Abstract
The SRO7/SOP1 encoded tumor suppressor homologue of Saccharomyces cerevisiae is required for maintenance of ion homeostasis in cells exposed to NaCl stress. Here we show that the NaCl sensitivity of the sro7Δ mutant is due to defective sorting of Ena1p, the main sodium pump in yeast. On exposure of sro7Δ mutants to NaCl stress, Ena1p fails to be targeted to the cell surface, but is instead routed to the vacuole for degradation via the multivesicular endosome pathway. SRO7-deficient mutants accumulate post-Golgi vesicles at high salinity, in agreement with a previously described role for Sro7p in late exocytosis. However, Ena1p is not sorted into these post-Golgi vesicles, in contrast to what is observed for the vesicles that accumulate when exocytosis is blocked in sec6-4 mutants at high salinity. These observations imply that Sro7p has a previously unrecognized role for sorting of specific proteins into the exocytic pathway. Screening for multicopy suppressors identified RSN1, encoding a transmembrane protein of unknown function. Overexpression of RSN1 restores NaCl tolerance of sro7Δ mutants by retargeting Ena1p to the plasma membrane. We propose a model in which blocked exocytic sorting in sro7Δ mutants, gives rise to quality control-mediated routing of Ena1p to the vacuole.
INTRODUCTION
Targeted delivery of secretory vesicles to discrete regions of the plasma membrane is essential for establishment and maintenance of cell polarity (Nelson and Yeaman, 2001; Pruyne et al., 2004). The tumor suppressor lethal(2)giant larvae (Lgl) has an important role in establishing cell polarity in developing Drosophila embryos (Wodarz, 2000; Humbert et al., 2003; Bilder, 2004). Homozygous inactivation of the lgl gene results in improper targeting of selected epithelial transmembrane proteins to their correct apical destination (Bilder et al., 2000; Hutterer et al., 2004; Wirtz-Peitz and Knoblich, 2006), leading to defects in apical-basal polarity. Lgl also plays an important role in asymmetric cell division by contributing to the polarization of cytosolic fate determinants in mitotic neuroblasts (Ohshiro et al., 2000; Peng et al., 2000; Betschinger and Knoblich, 2004). Lgl deficiency therefore results in improper cell fate specification and failure to differentiate correctly. The loss of cell polarity in lgl mutants is proposed to be a primary reason for disruption of larval tissue organization and for the massive overgrowth of the larval brain and the imaginal discs (Bilder, 2004). Hence, Lgl appears to be involved in a pathway that coordinates protein sorting, cell polarity, and growth control.
The Lgl protein is the founding member of a growing family of WD-40 repeat proteins conserved in eukaryotes ranging from yeast to man (Tomutsune et al., 1993; Strand et al., 1995; Koyama et al., 1996; Kagami et al., 1997; Larsson et al., 1998; Grifoni et al., 2004). Studies in yeast have provided clues to the molecular function of these proteins, implicating involvement in late stages of polarized secretion (Kagami et al., 1998; Lehman et al., 1999; Gangar et al., 2005; Zhang et al., 2005). Current models state that a conserved multisubunit complex, known as the exocyst, assembles as cargo vesicles arrive at the exocytic sites, establishing the primary connection between the plasma membrane and the secretory vesicle (TerBush et al., 1996; Guo et al., 2000; Wiederkehr et al., 2004). After this vesicle tethering, the coiled-coil SNARE (soluble N-ethylmalemide–sensitive factor attachment protein receptor) proteins, opposed on vesicle (v-SNAREs) and target membranes (t-SNAREs), form a complex that mediates a short-range interaction between the vesicle and plasma membranes. Studies in yeast have revealed that the two yeast Lgl homologues Sro7p and Sro77p (also named Sop1p and Sop2p) interact with myosins (Kagami et al., 1998, Gangar et al., 2005) components of the exocyst (Lehman et al., 1999; Zhang et al., 2005; Grosshans et al., 2006), and Sec9p (Lehman et al., 1999; Gangar et al., 2005), a t-SNARE protein involved in docking of post-Golgi transport vesicles with the plasma membrane. Deletion of both SRO7 and SRO77 leads to a cold-sensitive phenotype (Kagami et al., 1998) that is characterized by defective secretion and intracellular accumulation of post-Golgi vesicles (Lehman et al., 1999).
We isolated the yeast SOP1/SRO7 gene (hereafter referred to as SRO7) by complementation of a salt-sensitive Saccharomyces cerevisiae mutant (Larsson et al., 1998; Wadskog et al., 2004). Mutants having a deleted SRO7 gene are specifically sensitive to NaCl stress and exhibit increased intracellular levels of Na+ when exposed to high NaCl media. The sro7Δ sro77Δ double mutant is hypersensitive to NaCl, and this phenotype is partially rescued by ectopic expression of the Drosophila lgl gene, indicating functional conservation of the homologues (Larsson et al., 1998). The primary pathway for sodium export in S. cerevisiae occurs via the plasma membrane P-type ATPase encoded by the ENA1/PMR2 gene (Garciadeblas et al., 1993; Wieland et al., 1995). Expression of ENA1 is vital for maintenance of intracellular ion homeostasis in salt-stressed yeast cells and is subject to a complex network of regulatory interactions involving not only salt or pH stress but also general metabolic processes (Serrano et al., 1999; Wadskog and Adler, 2002). A second Na+ extrusion system in S. cerevisiae is the cation/proton antiporter encoded by NHA1 gene (Prior et al., 1996; Banuelos et al., 1998). This transporter becomes important for Na+ efflux only at acidic external pH values, whereas its expression does not seem to be controlled by salt stress or changes in pH.
In this article, we demonstrate that the salt sensitivity of sro7Δ mutants is due to failure of the ENA1-encoded sodium pump to be expressed on the cell surface. Instead, Ena1p is diverted for vacuolar degradation in a manner dependent on ubiquitin ligase and the vacuolar protease Pep4p. The observed mis-targeting of Ena1p in salt-stressed mutants is accompanied by accumulation of secretory vesicles. However, defective sorting only appears to involve a fraction of the vesicle-cargo-proteins. A number of plasma membrane proteins examined are correctly sorted in salt-stressed sro7Δ mutants. We have also identified a novel gene, RSN1, which functions as a multicopy suppressor of the salt-sensitive growth of sro7Δ mutants by restoring targeting of Ena1p to the cell surface. We postulate that Sro7p is involved in the molecular machinery required for targeting of Ena1p to the plasma membrane and that Ena1p is identified by the Golgi quality control system and rerouted to the vacuole for degradation in the absence of Sro7p.
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions
All strains and plasmids used in this study are listed in Tables 1 and 2. The wild-type strain used in most experiments was W303-1A and the congenic sro7Δ mutant was WKL-2A (Table 1). Unless otherwise stated, strains were grown at 30°C in rich YP media with 20 g/l of glucose, pH 6.5. Defined CBS medium was composed as previously described (Verduyn et al., 1992). For salt induction, appropriate volumes of medium containing 2 M NaCl were added to the cultures to obtain the final NaCl concentration.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| W303-1B | MATαade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| NY179 | MATa leu2-3,112 ura3-52 | P. Novick |
| BY4741 | MATa his3 leu2 ura3 | Euroscarf |
| WKL-2A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sro7::LEU2 | Larsson et al. (1998) |
| WKL-2B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sro7::LEU2 | Larsson et al. (1998) |
| Y05451 | BY4741 sro7:: kanMX4 | Euroscarf |
| Y00852 | BY4741 rsn1:: kanMX4 | Euroscarf |
| YN10 | MATa ade2oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ena1::HIS3 | Wieland et al. (1995) |
| WKL-210 | YN10 sro7::LEU2 | This study |
| YIW-2 | W303-1B ENA1-3xHA | This study |
| YIW-3 | W303-1B pep4::kanMX4 ENA1-3xHA | This study |
| YIW-4 | WKL-2A ENA1::3xHA | This study |
| YIW-5 | WKL-2A pep4::kanMX4 ENA1-3xHA | This study |
| YIW-18 | BY4741 end4::kanMX4 sro7::LEU2 | This study |
| YIW-19 | BY4741 vps27:: kanMX4 sro7::LEU2 | This study |
| YIW-20 | YIW-18 ENA1-3xHA | This study |
| YIW-21 | YIW19 ENA1-3xHA | This study |
| YIW-22 | NY786 sro7::LEU2 | This study |
| YIW-23 | YIW-22 ENA1-3xHA | This study |
| YCS-1 | W303-1B RSN1-CFP | This study |
| RSY255 | MATα ura3-52 leu2-3,112 | R. Sheckman |
| NY772 | MATa leu2-3,112 ura3-52 sec3-2 | P. Novick |
| NY774 | MATα leu2-3,112 ura3-52 sec4-8 | P. Novick |
| NY776 | MATα leu2-3,112 ura3-52 sec5-24 | P. Novick |
| NY778 | MATα leu2-3,112 ura3-52 sec6-4 | P. Novick |
| NY780 | MATα leu2-3,112 ura3-52 sec8-9 | P. Novick |
| NY782 | MATa leu2-3,112 ura3-52 sec9-4 | P. Novick |
| NY784 | MATa leu2-3,112 ura3-52 sec10-2 | P. Novick |
| NY786 | MATa leu2-3,112 ura3-52 sec15-1 | P. Novick |
| H1394 | NY179 sro7::URA3 | This study |
| H1295 | NY772 sro7::URA3 | This study |
| H1396 | NY774 sro7::URA3 | This study |
| H1296 | NY776 sro7::URA3 | This study |
| H1297 | NY778 sro7::URA3 | This study |
| H1298 | NY780 sro7::URA3 | This study |
| H1397 | NY782 sro7::URA3 | This study |
| H1299 | NY784 sro7::URA3 | This study |
| H1300 | NY786 sro7::URA3 | This study |
| 27061b | MATa ura3 trp1 | Galan et al. (1996) |
| 27064b | MATa ura3 trp1npi1 | Galan et al. (1996) |
| npi1sro7 | MATa ura3 trp1npi1 sro7::kanMX4 | This study |
Table 2.
