Abstract
In response to stress conditions (such as nutrient limitation or accumulation of damaged organelles) and certain pathological situations, eukaryotic cells use autophagy as a survival mechanism. During nutrient stress the main purpose of autophagy is to degrade cytoplasmic materials within the lysosome/vacuole lumen and generate an internal nutrient pool that is recycled back to the cytosol. This study elucidates a molecular mechanism for linking the degradative and recycling roles of autophagy. We show that in contrast to published studies, Atg22 is not directly required for the breakdown of autophagic bodies within the lysosome/vacuole. Instead, we demonstrate that Atg22, Avt3, and Avt4 are partially redundant vacuolar effluxers, which mediate the efflux of leucine and other amino acids resulting from autophagic degradation. The release of autophagic amino acids allows the maintenance of protein synthesis and viability during nitrogen starvation. We propose a “recycling” model that includes the efflux of macromolecules from the lysosome/vacuole as the final step of autophagy.
INTRODUCTION
Autophagy is a carefully orchestrated process responsible for the rapid degradation of large portions of cytoplasm in the lysosome/vacuole lumen (Kim and Klionsky, 2000). The identification of >20 conserved autophagy-related (ATG) genes in the yeast Saccharomyces cerevisiae (Klionsky et al., 2003) has provided some insight into the molecular basis behind autophagy. In this process, cytoplasm is nonselectively sequestered by a double-membrane vesicle, an autophagosome, which fuses with the lysosome/vacuole. The resulting single-membrane intravacuolar autophagic body is subsequently lysed, the cargos are typically degraded, and the resulting macromolecules are reused to synthesize essential cellular components. For ease of description, autophagy can be separated into several steps, including induction, vesicle formation, retrieval of Atg proteins, fusion with the lysosome/vacuole, and processing of the cargo (Klionsky, 2005).
It has long been assumed that during extreme conditions, such as nutrient shortage, autophagy provides an internal nutrient pool to maintain the metabolism essential for survival. One example of this function is illustrated by the observation that the survival of neonatal mice is dependent upon the amino acids produced by autophagy for the maintenance of energy homeostasis and viability (Kuma et al., 2004). Another example of the survival-promoting role of autophagy in mammalian cells is the recent evidence showing that in the absence of apoptosis, Bax- and Bak-deficient mice activate autophagy, maintain ATP production, and ultimately sustain viability for several weeks after growth factor withdrawal (Lum et al., 2005).
Although the accumulated evidence suggests that the recycling of degradation products generated by autophagy is a critical part of its function, no experimental data have ever demonstrated the existence of such a process. In fact, while virtually all reviews that discuss autophagy refer to breakdown in the lysosome/vacuolar lumen followed by recycling of the resulting macromolecules, none of the accompanying models include the recycling step as part of the autophagic process, and no mechanistic information has been published that directly connect autophagy with cytosolic amino acid levels. However, autophagy would be largely pointless, at least as a starvation response, without this final step.
One purpose of autophagy is to degrade cytoplasmic components and recycle the resulting macromolecules that are essential for cell survival when nutrients are scarce. Accordingly, it must break down the single-membrane autophagic body that results from fusion of the autophagosome with the vacuole. This breakdown event depends on the acidic pH of the vacuole lumen and Prb1. Two other proteins have also been reported to be involved in this step, Atg15/Cvt17 (Epple et al., 2001, 2003; Teter et al., 2001) and Atg22/Aut4 (Suriapranata et al., 2000). Atg15 is a putative lipase and seems likely to function directly in the intravacuolar lysis of autophagic bodies. In contrast, Atg22 is a putative integral membrane protein located in the limiting membrane of the vacuole, with limited homologies to permeases. It was suggested that the breakdown of autophagic bodies depends on Atg22, because starving atg22Δ/aut4Δ mutant cells exhibit a slight accumulation of autophagic bodies inside the vacuole, and they are partially defective in total protein turnover (Suriapranata et al., 2000). In contrast, the cytoplasm-to-vacuole targeting (Cvt) pathway, a type of specific autophagy involved in biosynthetic delivery to the vacuole (Kim and Klionsky, 2000), is normal in the atg22Δ mutant cells. In particular, the breakdown of the single-membrane intravacuolar Cvt bodies was unaffected by the absence of Atg22 (Suriapranata et al., 2000). These data imply a fundamental difference in the composition of the membranes that form the Cvt versus autophagic bodies and/or the breakdown process used in the Cvt and autophagy pathways.
In this article, we carefully examined the role of Atg22 in autophagy and the breakdown of autophagic bodies. In contrast to previously published data, we find that Atg22 is not directly required for the steps of autophagy up to and including vesicle breakdown. Instead, our results suggest that Atg22 functions as an amino acid effluxer on the vacuolar membrane. In addition, two other vacuolar amino acid effluxers, Avt3 and Avt4, which seem to be part of the same family as Atg22, were discovered to be also required for maintenance of viability under nitrogen starvation conditions in the absence of leucine in a leucine auxotrophic strain. Finally, these results support a model wherein the efflux of amino acids resulting from the breakdown of autophagic bodies within the vacuole lumen constitutes the final step of autophagy.
