Abstract
Structural studies on various domains of the ribonucleoprotein signal recognition particle (SRP) have not converged on a single complete structure of bacterial SRP consistent with the biochemistry of the particle. We obtained a three-dimensional structure for Escherichia coli SRP by cryoscanning transmission electron microscopy and mapped the internal RNA by electron spectroscopic imaging. Crystallographic data were fit into the SRP reconstruction, and although the resulting model differed from previous models, they could be rationalized by movement through an interdomain linker of Ffh, the protein component of SRP. Fluorescence resonance energy transfer experiments determined interdomain distances that were consistent with our model of SRP. Docking our model onto the bacterial ribosome suggests a mechanism for signal recognition involving interdomain movement of Ffh into and out of the nascent chain exit site and suggests how SRP could interact and/or compete with the ribosome-bound chaperone, trigger factor, for a nascent chain during translation.
INTRODUCTION
The mechanism of cotranslational protein targeting is conserved in most organisms. The initial step involves binding of the signal recognition particle (SRP) to both a ribosome and a signal sequence on the nascent polypeptide chain emerging from the ribosome. SRP targets the ribosome-nascent chain complexes to membranes via binding to the SRP receptor (Gilmore et al., 1982). Receptor binding and transfer of the nascent polypeptide from SRP to the translocation machinery in the membrane are regulated by the GTPase activities of SRP and SRP receptor (for review, see Millman and Andrews, 1997).
Depending on the organism, the components of SRP can vary in number and size. Bacteria have the simplest SRP, composed of a single polypeptide and a small RNA molecule. In Escherichia coli, these subunits are called “fifty-four homologue” (Ffh; for its homology to the SRP54 subunit of human SRP) and 4.5S RNA, respectively. Based on sequence comparisons, the primary sequence of Ffh has been arbitrarily divided into the N (amino-terminal), G (GTPase), and M (methionine-rich) regions.
Several structures have been determined for isolated domains of Ffh. Structures of the apo-form of the NG domain of Ffh (Freymann et al., 1997), and various guanine nucleotide-bound forms (Freymann et al., 1999) revealed that the N and G regions, although distinct based on sequence alignments, are structurally organized as a single tertiary domain. The M domain forms another distinct domain that binds 4.5S RNA and contains the putative signal sequence binding site. M isolated from T. aquaticus Ffh (Keenan et al., 1998) and human SRP54 (Clemons et al., 1999) have been crystallized in the apo form, and E. coli M was solved from a cocrystal with a fragment of the 4.5S RNA (Batey et al., 2000). A crystal that contained full-length Ffh from T. aquaticus (Keenan et al., 1998) revealed the structures of both NG and M from a single organism but the linker between them was not resolved, and the crystal packing precluded unambiguous determination of the relative orientation of the two domains.
Recently, a structure for the archaea Sulfolobus solfataricus full-length SRP54 bound to a fragment of SRP RNA was solved crystallographically (Rosendal et al., 2003). The complete backbone of the protein component was resolved, but a 19-kDa protein normally found in this SRP was not included. In addition, much of the archaeal SRP RNA was missing, including a region corresponding to nucleotide 84 of E. coli SRP RNA, a site that can be cross-linked to the ribosome (Rinke-Appel et al., 2002) and therefore is probably involved in binding SRP to the ribosome. Interestingly, in S. solfataricus SRP54 the relative orientation and position of M and NG were markedly different from all three of the possible arrangements deduced from the T. aquaticus Ffh structure. However, the domain structures were conserved, suggesting that the NG and M domains behave primarily as separate rigid bodies with high structural conservation across different species.
The length of the SRP RNA varies greatly among organisms from ∼300 nucleotides (nt) in mammals and archaea (for review, see Poritz et al., 1988) to 70 nt in Mycoplasma mycoides (Samuelsson and Guindy, 1990). In organisms with a longer SRP RNA, the particle generally contains multiple protein components; indeed, one proposed role of the RNA is to act as a scaffold for particle assembly. The E. coli SRP RNA is 114 nt of which nucleotides 39–64 have been implicated in binding to Ffh. Little is known about the function of the rest of the RNA in E. coli SRP; additional binding proteins have not been identified, and nucleotides 1–32 and 75–114 can be deleted without compromising viability in rich medium (Batey et al., 2000).
Electron spectroscopic imaging (ESI) of mammalian SRP suggested that much of the RNA functions as a scaffold for six SRP polypeptides (Andrews et al., 1985). Based on a structural model deduced from scanning transmission electron microscopy (STEM) micrographs, it was proposed that SRP could span the ribosome from the peptide bond formation site (for elongation arrest) to the peptide exit site (where SRP binds the emerging signal sequence). This model for SRP binding to the ribosome was recently verified and refined by cryo-electron microscopy of canine SRP bound to the 80S ribosome (Halic et al., 2004) to indicate direct binding of SRP to a region of the ribosome adjacent to the nascent chain exit site and to the elongation factor binding site near the peptide bond formation site. This structure of SRP also suggests that ribosome binding is another important function for the SRP RNA.
Further structural information is required for several forms of SRP to determine the molecular mechanisms of signal peptide binding, regulation, and release. We have visualized E. coli SRP by using cryo-STEM and calculated a three-dimensional (3-D) reconstruction. ESI in the STEM was used to locate the RNA molecule in three dimensions within the SRP reconstruction, and then we incorporated data from existing structures to generate a high-resolution model of E. coli SRP. This model is consistent with our measurements of interdomain distances by fluorescence resonance energy transfer (FRET) for free SRP in solution. Docking our model on the ribosome revealed a potential mechanism for signal peptide binding that involves rotation of the M domain of Ffh. Motion of M relative to NG is also predicted from FRET data for SRP. Our model also explains why SRP binding to its receptor, FtsY, and trigger factor (TF) binding on the ribosome may be mutually exclusive.
MATERIALS AND METHODS
Preparation of Bacterial Signal Recognition Particle
E. coli SRP was made by incubating a fivefold molar excess of recombinant poly-histidine-tagged Ffh (Zheng and Gierasch, 1997) and in vitro synthesized 4.5S RNA for 10 min at 0°C and then 10 min at 37°C (Freymann et al., 1997). RNA-containing complexes were separated from unbound protein by ion-exchange chromatography by using Sepharose-DEAE Fast-Flow (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) (adapted from Walter and Blobel, 1983). This eluate was then applied to a column containing nickel-nitrilotriacetic acid resin (QIAGEN, Valencia, CA) to remove unbound RNA. From an initial binding reaction containing 4.0 nmol of Ffh, the typical yield of SRP obtained by this purification procedure was 20% as 300 μl of 2.6 μM SRP (determined by protein quantification).
Electron Microscopy and Data Collection
Grids containing a carbon film were glow discharged. Purified complexes were adsorbed to the carbon films by injecting the sample into an equal volume of 2.5 mM magnesium acetate on the support. The grid was transferred, in succession, to a series of drops of 2.5 mM magnesium acetate and then was washed with droplets of 50 mM ammonium acetate. Residual ammonium acetate was wicked away and allowed to evaporate partially to form a thin layer across the grid, and then the grid was plunged into liquid ethane. Frozen grids were freeze-dried in a STEM (model HB601UX, Vacuum Generators, Hastings, United Kingdom) at a temperature of −130°C.
“Low-dose” dark field electron micrographs of the freeze-dried SRP were obtained at −130°C in the STEM at a magnification of 500,000× with a gun voltage of 100 kV, corresponding to an electron dose of 15 e/Å2. Images were acquired digitally by using a 3-Å electron beam at pixel spacings of 3.3Å. Particle images were converted to mass/pixel by calibrating the STEM image intensity to the carbon film thickness. Carbon film thickness was determined by analysis of the energy loss spectrum (Egerton, 1986) obtained on a Zeiss EM902 transmission electron microscope. The STEM images were filtered by a 1-pixel radius Gaussian blur and contrast enhanced for optimal viewing.