Plasmids
| Plasmid | Description/Source |
|---|---|
| pH/B316 | SRO7 cloned in pRS316 [CEN URA3] (Larsson et al., 1998) |
| YEpH/B | SRO7 cloned in YEplac195 [2μ URA3] (Larsson et al., 1998) |
| pJW11 | ENA1 behind PMA1-promotor in pRS316 (Wieland et al., 1995) |
| pMPY-HA | Plasmid for integrative 3xHA tagging (Schneider et al., 1995) |
| pNHA1-GFP | NHA1- GFP in pGRU1 (Kinclova et al., 2001) |
| pVR102 | SHO1-GFP (Reiser et al., 2000) |
| pVR28 | PBS2-GFP (Reiser et al., 1999) |
| pRS314STE2-GFP | STE2-GFP in pRS314 (Stefan and Blumer, 1999) |
| pDH3 | Plasmid containing CFP/KAN integration cassette (YRC, University of Washington) |
| pHR81 | [2μ URA3 LEU2-d] (Nehlin et al., 1989) |
| pRSN1 | RSN1 inserted in pHR81 (This study) |
| pUG35 | [CEN URA3] (Niedenthal et al., 1996) |
| pUG35-ENA1 | ENA1-GFP behind MET25 promoter (M. Cyert and V. Heath) |
| pUG35-GAP1 | GAP1-GFP behind MET25 promoter (This study) |
| pUG35-SUC2 | SUC2-GFP behind MET25 promoter (This study) |
| pRS423 | [2μ HIS3] (ATCC) |
| pRS423-RSN1 | RSN1 inserted in pRS423 (This study) |
| pRS426 | [2μ URA3] (ATCC) |
| pRS426-RSN1 | RSN1 inserted in pRS426 (This study) |
Growth Curves and Drop Tests
For growth curves, E-flasks containing 50 ml of defined CBS medium were inoculated to an OD610 of 0.05, using cells from a preculture grown to stationary phase. The cell cultures were incubated with shaking (180 rpm) at 30°C, unless otherwise stated. OD610 was measured once every hour and plotted versus time. For drop tests, overnight cultures were diluted as described, and drops of 8 μl were spotted onto solid agar media.
Northern Blot Analysis
For Northern analysis of ENA1 mRNA, cells were grown to midlog phase in basal medium before being subjected to NaCl stress. Cells were harvested on ice at time points indicated, and total RNA was isolated by standard procedures. The amount of RNA was determined by spectrophotometry and semiquantitatively using ethidium bromide stained agarose gels. Samples containing 15 μg of total RNA were denatured and run on low formaldehyde (2.5% vol/vol) agarose gels. Blotting onto positively charged nylon membranes was preformed by capillary transfer, using 10× SSC as transfer buffer. The filters were prehybridized for 4 h at 60°C in 5× SSC, 10 mM sodium phosphate (pH 6.5), 10× Denhardt's solution, 2% SDS, and 100 μg/ml herring sperm DNA. Hybridization was preformed at 60°C for 8–10 h in the same solution supplemented with 10% PEG4000. Probes were labeled and added at 5 ng/ml. For detection of the ENA1 transcript a 5′-AAT CAT GCA TGG CAA ACG AGA TTA CT-3′ probe, which shows little identity to other ENA genes, was used. As a loading control we used IPP1 mRNA detected using a 5′-TGT CTG GTA GTG TAG GTC ATT AGT-3′ probe. Signal intensities were quantified with a Phosphoimager (Molecular Dynamics, Sunnyvale, CA; 425E), using the manufacturer-supplied program, Image Quant.
Tagging of Ena1p
For epitope tagging of the genomic copy of ENA1, a three times–repeated hemagglutinin (HA) epitope was integrated in the ENA1 gene using PCR amplification of a tag-URA3-tag cassette in the pMPY-HA vector, kindly provided by Dr. B. Futcher (CSHL; Schneider et al., 1995). The PCR primers were designed such that 16 bases were complementary to pBluescript sequences of pMPY-HA, whereas flanking regions of ∼50 bases were complementary to and in frame with the 3′-end of the ENA1 gene. The PCR reaction was carried out using 50 ng of linearized pMPY-HA, 15 pmol of each primer, 0.5 mM dNTPs, 5 μl 10× PCR buffer 1 (Long template PCR kit, Roche, Indianapolis, IN) and 0.5 μl DNA polymerase mix (Long template PCR kit, Roche) in a total volume of 50 μl. The PCR reaction conditions were as follows: 5 min at 94°C, 1 min at 50°C, 3 min at 72°C for one cycle; 1 min at 94°C, 1 min at 50°C, and 3 min at 72°C for nine cycles. PCR products were purified by QIAquick PCR Purification kit, (QIAGEN, Chatsworth, CA), and 1 μg of purified DNA was used for transformation. After selection for stable transformants on medium lacking uracil, positive clones were cultured in YPD overnight to allow for URA3 gene loop out by recombination between the repeated epitope regions. Cells from this culture were transferred to 5-fluoro-orotic acid plates (5-FOA, 1.0 g/l), and resistant colonies were examined by PCR and Western analysis to confirm correct tagging of ENA1. The CEN plasmid pUG35-ENA1 containing ENA1-GFP behind the methionine repressible MET25 promoter was a generous gift from Drs. M. Cyert and Victoria Heath.
Quantitative PCR
To determine SRO77 expression 0.5 μg of total RNA was used for reverse transcription using Bio-Rad iScript First Strand Synthesis kit according to the manufacturers instructions (Richmond, CA). The reaction was scaled down to 10 μl, and each sample was reverse transcribed in duplicate. A pool of samples was used as control for genomic DNA contamination. For the detection of SRO77, the forward primer 5′-GGGCAAAAGCAAATAGAGGT-3′, and reverse primer 5′-CAATCAGCATCCAATCCAAG-3′, were used. For the detection of the control IPP1, the forward primer 5′-CGTAAGCCACCCAGAAACTAA-3′ and reverse primer 5′-ATCGGTCTCACCTTCATCCA-3′, were used. Quantitative PCR was performed on the Stratagene Mx3005p, using BD QTaq Polymerase mix with 0.4 μM of each primer and 0.25× SYBR Green I (Molecular Probes, Eugene, OR). cDNA corresponding to 25 ng of RNA was used in each PCR reaction. Denaturation for 2 min at 95°C was followed by 45 cycles of 20s at 95°C, 20s at 60°C, and 20s at 72°C. The fluorescence detection was performed at the 72°C step. After amplification dissociation curve analysis was performed to verify the product formed. Statistic analysis was performed using Excel 2003 (Microsoft, Redmond, WA). The IPP1 gene was used as a reference to calculate the relative expression of SRO77.
Western Blot Analysis
For immunoblot analysis cells cultured to midlog phase in YPD medium were subjected to a step increase in NaCl concentration, and cell samples harvested by centrifugation were frozen in liquid nitrogen. To lyse the cells, pellets were resuspended in 2 M NaOH (300 μl/OD610) and incubated on ice for 10 min. The cell suspensions were then neutralized by the addition of trichloroacetic acid to a final concentration of 40%. Before a subsequent 10 min incubation on ice, cells were centrifuged and washed twice in 1 M Tris-HCl, pH 8.0 and resuspended in 2× Laemmli buffer (100 mM Tris-HCl, pH 6.8, 8% SDS, 40% glycerine, 2% mercaptoethanol, 0.005% bromophenol blue). Samples corresponding to 10 ml of OD610 = 0.5 were incubated at 50°C for 5 min, loaded onto 10% polyacrylamide criterion gels (Bio-Rad), resolved by SDS-PAGE, and blotted onto nitrocellulose membranes. After transfer, the filters were stained for 5 min in Ponceau S and washed with distilled H2O to observe the quality of the transfer and check the total amount of protein. HA-tagged Ena1p was detected using anti-HA antibodies (Roche) at 1:2000 dilution as the primary antibody and anti-mouse HRP-conjugated antibody (Roche) at 1:4000 dilution as the secondary antibody. Blots were developed using Lumi-light chemiluminescence detection system (Roche). Immunoblot analysis of Bgl2p (Adamo et al., 1999) and Sso1/2p and Snc1/2p (Lehman et al., 1999) was performed as previously described.
Tagging of Rsn1p, Gap1p, and Suc2p with Fluorescent Tracers
Genomic tagging of RSN1 with the cyan fluorescent protein (CFP) was conducted according to the procedure described by the Yeast Resource Center, University of Washington (http://depts.washington.edu/∼yeastrc). The integration cassette from plasmid pDH3 was amplified by PCR with the following primers: 5′-GCG GAC CCA AAG TAT AAG GAA GAA GAG AGT CGT TCG GCA GTG GTC GAC GGA TCC CCG GG-3′ and 5′-AAA CAA ATA GGA ATA GAT AAT TTC TGA TAA TGT TTA ACC TTT AGA TCG ATG AAT TCG AGC-3′, which contain sequences from the 3′ end of the RSN1 gene (underlined) and sequences from the pDH3 plasmid. Wild-type cells were transformed with the PCR product and plated on selective YPD G418 medium. Positive clones were further verified by PCR. Finally the region of integration was sequenced to ascertain that the proper reading frame was used.
To tag Gap1p by green fluorescent protein (GFP) on its C-terminus, PCR was used to synthesize a fragment containing the GAP1 coding sequence lacking its stop codon. The primers used were 5′-AAA AAA CCC GGG ATG AGT AAT ACT TCT TCG TA and CGG AGT ATC GAT ACA CCA GAA ATT CCA GAT TC-3′, containing SmaI and ClaI restriction sites, respectively. After digestion, the fragment was cloned into the plasmid pUG35 (URA3 CEN plus gene encoding the yeast-enhanced GFP), which was cleaved with corresponding enzymes, to produce the GAP1-GFP fusion. The construct was confirmed by restriction enzyme analysis and fluorescence microscopy.
Tagging of Suc2p by GFP on its C-terminus was performed by the same procedure using the primers 5′-AAA AAA ACT AGT CTC TCA GAG AAA CAA GCA-3′ and 5′-AAA AAA GAA TTC CTT CCC TTA CTT GGA AC-3′ to generate a SUC2 fragment flanked by Spe1 and EcoR1 restriction sites. The fragment was cloned into the pUG35 plasmid as described for pUG35-GAP1.
Fluorescence Microscopy
Cells taken from exponential growth phase in defined media were subjected to conditions as described in Results and viewed with the Leica DMRXA microscope (Deerfield, IL) using a Fluotar lens and the Leica 513852 filter for GFP or CFP fluorescence. To obtain a representative picture of the appearance of the average cell in the culture >1000 cells were observed for each condition shown. Photographs were captured using a Hamamatsu ORCA cooled CCD camera (Bridgewater, NJ). For N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64, Molecular Probes) labeling of yeast vacuoles, exponentially growing cells were incubated at 30°C with 32 μM FM4-64 for 15 min, followed by washing and resuspension in fresh medium for 1–1.5 h (Vida and Emr, 1995), before expression of ENA1 was induced.
Confocal Microscopy
The CFP-tagged cells were imaged using a laser scanning confocal unit (MRC1024, Bio-Rad) mounted on the side port of an inverted microscope (TE-300, Nikon, Melville, NY). The CFP was excited using a fiber-coupled blue diode laser (405 nm). The samples were imaged using a 100× Plan Fluor objective (Nikon) with a numerical aperture of 1.3. A longitudinal resolution of 1.2 μm was obtained by means of the adjustable confocal pinhole. Because of the weak CFP signal a 405-nm Raman Notch Filter (FWHM 10 nm; Melles Griot, Rochester, NY) was used to prevent the excitation light of reaching the detector. Thus, no additional emission filters was used. The signal was collected by using a five-frame Kalman collection filter. Relative fluorescence was measured using LaserPix software (Bio-Rad).
Electron Microscopy
Wild-type and sro7Δ strains were grown to midlog phase in YPD at 25°C. Cells were shifted for 2 h in YPD or YPD/0.6 M NaCl media and processed for electron microscopy (EM) as described previously (Adamo et al., 1999).