MATERIALS AND METHODS
Strains, Plasmids, and Media
The S. cerevisiae strains used in this study are listed in Table 1. For gene disruptions, the entire coding region was replaced with either the Kluveromyces lactis HIS3, the Schizosaccharomyces pombe HIS5, or the S. cerevisiae TRP1, LEU2, or URA3 genes by using polymerase chain reaction (PCR) primers containing ∼60 bases of identity to the regions flanking the open reading frame.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4742 | MATα leu2-0, his3-1, lys2-0, ura3-0 | ResGen/Invitrogen |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 | Robinson et al., 1988 |
| lys2-801 suc2-Δ9 mel GAL | ||
| TN124 | MATα leu2-3,112 ura3-52 trp1 | Noda et al., 1995 |
| pho8Δ::PHO8Δ60 pho13Δ::LEU2 | ||
| WCG4a | MATα his3-11,15 leu2-3,112 ura3 | Thumm et al., 1994 |
| WHY1 | SEY6210 atg1Δ::HIS5 S.p. | Shintani et al., 2002 |
| YTS178 | SEY6210 vac8Δ::HIS5 S.p. | Cheong et al., 2005 |
| ZFY1 | WCG4a atg22Δ::HIS3 | This study |
| ZFY2 | WCG4a pep4Δ::LEU2 | This study |
| ZFY4 | WCG4a atg1Δ::URA3 | This study |
| ZFY6 | SEY6210 atg22Δ::TRP1 | This study |
| ZFY8 | SEY6210 atg22Δ::TRP1 vac8Δ::URA3 | This study |
| ZFY14 | TN124 atg22Δ::TRP1 | This study |
| ZFY15 | SEY6210 ATG22-GFP::HIS3 S.k. | This study |
| ZFY16 | BY4742 ATG22-GFP::HIS3 S.k. | This study |
| ZFY19 | SEY6210 avt3Δ::HIS3 | This study |
| ZFY20 | SEY6210 avt3Δ::HIS3 avt4Δ::URA3 | This study |
| ZFY22 | SEY6210 avt3Δ::HIS3 avt4Δ::URA3 atg22Δ::TRP1 | This study |
| ZFY32 | SEY6210 avt4Δ::HIS3 | This study |
| ZFY36 | SEY6210 ATG22-HA::TRP1 | This study |
| ZFY38 | SEY6210 ATG22-protein A::TRP1 pep4Δ::LEU2 | This study |
Cells were grown in rich (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal (SMD; 0.67% yeast nitrogen base, 2% glucose, amino acids and vitamins as needed) media. Starvation experiments were conducted in synthetic medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose).
Microscopy
For fluorescence microscopy, cultures were grown in SMD until early log phase. To label the vacuolar membrane, the cells were pelleted and resuspended in fresh SMD at OD600 = 1.0. FM 4-64 was added to a final concentration of 8 μM, and the culture was incubated at 30°C for 30 min. The cells were washed and resuspended in either SMD or SD-N at OD600 = 1.0. After a 2-h incubation, the samples were examined using a DeltaVision Spectris microscope (Applied Precision, Issaquah, WA) fitted with differential interference contrast optics and Olympus camera IX-HLSH100 with softWoRx software (Applied Precision). Electron microscopy was performed essentially as described previously (Kaiser and Schekman, 1990). Briefly, cells were fixed in potassium permanganate and embedded in Spurr's resin. After resin polymerization, 65- to 75-nm sections were mounted on nickel grids. The grids were stained with 1% uranyl acetate followed by lead citrate, and then they were imaged using a Philips CM-100 transmission electron microscope.
Yeast Vacuolar Amino Acid Analysis
The Cu2+ method (Ohsumi et al., 1988) was used for extraction of vacuolar amino acid pools from yeast cells. In summary, cells at OD600 = 1.6 were harvested, washed twice with distilled water, resuspended in AA buffer (2.5 mM potassium phosphate buffer, pH 6.0, 0.6 M sorbitol, 10 mM glucose, and 0.2 mM CuCl2), and incubated at 30°C for 10 min. Cell suspensions were collected by filtration on membrane filters (0.45 μm; Millipore, Billerica, MA) and washed five times with the AA buffer lacking 0.2 mM CuCl2. The cells retained on the filter were resuspended in distilled water and boiled for 15 min. Then, they were subjected to ultracentrifugation at 100,000 × g for 1 h. The supernatant was collected as the vacuolar fraction. The amino acid analysis was performed at the Protein Chemistry Laboratory (Texas A&M University, College Station, TX) by using a Hewlett Packard AminoQuant II system, and the data were analyzed and reviewed by Dr. Henriette A. Remmer (Protein Structure Facility, University of Michigan, Ann Arbor, MI).
Subcellular Fractionation
Cells were grown to mid-log phase in SMD medium and converted to spheroplasts as described previously (Kim et al., 1999). Supernatant and pellet fractions were generated by centrifugation at 13,000 × g for 5 min. All samples were collected by trichloroacetic acid precipitation and subjected to immunoblotting as described previously (Kim et al., 1999).
