln, NE).
Preparation of Samples for Mass Spectrometry
Silver staining was performed according to the modified Blum method, and samples were extracted by tryptic digestion (Blum et al., 1987; Mortz et al., 2001; Shibatani et al., 2005). Gels were fixed in 40% (vol/vol) ethanol and 10% (vol/vol) acetic acid, washed three times with 30% (vol/vol) ethanol for 20 min, and washed once with ddH2O. The gel was sensitized with 0.02% NaS2O3, washed three times with ddH2O, incubated with 0.1% silver nitrate for 20 min at 4°C, rinsed three times, and developed with 3% Na carbonate and 0.05% formalin. Protein bands were excised from silver-stained gels, dried, destained in 15 mM K ferricyanide and 50 mM NaS2O3 at 24°C for 15 min, and washed with 100 mM NH4HCO3. Destained gels fragments were dried, rehydrated with 0.01 mg/ml sequence-grade modified porcine trypsin (Promega, Madison, WI) in 50 mM NH4HCO3 and 5 mM CaCl2, and incubated at 37°C overnight. Trypsinized fragments were collected by bath sonication in 50 μl of 25 mM NH4HCO3 and again after adding 50 μl of 50% acetonitrile. The supernatant was collected, and the gel fragment was sonicated repeatedly in 50 μl of 5% formic acid and again after adding 50 μl of 50% acetonitrile. Supernatants were pooled. DTT was added to a final concentration of 1 mM. The sample was dried and stored at −80°C before analysis.
Mass Spectrometry
Samples were dissolved in 5% formic acid before liquid chromatography using a 10 cm × 180 μm Biobasic-C18 capillary column (Thermo Hypersil Keystone, West Palm Beach, FL) and analysis by electrospray ionization tandem mass spectrometry (MS/MS) using an LCQ Deca XP Plus ion trap mass spectrometer (ThermoElectron, San Jose, CA). Peptides were eluted using a mobile phase of 0.1% formic acid in water and either a 30-min 0–50% acetonitrile gradient or a 110-min 0–45% acetonitrile gradient. The instrument was set to trigger data-dependent MS/MS acquisition of the three most intense ions detected during the MS survey scan. Mass spectra were analyzed with the Sequest algorithm (ThermoElectron) as described by Eng and coworkers (Eng et al., 1994) using the UniProt/Swiss-Prot protein database (Uniprot release 5.1, URL: http://www.us.expasy.org/, Accessed 5/13/05). Search results were further analyzed by PeptideProphet (Keller et al., 2002) and ProteinProphet (Nesvizhskii et al., 2003) (Seattle Proteome Center, URL: http://tools.proteomecenter.org/windows.php). Proteins identified by ProteinProphet utilized a probability cutoff of p > = 0.99, which corresponded to an error of 0.002. Peptides were included in the coverage calculation when the adjusted PeptideProphet probability was greater than or equal to 0.9. All results were manually inspected.
Peptidase Assays
RRL proteasomes were preincubated for 30 min on ice in 50 mM BisTris-HCl, pH 7.0, containing 50 or 500 mM EACA or 0.01% of Coomassie G250. Reactions were then incubated for 30 min at 37°C with 100 μM N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (sLLVY-AMC) in buffer B (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 10 mM KCl, 1 mM DTT, 1 mM ATP) in the presence of 50 or 500 mM EACA or 0.0012% Coomassie G250. The reaction was stopped by adding 2 volumes of 100% ethanol, and AMC fluorescence was measured at 380 nm/440 nm excitation/emission (Hoffman et al., 1992; Oberdorf et al., 2001). For in-gel assays, BN- and CN-PAGE gel strips were incubated at 37°C for 30 min with 200 μM sLLVY-AMC in buffer B. The gel was exposed and photographed using a UV transiluminator (Gel Doc 1000, Bio-Rad, Hercules, CA).
Western Blotting
Proteins were transferred to PVDF membranes, blocked with 5% dried milk, incubated with rabbit PA28β antisera (1:2000 dilution) raised against peptides encoding C-terminal 21 amino acid residues of PA28β (Ahn et al., 1996) or anti-proteasome antisera raised against the recombinant murine α-3 20S subunit (Yang et al., 1995). Signals were obtained using horseradish peroxidase–conjugated goat anti-rabbit secondary antibody (1:5000 dilution; Bio-Rad) using SuperSignal (Pierce, Rockford, IL) according to the manufacturer's instructions.
Protein Degradation and Release Assay
Cystic fibrosis transmembrane conductance regulator (CFTR) was translated in vitro in RRL supplemented with [35S]Met and canine pancreas rough microsomal membranes (Carlson et al., 2005). Microsomes containing integrated CFTR were isolated by pelleting (180,000 × g for 10 min) through 0.5 M sucrose in buffer C (100 mM KoAc, 5 mM Mg(OAc)2, 1 mM DTT, and 50 mM HEPES-NaOH, pH 7.5) and resuspended in buffer C containing 0.1 M sucrose. In vitro degradation conditions were exactly as described elsewhere (Carlson et al., 2005). Samples were incubated for 2 h at 37°C in the presence of 100 μM MG132. Cytosolic CFTR fragments released during the reaction were recovered by pelleting membranes through 0.5 M sucrose in buffer C (180,000 × g for 10 min). Proteasomes in the supernatant were subsequently isolated by ultracentrifugation as above and separated by 2D BN-PAGE/SDS-PAGE. Gels were fixed, silver-stained, dried, and exposed to Kodak film (Eastman Kodak, Rochester, NY). 14C-methylated lysozyme (Sigma) was incubated in the identical RRL degradation system. Proteasomes were separated by BN-PAGE, and the gel was stained with silver or Coomassie G-250 and imaged by autoradiograpy.