ESI images were collected in the STEM by using a 6-Å electron beam at 3.3-Å pixel spacings. At each point, electrons were counted for four windows of spectroscopically separated energy loss values (centered at 0, 112.5, 125, and 150 eV) captured simultaneously in parallel.
Three-dimensional Reconstruction of SRP
Individual single molecule images of E. coli SRP were identified and extracted using the SPIDER/WEB image processing software (Frank et al., 1996) resulting in a set of 3236 particle images. After normalization of the images, they were randomly divided into 16 subsets of 201 particles. For each subset the relative angular orientations of the randomly oriented particles was determined by angular reconstitution by using iterative quaternion-based angular determination (IQAD) (Farrow and Ottensmeyer, 1993). The distributions of angles determined by IQAD were used to calculate completely independent 3-D reconstructions of SRP by back-projection of the individual subsets of original unfiltered images. The reconstructions generated for the individual subsets were refined by a projection matching algorithm (Penczek et al., 1994) created with SPIDER. The refined reconstructions were aligned in three dimensions by using EMAN (Ludtke et al., 1999) and combined into a single reconstruction by using a round of multireference projection matching. The reconstruction was further refined using a two-stage modified version of the projection matching technique (Penczek et al., 1994) created with SPIDER. Briefly, class images were generated by forward projection of the 3-D structure from the previous iteration. For the first stage of the refinement process, the class images are generated at isotropically distributed orientations at a relatively coarse angular interval (10–15°) and used in the projection matching for the initial alignment of each of the particle images. Next, a second set of class images is calculated for each particle image at a finer angular interval (0.3–1.5°) within a matrix defined by two of the top matched class images from the first stage. This two-stage process results in a very fine sampling of the angular space with a much shorter computation time than if the fine angular interval was used throughout the entire projection matching process.
Reconstructions of the STEM images were calculated on Silicon Graphics IndigoII and Origin2000 Workstations (Silicon Graphics, Mountain View, CA). Visualization of structure data was done using Insight II (Accelrys, San Diego, CA). Ffh structures were aligned using an alignment feature of Insight II.
Electronic Spectroscopic Imaging
For each set of ESI images of well separated SRP complexes, the average signals for the 112.5-, 125-, and 150-eV loss images (essentially carbon film) were used to calculate a correction factor to normalize the detectors to the electron energy loss spectrum curve approximation: I = A · E−R for carbon (where I is intensity, E is electron loss voltage, and A and R vary with respect to specimen thickness and chemical composition; Egerton, 1986). Individual particles of SRP were identified from low angle scattering images. Particle images were extracted from complementary regions of the corresponding 112.5-, 125-, and 150-eV loss images. For each pixel in the particle area of the 150 eV loss image, the background intensity due to the carbon film was extrapolated from the corresponding 112.5- and 125-eV loss pixel intensities. The extrapolated background was subtracted from the actual 150-eV loss pixel intensities, to generate an image from only those electrons that had interacted with and lost energy to the phosphorus L-shell electrons, referred to as the “net phosphorus signal.”
Three of the energy loss images (112.5, 125, and 150 eV) were summed together and projection matched to the SRP reconstruction generated using low-dose STEM images. The corresponding net phosphorus signal images were reconstructed into a 3-D structure by back-projection based on the angular orientation assigned from the summed energy loss images. The resulting reconstructions are inherently noisy due to both quantum noise and radiation damage; therefore, they are presented at an arbitrary threshold that illustrates a relatively contiguous signal representing a 3-D phosphorous localization map.
Fluorescence Resonance Energy Transfer
Ffh was expressed from the pGEV2 plasmid (kind gift of A. Gronenborn; Huth et al., 1997), resulting in a fusion protein with the “solubility-enhancing tag” GB1 from the streptococcal protein G at the N terminus and a hexa-histidine tag at the C terminus of Ffh. Mutant versions of the Ffh fusions were expressed in the BL21 (DE3) strain of E. coli cells and purified by ion exchange chromatography followed by nickel-affinity chromatography. The signal peptide binding activity was verified for each of the mutants (Cleverley and Gierasch, 2002). A 43-nt fragment of 4.5S RNA fragment was prepared as described in Zheng and Gierasch (1997).
For the generation of proteins with single tryptophan and cysteine residues for FRET experiments, a mutant version, referred to as Ffh*, was initially made in which the single tryptophan of the GB1 moiety was replaced with phenylalanine and Cys406 of the Ffh was mutated to serine (original Ffh amino acid numbering is retained for easy reference). The following proteins were created: Ffh* R11W/Q340C, Ffh* R11W/S366C, Ffh* R49W/Q340C, Ffh* R49W/S366C, and Ffh* R141C/M347W. Cysteines were externally labeled with the 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) fluorophore (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. The labeling efficiencies were in the 0.75–0.85 range for all mutants as determined from spectrophotometric measurements. These values were incorporated into the data analysis during calculation of the distance distributions.
Time-resolved FRET measurements were carried out on a custom-built single photon counting apparatus provided by the C. R. Matthews laboratory (University of Massachusetts School of Medicine, Worcester, MA). Excitation of fluorescence at 295 nm was accomplished by the triplet output of a mode-locked titanium:sapphire laser, and fluorescence decays were monitored at 350 nm (Falkovskaia and Gierasch, unpublished data). The analysis of time-resolved decay profiles and the distribution of distances between donor and acceptor fluorophores were carried out using the Savuka program (version 6.0.1; C. R. Matthews laboratory). Further details of the FRET experiments are presented in Supplemental Materials and will be published elsewhere.
All fluorometric measurements were carried out in 10 mM HEPES, pH 7.6, 300 mM sodium chloride, 10% glycerol, and 5 mM magnesium chloride. Protein was incubated with a 1.5 M excess of 4.5S RNA fragment to ensure complete formation of ribonucleoprotein particles.
RESULTS
SRP 3-D Reconstruction
We assembled SRP from its constituent protein and RNA. The reconstituted E. coli SRP exhibited the expected GTPase activity (our unpublished data), and the Ffh was shown to bind signal sequences (Cleverley and Gierasch, 2002); therefore, we conclude that the particles used for imaging were functional. To obtain sufficient signal-to-noise ratio to visualize the relatively small E. coli SRP (∼90 kDa), we used a freeze-dried, unstained sample in the dark-field imaging mode of a STEM. Mass determinations from the STEM images confirmed that the particles used for the reconstruction were monomers of SRP with a mass of 77 ± 15 kDa. The STEM images of SRP were as curved rods with one end larger than the other (Figure 1A). Angular assignment of the individual images revealed there was a slight preference for some orientations of SRP binding to the carbon support (see Supplemental Materials). Filtered back-projection included orientation density-dependent filtering for the calculation of the SRP reconstruction to ensure that the higher occurrence projections were not overweighted in the final reconstruction.
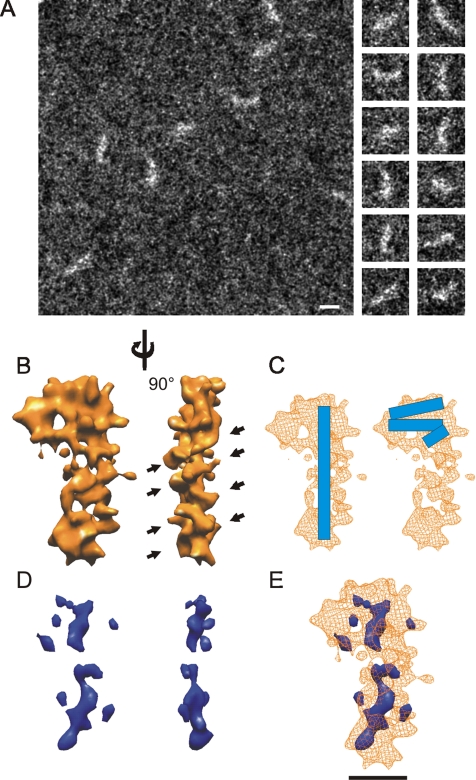
Figure 1.