Autophagy Assays
The alkaline phosphate assay to measure activity from Pho8Δ60 has been described previously (Noda et al., 1995; Abeliovich et al., 2003). The green fluorescent protein (GFP)-Atg8 processing assay was carried out as described previously (Cheong et al., 2005).
RESULTS
atg22Δ Cells Display Normal Autophagy
The Cvt pathway shares most of the components needed for autophagy. In both pathways, the basic mechanism of cytoplasm-to-vacuole transport involves the formation of a double-membrane vesicle that enwraps cytoplasmic components and delivers them into the vacuole for subsequent degradation. We began our analysis with an interest in what is typically viewed as the last step of autophagy, intravacuolar vesicle breakdown. The putative lipase Atg15 is needed for the disintegration of both Cvt and autophagic bodies (Teter et al., 2001). In contrast, it was reported that another protein, Atg22, is only needed for breakdown of autophagic bodies (Suriapranata et al., 2000). This finding was intriguing because it implied a difference between the membrane composition of Cvt vesicles and autophagosomes. The conclusion about the pathway specificity of Atg22 is based on the following observations: 1) electron microscopy revealed that Atg22-deleted cells accumulate a low level of autophagic bodies within the vacuole lumen, 2) the atg22Δ mutant displayed a partial reduction in total protein turnover under starvation conditions, and 3) processing of prApe1 seemed normal in this mutant (Suriapranata et al., 2000). However, other assays including the analysis of Pho8Δ60 (Noda et al., 1995), the most commonly used procedure for monitoring autophagy, were not used to examine the atg22Δ mutant. If the breakdown of autophagic vesicles really depends on Atg22, we should observe a general autophagy-defective phenotype in atg22Δ mutant cells.
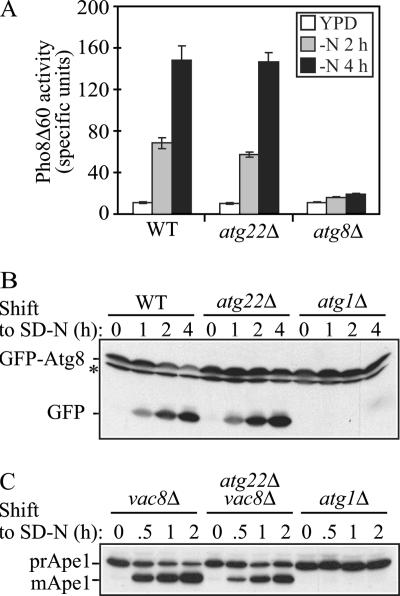
To make a quantitative measurement of autophagy in the atg22Δ mutant, we analyzed the truncated version of Pho8, Pho8Δ60, which resides in the cytosol and can only be delivered into the vacuolar lumen by autophagy. Once it enters into the vacuole, it is processed by removal of a C-terminal propeptide to generate the active form. As a nonspecific cargo, Pho8Δ60 serves as a protein marker for bulk autophagy. The Pho8Δ60 activity was measured in wild-type, atg22Δ, and atg8Δ cells under vegetative and starvation conditions (Figure 1A). As expected, wild-type cells showed Pho8Δ60-dependent alkaline phosphatase activity that increased between 2 and 4 h of starvation, whereas the atg8Δ autophagy-defective strain retained a background level of activity. The atg22Δ mutant demonstrated an increase of Pho8Δ60 activity similar to that observed in the wild-type strain, indicating normal breakdown of autophagic bodies.
Figure 1.
The atg22Δ mutant displays normal autophagy. (A) Pho8Δ60 activity, a marker for nonspecific autophagy, indicates this process is normal in atg22Δ cells. The Pho8Δ60-dependent alkaline phosphatase activity was measured before and 2 or 4 h after shifting from YPD to SD-N medium. The error bars indicate the SD of two independent experiments. (B) GFP-Atg8 processing is normal in the atg22Δ mutant. The wild-type (WT, SEY6210), atg1Δ (WHY1), and atg22Δ (ZFY6) strains expressing GFP-Atg8 were grown in SMD to mid-log phase and shifted to SD-N to induce autophagy. At the indicated times, aliquots were removed and examined by immunoblot using anti-GFP antibody. The position of free GFP, indicating autophagy-dependent processing of GFP-Atg8, is indicated. The asterisk indicates a nonspecific band. (C) In the atg22Δ mutant autophagic delivery of prApe1 is normal in the Cvt pathway-defective vac8Δ background. The vac8Δ (YTS178), atg22Δ vac8Δ (ZFY8), and atg1Δ (WHY1) strains were cultured as described above. At the indicated times, aliquots were taken and checked by immunoblot using anti-Ape1 antiserum. The positions of precursor and mature Ape1 are indicated.