RESULTS
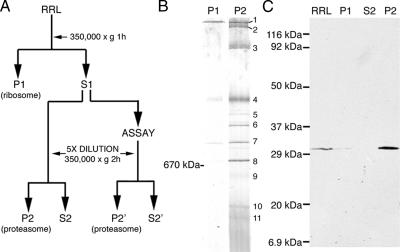
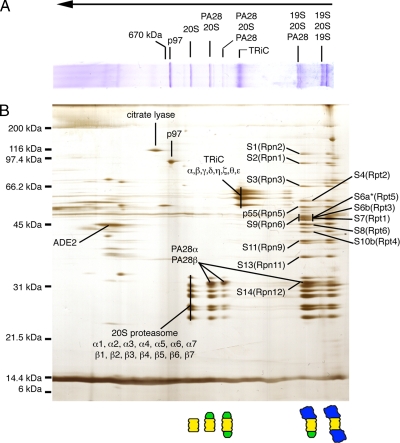
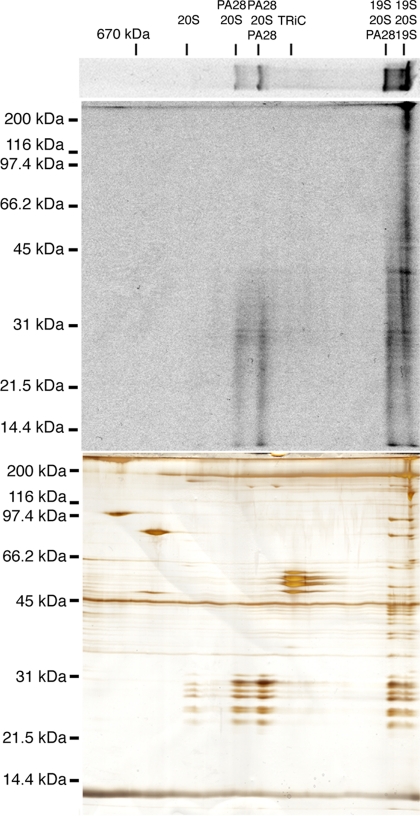
Cytosolic proteasomes were collected from RRL by differential sedimentation (Figure 1A), and crude pellets were resuspended and analyzed directly by BN-PAGE (Figure 1B) or SDS-PAGE (Figure 1C). Eleven distinct protein bands (>440 kDa in size) were readily detected by Coomassie staining (Figure 1B). Immunoblotting for the 20S α3-subunit (Figure 1C) and quantitation of Coomassie stained gels revealed that this process recovered ∼90% of total RRL proteasomes for analysis. BN-PAGE gel strips were subsequently analyzed in the second dimension by SDS-PAGE (Figure 2B), and silver-stained bands were then excised and subjected to tryptic digestion, liquid chromatography, and tandem mass spectrometry (LC-MS/MS). Uninterpreted MS/MS spectra were analyzed using the Sequest algorithm, and results were filtered using PeptideProphet and ProteinProphet as described in Materials and Methods. Major proteins identified in this analysis included all 14 α and β subunits of the 20S proteasome, PA28α and PA28β, 14 subunits of the 19S RC, all 8 subunits of the cytosolic chaperonin TRiC and p97(VCP/cdc48; Figure 2B and Table 1).
Figure 1.
(A) Strategy for isolation of RRL proteasomes by differential centrifugation. (B) The initial ribosome pellet (P1) and proteasome pellet (P2) were separated by BN-PAGE and stained with Coomassie G-250. A single major species predominated in P1 fraction, and 11 major bands were visualized in P2, several of which appeared as closely spaced doublets. (C) RRL, P1, S2, and P2 fractions were analyzed by SDS-PAGE and Western blotting using antisera raised against the 20S α3-subunit.
Figure 2.
(A) BN-PAGE gel from proteasome fraction (P2) was subjected to SDS-PAGE in the second (vertical) dimension. (B) Silver-stained 2D-BN/SDS-PAGE gel. Bands of interest were excised, trypsin-digested, and subjected to analysis by LC-MS/MS. Identities and corresponding locations of proteins are indicated. 19S RC subunits are based on nomenclature of human erythrocyte proteasomes (Dubiel et al., 1995) with corresponding yeast subunits (Saccharomyces cerevisiae) shown in parentheses (Finley et al., 1998). Identity and location of capped proteasome populations is diagrammed at the bottom. 20S proteasome is shown in yellow, PA28 in green, and 19S RC in blue. Migration of thyroglobulin marker (670 kDa) in BN-PAGE is indicated on the top.
Table 1.