STEM and ESI imaging of E. coli SRP. (A) A representative low-dose STEM micrograph of a carbon support containing E. coli SRP (left). Putative particles of SRP were extracted as individual images (right) for further structural analysis. Bar, 100 Å. (B) Two views of the reconstruction of E. coli SRP generated from low-dose cryo-STEM data. Black arrows indicate repeating ridges in the reconstruction. The reconstruction has been filtered to 12 Å and is displayed as a hard surface at a threshold corresponding to ∼90 kDa. This image display generates sharp edges not expected at the resolution obtained but produces a more interpretable image. (C) Two possible locations for the RNA (blue bar) in the reconstruction of SRP (wireframe). (D) Three-dimensional reconstruction representing the location of phosphorus atoms within SRP. The reconstruction is presented at the same two views as shown for the complete particle in B. (E) The overlap of the SRP reconstruction and the 3-D phosphorus map indicating the relative position of the phosphorus within SRP. Bar, 50 Å.
The STEM data converged on a 3-D structure for E. coli SRP resembling the numeral 9 from a “front” view (Figure 1B, left) with a globular “head” and a “tail” extending 90 Å from one side of the head. The tail gives the reconstruction an overall extended conformation with a maximal length of 150 Å, and width of 85 Å at the head and a minimal width of 42 Å along the tail in this orientation. The structure has a slight curvature along the tail, but it is otherwise flat (thickness 30 Å). The resolution of our SRP reconstruction was estimated to be 12 Å by Fourier shell correlation (FSC) by using the 3σ criteria (Orlova et al., 1997) (see Supplemental Materials for complete FSC plot). There are three possible locations for the RNA molecule in this reconstruction: first, the RNA could be completely localized to the tail, extending most of the length of the particle (Figure 1C, left); second, the RNA could be folded in such a way that it occupies the globular head of the reconstruction (Figure 1C, right); or, third, the RNA could be positioned so that part of it is in the head and part of it is in the tail (see Supplemental Materials for a schematic of all of these possibilities). The first two RNA locations are potentially compatible with the remainder of the particle being the protein component, Ffh (i.e., the head region or the tail region). The third position of the RNA would require that two relatively distant regions of the reconstruction (the distal end of the tail and the remaining head volume) would need to be filled by Ffh, and it is not readily apparent how this could be achieved in a biologically relevant/possible way (see Supplemental Materials for a description of model building). The length of the tail in the reconstruction is consistent with a highly base-paired double helix. Moreover, the repetitive features of an RNA helix can be discerned in the right-hand view of the particle tail (Figure 1B, arrowheads), and, although the tail deviates from the ribbon-like structure that would be expected for a perfect double-helical RNA molecule at the resolution of the reconstruction, it corresponds well with a rod-like RNA model that has the expected secondary structure of E. coli SRP RNA (see Supplemental Materials). However, chemical and enzymatic probing data of E. coli SRP and FRET measurements of SRP RNA bound to Ffh suggest that at least in solution the RNA can fold back on itself when in complex with Ffh (Buskiewicz et al., 2005a, b), which would be more consistent with the RNA being located in the head of the particle. To resolve this apparent discrepancy and to correctly assign the RNA and protein components in the reconstruction of E. coli SRP, it was therefore essential to localize the RNA in the structure directly.
SRP RNA Mapping
To locate the nucleic acid within E. coli SRP, the distribution of phosphorus in the particle was determined by ESI. Electrons inelastically scattered by L-shell electrons of a phosphorus atom predominantly lose 150 eV of energy (Bazett-Jones and Ottensmeyer, 1982) and can be identified with an electron spectrometer attached to the STEM. These electron spectroscopic images of the phosphorus distribution were used to generate a 3-D reconstruction of the phosphorus within SRP. The resolution in these maps is limited by radiation damage and quantum noise but is just sufficient to track double-stranded nucleic acids in particles, including canine SRP (Andrews et al., 1987). The 3-D ESI phosphorus map is quite noisy, as expected from the small number of phosphorus atoms imaged. Therefore, the reconstruction was filtered to 16 Å and shown at an arbitrary threshold that generates a mostly contiguous volume. Lowering the threshold further increases the size of the spurious signals (small structures not connected to the main signal) without changing the overall interpretation. Increasing the threshold leads to further fragmentation, but the main signals remain consistent with an extended RNA in SRP. Thus, we conclude that the map is sufficient to demonstrate that the RNA adopts an extended rod-like structure with a small but significant bend in the middle (Figure 1D). Although the phosphorus map provides little information about the RNA structure beyond that it forms an extended rod, it was calculated independently of the total structure. Therefore, the fact that when the SRP reconstruction was displayed together with the phosphorus map, the phosphorus map overlapped the SRP reconstruction (Figure 1E), confirmed that the tail of the SRP reconstruction contains RNA, whereas the head region of SRP is mostly protein. The size of the gap in the main axis of the phosphorus map and the spurious signals outside of this area suggests the limit to which the phosphorus map can be used to locate the RNA in the particle.
Generation of ec_SRP, a High-Resolution Model of SRP
The reconstructions in Figures 1 and 2 are displayed as having a hard shell that makes interpretation easier but does not permit easy assessment of the sharpness of the edges of the reconstruction. To take this into account and dock crystallographic data into the reconstruction, we examined the latitude for fitting by decreasing the viewing threshold (i.e., increasing the visualized volume) of the SRP reconstruction. When the threshold was reduced such that the reconstruction volume was increased by ∼50% (Figure 2A, black areas), it resulted in a “filling in” of the reconstruction without significantly changing the extent of the exterior boundaries. Thus, the edges of the reconstruction are quite sharp, and the framework/boundary for the docking of the individual domains of E. coli SRP into the reconstruction is quite obvious.
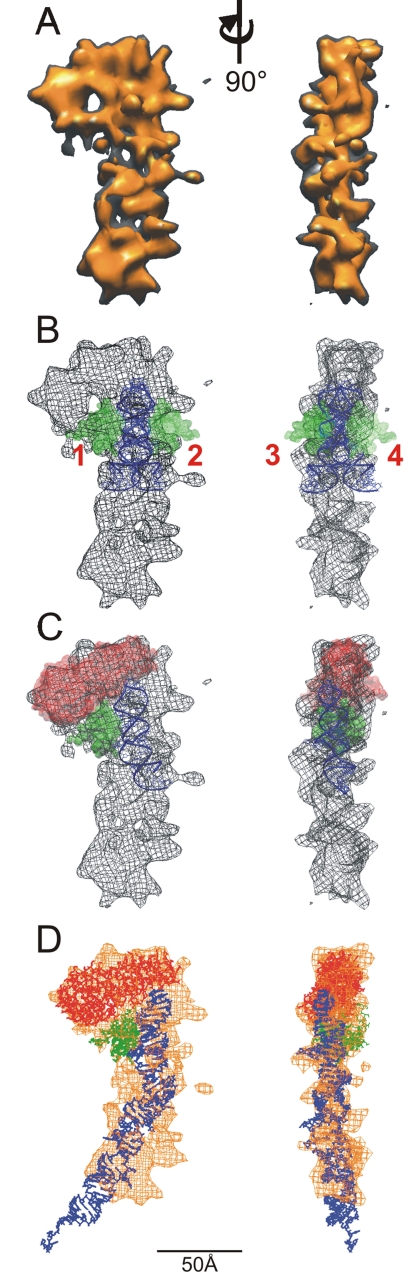
Figure 2.
Determination of the domain organization of the E. coli Ffh in our SRP reconstruction. (A) The SRP reconstruction visualized at a threshold that corresponds to the molecular mass of E. coli
Two factors affect the appearance of the reconstruction at a particular viewing threshold. First, using an average density to analyze a particle containing both RNA and protein should result in portions of the reconstruction containing solely protein to be underrepresented (or underestimated) and the RNA-only portions to be overrepresented. Second, the volume enclosed assumes that the reconstruction was calculated from a set of images of a particle adopting only one conformation. Areas of conformational heterogeneity in the image population result in lower density regions in the reconstruction due to misalignment of flexible regions when calculating the reconstruction (discussed below). Finally, the reconstruction will be dominated by the most prevalent structure on the grid with other structures optimally aligned to the dominant one such that a single unique reconstruction is generated that represents the most populous SRP structure on the grid (the effect of the reconstruction process on the final result is described in the Supplemental Materials).