In addition to Pho8Δ60, there are several other assays available to monitor autophagy, each one reflecting a different parameter of the process. For example, Atg8 is a ubiquitin-like protein that is conjugated to phosphatidylethanolamine and seems to be the only Atg protein, aside from the cargo receptor Atg19, that remains associated with the completed autophagosome and autophagic body. The population of GFP-tagged Atg8 that is within the lumen of these vesicles becomes trapped. When the autophagic bodies are broken down, Atg8 is degraded, whereas the GFP moiety remains relatively stable. Thus, the generation of free GFP reflects the delivery of autophagosomes, and in particular, the vesicle inner membrane, to the vacuole, and it can also be used to monitor the efficiency of lysis of the autophagic bodies (Cheong et al., 2005). Wild-type, atg22Δ, and atg1Δ strains were transformed with a plasmid encoding GFP-Atg8, grown in SMD with auxotrophic amino acids and shifted to SD-N to induce autophagy. At the indicated time, aliquots were removed and analyzed by Western blot by using antiserum against GFP. As shown in Figure 1B, wild-type cells displayed an increase in the level of free GFP over time in starvation conditions. In contrast, the atg1Δ mutant that is defective in autophagy accumulated only full-length GFP-Atg8. In the atg22Δ cells, free GFP was detected in increasing amounts as starvation proceeded, similar to the result seen with the wild-type strain. Thus, by this assay as well, autophagic bodies were broken down in the atg22Δ mutant.
To extend our analysis, we chose to examine the autophagy pathway by monitoring the maturation of prApe1, a marker for specific autophagy, in the Cvt pathway-defective vac8Δ background (Cheong et al., 2005). In this background, the maturation of prApe1 under starvation conditions is due to autophagy, allowing us to use a Western blot-based “pulse/chase” analysis. Cells were grown to early log phase in SMD medium, shifted to SD-N, and protein extracts were examined by Western blot. The vac8Δ strain accumulated only prApe1 during vegetative growth, but it was able to efficiently deliver the precursor to the vacuole by autophagy, where it was processed to the mature form (Figure 1C). The atg1Δ mutant again served as a negative control and accumulated only the precursor form, prApe1, under both conditions. As with vac8Δ cells, we observed the mature aminopeptidase I (Ape1) band by immunoblot in the vac8Δ atg22Δ double deletion mutant after shifting to starvation conditions, supporting our conclusion of a normal autophagy pathway in the absence of Atg22. We also examined the kinetics of prApe1 maturation through the Cvt pathway in the atg22Δ mutant by using a radioactive pulse–chase analysis and found essentially normal processing (our unpublished data) in agreement with the previously published results (Suriapranata et al., 2000). Taken together, these data suggested that Atg22 is not defective in either autophagy or the Cvt pathway.
Breakdown of Autophagic Bodies Is Kinetically Delayed in the atg22Δ Mutant
Suriapranata et al. (2000) observed the accumulation of autophagic bodies with 350 nm diameter inside the vacuole by electron microscopy in the atg22Δ mutant, which suggests a defective autophagy phenotype. However, based on our data, the atg22Δ mutant displayed normal autophagy, at levels similar to the wild-type strain. This finding suggested that the vacuolar lysis of autophagic bodies should also be normal. To address this apparent discrepancy, we examined the vacuolar accumulation of autophagic bodies by using electron microscopy. To minimize potential differences, we chose the same strain background, WCG4a, as used in the previous study. Autophagic bodies are single-membrane vesicles that result from the fusion of autophagosomes with the vacuole; once delivered into the vacuolar lumen, they are broken down in a Pep4-dependent manner. Therefore, we chose the wild-type and pep4Δ mutant strain as negative and positive controls, respectively.
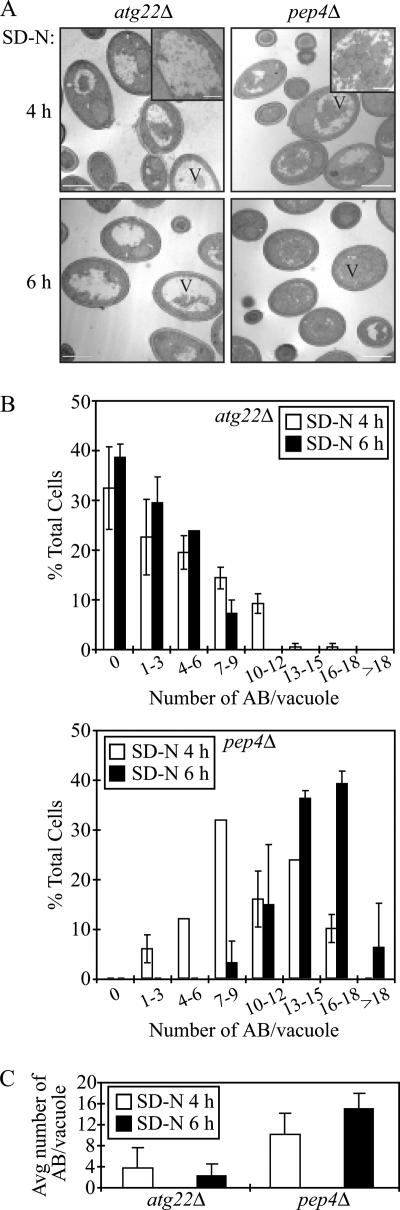
We first examined the cells after shifting to SD-N for 4 h. As shown in Figure 2A, the pep4Δ mutant accumulated autophagic bodies within the vacuole lumen as expected. In contrast, the wild-type strain did not accumulate autophagic bodies at all (our unpublished data), because they were rapidly broken down. As with the pep4Δ strain, we also observed the accumulation of some autophagic bodies inside the vacuole in the atg22Δ mutant. To compare the accumulation phenotype between the atg22Δ and pep4Δ mutants, we quantified the data (Figure 2B). We determined the number of autophagic bodies per vacuole, only counting those cells containing similar-sized vacuoles. In the atg22Δ mutant, the highest proportion of the vacuoles at the 4-h time point contained zero autophagic bodies, and the average number was 3.74 ± 3.87 (Figure 2C). However, in the pep4Δ mutant, the average number of accumulated autophagic bodies per vacuole was 10.14 ± 4.03, with seven to nine autophagic bodies in 32% of the vacuoles. Thus, the atg22Δ mutant accumulated far fewer autophagic bodies than the pep4Δ mutant. This result suggested two possibilities: either fewer autophagosomes were produced in the atg22Δ mutant, or the breakdown of the autophagic bodies was partially defective and/or kinetically delayed.