Major proteins identified in this analysis
| Complex | Subunits | Accession number |
MW | Peptides ID | Sequence coverage (% amino acids) | |
|---|---|---|---|---|---|---|
| Swissprot | NCBI | |||||
| 20S | α1 | PSA6_HUMAN | 46397659 | 27399 | 10 | 42.3 |
| α2 | PSA2_HUMAN | 130850 | 25767 | 5 | 21.5 | |
| α3 | PSA4_HUMAN | 130861 | 29484 | 6 | 21.8 | |
| α4 | PSA7_HUMAN | 12643540 | 27887 | 8 | 24.6 | |
| α5 | PSA5_HUMAN | 38258905 | 26411 | 6 | 34.4 | |
| α6 | PSA1_HUMAN | 130848 | 29556 | 9 | 35.4 | |
| α7 | PSA3_HUMAN | 130859 | 28302 | 8 | 27.6 | |
| β1 | PSB6_HUMAN | 20532407 | 25358 | 3 | 13.0 | |
| β2 | PSB7_HUMAN | 17380263 | 29965 | 5 | 17.3 | |
| β3 | PSB3_HUMAN | 20532411 | 22949 | 8 | 40.0 | |
| β4 | PSB2_HUMAN | 1709762 | 22836 | 6 | 31.3 | |
| β5 | PSB5_HUMAN | 1172607 | 22897 | 8 | 36.5 | |
| β6 | PSB1_HUMAN | 130853 | 26489 | 5 | 24.5 | |
| β7 | PSB4_HUMAN | 3915810 | 29192 | 2 | 13.3 | |
| PA28 | α | PSME1_HUMAN | 1170519 | 28723 | 3 | 14.5 |
| β | PSME2_HUMAN | 18203506 | 27230 | 3 | 12.6 | |
| 19S | S1 (Rpn2) | PSD1_HUMAN | 51704332 | 105836 | 3 | 3.9 |
| S2 (Rpn1) | PSD2_HUMAN | 6174930 | 100200 | 4 | 5.0 | |
| S3 (Rpn3) | PSD3_HUMAN | 20532405 | 60978 | 16 | 26.4 | |
| p55 (Rpn5) | PSD12_HUMAN | 20978544 | 52904 | 4 | 8.3 | |
| S4 (Rpt2) | PRS4_HUMAN | 49065817 | 49185 | 3 | 7.0 | |
| S6a (Rpt5) | PRS6A_HUMAN | 20532406 | 49204 | 1a | 2.1 | |
| S6b (Rpt3) | PRS6B_HUMAN | 20532409 | 47366 | 4 | 11.7 | |
| S7 (Rpt1) | PRS7_HUMAN | 547930 | 48503 | 8 | 21.5 | |
| S8 (Rpt6) | PRS8_HUMAN | 49065819 | 45626 | 7 | 19.0 | |
| S9 (Rpn6) | PSD11_HUMAN | 20978543 | 47333 | 12 | 32.8 | |
| S10a (Rpt4) | PRS10_HUMAN | 51702772 | 44173 | 5 | 14.1 | |
| S11 (Rpn9) | PSD13_HUMAN | 20978558 | 42918 | 7 | 17.8 | |
| S13 (Rpn11) | PSDE_HUMAN | 51701716 | 34577 | 2 | 6.5 | |
| S14 (Rpn12) | PSD8_HUMAN | 1346766 | 30005 | 2 | 7.0 | |
| p97 | TERA_HUMAN | 6094447 | 89191 | 20 | 30.3 | |
| TRiC | α | TCPA_HUMAN | 135538 | 60344 | 11 | 21.9 |
| β | TCPB_HUMAN | 6094436 | 57357 | 13 | 28.3 | |
| γ | TCPG_HUMAN | 66774185 | 60534 | 18 | 33.9 | |
| δ | TCPD_HUMAN | 52001478 | 57793 | 7 | 16.2 | |
| η | TCPH_HUMAN | 3041738 | 59367 | 18 | 39.0 | |
| ζ | TCPZ_HUMAN | 730922 | 57893 | 7 | 19.2 | |
| θ | TCPQ_HUMAN | 9988062 | 59489 | 18 | 35.8 | |
| ε | TCPE_HUMAN | 1351211 | 59671 | 11 | 20.3 | |
| Citrate lyase | ACLY_HUMAN | 20141248 | 120825 | 16 | 18.3 | |
| ADE2 | PUR6_HUMAN | 131628 | 46948 | 9 | 25.2 | |
a Only one peptide fragment was identified.
Based on the signature pattern of the 20S core subunits, 5 of the 11 protein complexes visualized by BN-PAGE were unequivocally identified as proteasomes. The smallest species was comprised exclusively of 20S α- and β-subunits and thus represented free 20S particles (∼750 kDa). Two slightly larger species contained an additional pair of proteins identified as PA28α and PA28β. The size of these complexes and the relative intensity of PA28 staining indicate that they represent singly and doubly PA28-capped proteasomes, respectively (∼900 and ∼1100 kDa). The two largest proteasome species contained 20S particles and at least 14 known subunits of the 19S RC. One of these contained 19S RC and PA28αβ and therefore represents hybrid (PA28-20S-19S) proteasomes. This complex migrated as a closely spaced doublet and also contained singly 19S-capped (19S-20S) proteasomes (see also Figure 4B). In contrast, the largest complex lacked PA28 and contained 19S and 20S subunits and constitutes doubly 19S-capped (19S-20S-19S) proteasomes. Thus, in a single-step purification, we show that unstimulated cytosol contains substantial amounts of 20S, PA28-20S, 19S-20S, PA28-20S-19S, and 19S-20S-19S proteasomes and relatively small amounts of PA28-20S-PA28 proteasomes.
Figure 4.
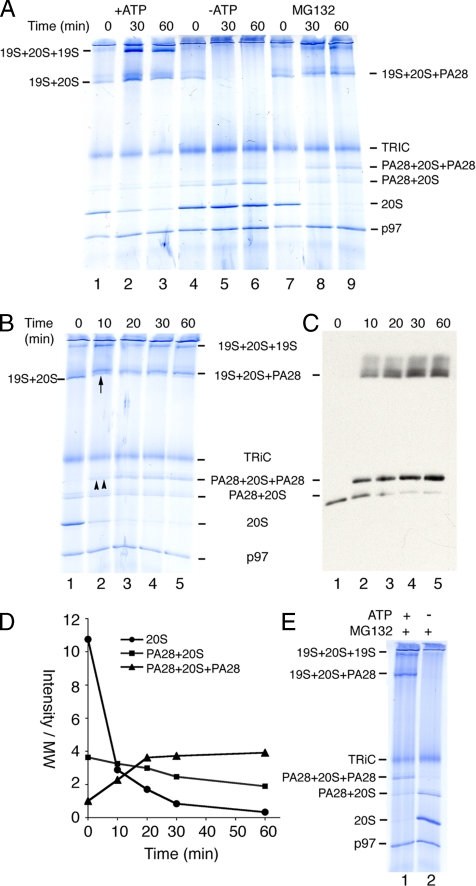
(A) Ribosome-free RRL was incubated at 37°C under the conditions indicated and proteasomes were pelleted, separated by BN-PAGE, and subjected to Coomassie staining. Migration of proteasomes and TRiC is indicated on right. (B and C) Time course of RRL incubation in 100 μM MG132. (B) Coomassie staining and (C) immunoblotting for PA28 revealed rapid formation of doubly capped PA28 and hybrid proteasomes (double and single upward arrows, respectively; lane 2). (D) Quantitation of Coomassie-stained gels showing time course of PA28 recruitment and corresponding decrease in free 20S and singly capped PA28 proteasomes. (E) ATP supplemented (lane 1) or ATP depleted (lane 2) RRL was incubated for 1 h with MG132, and proteasomes were then pelleted and analyzed as in A.