Secretion Assays
Bgl2p secretion assays were performed as described previously (Adamo et al., 1999). The examined strains were grown to midlog phase in YPD at 25°C and then shifted to YPD or YPD + 0.6 M NaCl media for 2 h. Invertase secretion assays were performed with cells transformed with the pUG35-SUC2 plasmid. Cells were grown to midlog phase in CBS medium at 25°C and then incubated for 2 h in methionine-free CBS or CBS + 0.6 M NaCl media to de-repress SUC2 expression. Invertase secretion was assayed as previously described (Lehman et al., 1999).
Suppressor Screen
To isolate multicopy suppressor genes of sro7Δ salt sensitivity, the mutant strain was transformed with the pHR81-based yeast genomic library (Nehlin et al., 1989). Transformants were first selected on YNB plates lacking uracil and then replica plated onto YPD containing 0.7 M NaCl. Positive clones, which contained plasmid inserts that restored growth of the sro7Δ mutant at high salinity, were isolated. After plasmid rescue and retransformation, library inserts were partially sequenced and compared against the yeast genome database via BLAST searches. For those inserts containing multiple open reading frames (ORFs), each ORF was subcloned into pRS426 and transformed into sro7Δ. A total of ∼12,000 transformants were screened.
Subcellular Fractionation
To localize Rsn1p by fractionation, wild-type W303-1B cells harboring CFP tagged Rsn1p were grown to OD610 0.5–1.0 in 200 ml YPD medium. Cells were washed by centrifugation and resuspended in twice the pellet volume of resuspension buffer (R-buffer), containing 50 mM Tris, pH 7.5, 5 mM EDTA, Complete protease inhibitor cocktail (Boehringer Mannheim, Germany, 1 tablet per 50 ml cell extract), and 0.1 M KCl. Four volumes of glass beads (0.5 mm ∅) were added, and the cells were then processed in a bead beater four times for 30 s. Unlyzed cells were removed by centrifugation at 500 × g for 5 min at 4°C. The lysate was divided into three aliquots and diluted with an equal volume of either R-buffer, R-buffer with 2.0 M KCl, or R-buffer with 2% Triton X-100. Centrifugation at 10,000 × g for 10 min resulted in pellets (P10) and supernatants (S10). P10 was resuspended in R-buffer, and S10 was further centrifuged at 100,000 × g for 1 h in a Beckman TL-100 ultracentrifuge (Berkeley, CA). The resulting pellets were resuspended in R-buffer. Aliquots containing equal amounts of cell material were boiled for 1 min in 2× Laemmli buffer (see above) and loaded to 10% Tris-HCl criterion gels (Bio-Rad). Immunoblot analysis was performed as previously described for Ena1p-HA using anti-GFP as the primary antibody (kindly provided by Dr. Pam Silver).
Post-Golgi vesicles were isolated as previously described (Brennwald et al., 1994; Lehman et al., 1999). Briefly, wild-type cells and sro7 mutants were cultured to midlog phase in YPD medium and shifted for 2 h in YPD or YPD + 0.6 M NaCl media at 25°C. Harvested cells were spheroplasted and lysed, and a P3 membrane fraction enriched in post-Golgi vesicles was prepared by differential centrifugation (Walworth and Novick, 1987). This fraction was resuspended and layered onto a 20–40% sorbitol velocity gradient that was fractioned as described previously (Brennwald et al., 1994). The presence of the vesicle marker Snc1/2p and Ena1p-HA were determined by immunoblotting followed by quantitation on a LI-COR Odyssey Infrared imaging system (Cincinnati, OH).
RESULTS
The NaCl Sensitivity of the sro7Δ Mutant Is Due to Defective Ena1p Stability Rather than Defective ENA1 Expression
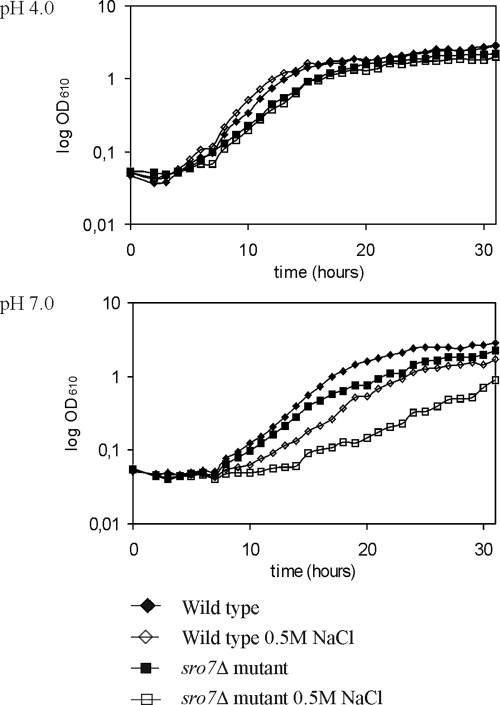
We have previously observed that mutants having a deleted SRO7 gene show increased sensitivity to NaCl (Larsson et al., 1998). As demonstrated in Figure 1, this sensitivity is strongly dependent on the pH of the growth medium. At acidic pH, the sro7Δ mutant showed wild-type growth behavior both in saline and nonsaline medium. At neutral pH, however, the presence of 0.5 M NaCl caused a significantly stronger growth inhibition of the mutant strain. S. cerevisiae uses two different transport systems to export Na+, the NHA1-encoded Na+/H+ antiporter (Banuelos et al., 1998) and the ENA1-encoded Na+-extruding ATPase (Garciadeblas et al., 1993; Wieland et al., 1995). Because Nha1p is active at low pH, whereas Ena1p has its optimum activity around neutral pH, we anticipated that ENA1 might be a prime target for the effects of the SRO7 deletion. We therefore analyzed ENA1 expression by Northern blot analysis (Figure 2A). Total RNA was extracted from wild-type and sro7Δ mutant cells at various time points after addition of NaCl to 0.5 M concentration to cultures growing exponentially in YPD medium. In wild-type cells there was a rapid, transient transcriptional induction of the ENA1 gene, reaching its maximum 30 min after the salt shock, followed by a return to low expression levels. The expression profile of the sro7Δ mutant was very similar to that of wild-type cells, indicating that the salt-sensitive phenotype is not a consequence of defective ENA1 expression.
Figure 1.
Growth of wild-type and sro7Δ strains in the presence and in the absence of NaCl stress. E-flasks containing complete minimal CBS media supplemented with 0 or 0.5 M NaCl at pH 4.0 or pH 7.0 were inoculated from overnight cultures of wild-type (W303-1A) or sro7Δ (WKL-2A) cells to an OD610 of 0.05. Growth was monitored by following OD610.
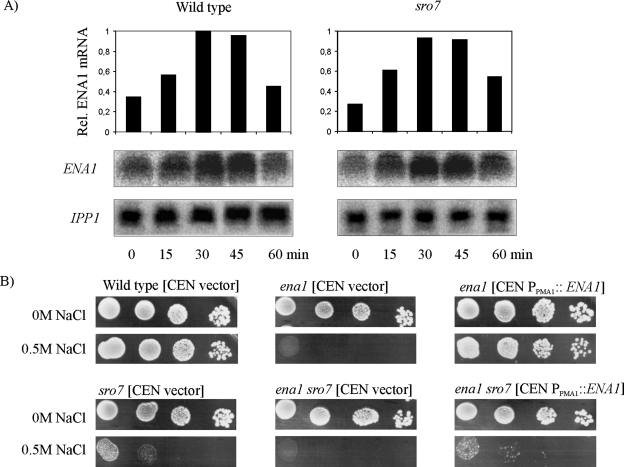
Figure 2.
The sro7Δ mutant shows wild-type expression of ENA1, and its salt tolerance is not improved by overexpression of ENA1. (A) Northern blot analysis of ENA1 expression after shift of exponentially growing wild-type (W303-1A) or sro7Δ (WKL-2A) strains to medium containing 0.5 M NaCl. Total RNA was extracted from cells and harvested at time points indicated. Each sample contains extract from the same number of cells. Transcript levels were first normalized to control mRNA (IPP1) and then presented relative the wild-type value at 30 min. (B) Drop-test monitoring growth of ena1Δ and ena1Δ sro7Δ mutants, carrying the pRS316 empty control plasmid and the same plasmid containing the ENA1 gene behind the strong PMA1 promoter (Wieland et al., 1995). Tenfold dilutions of CBS cultures of OD610 1.0 were spotted onto CBS and CBS + 0.5 M NaCl plates. Plates were photographed after incubation for 48 h at 30°C.
To further explore whether enhanced expression of ENA1 might overcome growth inhibition of sro7Δ in saline media, a centromeric plasmid, pJW11, driving ENA1 expression from the strong constitutive PMA1 promoter (Wieland et al., 1995), was introduced into ena1Δ and ena1Δ sro7Δ mutants. Serial dilutions of overnight cultures of these strains and control strains transformed with empty plasmids were spotted onto solid media with and without salt (Figure 2B). Notably, the ena1Δ sro7Δ strain overexpressing ENA1 remained as sensitive to 0.5 M NaCl as the sro7Δ strain expressing ENA1 from its native promoter. Similarly, the salt tolerance of sro7Δ cells was not enhanced after transformation with a centromeric plasmid carrying ENA1 controlled by its own promoter or alternatively expressing a functional ENA1-GFP construct from the strong MET25 promoter (unpublished data). Thus, salt sensitivity of the sro7Δ mutant cannot be rescued by increased expression of the ENA1 gene.
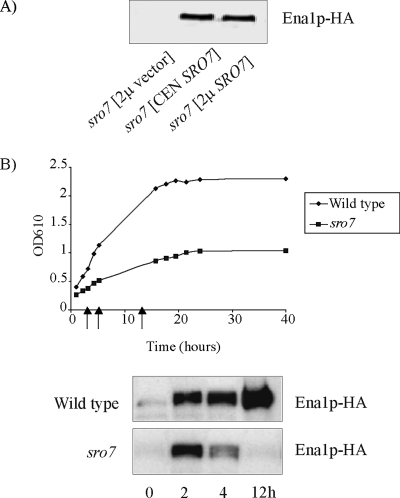
Having observed wild-type expression of ENA1 in sro7Δ mutants, we went on to examine the fate of the ENA1 encoded protein. The genomic copy of ENA1 in the wild-type and the sro7Δ mutant therefore was modified to encode three HA epitopes at the C-terminus of the protein (see Materials and Methods). Yeast cells expressing this tagged protein produced a protein detectable with HA antibodies on immunoblots, with an apparent molecular weight of ∼130 kDa. The tagged wild-type strain maintained its original salt tolerance, and the HA tagged sro7Δ mutant showed wild-type salt tolerance after reintroduction of the SRO7 gene on a plasmid, indicating that the tagged protein was functional in both strains (unpublished data). A first indication that lack of the SRO7 gene affects the stability of Ena1p came from immunoblot analysis of extracts of the sro7Δ mutant (Figure 3A). This analysis failed to detect any HA-tagged Ena1p 5 h after the mutant had been shifted to medium containing 0.5 M NaCl. In similar experiments with mutants transformed to contain the SRO7 gene on either a centromeric or a 2μ plasmid, tagged Ena1p was clearly detectable. We therefore performed a time-course study of the Ena1p fate in exponentially growing wild-type and sro7Δ strains shifted to medium of increased salinity. In wild-type cells the Ena1p levels increased after the shift to 0.5M NaCl, and after long-time incubation (12 h) there was a massive accumulation of Ena1p (Figure 3B). However, in sro7Δ mutants the Ena1p level initially increased as in wild-type cells, but then declined to undetectable levels >4 h after the salinity shift (Figure 3B). These observations suggest that maintained stability of Ena1p is dependent on a functional Sro7p.