Figure 2.
The atg22Δ mutant cells display a kinetic delay in the breakdown of autophagic bodies. (A) The atg22Δ (ZFY1) and pep4Δ (ZFY2) strains were grown to mid-log phase in YPD and shifted to SD-N medium for 4 and 6 h. Cells were fixed and then examined by electron microscopy as described in Materials and Methods. The bars in the main images (X7,900) and insets (X19,000) represent 2 and 0.5 μm, respectively. (B) Quantification of autophagic body (AB) accumulation. The number of autophagic bodies in ∼100 or 50 cells containing vacuoles of similar size in atg22Δ and pep4Δ cells, respectively, were quantified. The error bars represent the SD (C) The average number of vacuoles from the quantification in B is depicted.
The normal autophagy phenotype observed in the atg22Δ mutant excluded the first possibility. To test the second possibility, we extended our electron microscopy analysis by observing the accumulation of autophagic bodies in cells starved for 6 h. As shown in Figure 2, A and B, atg22Δ mutant cells accumulated fewer autophagic bodies at 6 h compared with cells that had been starved for 4 h. The percentage of vacuoles containing zero autophagic bodies increased from 31.5% at 4 h to 38.9% at 6 h. Similarly, the vacuoles containing one to three autophagic bodies increased from 22 to 29.6%. The average number of autophagic bodies per vacuole decreased to 2.24 ± 2.31 (Figure 2C). In contrast, in the pep4Δ mutant, it was difficult to discern the vacuole boundary because the organelles were essentially filled with autophagic bodies at this time point. The average number of autophagic bodies increased to 15.01 ± 2.86, and >75% of the cells contained 13–18 autophagic bodies. Taken together, this result strongly supported our hypothesis that autophagic bodies in the atg22Δ mutant were indeed gradually broken down, with the rate of degradation much slower than in the wild-type strain. Thus, partially accumulated autophagic bodies seen in this mutant might be due to a kinetic delay in the breakdown process.
Atg22 Is a Vacuolar Integral Membrane Protein
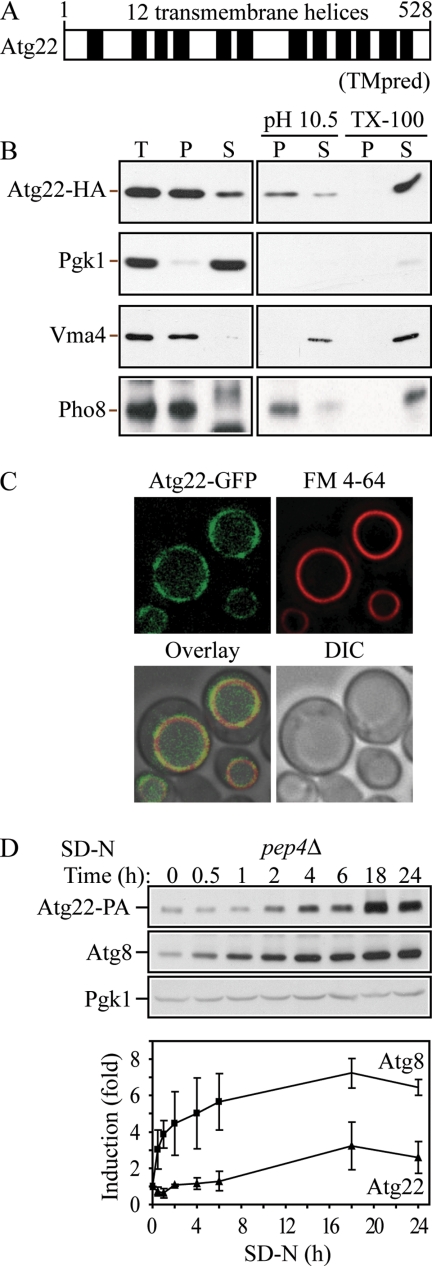
To identify the molecular function of Atg22, we searched databases, but we found that Atg22 does not show significant similarity with characterized proteins in other organisms. ATG22 encodes a protein of 528 amino acids with a predicted molecular mass of 58 kDa. Based on TMpred hydrophobicity analysis (http://www.ch.embnet.org/software/TMPRED_form.html), Atg22 is predicted to contain 11–12 transmembrane helices (Figure 3A). A computer-based analysis was carried out to identify all members of the major facilitator superfamily (permeases) in S. cerevisiae that are characterized by two structural units of six transmembrane-spanning α-helical segments connected by a cytoplasmic loop (Nelissen et al., 1997). These parameters allowed the consideration of proteins with a length of 500–600 amino acids, making up a total of 12 transmembrane-spanning segments. Within this study, Atg22 is characterized as a predicted permease with unknown function. Additionally, Atg22 shows limited similarity with some putative transporters, such as the multidrug-efflux transporter Bmr3 from B. subtilis (Ohki and Murata, 1997; Suriapranata et al., 2000). To gain more insight about Atg22, we performed several experiments to examine the biosynthesis of the protein.