Purified Proteasomes Retain Peptidase Activity
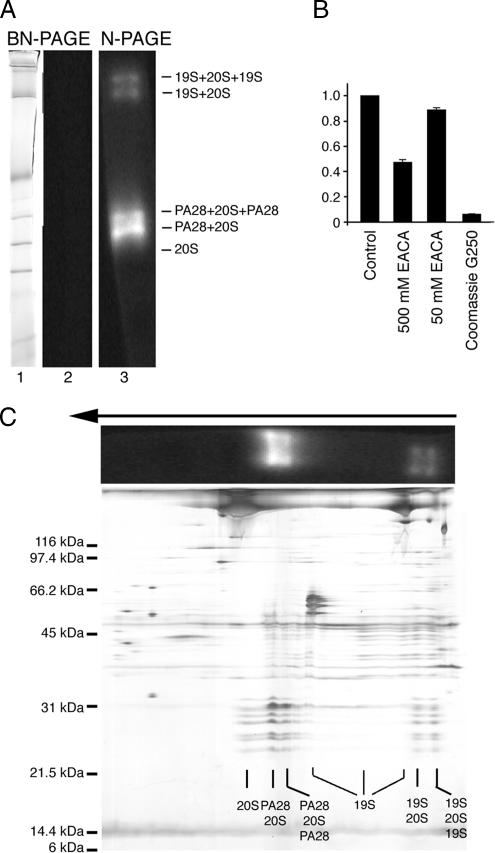
To determine whether isolated proteasomes retained function, in-gel assays were performed using the fluorogenic peptide substrate sLLVY-AMC. Initial experiments using BN-PAGE gels failed to reveal peptidase activity (Figure 3A, lanes 1 and 2), suggesting that components in the gel might inhibit proteasomes. Consistent with this, we showed that Coomassie G-250 was a potent inhibitor of chymotrypsin-like activity (>90% inhibition), whereas a high concentrations of ε-aminocaproic acid (500 mM) produced a mild reduction (50%) in substrate cleavage (Figure 3B). RRL was therefore subjected to a variation of BN-PAGE (previously termed Clear-Native PAGE; Schägger et al., 1994; Wittig and Schägger, 2005) using similar gel conditions conditions but omitting Coomassie G-250 dye in the cathode buffer and reducing the EACA concentration to 50 mM. As previously reported (Wittig and Schägger, 2005), native-PAGE resulted in less resolution and a more complex staining pattern than BN-PAGE (Figure 3C). However, five distinct 20S proteasome complexes were identified, and in-gel assays demonstrated that four of these retained peptidase activity. These active complexes also comigrated with PA28-capped and 19S-capped proteasomes (Figure 3, A, lane 3, and C). Given the reduced resolution of native-PAGE, however, we could not conclusively determine whether hybrid and singly 19S-capped proteasomes were both present in the second largest band. As expected, isolated 20S particles lacked proteolytic activity due to the native closed conformation of α-subunit N-termini (Groll et al., 2000). Several heterogeneous bands containing 19S RC subunits were also visualized by native PAGE. This confirmed the presence of 19S RC in RRL, which was likely partially dissociated during electrophoresis due to the increased lability of free particles. Intact 19S RC was not visualized by BN-PAGE, suggesting that it is also sensitive to the harsher gel conditions.
Figure 3.
(A) RRL proteasomes were separated by BN-PAGE or CN-PAGE. Gel strips were incubated with sLLVY-AMC and Coomassie stained (lane 1) or photographed using a UV transilluminator Gel-doc (Bio-Rad; lanes 2 and 3). (B) Peptidase activity of RRL proteasomes was determined using sLLVY-AMC in the presence of 0 mM (control), 50 mM, and 500 mM EACA or 0.01% Coomassie G-250. Values were normalized to control conditions. (C) Proteasomes separated by CN-PAGE (A) were subsequently separated by SDS-PAGE to verify the location and composition of active proteasomes.
Proteasome Inhibition Stimulates PA28 Binding to 20S Particles
We next investigated whether RRL proteasomes represented static or dynamic populations. Incubation at 37°C in the presence of ATP substantially increased the fraction of 19S capped proteasomes and correspondingly decreased free 20S particles (Figure 4A, lanes 1–3). In contrast, ATP depletion caused rapid dissociation of 19S-capped complexes as predicted (Orino et al., 1991) and resulted in a slight increase in singly PA28-capped proteasomes, presumably due to release of 19S RC from hybrid proteasomes (Figure 4A, lanes 4–6). Surprisingly, β-subunit inhibition also had a dramatic effect on the proteasome profile. MG132, a potent inhibitor that blocks all three catalytic β-subunit activities (Rock et al., 1994; Bogyo et al., 1997; Oberdorf et al., 2001), markedly stimulated PA28 recruitment and rapidly converted nearly all 20S proteasomes to doubly PA28-capped proteasomes (Figure 4, A, lanes 7–9, and B). Although a mild increase in 19S binding was observed for some experiments, this effect was not reproducible. MG132 also stimulated the formation of hybrid proteasomes as demonstrated by a slight increase in size of the 19S-20S proteasome complex (Figure 4B) and PA28 immunoblotting (Figure 4C). Within minutes of adding inhibitor, PA28 was bound to essentially all free ends of exposed α-subunits (Figure 4, C and D). When MG132 was added after ATP depletion, however, 19S caps were released but no additional doubly capped PA28 or hybrid proteasomes were formed (Figure 4E). Thus, even though PA28 does not require ATP to bind the 20S core in vitro and addition of cytosolic ATP by itself did not appreciably stimulate PA28 association (Figure 4A), PA28 recruitment in cytosol is controlled by ATP-dependent mechanisms (discussed further below).