To construct a high-resolution model of SRP, we first assigned the approximate position of the RNA in the particle as shown in Figure 1E. This position of the RNA restricts the possible locations for the M domain that was modeled using the structure from the cocrystal of E. coli M and a fragment of SRP RNA (PDB: 1DUL). Various possible locations of M were examined by rotating the RNA. During the modeling, the RNA was rotated in small increments however, for illustrative purposes, the results of rotation at 90° intervals are shown in Figure 2B. Even when the SRP reconstruction is visualized at a conservative threshold (150% of the expected molecular mass), only one orientation (Figure 2B, 1) has M positioned entirely within the volume of the reconstruction. When rotated 180° (Figure 2B, 2), the M domain extends into an invagination in the SRP structure. Similarly, in the other two orientations shown (±90°; Figure 2B, 3 and 4) M extended significantly beyond even the most conservative border of the SRP reconstruction. The relatively low resolution of the 3-D phosphorus map allows for some latitude in the vertical positioning of the RNA molecule (see Supplemental Materials). Therefore the fit of the M-domain in the reconstruction was examined after vertical translation of the RNA-bound M structure. An upward shift within the reconstruction positioned the M domain either in the low density region (the “hole”) located just above position 1 or outside the mass distribution (similar to positions 3 and 4), or, if it resulted in an acceptable fit of the M domain within the particle mass there was too little space left to fit NG within the head of the reconstruction (see Supplemental Materials). Shifting the RNA downward moves M below the mass of the head region and positions it too far from the NG domain to be compatible with an appropriate interdomain linker (see Supplemental Materials).
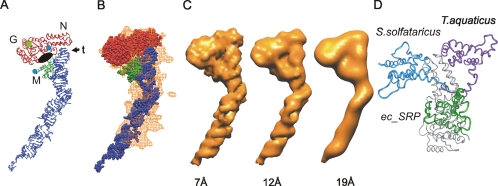
Figure 3.
Comparison of the ec_SRP model with the reconstruction of E. coli SRP. (A) Ribbon diagram representation of ec_SRP (colored as in Figure 2D), highlighting potential regions of interest, including the N, G, and M regions as well as the tetraloop (t) of the SRP RNA. The amino terminus of NG and the carboxyl terminus of M are indicated by blue circles. A black oval indicates the approximate location of the proposed signal sequence binding region of the M domain and the guanine nucleotide binding site of NG is indicated by a green square. (B) Alignment of the ec_SRP model into the reconstruction of E. coli SRP, from Figure 2D. (C) The ec_SRP model was filtered to three different resolutions (left, 7 Å; middle, 12 Å; and right, 19 Å) and displayed at respective threshold values to generate hard surface representations corresponding to ∼90 kDa (see Supplemental Materials for additional views and direct comparison with the reconstruction). (D) The relative orientations of M in ec_SRP (green), SRP54 from S. solfataricus (PDB: 1QZW, blue), and the B/A configuration of T. aquaticus apo-Ffh (adapted from PDB: 2FFH, purple). The slight differences in the respective NG domains are similar to the amount of variation seen in the NG domain from a single organism (T. aquaticus) depending on guanine nucleotide occupancy of the G region. For simplicity, only one of the aligned NG domains is shown (gray). The major difference between the three structures is the relative locations of the M domain. To represent a complete polypeptide backbone for the T. aquaticus structure, an energy minimized 11 amino acid NG-M linker has been modeled. To show the approximate location of the NG-M linker in the Ffh component of ec_SRP residues 298–318 of S. solfataricus SRP54 were positioned between NG and M of ec_SRP.
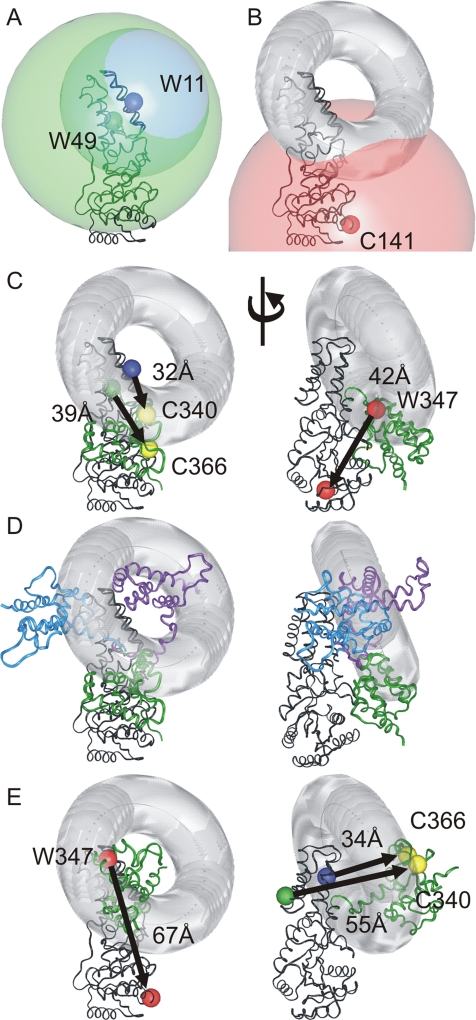
Figure 4.
Three-dimensional analysis of FRET-determined interdomain distances. (A) Distances obtained by FRET analysis represented as spheres centered at residue 11 (radius 30 Å, large blue sphere) and residue 49 (radius 44 Å, large green sphere) of the NG domain. The smaller colored spheres indicate the positions of tryptophan donors at residues 11 and 49. (B) A circle representing the intersection of the two spheres from A was calculated and is displayed as a gray toroid. A sphere of radius 50 Å (red) was centered on the residue corresponding to 141 in E. coli Ffh to represent prohibited space for residue 347 implied from the lack of FRET between these two positions. (C) The NG-M configuration defined by ec_SRP is shown with the distances measured from the α-carbon atoms of the residues corresponding to the donors and acceptors (Table 1). The arrow is displayed going from the donor position to the acceptor position. The respective residues in M colocalize well with the intersection of the FRET-defined interdomain distances (gray toroid). Left, same view as that in Figure 3D. Right, after a
Positioning the M domain as shown in Figure 2B, 1 left a relatively constrained space in the head of the SRP reconstruction for the NG domain. The thickness of the particle constrains the NG domain to an orientation with the long axis aligned with the plane of the particle. Therefore, the NG domain could be docked into the remaining volume of the globular head in only two main orientations; one orientation with the N region above the RNA molecule and the G region next to the M domain; and the other orientation with the N region near the M domain and the G region above the RNA molecule (see Supplemental Materials). Also, crystallographically determined structures available for the NG domain have a high degree of rotational symmetry. To set the rotational orientation of the NG domain, the length of an NG-M linker from the structure of S. solfataricus SRP54 (∼40 Å) was used to constrain the distance between the carboxy terminus of the NG domain and the amino terminus of the M domain. Surprisingly, there was a fairly limited region of rotational space that was compatible with this linker length, for either of the two N-G orientations (see Supplemental Materials). Therefore, the T. aquaticus NG (PDB: 2FFH) was docked into the reconstruction in the two N-G orientations by using the optimal rotation determined from the linker length analysis (see Supplemental Materials). The N-G orientation with the N region above the RNA gave a more optimal visual fit (see Supplemental Materials), and there were no significant molecular clashes of NG with either M or RNA. This orientation of NG was also compatible with docking SRP to FtsY and to the ribosome (see below). Finally, this orientation of NG in Figure 2C (but not the other NG orientation) results in a positioning of NG relative to M that is compatible with FRET data for the particle in solution (see below). Once the position of the individual domains was assigned, we replaced the structure for the E. coli M domain, in which several amino acids are missing from a flexible loop thought to be involved in signal sequence binding, with the complete M structure from S. solfataricus. Finally, the remaining RNA fragment was constructed using a portion of a model of extended E. coli SRP RNA obtained from the SRP database (Rosenblad et al., 2003).