Figure 3.
Atg22 is an integral vacuolar membrane protein. (A) Hydrophobicity analysis based on TMpred (Hofmann and Stoffel, 1993) predicts 12 transmembrane helices, which are indicated schematically. (B) Atg22 is an integral membrane protein. Cells expressing Atg22-3xHA (ZFY36) were grown in YPD, converted to spheroplasts, and osmotically lysed and fractionated into total (T), supernatant (S), and pellet (P) fractions, as described in Materials and Methods. The resulting pellet was subjected to the indicated treatment: 0.5 mM Na2CO3, pH 10.5, or 1% Triton X-100. After centrifugation, S and P fractions were collected and analyzed by immunoblots with anti-hemagglutinin (HA) antibody or the indicated antisera. The positions of Atg22-HA, and the markers Pgk1 (cytosolic), Vma4 (peripheral membrane), and Pho8 (integral membrane) are indicated. (C) Atg22 is localized on the vacuolar membrane. Cells expressing Atg22-GFP (ZFY16) were grown in YPD, treated with FM 4-64 to label vacuoles, and analyzed by fluorescence microscopy as described in Materials and Methods. DIC, differential interference contrast. (D) The expression level of Atg22 seemed constant under vegetative growth and was induced under nitrogen starvation conditions. Cells expressing Atg22-protein A (ZFY38) in the background of pep4Δ were grown in YPD and shifted to SD-N. At the indicated times, aliquots were removed and analyzed by immunoblot using anti-protein A (PA) antibody and antisera to Atg8 and Pgk1; Pgk1 was used as a loading control. The blot is shown for a representative experiment and the graph plots the data for three independent experiments. The error bars represent the SD.
First, to assess whether Atg22 is an integral membrane protein, we carried out subcellular fractionation. We generated a functional C-terminal 3xHA fusion at the chromosomal ATG22 locus. Lysed spheroplasts of the strain expressing Atg22-3xHA (ZFY36) were separated into soluble and pelletable fractions. As shown in Figure 3B, Atg22 was found predominantly in the pellet fraction when spheroplasts were lysed in the presence of buffer alone. Similarly, 1.0 M KCl did not extract Atg22 into the supernatant fraction (our unpublished data), and Na2CO3 at pH 10.5 solubilized only a small amount of the protein, indicating that Atg22 is not a peripheral membrane protein. In contrast, extraction with Triton X-100 completely solubilized Atg22 into the supernatant fraction. Cytosolic Pgk1 was almost completely absent from the pellet fraction, indicating the successful lysis of spheroplasts and separation of soluble and pelletable fractions. Vma4, a peripheral vacuole membrane protein, was extracted into the supernatant fraction in the presence of KCl (our unpublished data) and Na2CO3. Pho8, an integral membrane protein, served as a positive control, and displayed a similar fractionation pattern as seen with Atg22. Thus, combined with the hydrophobicity analysis, this result strongly indicated that Atg22 is an integral membrane protein.
Suriapranata et al. (2000) visualized the intracellular localization of Atg22 at the vacuolar membrane, by examining a plasmid-based fusion protein consisting of GFP fused at the N terminus of Atg22, under the control of the MET25 promoter. Because of the differences in the phenotype of the atg22Δ mutant in our current studies compared with the previously published data, we also examined the subcellular localization of Atg22. The gene encoding GFP was integrated in-frame at the 3′ end of ATG22 at the chromosomal locus, under the control of its native promoter. Cells expressing Atg22-GFP were grown to mid-log phase, treated with FM 4-64, and imaged with a fluorescence microscope. As shown in Figure 3C, Atg22 localized primarily to the vacuolar membrane, and it overlapped with the FM 4-64–labeled vacuole. Thus, our result was consistent with that in the previous study. Taken together, Atg22 is a vacuolar integral membrane protein.