19S and PA28 Caps Cooperate in Degrading Soluble and Membrane-bound Substrates
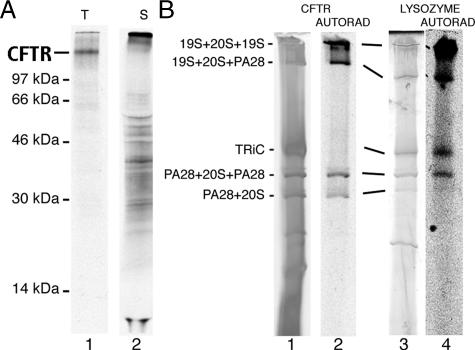
The hallmark of proteasome-mediated degradation is ATP-dependent conversion of substrate into small, trichloroacetic acid (TCA)-soluble peptides ∼8–20 aa in length. However, we previously showed that when proteolytic β-subunits are severely compromised (e.g., by MG132), substrates continue to be unfolded, and large degradation intermediates accumulate and remain bound to proteasomes (Oberdorf et al., 2006). These findings prompted us to investigate which population(s) of proteasomes participate in the degradation (and sequestration) of specific substrates. Two well-defined substrates were examined, the polytopic membrane protein CFTR and the soluble protein lysozyme, both of which are efficiently degraded into TCA soluble fragments by the RRL ubiquitin-proteasome pathway (Hershko et al., 1984; Xiong et al., 1999; Oberdorf et al., 2001).
To identify proteasome species involved in degradation, we used an in vitro system previously developed in our laboratory that reconstitutes biosynthesis and ER-associated degradation (ERAD) of misfolded integral membrane proteins (Xiong et al., 1999; Oberdorf et al., 2001; Carlson et al., 2005). In this system, in vitro–translated CFTR generates full length (∼160 kDa) 35S-labeled protein that is cotranslationally glycosylated and integrated into ER microsomal membranes (Figure 5A, lane 1, and Xiong et al., 1999). When microsomes were isolated and incubated in RRL in the presence of MG132, large heterogeneous CFTR-derived fragments were released from the membrane and accumulated in the cytosol (Figure 5A, lane 2). Previous analyses has shown that these cytosolic fragments originate from membrane-bound, newly synthesized CFTR as a result of ATP-dependent retrotranslocation (Carlson et al., 2006; Oberdorf et al., 2006). When analyzed by BN-PAGE, cytosolic CFTR degradation intermediates specifically comigrated with four distinct complexes that were identified as singly and doubly PA28-capped, hybrid, and doubly 19S-capped proteasomes (Figure 5B). Similarly, 14C lysozyme was also recovered together with PA28-capped, hybrid, and 19S-capped proteasomes in the presence of MG132 (Figure 5B, lanes 3 and 4). Thus sequestration of degradation intermediates by inhibited, capped proteasomes is a general process that is not restricted to ERAD substrates. Interestingly, lysozyme was also recovered with the cytosolic TRiC complex. This suggests that a subset of lysozyme molecules may also be involved in refolding and that both proteasomes and TRiC maintain their native structure and substrate-binding properties during BN-PAGE purification.
Figure 5.
(A) Autoradiogram of SDS-PAGE gel showing translation products (t) for in vitro–translated CFTR (lane 1) and cytosolic degradation intermediates released into the supernatant (s) in the presence of MG132 (lane 2). Input for lane 2 is six times that of lane 1 to better visualize the heterogeneous distribution of degradation products. (B) BN-PAGE analysis of cytosolic fragments of [35S]-CFTR (lanes 1 and 2) and 14C-lysozyme (lanes 3 and 4) after degradation in RRL in the presence of MG132. Migration of proteasomes was determined by silver or Coomassie staining (lanes 1 and 3, respectively), and gels were then subjected to autoradiography (lanes 2 and 4). Migration of hybrid proteasomes varied somewhat because of gel conditions. CFTR fragments comigrated precisely with four distinct proteasome species (lane 2), whereas lysozyme was bound to both proteasomes and the TRiC complex (lane 4).
19S and PA28 Capped Proteasomes Bind Different Cohorts of Degradation Intermediates
Finally, we tested whether different proteasome species sequester different distributions of degradation products by comparing autoradiograms and silver-stained 2D BN/SDS-PAGE gels (Figure 6). These results confirmed that CFTR fragments comigrated precisely with specific proteasome species. Doubly 19S-capped proteasomes remained bound to the entire range of CFTR degradation products including fragments larger than CFTR (>160 kDa), which presumably contain polyubiquitin chains that have not yet been removed. In contrast, singly and doubly PA28-capped proteasomes bound primarily to smaller fragments less than ∼40 kDa, whereas hybrid proteasomes contained small and intermediate sized fragments (<40 to ∼60 kDa). Thus each capped proteasome subtype sequestered a distinct but overlapping subpopulation of degradation intermediates that were derived from a single full-length substrate. Interestingly, no CFTR fragments were recovered with the chaperonin complex TRiC or with p97, an AAA-ATPase that specifically stimulates CFTR degradation by facilitating extraction of its transmembrane domains (Gnann et al., 2004; Carlson et al., 2006; Oberdorf et al., 2006). Thus although p97 likely interacts with CFTR as degradation is initiated, it appears to be released after the substrate is transferred to the proteasome.
Figure 6.
BN-PAGE gel strip from Figure 5 containing cytosolic CFTR fragments was separated by 2D SDS-PAGE and analyzed by autoradiography (top) and silver staining (bottom).