In the final model, referred to as ec_SRP, the α-helical bundle of the N region is situated above the hairpin tetraloop of the RNA and M is adjacent to the GTPase region of NG (Figures 2D and 3A). The SRP RNA adopts a rod-like conformation with a significant bend at the approximate midpoint of the rod. There are approximately eight “base pairs” of RNA (∼20 Å) that extend beyond the SRP reconstruction volume. In our model, the spacing between nucleotides in the distal half of the RNA is the same as for theoretical double-stranded, helical RNA with a few nucleotides left unpaired to match loops and bulges from the predicted secondary structure. Using the crystallized portion of the E. coli SRP RNA (nucleotides 33–74) as a molecular “measuring stick,” 22 nucleotide pairs (which includes a symmetric loop and an asymmetric loop) is ∼50 Å. Extending this to the full-length SRP RNA (∼56 nucleotide pairs predicted to have similar occurrences of bulges and loops) generates an expected length of 128 Å, which, at ∼12 Å shorter than the RNA model shown is consistent with the length of the RNA in our reconstruction. It is also possible that the tertiary structure of this region of the SRP RNA adopts a more irregular, complex configuration, such as side loops extending from the main helix or a bending back of the distal end, which would result in an overall length that is consistent with our reconstruction. Alternatively, it is possible that this region of the RNA is so mobile that it is not visible due to the averaging that occurs during the reconstruction process.
The diameter of the tail of the reconstruction is elliptical with dimensions of ∼42 Å by 30 Å (±∼12 Å) compared with 25 Å for the diameter of the RNA model. The 30-Å dimension is consistent with a largely helical RNA at the resolution of our reconstruction. The larger dimension in the reconstruction suggests conformational flexibility in the RNA that could come from mobile loops that extend away from the main RNA helix and/or from movement of the RNA in one dimension. Consistent with these possibilities, the RNA exhibits lower density than expected (see above). At a resolution around 10 Å it should be possible to identify the characteristic shape of a double-stranded RNA, although, for a 3-D reconstruction of the E. coli 50S ribosomal subunit, with a much better resolution (7.5 Å by FSC at 3σ threshold), a ribbon-like structure for the rRNA was not always readily apparent (Matadeen et al., 1999). Furthermore, the SRP RNA is not continuously double-stranded over the length of the molecule. Remarkably, it is still possible to recognize a repeating feature in the reconstruction (Figure 1B, right, indicated by arrows), with an average spacing of 25 Å, consistent with the helical repeat distance (e.g., the spacing between minor grooves) in the SRP RNA model. This repeat pattern more closely resembles the predicted structure of SRP RNA than it does a strict RNA double helix consistent with the resolution obtained for our reconstruction (see Supplemental Materials).
The possibility that there is some conformational heterogeneity within the SRP imaged means that the resolution obtained may not be the same for all areas of the image. For example, the lower than expected mass in the tail region and the elliptical diameter of the tail suggest that in the long dimension the resolution may be lower than other areas of the reconstruction. Filtering the ec_SRP model to different resolutions provides an alternate method for estimating the resolution of the SRP reconstruction and the extent to which the model represents reconstruction. The SRP reconstruction (Figure 1B) clearly has more fine structural detail than a 19-Å resolution version of ec_SRP (Figure 3C, right). Also, the decreased density and increased width of the tail of the reconstruction and the lack of correspondence of other high-resolution features preclude selecting the 7-Å resolution reference (Figure 3C, left) as the model that most closely resembles the reconstruction. Therefore, the resolution of the model that most closely resembles the SRP reconstruction is one that has been uniformly reduced to ∼12Å (Figure 3C, middle), consistent with the value obtained by FSC at the 3σ threshold. However, the width of the tail of the reconstruction is not consistent with this model, but it is consistent with mobility of this region in the alignment used for the reconstruction. A more detailed comparison with different views of the filtered models is consistent with this conclusion (see Supplemental Materials) and supports the relative domain organization in our model (Figure 3A). The main structural features that constrained domain organization were the phosphorus map and an ∼40-Å linker between NG and M. Other features were secondary to these constraints as described above and in the Supplemental Materials. During the modeling process, the domain organization reported in other structural analyses of SRP was not used as a constraint.
We compared ec_SRP to the domain organization reported for structures of full-length SRP54 from S. solfataricus (56% similarity to E. coli Ffh) and Ffh from T. aquaticus (65% similarity to E. coli Ffh) post hoc. The structures of the NG and M domains are all similar with the main differences being their relative positions allowing superposition of the three models (Figure 3D). The structure of S. solfataricus RNA-bound SRP54 revealed that NG and M adopted an L-shaped orientation, with the SRP RNA fragment extending from the M domain, giving the whole particle a U-shape. Due to the unresolved NG-M linker of the T. aquaticus apo-Ffh structure (PDB: 2FFH), three potential configurations are generated that could represent full-length T. aquaticus Ffh: A/A (composed of A chain NG and A chain M from the PDB coordinates, respectively), B/A and C/A. In Figure 3D, the B/A domain organization for T. aquaticus is shown, as it is the most consistent with the structure of S. solfataricus SRP54. Comparison of our model for Ffh to the S. solfataricus and T. aquaticus structures (Figure 3D) demonstrates that the organization of M is different in each of the three models, but all three can be rationalized by movement of the M domain as a rigid body on a relatively rigid linker arm that is flexible at each end. This is in agreement with a previous proposal that there is flexibility within the NG-M linker (Rosendal et al., 2003) but significantly expands the range of movement to accommodate the S. solfataricus and T. aquaticus configurations. Although the range of motion seems large, it is confined to one area on one side of the NG domain (see below).
FRET Measurements Suggest Flexibility in the Spatial Arrangement of NG and M of Ffh in Solution
Fluorescence resonance energy transfer represents an independent method to explore the relative positioning of the NG and M domains of Ffh. Two residues within the N region, R11 and R49, and M347 in the M domain were substituted with a tryptophan (one at a time) to serve as a donor fluorophore. Residues Q340 and S366 in the M domain and R141 in the G region were mutated to cysteine for labeling with CPM, a fluorophore that acted as an acceptor moiety. The protein was bound to 43 nt of SRP RNA, and the extent of FRET was calculated for the various donor-acceptor pairs by measuring the time-resolved intensity decays of Trp11 and Trp49 (as described in Supplemental Materials).
The average spatial separation between two sites was evaluated by fitting the lifetime data to a FRET model that assumed a Gaussian distribution of distances. The goodness of fit was estimated by χR2 and was within the range of 1.2–1.4. Surprisingly, the Gaussian width was broad with an average value of 22–25 Å, strongly suggesting that the distance between M and NG is not fixed in solution. The distance measurements obtained between the respective residues within ec_SRP are in good agreement with the FRET data (Table 1 and Figure 4C).
Table 1.
Spatial separation between different donor-acceptor pairs of E. coli Ffh as found in free SRP in solution compared with measurements of various Ffh models
| Protein | Donor (residue no.) | Acceptor (residue no.) | Distance, Å |
|||
|---|---|---|---|---|---|---|
| From FRETa | ec_SRPb | S. solfataricusb | T. aquaticusb | |||
| Ffh* R11W/Q340C-CPMc | 11 | 340 | 30 | 32 | 28 | 29 |
| Ffh* R11W/S366C-CPM | 11 | 366 | 29 | 32 | 38 | 36 |
| Ffh* R49W/Q340C-CPM | 49 | 340 | 43 | 46 | 35 | 48 |
| Ffh* R49W/S366C-CPM | 49 | 366 | 45 | 39 | 44 | 56 |
| Ffh* M347W/R141C-CPM | 347 | 141 | N/Ad | 42 | 65 | 73 |
a There is an uncertainty of ±3 Å in the FRET-determined distance measurements due to an approximate 10% uncertainty in the orientation factor (see Supplemental Materials).
b Distance measurements for the Ffh structures/models were made from the α-carbon of the respective residues by using InsightII. This simplification introduces an uncertainty in the measurement of approximately ±3 Å.
c Amino-terminal fusions of Ffh with GB1. Residues are numbered as in wild-type E. coli Ffh.
d No FRET was detected. Distances of >50 Å are not resolved by this method.