In addition, we also examined the expression level of Atg22 under both vegetative growth and nitrogen starvation conditions. We generated a functional C-terminal 3xProtein A fusion at the chromosomal ATG22 locus. To eliminate the cleavage of protein A by vacuolar hydrolases, we knocked out the gene encoding vacuolar proteinase A, PEP4. The pep4Δ cells expressing Atg22-3xProtein A were grown to mid-log phase and then incubated in nitrogen starvation medium. At the indicated time, aliquots were removed and analyzed by Western blot by using antiserum against protein A. As shown in Figure 3D, Atg22 displayed an approximately threefold increase after 18-h starvation, relative to the loading control Pgk1, which displayed a constant level over time. This finding is in agreement with published data indicating a 2.7-fold increase in expression of Atg22 after 1 d of nitrogen depletion (Gasch et al., 2000). As a comparison, we examined Atg8, which is induced by nitrogen starvation (Huang et al., 2000). This protein increased its accumulation in the pep4Δ strain immediately when cells were shifted to SD-N, whereas the increase in Atg22 was apparent only after starvation for 2 h. Atg8 also showed a substantially higher level of induction, but both proteins seemed to reach a maximum after 18 h.
The atg22 Mutant Is Defective in Recycling Amino Acids
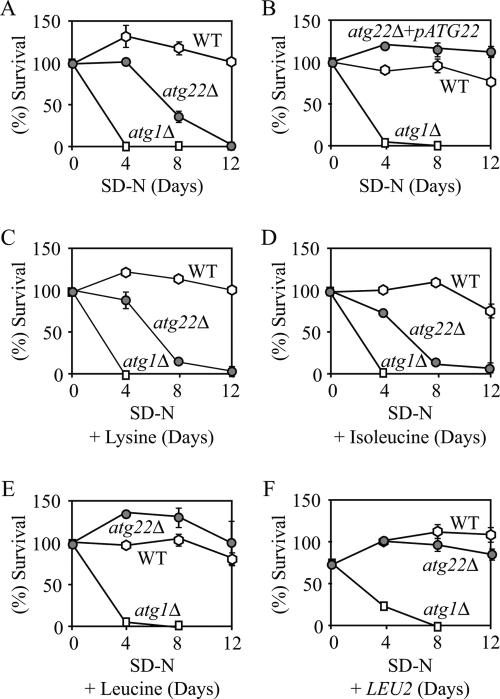
Autophagy genes are dispensable for vegetative growth, but they are required for survival during nutrient starvation conditions (Levine and Klionsky, 2004). It has been assumed that an internal nutrient pool provided by autophagy helps yeast cells sustain viability to survive nutrient deprivation. Accordingly, testing for viability during nitrogen starvation has been used as an assay to assess autophagic capacity. Although atg22Δ cells were not autophagy-defective by most assays, electron microscopy revealed that deletion of Atg22 caused the partial accumulation of autophagic bodies. Because breakdown of autophagic bodies is required for recycling the cargo contained within, deletion of Atg22 might also cause a loss of viability during nitrogen starvation. To test this possibility, we examined the viability of atg22Δ cells incubated in SD-N, by using wild-type and atg1Δ cells as positive and negative controls, respectively. As expected, atg1Δ cells exhibited a dramatic decrease in cell viability after 4-d starvation, whereas wild-type cells maintained robust viability even under prolonged nitrogen starvation (Figure 4A). The atg22Δ cells gradually lost viability through 12 d of starvation. The ATG22 gene expressed on a plasmid restored the viability of the atg22Δ cells to the wild-type level (Figure 4B), indicating that the loss of viability was due to the absence of Atg22. Thus, there was a certain discrepancy in the data: atg22Δ cells displayed normal autophagy by most criteria, but they were starvation sensitive, a characteristic of atg mutants.
Figure 4.
atg22Δ mutant cells displayed a loss of viability in starvation conditions. The wild-type (SEY6210), atg22Δ (ZFY6), and atg1Δ (WHY1) cells were grown in SMD containing auxotrophic amino acids and nucleosides to OD600 = 1.0, and then they were shifted to SD-N. The SD-N included no addition (A), 30 μg/ml lysine (C), 30 μg/ml isoleucine (D), and 50 μg/ml leucine (E). Cells harbored the plasmid pCu-ATG22, expressing Atg22 from the CUP1 promoter (B), or the pRS425 (LEU2) plasmid (F). At the indicated day, viability was determined by removing aliquots, plating on YPD in triplicate, and counting the number of colonies per plate after 2–3 d of growth. The addition of leucine, or the LEU2 or ATG22 genes restored viability of atg22Δ cells to wild-type levels. The addition of histidine or uracil resulted in essentially the same loss of viability as seen with lysine. The error bars indicate the SD of two independent experiments.