DISCUSSION
This study describes the first simultaneous isolation and characterization of total mammalian cytosolic proteasome pools in an intact, native and functional form. 2D BN/SDS-PAGE provides a relatively simple, high-resolution method for single-step isolation that bypasses the need for multiple column chromatography, proteasome antibodies, or proteasome subunit modification. Together with LC-MS/MS, this approach unambiguously identified six different proteasome species as 20S, singly and doubly PA28-capped, singly 19S-capped, hybrid, and doubly 19S-capped proteasomes. With the exception of doubly PA28-capped proteasomes, which were present in relatively low quantities, substantial amounts all other proteasome species were readily identified in freshly thawed, untreated RRL (Figure 2). Native-PAGE confirmed proteolytic activity in at least four proteasome species. Although native-PAGE has been widely used to study the composition, assembly, and functional characteristics of purified 20S and 19S RC particles (Leggett et al., 2002, 2005; Elsasser et al., 2005), the improved resolution of BN-PAGE now allows similar analyses to be performed directly from crude cellular lysates (Camacho-Carvajal et al., 2004). This enabled us to demonstrate that cellular proteasomes are remarkably dynamic and undergo rapid exchange and/or redistribution of regulatory caps in response to changes in the cellular environment. Interestingly, PA28-capped proteasomes constitute a significant fraction of the proteasome pool and appear to participate in degrading diverse substrates. These studies thus establish a powerful tool to monitor the physiological composition and organization of the global proteasome pool and to investigate how different regulatory caps carry out proteolytic activities in cells.
Although the existence of differentially capped proteasomes was established more than a decade ago (Hoffman et al., 1992), little is known regarding the in vivo relationships between different proteasome species, in part because of technical difficulties in isolation and the lability of native complexes (Tanaka et al., 1988; Driscoll and Goldberg, 1990; Udvardy, 1993; Shibatani and Ward, 1995; Hendil et al., 1998; Tanahashi et al., 2000). BN-PAGE now overcomes this constraint by allowing isolation of proteasomes under mild conditions that have little apparent effect on rearrangement or loss of bound caps. Consistent with this, modulation of ATP levels before BN-PAGE recapitulated the known ATP-dependence of 19S RC binding and yielded predictable changes in the fraction of 26S and 20S proteasomes. Similarly, the pool of free 20S particles was almost completely converted into PA28 capped proteasomes upon incubation with MG132. The rapid recruitment and exchange of 19S RC and PA28 complexes suggests that cytosolic proteasome populations reflects a steady state distribution governed by the relative rates of cap binding and release, which in turn is likely controlled by the specific needs of the cell.
A particularly striking finding was that virtually all exposed ends of 20S subunits were capped by PA28 within minutes of MG132 treatment. Although the exact mechanism of PA28 recruitment is not yet established, several possibilities can be envisioned. MG132 binding to β1-, β2-, and/or β5-subunits in the 20S cylinder could induce an allosteric conformational change that increases the affinity of free α-subunit ends for the PA28 heptamer. This would be consistent with a functional connection between 20S α- and β-subunits as suggested by the ability of PA28 to modulate β-subunit cleavage specificity (Li et al., 2001; Cascio et al., 2002). Alternatively, MG132 could exert indirect effects on RRL that might lead to PA28 association.
A third and intriguing possibility is that PA28 binding might be stimulated by the presence of substrate within the catalytic 20S core. PA28 is proposed to bind the free end of 19S-20S proteasomes where it induces a conformational change in α-subunits that gates open the chamber and facilitates the release of small peptide fragments (Whitby et al., 2000; Forster et al., 2005). However, when cleavage is prevented by MG132, partially degraded fragments derived from both soluble and transmembrane substrates remained bound to PA28-capped proteasomes (Figure 5 and Oberdorf et al., 2006). Importantly, PA 28 recruitment was not significantly affected by ATP levels but was completely abolished when 19S caps were released by ATP depletion before MG132 addition. These findings are consistent with previous reports that ATP stimulates formation of PA28-capped proteasomes in Hela cell extracts (Tanahashi et al., 2000). Moreover, they suggest that substrate translocation (and/or accumulation), rather than MG132 binding, might allosterically increase the affinity of exposed α-subunit N-termini for PA28. If this were the case, then PA28 could be mechanistically coupled to the proteolytic cycle, binding 20S as fragments accumulate and releasing as the core is emptied. Trapped substrates might also maintain the free 20S end in a relatively open conformation, thus favoring PA28 binding. Although speculative, such a model would explain why relatively few doubly capped PA28 proteasomes are present in resting RRL and why it has been difficult to isolate native PA28-20S complexes from cells where functional proteasomes generate small peptide fragments that are readily released (Tanahashi et al., 2000; Cascio et al., 2002).
Our results also revealed that inhibited proteasome species trap fragments of different lengths. Because all of these fragments were derived from a single substrate (CFTR), they likely represent different stages of the degradation cycle. Doubly 19S-capped proteasomes remained associated with the entire range of cytosolic CFTR fragments, consistent with their ability to recognize ubiquitinated substrates and degrade intact proteins and small peptides. In contrast, PA28-capped proteasomes primarily contained peptides less than ∼40 kDa in size, and hybrid proteasomes contained only slightly larger fragments. Although it is difficult to precisely extrapolate from inhibited to fully functional proteasomes, these results suggest that doubly 19S-capped proteasomes initiate degradation of intact ubiquitinated substrates and then recruit PA28 as degradation proceeds, perhaps as a consequence of ATP-dependent 19S RC dissociation (Babbitt et al., 2005). Such a process could initially form hybrid proteasomes and lastly PA28-capped proteasomes that complete degradation of deubiquitinated smaller polypeptides (Ma et al., 1992). Alternatively, our results do not rule out the possibility that substrates and/or degradation intermediates could be transferred to different proteasome subtypes as they are de-ubiquitinated and progressively cleaved. In either case the relative distribution of proteasome species would likely reflect total complement of ongoing degradation cycles in the cell at any given time.