Figure 3.
The stability of Ena1p is dependent on SRO7. (A) Immunoblot analysis of Ena1p-HA expressing sro7Δ mutants transformed with and empty plasmid (pRS316), a CEN-SRO7 plasmid (pH/B316), and a 2μ-SRO7 plasmid (YEpH/B). Cells were cultured to midlog phase, exposed to 0.5 M NaCl for 5 h, harvested, and analyzed for the presence of Ena1p-HA. (B) A time-course study of the appearance of Ena1p-HA in wild-type and sro7Δ cells, after exposure to medium containing 0.5 M NaCl. Cell samples were withdrawn at stages indicated in the growth curve (arrows), and total protein extracts were analyzed for presence of Ena1p-HA. For immunoblot analysis protein extracts were subjected to SDS-PAGE in 10% polyacrylamide gels. All lanes were loaded with identical amounts of total protein. Blotted filters were analyzed for Ena1-HA by primary mouse anti-HA antibodies and secondary HRP-conjugated anti-mouse antibodies (Roche).
We also examined the possibility that salt stress might perturb expression of the iso-gene SRO77 such that sro7Δ plus salt generates a sro7Δ sro77Δ phenotype. The SRO77 expression was, however, little affected by shifting wild-type cells or sro7Δ mutants to medium containing 0.6M NaCl (Supplementary Figure S1).
Ena1p Is Sorted in a MVE-dependent Way from Internal Compartments to the Vacuole in Salt-stressed sro7Δ Mutants
Most major pathways for degradation of membrane proteins terminate in the vacuole (Katzmann et al., 2002; Horak, 2003). To investigate the fate of Ena1p in the sro7Δ mutant, we therefore constructed the sro7Δ pep4Δ double mutant, in which vacuolar protease activity is diminished due to deletion of the gene for the master protease, Pep4p (Jones, 1991). If SRO7 deficiency results in sorting of Ena1p to the vacuole for degradation, the levels of Ena1p would be expected to remain unchanged in the sro7Δ pep4Δ mutant lacking active vacuolar hydrolases. This prediction was supported by immunoblot analysis (Figure 4A). After shift to increased salinity, the sro7Δ pep4Δ mutant maintained wild-type levels of Ena1p at time points when levels were clearly reduced in the sro7Δ single mutant. We conclude that Ena1p in salt-stressed sro7Δ mutants is diverted to the vacuole and subjected to Pep4p-dependent protease degradation.
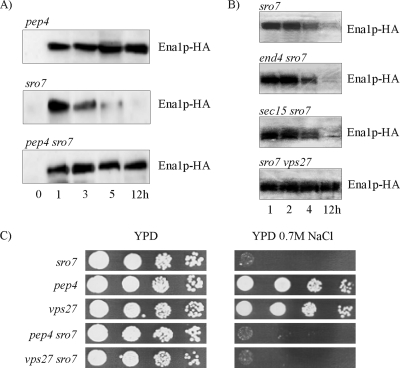
Figure 4.
Inactivation of the vacuolar protease Pep4p stabilizes Ena1p, but does not rescue growth of sro7Δ mutants on NaCl media. Time-course study of Ena1p-HA levels in (A) pep4Δ, sro7Δ and pep4Δ sro7Δ mutants and (B) sro7Δ, end4Δ sro7Δ, sec15-1 sro7Δ and vps27Δ sro7Δ mutants after exposure of cells to medium containing 0.5 M NaCl. Cell samples were withdrawn at time points indicated, and total protein extracts were analyzed for presence of Ena1p-HA by immunoblot analysis. The sec15-1 sro7Δ (ts) mutant was incubated at 34°C during salt treatment to inactivate Sec15-1p without causing total growth arrest. Immunoblot analysis was performed as described in Figure 3 and Materials and Methods. (C) Drop-test monitoring growth of sro7Δ, pep4Δ, vps27Δ, pep4Δ sro7Δ, and vps27Δ sro7Δ on YP and YP + 0.7 M NaCl plates. Tenfold dilutions of YP cultures of OD610 1.0 were spotted onto the plates, which were photographed after incubation for 48 h at 30°C.
S. cerevisiae has multiple secretory pathways leading to the vacuole, which originate from the endoplasmic reticulum (ER), Golgi, or the plasma membrane (Wendland et al., 1998). To delineate the pathways by which Ena1p is diverted to the vacuole in cells lacking SRO7, we constructed sro7Δ mutants that were blocked in specific steps in secretory or degradation pathways. If Ena1p is sorted to the vacuole via the cell surface, the sodium transporter should be stabilized in an end4Δ mutant, because of inhibited internalization from the plasma membrane. However, immunoblot analysis revealed equal instability of Ena1p in salt-stressed end4Δ sro7Δ mutants as in sro7Δ mutants (Figure 4B), suggesting sorting to the vacuole occurs independent of the plasma membrane. Similar results were obtained with the sec15-1 sro7Δ mutant, which is conditionally blocked in a late stage of the secretory pathway. The experiments were performed at 34°C where the temperature-sensitive sec15-1 mutant displays growth inhibition, but not a complete growth arrest. After 3–5-h incubation at 0.5 M NaCl Ena1p disappeared from the immunoblots of the sec15-1 sro7Δ mutant (Figure 4B), as for the sro7Δ single mutant, indicating that Ena1p is routed to the vacuole at steps preceding the Sec15p-mediated tethering of post-Golgi vesicles for subsequent membrane fusion. By contrast, Ena1p remained as stable in a sro7Δ vps27Δ mutant (Figure 4B), as in a sro7Δ pep4Δ mutant. Mutants defective in VPS27 are blocked in exit from prevacuolar/endosomal compartments and trap ubiquitinated vacuolar and recycling proteins in endosomes of the so-called MVE (multivesicular endosomes; Bilodeau et al., 2002) pathway. Taken together, our results suggest that in NaCl-stressed sro7Δ mutants Ena1p is sorted from Golgi to the vacuole via the MVE pathway and does not pass via the plasma membrane.
Missorting in sro7Δ Mutants Is Exacerbated by NaCl Stress
Despite the stabilization of Ena1p in the sro7Δ pep4Δ and sro7Δ vps27Δ mutants no obvious differences in salt tolerance were observed between these strains and the sro7Δ mutant (Figure 4C). Hence, stabilization of Ena1p was not sufficient per se to restore the salt tolerance of sro7Δ mutants. This suggests the cell is incapable of sorting Ena1p correctly to the plasma membrane in the absence of SRO7, even when Ena1p remains stable. This result agrees with the previous observation that overexpression of the ENA1 gene does not improve salt tolerance of the sro7Δ mutant (Figure 2B).
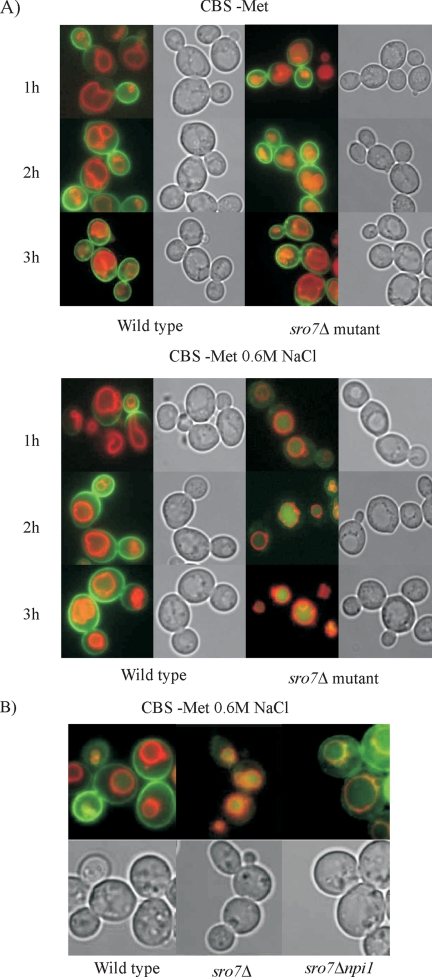
To further study the fate of Ena1p in wild-type cells and sro7Δ mutants, we used the pUG35-ENA1 plasmid (gift from M. Cyert) containing an ENA1-GFP insert driven by the methionine-repressible MET25 promoter. The expression of ENA1-GFP from this plasmid restores wild-type NaCl tolerance of ena1Δ mutants (unpublished data), demonstrating that the Ena1p-GFP fusion is functional. This construct allowed us to examine the influence of salt stress on Ena1p secretion, because ENA1 expression can be induced without NaCl exposure. Transformants of wild-type and sro7Δ cells cultured at repressing methionine concentration (0.5 mM) were washed free of methionine to de-repress ENA1-GFP expression and incubated in CBS medium with and without 0.6 M NaCl. Fluorescence microscopy of these transformants showed that Ena1p-GFP, independent of salinity, gradually accumulated in the periphery of wild-type cells, first appearing around the rim of the bud or in the bud neck of the cells (Figure 5A). A similar pattern was observed in nonstressed sro7Δ mutants. However, in sro7Δ mutants subjected to 0.6 M NaCl stress, the GFP staining was coincident with the lumen of the vacuoles, as visualized by costaining with the vital dye FM4-64, which stains the delimiting vacuolar membrane (Vida and Emr, 1995). These experiments confirmed routing of Ena1p to the vacuolar lumen in sro7Δ mutants and also demonstrate that the sorting into the endosomal pathway for vacuolar degradation is dependent on NaCl stress.
Figure 5.
Mis-sorting of Ena1p in sro7Δ mutants is exacerbated by NaCl stress and is dependent on the ubiquitin ligase Rsp5p. (A) Wild-type cells and sro7Δ mutants transformed with the pUG35-ENA1 plasmid, expressing Ena1p-GFP from the MET25 promoter, were cultured overnight in CBS medium containing 0.5 mM methionine. These cultures were used to inoculate identical fresh medium, and cells were cultured to midlog phase before being stained for 15 min with the vital dye FM4-64, to visualize vacuolar membranes. The cells were then washed, resuspended in fresh medium, and incubated for another 1.5 h before being washed again and transferred to methionine-free medium with and without 0.6 M NaCl. Removal of methionine induces expression of Ena1p-GFP from the MET25 promoter. Samples were removed and photographed at the times indicated. (B) Wild-type cells, sro7Δ single mutants, and sro7Δnpi1 double mutants, which express low levels of RSP5, were treated as in A and incubated for 2 h in methionine-free medium at 0.6 M NaCl. Yellow regions indicate Ena1p-GFP and FM4-64 colocalization.