Because multiple positions within Ffh were analyzed by FRET, the distance measurements can be used to provide more complex constraints than one-dimensional measurements. To evaluate the FRET data in three dimensions, individual distances were represented by spheres centered at the α-carbons of the appropriate residues within the NG domain (Figure 4). For simplicity, the distances between the donor at residue 11 and the acceptors at residues 340 and 366 were approximated as being equidistant, generating a single sphere centered at residue 11 (Figure 4A, blue sphere). The same assumption was made for the distances between residue 49 and residues 340 and 366 (Figure 4A, green sphere). The intersection of two spheres is a circle whose equation can be solved. This intersection circle was displayed as a toroid (i.e., a circle with 3-D thickness) to account for various sources of uncertainty in the distance measurements such as the broad Gaussian width distribution obtained for the FRET data, the simplification of the distances between the donor residues and residues 340 and 366 and measurements of the Ffh structure from α-carbon atoms. This toroid (Figure 4B, gray) designates the potential space in which residues 340 and 366 of the M domain are localized. The lack of FRET between residues 141 and 347 defines a third sphere (Figure 4B, red sphere) of prohibited space for residue 347 of the M domain. The distances between the residues used for the FRET measurements are in good agreement with ec_SRP (Figure 4C). Significantly, the relative orientations of M in other Ffh homologues (described above) are located close to the gray toroid defined by the FRET data (Figure 4D). This result is consistent with movement of M relative to NG being confined to this region. The T. aquaticus configuration fits less well, with the majority of the M domain positioned in the hole of the toroid, so the S. solfataricus and ec_SRP configurations could define the minimal region of movement, but this region could be expanded to include the T. aquaticus configuration.
DISCUSSION
SRP is a key “adaptor” molecule, performing many linked functions: signal sequence recognition, ribosome binding, and regulated release of a nascent chain to the membrane translocation machinery. To understand these roles requires knowledge of the structural states of SRP and identification of the sites of interaction with its binding partners. Here, we fit the structures of domains and fragments of SRP from different organisms to the reconstruction of SRP calculated from STEM micrographs to obtain a model for the complete structure of E. coli SRP.
Consistent with other structural analyses of SRP, the different domains of SRP could be fit into our reconstruction for E. coli SRP only if the relative orientation of NG and M was changed (Figure 3D). Thus, the relative positioning of NG and M is different in every structure for Ffh and SRP yet solved. These differences may reflect bona fide differences between species, or they may indicate a common flexibility among all SRP exacerbated (or revealed) by the different specimen preparations, such as crystal packing, freeze drying on carbon or other conditions. Although there are clearly differences between SRPs from different organisms, the latter case is strongly supported by the broad FRET distance distributions between two fluorophores on the different domains of Ffh. The distance distributions imply that a dynamic range of Ffh conformations, possibly multiple discrete conformations, was sampled, which could be conferred by a flexible interdomain linker that permits motion of the M domain within a limited region (Figure 4). Flexibility is also consistent with recent observations suggesting that the M and NG domain organization changes for E. coli Ffh when it binds RNA (Buskiewicz et al., 2005a). Assuming conformational flexibility exists, the different crystal structures might result from the different specimen preparations or crystallography conditions used for structure determination. Consistent with this possibility, a cross-linking study of the binding of Ffh and FtsY in T. aquaticus suggests an NG-M domain organization that is not compatible with that obtained by crystallography (Chu et al., 2004). These results confirm that M can move relative to NG in SRP, a conclusion that could only be drawn by comparing several different SRP structures obtained from a variety of environments and using different techniques.
Some features of the SRP reconstruction suggest that the RNA has different conformations as well. The tail of the SRP reconstruction is wider than the SRP RNA in ec_SRP (Figure 3B, see Supplemental Materials for further discussion). Also, the density in the tail seems lower than in the head region (Figure 2A), even though double-stranded nucleic acid should be more dense than protein. Washout from the averaging of slightly different conformations due to local flexibility of the RNA tail of SRP would account for the wider and less dense tail of the SRP reconstruction. Quantum noise from the fewer number of images from the direction depicted in Figure 1B, left, could reduce the accuracy of the alignment of this region of the image and thereby may also contribute to the lower recorded density in the tail of the reconstruction. Nevertheless, the pattern of intensity expected for double-stranded RNA was discernible in the narrow view (Figure 1B, right), suggesting that although there was conformational variation within the images there was a dominant conformation (see Supplemental Materials for further discussion). Consistent with the interpretation that the RNA changes conformation, the accessibility of nucleotide 27 (located in the kinked region of ec_SRP) to chemical reagents changes when Ffh binds RNA (Buskiewicz et al., 2005a). The carbon film specimen support for the STEM imaging had been treated by glow discharge and incubated with a magnesium-containing buffer before SRP addition to increase the hydrophilicity of the carbon film. The additional charges on the carbon film may have favored certain conformations of the SRP complex that optimized its contact with the charged surface. Thus the conformational flexibility of the RNA, especially around nucleotide 27, may be even greater in solution than recorded in our micrographs. Although it is also possible that adsorption of the SRP molecules to the carbon film may have induced conformational changes within the complex, previous studies of electron micrographs of protein molecules of known structures on carbon film demonstrated that such perturbations were minimal (Andrews and Ottensmeyer, 1982; Harauz et al., 1983).
It is somewhat counterintuitive that a molecule with such a dynamic range of motion would result in a 3-D reconstruction that represents a single, discrete conformation when displayed as a hard surface. The major indication that the reconstruction comes from a heterogeneous series of particles is the altered dimension and density of the RNA tail. That even this region has well defined edges indicates that the particles bound the carbon film as a series of discrete structures. The refinement algorithm used to reconstruct the SRP volume (primarily projection matching) causes the reconstruction to converge on the most common structure present within the population of STEM images. By aligning common elements within the structures of the various particles a conformation that is more highly represented in the image population dominates the reconstruction and mobile structural elements will be retained at their positions in the dominant structure, whereas at other locations, the same structures are averaged out and disappear. Consistent with this, detailed analysis of STEM micrographs of the Ffh protein in the absence of SRP RNA by using a variety of reconstruction algorithms revealed an Ffh conformation, similar to the T. aquaticus A/A configuration, in almost 60% of the images, whereas a structure resembling the S. solfataricus NG-M configuration was present in 30% of the images (our unpublished data). Nevertheless, reconstruction of the entire image population resulted in a single final reconstruction similar to the more abundant one (Mainprize et al., unpublished data). Therefore, the structure that we have obtained for E. coli SRP represents the most abundant conformation present within our image population, and other conformations present are either lost or evidenced only by areas of lower than expected density due to the averaging effect of the refinement procedure (see Supplemental Materials for further discussion).
In contrast to our finding that the RNA in SRP extends away from the particle in a rod shape, recent FRET data for E. coli SRP in solution suggests that the distal half of the RNA folds back onto the protein (Buskiewicz et al., 2005b). It has already been shown by cross-linking to the large ribosomal subunit that the RNA of E. coli SRP adopts an extended conformation when bound to the ribosome (Rinke-Appel et al., 2002); therefore, previous FRET studies of SRP in solution have revealed a conformation inconsistent with ribosome-bound SRP. Nevertheless, because this region of SRP RNA (nucleotides 1-32 and 75-114) has been demonstrated to be nonessential (Batey et al., 2000) and contributes at most an accessory function, the precise localization of the distal half of the RNA is unlikely to change the structure of the rest of the particle in a functionally significant way.