Finally, a significant body of evidence has implicated hybrid proteasomes in MHC class I antigen presentation (Hendil et al., 1998; Tanahashi et al., 2000; Cascio et al., 2002), wherein PA28αβ is proposed to modulate peptide cleavage and preferentially generate antigenic peptides (Hendil et al., 1998; Cascio et al., 2002). Reticulocytes lack intracellular organelles and thus do not present MHC-restricted antigens (Hoffman et al., 1992). It seems likely therefore that reticulocyte PA28 is primarily involved in protein degradation during the various stages of erythrocyte maturation (Etlinger and Goldberg, 1977; Wefes et al., 1995). It is unknown whether PA28 might produce a specific subset of peptides needed for erythrocyte differentiation. However, the high levels of PA28 relative to 20S, the robust induction of hybrid proteasomes after MG132 treatment, and their relative promiscuity in binding to diverse substrates suggests that PA28 likely plays a general role in protein turnover. On the other hand, the distribution of proteasomes observed here may also reflect specialized features of the highly active ubiquitin– proteasome system utilized by reticulocytes during final stages of degrading unneeded residual proteins. Although further investigations are clearly required to delineate the precise role of different proteasome species in different cell types and under different growth conditions, our data demonstrate that multiple proteasomes can participate in the degradation of a single substrate and provide a new approach for investigating physiological relationships between proteasome populations.
ACKNOWLEDGMENTS
The authors thank Dr. L. Musil and members of the Skach lab for valuable advice and comments. This work was supported National Institutes of Health Grants GM53457 (W.S.), DK51818 (W.S.), and T32HL07781 (T.S.) and Core Grant EY10572 (K.F.) and by the Cystic Fibrosis Foundation Therapeutics (W.S.).
Abbreviations used:
- 19S RC
19S regulatory subunit
- ER
endoplasmic reticulum
- PA28
proteasome activator, 28 kDa
- RRL
rabbit reticulocyte lysate
- BN-PAGE
blue native-polyacrylamide gel electrophoresis.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0311) on September 20, 2006.
REFERENCES
- Ahn K., Erlander M., Leturcq D., Peterson P. A., Früh K., Yang Y. In vivo characterization of the proteasome regulator P.A28. J. Biol. Chem. 1996;271:18237–18242. doi: 10.1074/jbc.271.30.18237. [DOI] [PubMed] [Google Scholar]
- Babbitt S. E., et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Blum H., Beier H., Gross H. J. Improved silver staining of plant proteins, RNA, and DNA in polyacrylamide gels. Electropheresis. 1987;8:93–99. [Google Scholar]
- Bogyo M., McMaster J. S., Gaczynska M., Tortorella D., Goldberg A. L., Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliebe A., Grimm R., Kaiser G., Lubeck J., Soll J., Heins L. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Carvajal M., Wollscheid B., Aebersold R., Steimle V., Schamel W. Two dimensional Blue Native/SDS gel electrophoresis of multi-protein complexes from whole cellular lysates. Mol. Cell. Proteomics. 2004;3:176–182. doi: 10.1074/mcp.T300010-MCP200. [DOI] [PubMed] [Google Scholar]
- Carlson E., Bays N., David L., Skach W. R. Reticulocyte lysate as a model system to study endoplasmic reticulum membrane protein degradation. Methods Mol. Biol. 2005;301:185–205. doi: 10.1385/1-59259-895-1:185. [DOI] [PubMed] [Google Scholar]
- Carlson E., Pitonzo D., Skach W. p97 functions as a non-essential auxiliary factor to facilitate TM-domain extraction during CFTR ER-associated degradation. EMBO J. 2006;25:4557–4566. doi: 10.1038/sj.emboj.7601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio P., Call M., Petre B. M., Walz T., Goldberg A. L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., Slaughter C. A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J. Biol. Chem. 1990;265:4789–4792. [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Rechsteiner M. Subunits ofthe regulatory complex of the 26S protease. Mol. Biol. Rep. 1995;21:27–34. doi: 10.1007/BF00990967. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Pratt G., Ferrell K., Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- Eng J. K., McCormack A. L., Yates J. R., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Etlinger J. D., Goldberg A. L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. USA. 1977;74:54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S., Schmidt M., Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- Finley D., et al. Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- Forster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Früh K., Yang Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Curr. Opin. Immunol. 1999;11:76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- Glickman M., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Fu H., Larsen C. N., Coux O., Wefes I., Pfeifer G., Cjeka Z., Vierstra R., Baumeister W., Fried V., Finley D. Functional analysis of the proteasome regulatory particle. Mol. Biol. Rep. 1999;26:21–28. doi: 10.1023/a:1006928316738. [DOI] [PubMed] [Google Scholar]
- Gnann A., Riordan J., Wolf D. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htmm1p/EDEM and the cdc48 complex in yeast. Mol. Biol. Cell. 2004;15:4125–4135. doi: 10.1091/mbc.E04-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Bajorek M., Kohler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. A. gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Structure of 20S. proteasome from yeast at 2.4 A. resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Harris J. L., Alper P. B., Li J., Rechsteiner M., Backes B. J. Substrate specificity of the human proteasome. Chem. Biol. 2001;8:1131–1141. doi: 10.1016/s1074-5521(01)00080-1. [DOI] [PubMed] [Google Scholar]
- Hendil K. B., Khan S., Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20 S proteasomes. Biochem. J. 1998;332(Pt 3):749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Leshinsky E., Ganoth D., Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc. Natl. Acad. Sci. USA. 1984;81:1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kloetzel P. M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Kloetzel P. M. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Kloetzel P. M., Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Kopp F., Dahlmann B., Kuehn L. Reconstitution of hybrid proteasomes from purified PA700–20 S complexes and PA28alphabeta activator: ultrastructure and peptidase activities. J. Mol. Biol. 2001;313:465–471. doi: 10.1006/jmbi.2001.5063. [DOI] [PubMed] [Google Scholar]