Because sorting of Ena1p from Golgi to the vacuole appears dependent on the MVE pathway, we speculated that this trafficking requires Rsp5p-mediated ubiquitinylation, as reported for transporters such as Fur4p (Galan et al., 1996), Gap1p (Helliwell et al., 2001), and Tat2p (Beck et al., 1999). In a sro7Δnpi1 double mutant, which expresses only low levels of the essential ubiquitin-protein (E3) ligase Rsp5p (Hein et al., 1995), Ena1p-GFP was mis-sorted at high salinity to the limiting membrane of the vacuole rather than to the vacuolar lumen (Figure 5B). Because ubiquitinylation is a signal for delivery of cargo to the internal vesicles of MVEs, inhibited ubiquitinylation may lead to cargo accumulation in the MVE-limiting membranes, which ultimately fuses with the vacuolar membrane (Katzmann et al., 2002). Thus, the observed Ena1p-GFP distribution indicates requirement of ubiquitinylation for correct sorting of Ena1p to the vacuole.
Genetic Interactions between SRO7 and Late Secretory Mutants
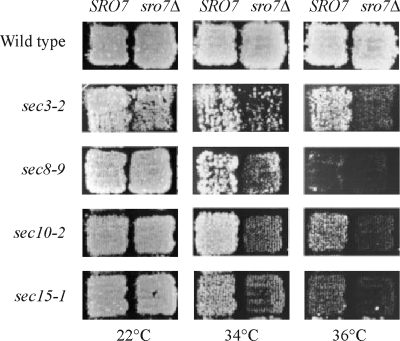
Lehman et al. (1999) reported that overexpression of SRO7 partially suppressed the temperature-sensitive phenotypes of mutations in SEC3, SEC8, SEC10, and SEC15, all of which encode subunits of the exocyst. However, high copy number suppression can also be the result of rather indirect interactions. We therefore tested if the sro7Δ knockout shows synthetic interactions with temperature-sensitive sec3-2, sec4-8, sec5-24, sec6-8, sec8-9, sec9-4, sec10-2, and sec15-1 mutations, by disrupting the SRO7 gene in the corresponding sec strains and testing growth at different temperatures. As shown in Figure 6, we saw clear evidence of synthetic interactions between sro7Δ and sec3-2, sec8-9, sec10-2, and sec15-1 at both 34 and 36°C, which, interestingly, are the same mutations that were suppressed by overexpression of SRO7 (Lehman et al., 1999). For sec4-8, we saw a very weak synthetic interaction with sro7Δ at 30°C, whereas no interactions were detected between sro7Δ and sec5-24, sec6-8, or sec9-4 (unpublished data). It should be noted, though, that the stronger phenotypes of these sec mutations, which prevent all growth at 34°C and above, make it more difficult to detect any synthetic interactions.
Figure 6.
Genetic interaction between SRO7 and various SEC genes. The SRO7 gene was disrupted in various sec mutants, and temperature sensitivity was monitored at different temperatures. Top row represents growth of wild-type (NY179) and sro7Δ (H1394) strains at 22, 34, and 36°C. The following rows display growth of different sec single mutants (left column) and the corresponding sro7Δ sec double mutants (right column). Genetic interactions are revealed as increased temperature sensitivity of double mutants. Plates were photographed after incubation for 48 h at the indicated temperatures.
The Salt Sensitivity of sro7Δ Mutants Is Accompanied by a Late Secretory Defect and the Accumulation of Post-Golgi Secretory Vesicles
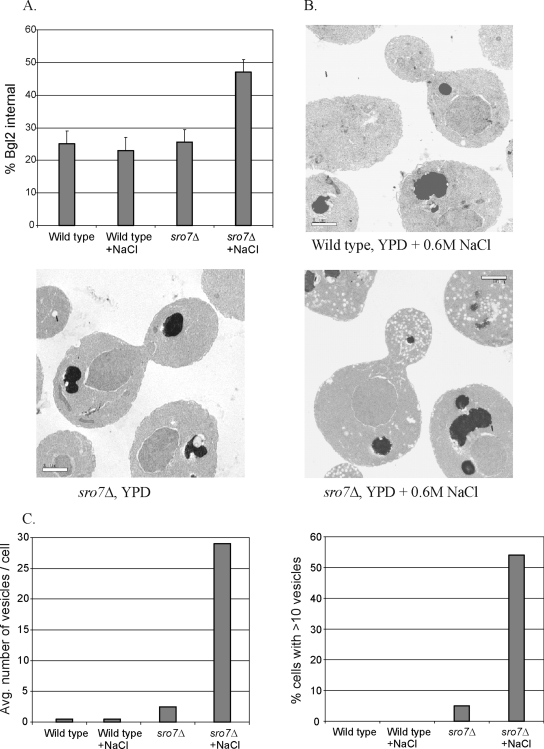
The genetic interactions described above as well as previous studies with sro7Δ sro77Δ mutants have shown that these proteins have a critical function in Rab-SNARE–mediated vesicle docking/fusion with the plasma membrane (Lehman et al., 1999; Grosshans et al., 2006). However, none of these studies examined the effect of high salt on the in vivo function of Sro7p in the cell. To explore this directly, we examined the secretory capacity of sro7Δ after exposure to 0.6 M NaCl. We first examined the secretion of the exoglucanase Bgl2p, a marker for one class of post-Golgi vesicles that delivers proteins to the cell surface (Harsay and Bretscher, 1995). Although the secretory capacity of wild-type cells is unaffected by exposure to salt, we find a dramatic defect in the surface delivery of Bgl2p (seen as a rise in internal pools) in sro7Δ mutants exposed to 0.6 M NaCl medium, whereas unstressed mutants showed a secretory capacity similar to that of wild-type cells (Figure 7A). Several attempts were made to determine the effect of salt on invertase secretion in sro7Δ cells. Unfortunately the presence of 0.6 M NaCl completely blocked the ability of sro7Δ mutants to de-repress invertase expression in low-glucose medium. We therefore inserted SUC2 behind the de-repressible MET25 promoter in a pUG35 plasmid that was transformed into wild-type cells and sro7Δ mutants. Assays for invertase secretion were then performed with cells incubated for 2 h at 25°C in methionine-free medium containing 0.6 M NaCl. These experiments revealed no significant difference in invertase secretion between sro7Δ and isogenic wild-type cells (unpublished data). A cargo-specific secretory defect is not unprecedented because we have previously shown that the cdc42-6 mutant also demonstrates a pronounced defect in Bgl2p secretion, whereas invertase secretion is normal (Adamo et al., 2001).
Figure 7.
The sro7Δ mutant exhibits secretion defects and accumulation of post-Golgi vesicles at high salinity. (A). Secretion of Bgl2p. Assays were performed on isogenic wild-type and sro7Δ stains after a 2-h shift to YP medium with 0.6 M NaCl (+NaCl) or YP medium without added NaCl. The results show the internal accumulation of Bgl2p as a percentage of the total Bgl2 in the cell. (B) Electron microscopy of cells from WT and sro7Δ strains after a 2-h shift to YP medium with 0.6 M NaCl show that sro7Δ mutant cells accumulate 80–100-nm post-Golgi vesicles in the bud. This accumulation depends on the addition of salt, because no vesicles accumulate in sro7Δ cells in the absence of a salt stress. (C) Quantitation of the secretory vesicle accumulation in sro7Δ cells after a 2-h shift to 0.6 M NaCl/YP. Cells were scored for severity and penetrance of the secretory defects. The bar graph on the left shows the accumulation of vesicles in the indicated strains, expressed as vesicles per cell, and was determined by the number of 80–100-nm vesicles observed per cell per thin section. The graph on the right shows the penetrance of the vesicle accumulation expressed as the number of cells containing more than 10 vesicles per cell. A minimum of 100 cells were used for the quantitation of each strain.
To determine more precisely the step in secretion effected by the exposure of sro7Δ mutants to high salt, we examined wild-type cells and sro7Δ mutants by thin-section electron microscopy (Adamo et al., 1999), after incubation for 2 h in YPD medium with and without 0.6 M NaCl. Cells showing a defective Golgi-to-plasma membrane delivery are known to accumulate 80–100-nm secretory vesicles (Novick et al., 1980). As expected, wild-type cells displayed few post-Golgi vesicles in the presence or absence of exposure to 0.6 M NaCl (Figure 7B). Although sro7Δ mutants show only a small increase in vesicles in normal media, when shifted to 0.6 M NaCl, sro7Δ mutants demonstrate a dramatic increase in the accumulation of 80–100-nm vesicles (Figure 7, B and C). This phenotype was highly penetrant (>55% of cells with >10 vesicles) and, unlike the cdc42-6 mutant (Adamo et al., 2001), there was no obvious correlation between vesicle accumulation and bud size (G. Rossi and P. Brennwald, unpublished observation).
To further characterize these vesicles as post-Golgi vesicles, velocity sedimentation analysis was performed. Lysates derived from spheroplasts of wild-type cells and sec6-4 and sro7Δ mutants having the chromosomal ENA1 gene modified to carry the 3xHA tag were layered onto sorbitol gradients as described previously (Adamo et al., 2001). All strains were shifted into 0.6 M NaCl media for 2 h to induce expression of Ena1p and the secretory defect associated with sro7Δ cells. As a positive control for a late secretory block, we shifted sec6-4 cells (containing Ena1-HA) into 0.6 M NaCl (to induce Ena1-HA) and then shifted the cells to 37°C for 2 h to impose the late secretory block. After centrifugation fractions were analyzed for Ena1p-HA, the vesicle marker Snc1/2p, and Bgl2p (unpublished data) by immunoblot analysis. The results, shown in Figure 8A (top graphs), strongly support the observations seen by EM and demonstrate sro7Δ mutants have a pronounced accumulation of post-Golgi vesicles (marked by Snc1/2p) in response to 0.6 M NaCl. We find these vesicles have a mobility (compare top graphs left and right) in the gradients that are identical to those that accumulate in sec6-4 cells, which is consistent with the EM and secretion results above. Finally we find that a peak of the periplasmic enzyme Bgl2p appears in the sro7Δ gradients (but not in wild-type gradient), which is coincident with the peak of Snc1/2p that appears in sro7Δ cells (unpublished data)—demonstrating that the accumulation of internal pools of Bgl2p in this mutant is due to a post-Golgi secretory defect.
Figure 8.