The NG-M configuration, deduced from other FRET studies that indicated a folding back of the RNA of SRP in solution, was a more opened version of the T. aquaticus A/A configuration. Interestingly, the RNA in this NG-M configuration occludes the FtsY binding site so that further interdomain movement must occur to enable SRP to bind to its receptor. Thus, it is possible that the RNA and the M domain are repositioned relative to NG in response to different stages of the SRP pathway. Consistent with this possibility, results from localized hydroxyl radical cleavage of E. coli SRP RNA indicated that there was at least one region of Ffh that was closer to the RNA tetraloop when SRP was bound to FtsY than in free SRP (Spanggord et al., 2005). Although the sites of cleavage that were obtained for SRP alone are not inconsistent with our model, there are two sites in the SRP/FtsY complex where they did not see cleavage that are close enough in our model that they might have obtained cleavage. As most of the radical induced cleavage experiments included both SRP and FtsY the model they generated may be more relevant to the complex. Nevertheless the data are not inconsistent with the structure we obtained. The main difference between the ec_SRP model and the model they generated is the orientation of the NG domain, which is rotated to a position similar to that in Supplemental Materials (Figure 4G, right).
SRP–Ribosome Interactions
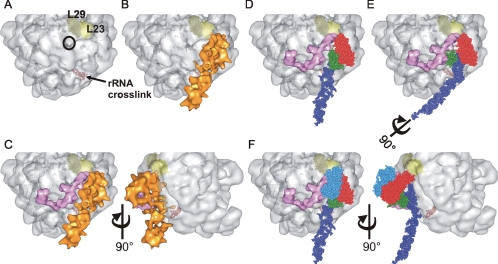
To explore a possible mode of interaction of SRP with the ribosome, ec_SRP was docked onto the structure of the H. marismortui ribosome by using published cross-linking data for bacterial SRP on the large ribosomal subunit. The RNA was positioned using the location of an RNA cross-link between nucleotide U84 of E. coli SRP RNA and nucleotides 2828–2837 of 23S rRNA (Rinke-Appel et al., 2002) (Figure 5A, red spheres). The binding site for the N region of Ffh was identified based on the cross-linking between two residues in the N region and the L23 protein of the 50S subunit by using a cross-linker with a 10-Å spacer arm (Gu et al., 2003) (Figure 5A, light yellow). These two sets of cross-linking data provide constraints that align our reconstruction onto the large ribosomal subunit (Figure 5B) such that the particle runs alongside the nascent chain exit site (Figure 5A, black circle) with M aligned at the location of closest approach to this site. These points clearly fix the location and orientation of the long axis of SRP on the ribosome. To our surprise, it was only possible to position SRP on the ribosome such that the Ffh M domain is located on the side of SRP opposite from the ribosome (Figure 5D). Furthermore, the majority of the potential contacts between E. coli SRP and the ribosome seem to be nucleic acid based. Unexpectedly, the kink in the RNA tail of SRP extends the distal half of SRP RNA away from the ribosome (Figure 5C) where it may recruit nonessential accessory factors. Although the model is placed at a precise angle with respect to the ribosome, there is considerable latitude possible in this angle that would still be consistent with our model and the known biology of SRP. The primary constraint used was not occluding the FtsY binding site on SRP while minimizing the distance between SRP and the nascent chain exit site on the ribosome.
Figure 5.
Docking SRP on the ribosome. (A) Surface of the 50S ribosomal subunit from H. marismortui (PDB: 1W2B) highlighting subunits L23 (light yellow) and L29 (dark yellow) and the nascent chain exit site (black circle). The nucleotides which correspond to a region of 23S rRNA found to cross-link to bacterial SRP RNA are displayed as red spheres. (B) Alignment of the SRP reconstruction (orange) onto the 50S ribosomal subunit maximizing the overlap of regions of SRP with respective cross-linking sites on the ribosome (L23 and rRNA). (C) The structure of E. coli TF (pink) modeled onto the ribosome along with the SRP reconstruction. TF and SRP are in very close proximity to each other and flank the nascent chain exit site. A second view related to the first by rotation of 90° illustrates the effect of the kink in the RNA structure. (D) The SRP reconstruction was replaced with the ec_SRP model and demonstrates the orientation of M (green) with respect to the other domains of SRP, the large ribosomal subunit and TF. The SRP RNA, NG and M domains are colored as in Figure 3B. (E) Rotation of the SRP RNA by 90° moves the M domain of SRP in toward the nascent chain exit site where it can effectively compete with TF for sampling nascent chains as they emerge from the ribosome. (F) The structural data of the binding of the NG domain of the SRP receptor, FtsY (PDB: 1RJ9, blue), to E. coli Ffh NG was incorporated onto the ribosomal subunit. This location of FtsY is likely to result in the displacement of TF from the ribosome explaining mutually exclusive binding of TF and FtsY to ribosome–SRP complexes. Two views related by rotation of 90° are shown.
To obtain insight into the possible functional significance of the interdomain movement of SRP, the structure of E. coli TF, another protein known to bind the ribosome adjacent to the nascent chain exit site, was added to our model of SRP docked to the ribosome. The model of ribosome-bound TF was generated as explained previously (Raine et al., 2004). Significantly, the model that resulted from docking the structures of TF and SRP independently explains how both TF and SRP can bind ribosomes simultaneously, such that both can access the nascent chain exit site of the ribosome (Figure 5C).
Consistent with the model in Figure 5C, cross-linking between TF and Ffh was seen only when both were bound to ribosomes (Buskiewicz et al., 2004). Furthermore, the cross-linker in TF was positioned at a site on the side of TF closest to Ffh in our ec_SRP model from which the 10-Å spacer arm of the cross-linker could easily bridge between the two proteins. The observation that the cross-linking pattern between Ffh and L23 of the large ribosomal subunit was altered upon TF binding to the ribosome is also consistent with the two binding sites being in proximity.
In the absence of TF, E. coli SRP has been shown to bind virtually any nascent chain as it emerges from the ribosome (Beck et al., 2000), suggesting that SRP may sample all nascent chains not bound by TF. Although the location of SRP on the ribosome is consistent with access to nascent chains, SRP binding to nascent chains in the absence of TF is somewhat artificial as there is good evidence that most, if not all, cytoplasmic ribosomes include bound TF. Moreover, most evidence suggests that only a subset of nascent chains (e.g., primarily polytopic membrane proteins) is bound by SRP. Nevertheless, the implication of these findings is that SRP and TF must both access the nascent chain exit site to sample nascent chains as they emerge from the ribosome.
A Model for Nascent Chain Sampling by TF and SRP
The ribosome-docked ec_SRP suggests that the M domain of Ffh, containing the primary signal sequence binding site, is relatively far from the nascent chain exit site. Thus, nascent chains emerging from the ribosome would first encounter TF and any chains with an affinity for TF would be drawn away from SRP. For SRP to sample nascent chains with low affinity for TF, we speculate that interdomain movement within SRP repositions M closer to the nascent chain exit site. Movement of M into and out of the nascent chain exit site would permit SRP to compete with TF for nascent chains containing hydrophobic segments and may also account for why nascent chain binding by SRP and TF often seems mutually exclusive because movement of M back to the position seen in Figure 5D would draw the nascent chain away from TF.
To move M into the nascent chain exit region requires only that the RNA molecule be rotated 90° about the long axis as shown in Figure 5E. This movement of M is consistent with the FRET data because the movement of the M domain is within the area permitted by the toroid (Figure 4C). More efficient rotation of M may be the elusive role for the RNA in E. coli SRP. If the primary function of the SRP RNA in E. coli is to move M relative to NG, then our model showing interactions of SRP to the ribosome for only one-half of the RNA may also explain why E. coli can survive with a shortened SRP RNA. Although the SRP RNA is positioned along the ribosome surface it makes relatively few contacts, therefore rotation might be accomplished by the conformational change that occurs in ribosomes translating hydrophobic sequences (Woolhead et al., 2004). Rotation of M may also result from or regulate GTP binding by SRP.