- Leggett D. S., Glickman M. H., Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 2005;301:57–70. doi: 10.1385/1-59259-895-1:057. [DOI] [PubMed] [Google Scholar]
- Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R., Walz T., Ploegh H., Finley D. Multiple associated proteins regulate proteasome structure and function. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li J., Gao X., Ortega J., Nazif T., Joss L., Bogyo M., Steven A. C., Rechsteiner M. Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta. EMBO J. 2001;20:3359–3369. doi: 10.1093/emboj/20.13.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. W., Strickland E., Demartino G. N., Thomas P. J. Recognition and processing of misfolded proteins by PA700, the 19S regulatory complex of the 26S proteasome. Methods Mol. Biol. 2005;301:71–81. doi: 10.1385/1-59259-895-1:071. [DOI] [PubMed] [Google Scholar]
- Lowe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Ma C. P., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J. Biol. Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- Mortz E., Krogh T. N., Khenrik V., Gorg A. Improved silver staining protocols compatible with large-scale protein identification using matrix assisted laser desorption/iionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Oberdorf J., Carlson E. J., Skach W. R. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry. 2001;40:13397–13405. doi: 10.1021/bi011322y. [DOI] [PubMed] [Google Scholar]
- Oberdorf J., Carlson E. J., Skach W. R. Uncoupling proteasome peptidase and ATPase activities results in cytosolic release of an ER polytopic protein. J. Cell Sci. 2006;119:303–313. doi: 10.1242/jcs.02732. [DOI] [PubMed] [Google Scholar]
- Orino E., Tanaka K., Tamura T., Sone S., Ogura T., Ichihara A. ATP-dependent reversible association of proteasomes with multiple protein components to form 26S complexes that degrade ubiquitinated proteins in human HL-60 cells. FEBS Lett. 1991;284:206–210. doi: 10.1016/0014-5793(91)80686-w. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Cohen R. E. Proteasomes and their kin: proteases inthe machine age. Nat. Rev. Mol. Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Hill C. P. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Realini C., Ustrell V. The proteasome activator 11 SREG (P.A28) and class I antigen presentation. Biochem. J. 2000;345(Pt 1):1–15. [PMC free article] [PubMed] [Google Scholar]
- Reguenga C., Oliveira M. E., Gouveia A. M., Sa-Miranda C., Azevedo J. E. Characterization of the mammalian peroxisomal import machinery: Pex2p, Pex5p, Pex12p, and Pex14p are subunits of the same protein assembly. J. Biol. Chem. 2001;276:29935–29942. doi: 10.1074/jbc.M104114200. [DOI] [PubMed] [Google Scholar]
- Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Toh-e A., Yokosawa H. Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem. Biophys. Res. Commun. 2000;273:509–515. doi: 10.1006/bbrc.2000.2980. [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W. A., von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Shibatani T., David L. L., McCormack A. L., Frueh K., Skach W. R. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- Shibatani T., Ward W. F. Sodium dodecyl sulfate (SDS) activation of the 20S proteasome in rat liver. Arch. Biochem. Biophys. 1995;321:160–166. doi: 10.1006/abbi.1995.1381. [DOI] [PubMed] [Google Scholar]
- Strickland E., Hakala K., Thomas P. J., DeMartino G. N. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26 S proteasome. J. Biol. Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- Tanahashi N., Murakami Y., Minami Y., Shimbara N., Hendil K. B., Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- Tanaka K. Molecular biology of the proteasome. Biochem. Biophys. Res. Commun. 1998;247:537–541. doi: 10.1006/bbrc.1998.8617. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Tsurumi C. The 26S proteasome: subunits and functions. Mol. Biol. Rep. 1997;24:3–11. doi: 10.1023/a:1006876904158. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Yoshimura T., Kumatori A., Ichihara A., Ikai A., Nishigai M., Kameyama K., Takagi T. Proteasomes (multi-protease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J. Biol. Chem. 1988;263:16209–16217. [PubMed] [Google Scholar]
- Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26S (1500 kDa) proteolytic complex. J. Biol. Chem. 1993;268:9055–9062. [PubMed] [Google Scholar]
- Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang C. C., Bozdech Z., Liu C. L., Shipway A., Backes B. J., Harris J. L., Bogyo M. Biochemical analysis of the 20S proteasome of Trypanosoma brucei. J. Biol. Chem. 2003;278:15800–15808. doi: 10.1074/jbc.M300195200. [DOI] [PubMed] [Google Scholar]
- Wang L., Dobberstein B. Oligomeric complexes involved in translocation of proteins across the membrane ofthe endoplasmic reticulum. FEBS Lett. 1999;457:316–322. doi: 10.1016/s0014-5793(99)01075-3. [DOI] [PubMed] [Google Scholar]
- Wefes I., Mastrandrea L. D., Haldeman M., Koury S. T., Tamburlin J., Pickart C. M., Finley D. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc. Natl. Acad. Sci. USA. 1995;92:4982–4986. doi: 10.1073/pnas.92.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Wittig I., Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- Wolf D. H., Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim. Biophys. Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Xiong X., Chong E., Skach W. R. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Yang Y., Früh K., Ahn K., Peterson P. A. In vivo assembly of the proteasomal complexes, implications for antigen processing. J. Biol. Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Kameyama K., Takagi T., Ikai A., Tokunaga F., Koide T., Tanahashi N., Tamura T., Cejka Z., Baumeister W., Tanaka K., Ichihara A. Molecular characterization of the “26S” proteasome complex from rat liver. J. Struct. Biol. 1993;111:200–211. doi: 10.1006/jsbi.1993.1050. [DOI] [PubMed] [Google Scholar]