(A) Accumulation of post-Golgi vesicles marked with Snc1/2p but not Ena1p in sro7Δ mutants shifted to high salt. Vesicle gradients of 20–40% sorbitol were performed on sro7Δ and isogenic wild-type strains after 2 h of growth in media with 0.6 M NaCl to induce both expression of HA-tagged Ena1 as well as the sro7Δ growth defect. As a positive control HA-Ena1p was introduced into a sec6-4 strain, which was grown in 0.6 M NaCl for 2 h (to induce HA-Ena1p) before shifting temperature to 37°C to induce the late secretory block. Normalized lysates from each strain were then layered onto the velocity gradients. Post-Golgi vesicles were detected by immunoblotting with rabbit anti-Snc1/2p antisera. HA-tagged Ena1p was detected with mAb 12CA5. Western blots were quantitated using an Odyssey InfraRed imaging system. (B) Ena1p is not retargeted to the vacuole after blocked exocytosis in late sec mutants. Wild-type (RSY255) cells and sec6-4 and sec4-8 mutants transformed with the pUG35-ENA1 plasmid, expressing Ena1p-GFP from the MET25 promoter, were cultured overnight in CBS medium and treated as described in legend to Figure 5 except that transferred cells were incubated at 25 or 37°C in the methionine-free medium with and without 0.6 M NaCl. Samples were removed and photographed 2 h after start of the final incubation.
To examine the effect of sro7Δ on sorting of Ena1p into post-Golgi vesicles, we first determined if salt-induced Ena1p-HA was found associated with the more conventional sec6-4 vesicles. As is shown in Figure 8A (bottom graphs), we find a major peak of Ena1p-HA that comigrates with the peak of Snc1/2p in these gradients. This peak depends on a late-secretory block because it disappears in an isogenic wild-type strain. We then examined whether Ena1p-HA was associated with the post-Golgi vesicles that accumulate in response to salt in a sro7Δ strain. In this case we found that Ena1p-HA was not detectable in the peak of Snc1/2p- and Bgl2p-containing vesicles, which accumulate in this strain. Therefore, although Ena1p-HA is properly sorted into post-Golgi vesicles accumulated in response to a sec6-4 mutant (shifted to high temperature), Ena1p-HA is not detectably sorted in post-Golgi vesicles that accumulate in response to a sro7Δ mutant (shifted to high salt). Therefore sro7Δ cells show two salt-sensitive defects, one that results in accumulation of post-Golgi vesicles and a second (earlier) defect that prevents the proper sorting of Ena1p-HA into these vesicles.
We next examined the effect of Ena1p sorting by a late block in secretion by following the distribution of Ena1p-GFP in wild-type and late sec mutants. Wild-type (RSY255), sec4-8 and sec6-4 strains were transformed with the pUG35-ENA1 plasmid and incubated in basal medium at permissive (25°C) and restrictive (37°C) temperature after de-repression of ENA1-GFP expression (Figure 8B). Restrictive conditions clearly blocked delivery of Ena1p-GFP to the cell surface in the sec mutants, as opposed to the seemingly intact secretion at the permissive temperature. However, the blocked secretion did not result in any obvious vacuolar sorting of accumulated Ena1p-GFP, instead GFP fluorescence remained as a punctate haze in the cytosol, indicating stalled Ena1p traffic in the secretory system. A similar distribution of Ena1-GFP fluorescence was recorded for sec mutants when experiments were repeated at 0.6 M NaCl. Hence, a block at a late stage in exocytosis does not promote vacuolar targeting of Ena1p even when cells are exposed to high salinity.
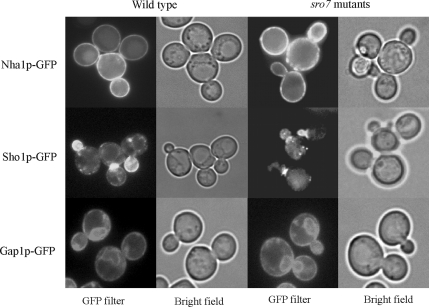
We were unable to identify any obvious mis-targeting in NaCl-exposed sro7Δ mutants for a number of other GFP-tagged membrane proteins selected for analysis by microscopy. This analysis revealed an apparently correct targeting of the constitutively expressed sodium-proton anti-porter Nha1p in NaCl-stressed sro7Δ mutants (Figure 9). Similarly, a GFP-tagged version of Gap1p, expressed from the heterologous MET25 promoter, showed a distribution in NaCl-stressed sro7Δ mutants similar that of wild-type cells. Gap1p is thought to be carried by a class of post-Golgi vesicles distinct from the high-density Bgl2p carrying vesicles (Roberg et al., 1997). Similarly, other GFP tagged proteins examined, i.e., the polarly localized Sho1p and Ste2p, as well as the membrane associated Pbs2p (Stefan and Blumer, 1999; Reiser et al., 2000), maintained a typical wild-type-like distribution in sro7Δ mutants even during salt stress (unpublished data for the Ste2p and Pbs2p localization). We conclude that sro7Δ mutants exposed to salt stress accumulate post-Golgi vesicles and restrict Ena1p delivery to the plasma membrane, but that the surface delivery of other proteins examined is partly or insignificantly inhibited. Thus, loss of SRO7 affects transport of only specific proteins, which is in agreement with the observation that alternative pathways exist for the transport of membrane proteins to the plasma membrane (Harsay and Bretscher, 1995; Roberg et al., 1997; Harsay and Schekman, 2002).
Figure 9.
Localization of selected GFP-tagged transmembrane proteins in wild-type and sro7Δ cells after a shift to high salinity. Cells were grown to midlog phase and then exposed to medium containing 0.5 M NaCl for 3 h before photography. Top, Nha1p-GFP, which localizes to the plasma membrane (Banuelos et al., 1998), shows a similar distribution in salt-stressed wild-type cells (left) and sro7Δ mutant cells (right). To express Nha1p-GFP, cells were transformed with the pGRU1 plasmid (Kinclova et al., 2001). Middle, Sho1p-GFP displays a similar pattern of polarized localization in NaCl-stressed wild-type and sro7Δ mutant cells, being concentrated in the growing bud and at the mother-bud neck as previously reported (Reiser et al., 2000). Bottom, Gap1p-GFP expressed from the MET25 promoter in the pUG35-GAP1 plasmid exhibits similar distribution in salt-stressed wild-type and sro7Δ mutant cells. Gap1p shows distribution to ER and the plasma membrane as reported for wild-type cells growing under nonstress conditions (Roberg et al., 1997).
Overexpression of the RSN1 Gene Suppresses Salt Sensitivity of the sro7Δ Mutant
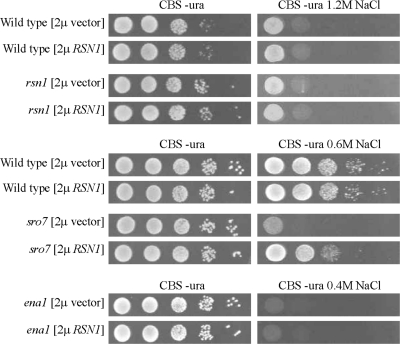
To find new genes that could fully or partially substitute for SRO7, sro7Δ mutants were used to select multicopy suppressors that restored growth at high salinity. To this end sro7Δ mutants were transformed with the pHR81-based yeast genomic library (Nehlin et al., 1989), plated on selective medium and after incubation replica-plated to high-salinity media. By this screen we identified three genes, including the SRO7 gene, which restored sro7Δ salt tolerance when retransformed into the mutant cells. The other two genes are so far uncharacterized and were named RSN-genes, for rescue of sro7 at high NaCl. Four independently isolated plasmids had inserts containing the ORF YMR266w, henceforth referred to as RSN1. This ORF encodes a protein of 953 amino acids containing 11 putative transmembrane regions (http://www.yeastgenome.org/) and belongs to a family of membrane-bound or -associated proteins of unknown function. Multicopy plasmids (pHR81 and pRS426) containing RSN1 and the corresponding empty plasmids were transformed into wild-type, sro7Δ, rsn1Δ, and ena1Δ strains, and growth was scored on selective agar plates with different salt concentrations (Figure 10). The rsn1Δ mutant displayed tolerance to NaCl similar to that of wild-type cells, whereas overexpression of RSN1 specifically rescued growth of the sro7Δ mutant at high salinity. This suppression was obviously not due to any Na+-protecting activity by Rsn1p itself, because multicopy expression of RSN1 in ena1Δ cells did not improve salt tolerance of these highly NaCl sensitive mutants (Figure 10).
Figure 10.
The RSN1 gene rescues growth of the sro7Δ mutant at high salinity. Cultures of wild-type (BY 4741), rsn1Δ, sro7Δ, and ena1Δ mutant strains transformed with the 2μ plasmid pRSN1 or the empty vector pRS423 were spotted in 10-fold dilutions onto defined CBS medium with or without NaCl as indicated, the first spot representing OD610 0.1. Plates were incubated for 48 h at 30°C before photography.
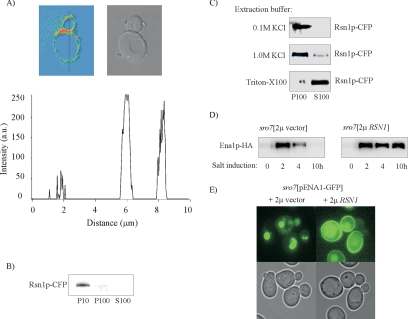
For further characterization of Rsn1p, we modified the chromosomal gene to encode a cyan fluorescent protein (CFP) at the carboxy terminus of the protein. Confocal microscopy demonstrated that the fluorescence localized along the cell perimeter (Figure 11A). The intensity distribution of the fluorescence also demonstrates a clear concentration of the tagged protein to the bud neck region. Consistent with the microscopic observations, subcellular fractionation detected the majority of Rsn1p-CFP in the 10,000 × g fraction and the remainder sedimented in the 100,000 × g fraction (Figure 11B). To investigate the association of Rsn1p-CFP with the sedimenting fractions, protein extracts were incubated with 1.0 M KCl or 1% Triton X-100 before centrifugation at 100,000 × g. As shown in Figure 11C, Rsn1p-CFP could not be extracted from the pellet with KCl, whereas treatment with detergent resulted in an almost complete recovery of Rsn1p-CFP in the soluble fraction. These results confirm a membrane location for the protein and suggest that Rsn1p is an integral membrane protein in agreement with the existence of several putative transmembrane domains in its sequence.
Figure 11.
Rsn1p is a plasma membrane protein and multicopy expression of RSN1 reroutes Ena1p to the cell surface in NaCl-stressed sro7Δ mutants. (A) Transmission image and fluorescence intensity distribution of CFP-tagged Rsn1p in wild-type cells as analyzed by confocal microscopy. The graph shows a line profile of the fluorescence intensity measured along the black line indicated in the figure. The tagged protein is clearly concentrated in the bud neck region (corresponding to the peak at 6 μm). (B) Subcellular fractionation of cells expressing Rsn1p-CFP. Total protein extract was centrifuged at 10,000 × g and 100,000 × g to separate soluble cytosolic proteins from pelletable structures. P10, 10,000 × g pellet; P100, 100,000 × g pellet; S100, supernatant from 100,000 × g centrifugation. (C) Rsn1p-CFP localizes to a membrane fraction. Cell lysates were treated with 0.1 M KCl, 1.0 M KCl, or 1% Triton X-100. Pellet and supernatant fractions were then separated by centrifugation at 100,000 × g. Rsn1p-CFP was detected by SDS-PAGE followed by immunoblotting using an anti-GFP antibody. (D) Immunoblot analysis of Ena1p-HA levels in sro7Δ mutants containing a control plasmid or a multicopy RSN1 plasmid. Cells were grown to exponential phase and shifted to medium containing 0.6 M NaCl before sampling at indicated time points. Immunoblot analysis was preformed as described in Figure 3 and Materials and Methods. Total proteins were extracted by alkaline cell lysis, and Ena1p-HA was detected by SDS-PAGE and immunoblotting with anti-HA antibody. Each sample represents an equal amount of total protein. (E) Fluorescence images of sro7Δ mutants transformed with the pUG35-ENA1 plasmid expressing Ena1p-GFP from the MET25 promoter and the pRS423 empty vector (left) or the pRS423-RSN1 vector (right) Cells cultured on defined medium containing 1 mM methionine and washed before transfer to methionine-free 0.5 M NaCl medium. Photographs were taken after 3-h incubation in the saline medium.