Alternatively (or in addition), this conformational change within SRP could act as a switch to regulate downstream events in the SRP pathway, such as the binding of SRP to FtsY. The binding site of TF on the ribosome is the same as that implicated for binding of the ribosome to the translocation machinery, so TF must be removed to clear the translocon binding site. Consistent with one function of FtsY being the displacement of TF from the ribosome (Buskiewicz et al., 2004), incorporating FtsY into our ec_SRP-ribosome model locates the SRP-bound FtsY such that it could directly displace TF (Figure 5F). Interestingly, movement of the G region relative to the N region of Ffh, which is implied by the variation between the structure of apo-Ffh NG and the structure of Ffh NG bound to FtsY (Freymann et al., 1997;Focia et al., 2004), would place the maximal displacement of the G region in a direction such that the proposed signal sequence binding site in SRP would be opened up upon FtsY binding (see Supplemental Materials).
One of the main strengths of our ec_SRP model is that it can be used to generate novel, yet testable, predictions regarding the interaction of SRP with both the ribosome and TF. Similar STEM and ESI data were used to predict a mechanism of binding of canine SRP to eukaryotic ribosomes that almost 20 yr later was shown to be largely correct (Andrews et al., 1985, 1987; Halic et al., 2004). Therefore, to complete our understanding of how the various components of the SRP pathway interact and function, additional SRP structures will be required. However, because our model is consistent with structural data currently available, it will be an important tool for analyzing the results of future biochemical and structural investigations of bacterial SRP and for identifying the ligand-induced conformational changes that are thought to be the basis for the targeting function of SRP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Doug Freymann for suggesting the modeling experiment comparing the relative domain positions in Ffh and the Ffh–Ftsy heterodimer shown in Supplemental Materials. We are grateful to J. Ortega for critical reading of the manuscript. This work was supported by Canadian Institutes of Health Research Grant FRN10490 to D.W.A. and National Institutes of Health Grant GM-34962 to L.M.G. D.W.A. holds the Canada Research Chair in Membrane Biogenesis.
Abbreviations used:
- 3-D
three-dimensional
- CPM, 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin
- FRET
fluorescence resonance energy transfer
- IQAD
iterative quaternion-based angular determination
- SRP
signal recognition particle
- STEM
scanning transmission electron microscopy
- TF
trigger factor.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0384) on September 20, 2006.
REFERENCES
- Andrews D. W., Ottensmeyer F. P. Electron microscopy of the poly-L-lysine alpha-helix. Ultramicroscopy. 1982;9:337–348. doi: 10.1016/0304-3991(82)90094-8. [DOI] [PubMed] [Google Scholar]
- Andrews D. W., Walter P., Ottensmeyer F. P. Structure of the signal recognition particle by electron microscopy. Proc. Natl. Acad. Sci. USA. 1985;82:785–789. doi: 10.1073/pnas.82.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D. W., Walter P., Ottensmeyer F. P. Evidence for an extended 7SL RNA structure in the signal recognition particle. EMBO J. 1987;6:3471–3477. doi: 10.1002/j.1460-2075.1987.tb02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R. T., Rambo R. P., Lucast L., Rha B., Doudna J. A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D. P., Ottensmeyer F. P. DNA organization in nucleosomes. Can. J. Biochem. 1982;60:364–370. doi: 10.1139/o82-043. [DOI] [PubMed] [Google Scholar]
- Beck K., Wu L. F., Brunner J., Muller M. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 2000;19:134–143. doi: 10.1093/emboj/19.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Deuerling E., Gu S. Q., Jockel J., Rodnina M. V., Bukau B., Wintermeyer W. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl. Acad. Sci. USA. 2004;101:7902–7906. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Kubarenko A., Peske F., Rodnina M. V., Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005a;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Peske F., Wieden H. J., Gryczynski I., Rodnina M. V., Wintermeyer W. Conformations of the Signal Recognition Particle Protein Ffh from Escherichia coli as Determined by FRET. J. Mol. Biol. 2005b;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Chu F., Shan S. O., Moustakas D. T., Alber F., Egea P. F., Stroud R. M., Walter P., Burlingame A. L. Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:16454–16459. doi: 10.1073/pnas.0407456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons W. M., Jr, Gowda K., Black S. D., Zwieb C., Ramakrishnan V. Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding. J. Mol. Biol. 1999;292:697–705. doi: 10.1006/jmbi.1999.3090. [DOI] [PubMed] [Google Scholar]
- Cleverley R. M., Gierasch L. M. Mapping the signal sequence-binding site on SRP reveals a significant role for the NG domain. J. Biol. Chem. 2002;277:46763–46768. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]
- Egerton R. F. New York: Plenum Press; 1986. Electron Energy-Loss Spectroscopy in the Electron Microscope. [Google Scholar]
- Farrow N. A., Ottensmeyer F. P. Automatic 3D alignment of projection images of randomly oriented objects. Ultramicroscopy. 1993;52:141–156. [Google Scholar]
- Focia P. J., Shepotinovskaya I. V., Seidler J. A., Freymann D. M. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Freymann D. M., Keenan R. J., Stroud R. M., Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Freymann D. M., Keenan R. J., Stroud R. M., Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP. Nat. Struct. Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Walter P., Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. Q., Peske F., Wieden H. J., Rodnina M. V., Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Becker T., Pool M. R., Spahn C. M., Grassucci R. A., Frank J., Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Harauz G., Andrews D. W., Ottensmeyer F. P. Electron microscopic visualization of the sidechains of the poly-L-lysine alpha-helix. Ultramicroscopy. 1983;12:59–64. doi: 10.1016/0304-3991(83)90305-4. [DOI] [PubMed] [Google Scholar]
- Huth J. R., Bewley C. A., Jackson B. M., Hinnebusch A. G., Clore G. M., Gronenborn A. M. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan R. J., Freymann D. M., Walter P., Stroud R. M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Ludtke S. J., Baldwin P. R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Matadeen R., Patwardhan A., Gowen B., Orlova E. V., Pape T., Cuff M., Mueller F., Brimacombe R., van Heel M. The Escherichia coli large ribosomal subunit at 7.5 A resolution. Structure. 1999;7:1575–1583. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- Millman J. S., Andrews D. W. Switching the model: a concerted mechanism for GTPases in protein targeting. Cell. 1997;89:673–676. doi: 10.1016/s0092-8674(00)80248-2. [DOI] [PubMed] [Google Scholar]
- Orlova E. V., Dube P., Harris J. R., Beckman E., Zemlin F., Markl J., Van Heel M. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 A resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- Penczek P. A., Grassucci R. A., Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Strub K., Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Raine A., Ivanova N., Wikberg J. E., Ehrenberg M. Simultaneous binding of trigger factor and signal recognition particle to the E. coli ribosome. Biochimie. 2004;86:495–500. doi: 10.1016/j.biochi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel J., Osswald M., von Knoblauch K., Mueller F., Brimacombe R., Sergiev P., Avdeeva O., Bogdanov A., Dontsova O. Crosslinking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh. RNA. 2002;8:612–625. doi: 10.1017/s1355838202020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad M. A., Gorodkin J., Knudsen B., Zwieb C., Samuelsson T. SRPDB: signal recognition particle database. Nucleic Acids Res. 2003;31:363–364. doi: 10.1093/nar/gkg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal K. R., Wild K., Montoya G., Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. Nucleotide sequence of a Mycoplasma mycoides RNA which is homologous to E. coli 4.5S RNA. Nucleic Acids Res. 1990;18:4938. doi: 10.1093/nar/18.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggord R. J., Siu F., Ke A., Doudna J. A. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat. Struct. Mol. Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983;34:525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- Woolhead C. A., McCormick P. J., Johnson A. E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Zheng N., Gierasch L. M. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol. Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.