Increased Expression of RSN1 Enhances Ena1p Stability in sro7Δ Mutants and Retargets the Protein to the Plasma Membrane
Because extra copies of RSN1 restore salt tolerance of the sro7Δ mutant (Figure 10), we wanted to examine the effects of the suppression on the fate of Ena1p. Therefore sro7Δ mutants carrying a HA-tagged Ena1p were transformed with a multicopy plasmid containing RSN1 and a corresponding empty plasmid. Immunoblot analysis of extracts from NaCl-exposed transformants demonstrated that Ena1p levels are stably maintained in sro7Δ mutants overexpressing RSN1, whereas Ena1p becomes gradually degraded in transformants carrying the empty plasmid (Figure 11D). Multicopy expression of RSN1 in NaCl-stressed sro7Δ mutants carrying the ENA1-GFP plasmid pUG35-ENA1 showed redistribution of Ena1p from the interior to the cell perimeter, similar to the distribution seen in wild-type cells (Figure 11E). These results strongly suggest that the basis for suppression of the NaCl sensitivity of sro7Δ mutants is retargeting of Ena1p to the plasma membrane and indicate a Sro7p-like role for Rsn1p in membrane traffic.
DISCUSSION
Previous work has shown that deletion of the yeast SRO7 gene produces a NaCl-sensitive phenotype and a disrupted Na+/K+ balance in cells subjected to NaCl stress (Larsson et al., 1998). Here we demonstrate that the NaCl sensitivity of sro7Δ mutants is due to defective targeting of the ENA1 gene product to its final destination in the plasma membrane. The ENA1 gene encodes a salinity-induced sodium transporter that belongs to the family of ion-transporting P-type ATPases and constitutes the main pathway for sodium export in yeast (Haro et al., 1991; Haro et al., 1993; Wieland et al., 1995). In cells lacking Sro7p the salt-induced sodium pump becomes degraded before the arrival at the plasma membrane, because additional mutations in the sro7Δ background that blocked Golgi to plasma membrane transport (sec15) or inhibited internalization from the cell surface (end4), did not prevent Ena1p degradation. The loss of Ena1p in sro7Δ mutants is caused by retargeting to the vacuole (Figures 4A and 5), probably via the MVE pathway because degradation of Ena1p does not occur in sro7Δ vps27Δ mutants (Figure 4B). Vps27p mediates, in conjunction with the ESCRT complexes (endosomal sorting complex required for transport), sorting of ubiquitinylated membrane proteins destined for degradation in the vacuole (Bilodeau et al., 2002; Katzmann et al., 2003). The fact that Ena1p remains improperly targeted in sro7Δ mutants even when the ENA1 gene is overexpressed, strongly suggests that trafficking of Ena1p to the cell surface involves Sro7p-dependent sorting and does not occur by default mechanisms. Although it is still an open question whether yeast plasma membrane proteins can be targeted to the vacuole by a default mechanism, studies have shown that ubiquitinylation mediates sorting of specific plasma membrane proteins from Golgi into the MVE pathway in response to nutritional signals (Helliwell et al., 2001; Umebayashi and Nakano, 2003). Ubiquitinylation defective sro7Δ mutants carrying the npi1/rsp5 mutation diverted Ena1p-GFP to the vacuole limiting membrane, whereas sro7Δ single mutants effectively sorted Ena1p-GFP to the vacuole lumen (Figure 5B). Rsp5p belongs to a family of evolutionarily conserved ubiquitin ligases with multiple functions (Shearwin-Whyatt et al., 2006), one of which being the tagging of selected membrane proteins for entry into the intralumenal MVEs (Katzmann et al., 2002). Hence, our results are consistent with a requirement for ubiquitinylation to direct Ena1p from Golgi into the MVE pathway. There is also evidence for a Golgi-based quality control mechanisms playing a role for rerouting of mis-folded or defective plasma membrane proteins into the endosomal/vacuolar pathway for degradation (Chang and Fink, 1995; Jenness et al., 1997; Zhang et al., 2001; Arvan et al., 2002). Mis-sorting of Ena1p may result from a defect in a number of steps from Golgi to the cell surface, ranging from defective cargo packaging or vesicle budding to a blocked recognition of cargo determinants at the site of exocytosis. Interestingly, salt-stressed sro7Δ mutants accumulate bona fide post-Golgi vesicles, suggesting that inhibited or restricted exocytosis may be a reason for the rerouting of Ena1p into the MVE pathway. Previous work has shown that a fraction of Sro7p is attached to the plasma membrane (Larsson et al., 1998; Lehman et al., 1999), and Sro7p interacts physically with Sec9p, a plasma membrane t-SNARE protein (Lehman et al., 1999). SRO7 interacts furthermore genetically with genes encoding subunits of the exocyst (Figure 6 and Lehman et al., 1999), and Sro7p was recently shown to interact directly with the Exo84p component of the exocyst complex (Zhang et al., 2005) and to serve as an effector of the Sec4p GTPase (Grosshans et al., 2006). Hence, there is strong evidence for a role of Sro7p in late exocytosis. Although cellular processes that identify targeting determinants at the plasma membrane remain uncharacterized (Mayor and Riezman, 2004), exocyst- and SNARE-interacting proteins might well play a role. It is likely that sites of active exocytosis require mechanisms to ensure that the correct cargo molecules are destined to the proper membrane sites. In fact, SNAREs are implicated in cargo sorting functions in the ER, where they might serve as primers for cargo capture (Springer and Schekman, 1998; Morsomme et al., 2003). However, absence of Ena1p in the post-Golgi vesicles that accumulate in salt-stressed sro7Δ mutants (Figure 8A) suggests that Ena1p is routed to the vacuole before being sorted into late secretory vesicles. This conclusion is also supported by the finding that a temperature-induced block of late exocytosis in sec6-4 mutants does not prevent Ena1p from being sorted into post-Golgi vesicles, and the imposed block does not provoke vacuolar routing of Ena1p (Figure 8B). These data clearly suggest that Sro7p fulfils a so far unrecognized role for delivery of Ena1p into post-Golgi vesicles and that loss of SRO7 leads to identification of this cargo protein for transport to and destruction in the vacuole. Exclusion from the normal sorting machinery may affect conformational characteristics and/or expose targeting signals that lead to recognition by the Golgi quality control system. Such detection may in turn initiate tagging of the protein for subsequent degradation via the MVE pathway. This scenario is in agreement with the transient appearance of Ena1p in NaCl-stressed sro7Δ mutants (Figure 3B). The fact that both mis-sorting of Ena1p and accumulation of post-Golgi vesicles are restricted to sro7Δ mutants exposed to NaCl stress suggests that Sro7p, both at the cell surface and in its proposed Golgi role, is involved in mechanisms that become destabilized and sensitive to ion-mediated stress by Sro7p deficiency.
Analysis of secretory vesicles from exocytic (Harsay and Bretscher, 1995) and vacuolar protein-sorting mutants (Harsay and Schekman, 2002) has identified vesicle populations with different physical characteristics and carrying different cargo molecules, indicating that exocytosis can follow at least three different routes in yeast. The only obvious phenotype of the sro7Δ mutant is its sensitivity to NaCl stress (Larsson et al., 1998), suggesting vacuolar mis-sorting is highly specific for Ena1p. This view is supported by the observation that plasma membrane–associated proteins representing both a constitutive plasma membrane localization (Nha1p-GFP; Banuelos et al., 1998) and a polarized distribution that changes during the cell cycle (Sho1p-GFP; Reiser et al., 2000) appear to show proper membrane targeting in sro7Δ mutants exposed to salt stress (Figure 9). Similar results were observed for Gap1p-GFP when expression was induced simultaneously with the NaCl stress. However, the significant accumulation of post-Golgi vesicles in salt-stressed sro7Δ mutants and the strong secretory defect for Blg2p, a marker for light density vesicles, demonstrates a defect in the docking and fusion of transport vesicles with the cell surface (Figures 7A and 8A). These findings suggest that there is no general sorting defect for plasma membrane proteins in salt-stressed sro7Δ mutants but that delivery of specific cargo molecules becomes restricted dependent on cargo characteristics or the pathway used.
The Drosophila Sro7p counterpart, the Lgl tumor suppressor has been suggested to fulfill a role similar to Sro7p in late exocytosis (Bilder et al., 2000; Bilder, 2004; Wirtz-Peitz and Knoblich, 2006). Lgl associates with the lateral membrane of polarized epithelia and is required to properly localize apical proteins and adherence junctions to organize the epithelial architecture in embryos (Manfruelli et al., 1996; Bilder, 2004). Müsch et al. (2002) showed that a Lgl homologue in Madin-Darby canine kidney cells (Mlgl) becomes associated with the lateral membrane as the cells make cell-to-cell contacts and establish a polarized phenotype. The membrane-bound Mlgl coimmunoprecipitated with the basolateral t-SNARE syntaxin 4, suggesting a specific role for Mlgl in basolateral exocytosis in polarized epithelial cells. Mlgl (Gangar et al., 2005) and Lgl (I. Wadskog, unpublished results and Larsson et al., 1998) substitute for Sro7p when expressed in sro7Δ or sro7Δ sro77Δ mutants, indicating the homologues participate in a highly conserved function. The ability to rescue the salt sensitivity of these mutants along with the ability to bind SNARE proteins may indicate that metazoan Lgl proteins have retained these dual functions in Golgi and endosomal cargo sorting and exocytic vesicle fusion.
Our search for multicopy suppressors of the sro7Δ salt sensitivity identified the previously uncharacterized RSN1 gene. Overexpression of RSN1 stabilizes Ena1p (Figure 11) restores its sorting to the cell surface and also restores NaCl tolerance. RSN1 encodes a 108-kDa protein, belonging to the yeast facilitator superfamily. Deletion of RSN1 does not produce any obvious phenotype and overexpression of RSN1 does not improve salt tolerance of ena1Δ mutants, indicating that Rsn1p per se cannot substitute for mis-targeted Ena1p. According to the localization experiments (Figure 11), Rsn1p is an integral membrane protein localized to the cell periphery, in particular to the bud-neck region. This distribution is reminiscent of that of Sro7p, which is along detected along the cell perimeter (Larsson et al., 1998; Lehman et al., 1999). Sro7p also coimmunoprecipitates with Myo1p (Kagami et al., 1998), a type II myosin that colocalizes with the actin ring in the bud neck (Lippincott and Li, 1998). Hence, the distribution of Rsn1p is consistent with a role in late exocytosis and in agreement with its ability to substitute for the Sro7p function. How exactly Rsn1p contributes to the reestablished cell surface targeting of Ena1p vesicles remains to be determined.
We have here presented evidence for a novel function of the yeast tumor suppressor homologue Sro7p that is required for sorting of Ena1p into Golgi-derived vesicles destined for the cell surface. The precise mechanism by which Sro7p exerts this function is at present unknown; however, identification and characterization of proteins regulating the functions of Sro7p may help to elucidate these mechanisms. These studies on the roles of Lgl proteins in protein sorting and polarized vesicle docking or fusion, may provide important information on how polarity is established and how cells sort plasma membrane proteins to specific domains on the cell surface or within the cell.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge kind gifts of plasmids containing Nha1p-GFP from H. Sychrova, Sho1p-GFP and Pbs2p-GFP from G. Ammerer, Ste2p-GFP from J. Blumer, and ENA1 driven by the PMA1 promoter from H. Rudolph. Plasmids for CFP or yellow fluorescent protein tagging of proteins were kindly provided by the Yeast Resource Center at University of Washington (T. Davis), and GFP antibodies were generously provided by P. Silver. We are also indebted to M. Cyert and V. Heath for making available their pUG35-ENA1 plasmid and to R. Haguenauer-Tsapis and P. Ljungdahl for providing strains. This work was supported by grants from the Swedish Research Council, Magnus Bergvalls Stiftelse, and the EU programs BIO-CL 950161 and ERB4061 PL95-0014 to L.A. and by a grant from the Swedish Cancer Society to H.R. Parts of this study were conducted at the SWEGENE Center for Biophysical Imaging.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0798) on September 27, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adamo J. E., Moskow J. J., Gladfelter A. S., Viterbo D., Lew D. J., Brennwald P. J. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo J. E., Rossi G., Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P., Zhao X., Ramos-Castaneda J., Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- Banuelos M. A., Sychrova H., Bleykasten-Grosshans C., Souciet J.-L., Potier S. The NHA1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology. 1998;144:2749–2758. doi: 10.1099/00221287-144-10-2749. [DOI] [PubMed] [Google Scholar]
- Beck T., Schmidt A., Hall M. N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Knoblich J. A. Dare to be different: asymmetric cell division in Drosophila, C. elegans, and vertebrates. Curr. Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Bilder D., Li M., Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Brennwald P., Kearns B., Champion K., Keranen S., Bankaitis V., Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Chang A., Fink G. R. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Moreau V., Andre B., Volland C., Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- Gangar A., Rossi G., Andreeva A., Hales R., Brennwald P. Structurally conserved interaction of Lgl family with SNAREs is critical to their cellular function. Curr. Biol. 2005;15:1136–1142. doi: 10.1016/j.cub.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Garciadeblas B., Rubio F., Quintero F. J., Banuelos M. A., Haro R., Rodriguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- Grifoni D., Garoia F., Schimanski C. C., Schmitz G., Laurenti E., Galle P. R., Pession A., Cavicchi S., Strand D. The human protein Hugl-1 substitutes for Drosophila lethal giant larvae tumour suppressor function in vivo. Oncogene. 2004;23:8688–8694. doi: 10.1038/sj.onc.1208023. [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Andreeva A., Gangar A., Niessen S., Yates J. R., 3rd, Brennwald P., Novick P. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J. Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Sacher M., Barrowman J., Ferro-Novick S., Novick P. Protein complexes in transport vesicle targeting. Cell Biol. 2000;10:251–255. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- Haro R., Banuelos M. A., Quintero F. J., Rubio F., Rodriguez-Navarro A. Genetic basis of sodium exclusion and sodium tolerance in yeast. A model for plants. Physiol. Plant. 1993;89:868–874. [Google Scholar]
- Haro R., Garciadeblas B., Rodriguéz-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- Harsay E., Bretscher B. Parallel pathways to the cell surface in yeast. J. Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., Andre B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- Helliwell S. B., Losko S., Kaiser C. A. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 2001;153:649–662. doi: 10.1083/jcb.153.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim. Biophys. Acta. 2003;1614:139–155. doi: 10.1016/s0005-2736(03)00195-0. [DOI] [PubMed] [Google Scholar]
- Humbert P., Russell S., Richardson H. Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays. 2003;25:542–553. doi: 10.1002/bies.10286. [DOI] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., Knoblich J. A. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 2004;6:845–854. doi: 10.1016/j.devcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Li Y., Tipper C., Spatrick P. Elimination of defective alpha-factor pheromone receptors. Mol. Cell. Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- Kagami M., Toh-e A., Matsui Y. SRO9, a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics. 1997;147:1003–1016. doi: 10.1093/genetics/147.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M., Toh-e A., Matsui Y. Sro7p, a Saccharomyces cerevisiae counterpart of the tumor suppressor l(2)gl protein, is related to myosins in function. Genetics. 1998;149:1717–1727. doi: 10.1093/genetics/149.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Stefan C. J., Babst M., Emr S. D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinclova O., Ramos J., Potier S., Sychrova H. Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol. Microbiol. 2001;40:656–668. doi: 10.1046/j.1365-2958.2001.02412.x. [DOI] [PubMed] [Google Scholar]
- Koyama K., Fukushima Y., Inazawa J., Tomotsune D., Takahashi N., Nakamura Y. T. The human homologue of the murine Llglh gene (LLGL) maps within the Smith-Magenis syndrome region in 17p11.2. Cytogenet. Cell Genet. 1996;72:78–82. doi: 10.1159/000134167. [DOI] [PubMed] [Google Scholar]
- Larsson K., Böhl F., Sjöström I., Akhtar N., Strand D., Mechler B., Grabowski R., Adler L. The Saccharomyces cerevisiae SOP1 and SOP2 genes, which act in cation homeostasis, can be functionally substituted by the Drosophila lethal(2)giant larvae tumor suppressor gene. J. Biol. Chem. 1998;273:33610–33618. doi: 10.1074/jbc.273.50.33610. [DOI] [PubMed] [Google Scholar]
- Lehman K., Rossi G., Adamo J. E., Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfruelli P., Arquier N., Hanratty W. P., Semeriva M. Ther tumor suppressor lethal(2)gigant larvae (l(2)gl), is required for cell shape change of epithelial cells during Drosophila development. Development. 1996;122:2283–2294. doi: 10.1242/dev.122.7.2283. [DOI] [PubMed] [Google Scholar]
- Mayor S., Riezman H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Morsomme P., Prescianotto-Baschong C., Riezman H. The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J. Cell Biol. 2003;162:403–412. doi: 10.1083/jcb.200212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsch A., Cohen D., Yeaman C., Nelson W. J., Rodriguez-Boulan E., Brennwald P. Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in Madin-Darby canine kidney cells. Mol. Biol. Cell. 2002;13:158–168. doi: 10.1091/mbc.01-10-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene. 1989;85:313–319. doi: 10.1016/0378-1119(89)90423-x. [DOI] [PubMed] [Google Scholar]
- Nelson W. J., Yeaman C. Protein trafficking in the exocytic pathway of polarized epithelial cells. Trends Cell Biol. 2001;11:483–486. doi: 10.1016/s0962-8924(01)02145-6. [DOI] [PubMed] [Google Scholar]
- Niedenthal R., Riles L., Johnston M., Hegemann J. H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ohshiro T., Yagami T., Zhang C., Matsuzaki F. Role of cortical tumour-suppressor protein in assymmetric division of Drosophila neuroblasts. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- Peng C. Y., Manning L., Albertson R., Doe C. Q. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- Prior C., Potier S., Souciet J.-L., Sychrova H. Characterization of the NHA1 gene encoding the Na+/H+ antiporter pf the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;387:89–93. doi: 10.1016/0014-5793(96)00470-x. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Reiser V., Ruis H., Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:1147–1161. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Salah S. M., Ammerer G. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2000;2:620–627. doi: 10.1038/35023568. [DOI] [PubMed] [Google Scholar]
- Roberg K. J., Rowley N., Kaiser C. A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B. L., Seufert W., Steiner B., Yang Q. H., Futcher A. B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Serrano R., et al. A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 1999;50:1023–1036. [Google Scholar]
- Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- Springer S., Schekman R. Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- Stefan C. J., Blumer K. J. A syntaxin homolog encoded by VAM3 mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 1999;274:1835–1841. doi: 10.1074/jbc.274.3.1835. [DOI] [PubMed] [Google Scholar]
- Strand D., et al. A human homologue of the Drosophila tumour suppressor gene l(2)gl maps to 17p11.2–12 and codes for a cytoskeletal protein that associates with non muscle myosin II heavy chain. Oncogene. 1995;11:291–301. [PubMed] [Google Scholar]
- TerBush D. R., Maurice T., Roth D., Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tomutsune D., Shoji H., Wakamutsu Y., Kondoh H., Takahashi N. A mouse homologue of the Drosophila tumour-suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- Umebayashi K., Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadskog I., Adler L. Ion homeostasis in Saccharomyces cerevisiae under NaCl stress. In: Hohmann S., Mager P., editors. Yeast Stress Responses. Vol. 1. Heidelberg: Springer-Verlag; 2002. pp. 201–239. [Google Scholar]
- Wadskog I., Maldener C., Proksch A., Madeo F., Adler L. Yeast lacking the SRO7/SOP1-encoded tumor suppressor homologue show increased susceptibility to apoptosis-like cell death on exposure to NaCl stress. Mol. Biol. Cell. 2004;15:1436–1444. doi: 10.1091/mbc.E03-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N. C., Novick P. Purification and characterization of constitutive secretory vesicles from yeast. J. Cell Biol. 1987;105:163–174. doi: 10.1083/jcb.105.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., Emr S. D., Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr. Opin. Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W. A., Van Dijken J. P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., De Craene J. O., Ferro-Novick S., Novick P. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 2004;167:875–887. doi: 10.1083/jcb.200408001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland J., Nietsche A. M., Strayle J., Steiner H., Rudolph H. K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Knoblich J. A. Lethal giant larvae take on a life of their own. Trends Cell Biol. 2006;16:234–241. doi: 10.1016/j.tcb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Wodarz A. Tumor suppressors: linking cell polarity and growth control. Curr. Biol. 2000;10:R624–R626. doi: 10.1016/s0960-9822(00)00658-8. [DOI] [PubMed] [Google Scholar]
- Zhang B., Chang A., Kjeldsen T. B., Arvan P. Intracellular retention of newly synthesized insulin in yeast is caused by endoproteolytic processing in the Golgi complex. J. Cell Biol. 2001;153:1187–1198. doi: 10.1083/jcb.153.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang P., Gangar A., Zhang J., Brennwald P., Terbush D., Guo W. Lethal giant larvae proteins interact with the exocyst complex and are involved in polarized exocytosis. J. Cell Biol. 2005;170:273–283. doi: 10.1083/jcb.200502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.