To resolve this discrepancy, we examined the reason for the starvation sensitivity of the atg22Δ strain. Atg22 is an integral vacuolar membrane protein, and its protein sequence places it in a family of permeases. Accordingly, we decided to test whether it was a vacuolar permease. We hypothesized that the absence of a permease would cause a loss of viability in strains that lack the biosynthetic capacity for the substrate of the permease under starvation conditions. To test our hypothesis, we decided to identify the substrate for the putative permease Atg22. The atg22Δ strain was auxotrophic for histidine, leucine, lysine, and uracil. Thus, we decided to test whether one of these four components could rescue the starvation-sensitive phenotype seen in atg22Δ cells. Wild-type, atg1Δ, and atg22Δ cells were starved in SD-N containing each of these components separately (Figure 4, C and D; our unpublished data). The addition of histidine, lysine, or uracil was not able to restore viability to the atg22Δ strain in SD-N. Similarly, the addition of other amino acids such as isoleucine that could be synthesized by the cell did not affect viability. In contrast, the addition of leucine, or complementation of the leu2 defect by transformation with a plasmid carrying the LEU2 gene, restored viability to the wild-type level (Figure 4, E and F). These results led us to conclude that Atg22 was a leucine effluxer on the vacuolar membrane.
To obtain additional evidence for the role of Atg22 as a vacuolar leucine effluxer, we decided to examine vacuolar levels of leucine. In addition, we considered the possibility that Atg22 might share substrates with two other vacuolar amino acid effluxers, Avt3 and Avt4, which belong to a family of yeast vacuolar amino acid transporters, designated as Avt1-7, in the yeast S. cerevisiae (Russnak et al., 2001). Among this family, Avt1 is required for uptake of neutral amino acids including tyrosine, isoleucine, and glutamine; leucine and asparagine might also be substrates, because competitive inhibition studies by Sato et al. (1984) have shown that leucine/isoleucine and asparagine/glutamine use the same vacuolar uptake system. Avt3 and Avt4, two closely related proteins, display the same specificity as Avt1, but they are synergistically involved in the efflux of amino acids from the vacuole into the cytosol, whereas Avt6 is responsible for the efflux of aspartate and glutamate. Thus, we decided to extend our analysis to Avt3/Avt4. Accordingly, we generated avt3Δ avt4Δ double mutant and atg22Δ avt3Δ avt4Δ triple mutant strains, and then we carried out amino acid analysis to examine vacuolar levels of various amino acids. The vacuolar amino acid fraction was extracted from log phase-grown cells, as described in Materials and Methods. We normalized the concentrations of the analyzed amino acids to that for arginine, which is stored in the vacuole, but is not a substrate for the Avt3/Avt4 permeases.
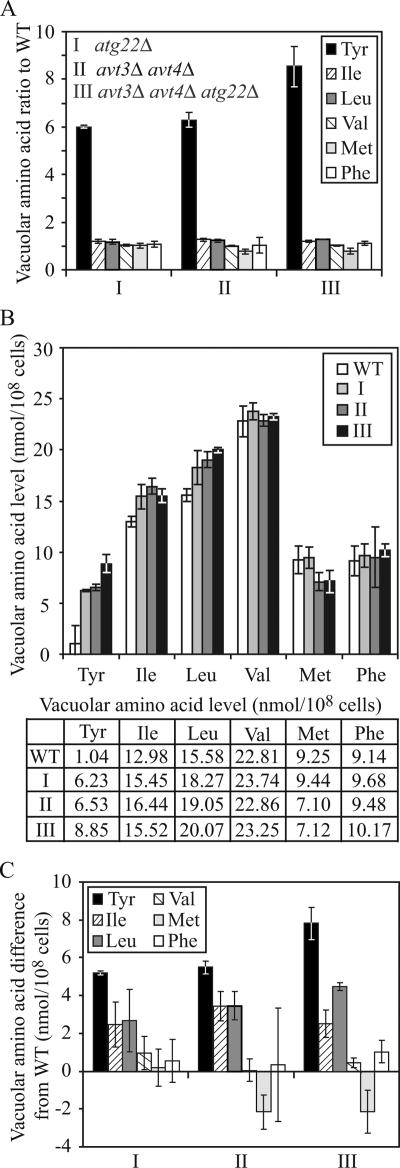
As shown in Figure 5, the most striking accumulation in all three mutant strains was seen with tyrosine, which showed a sixfold accumulation in the atg22Δ cells relative to the wild-type strain. A similar result was seen with the avt3Δ avt4Δ double mutant, whereas the triple mutant displayed an even higher level of accumulation, ∼8.5-fold higher than wild type. This result confirmed the validity of the analysis because tyrosine is a known substrate of the Avt3/Avt4 effluxers and should accumulate within the vacuole in the corresponding mutant strain. This finding also implicated Atg22 as a vacuolar effluxer for tyrosine.
Figure 5.
Yeast vacuoles accumulated a high level of tyrosine, isoleucine, and leucine in the absence of Atg22 and/or Avt3/Avt4. The wild-type (SEY6210), atg22Δ (ZFY6), avt3Δ avt4Δ (ZFY20), and avt3Δ avt4Δ atg22Δ (ZFY22) cells were grown in YPD medium and harvested at log phase. The preparation of vacuolar amino acid pools and analysis of amino acid composition are described in Materials and Methods. The results represent the mean and SD of three experiments. The amino acid concentration was normalized to the highest concentration of arginine among three experiments. The results are displayed as the ratio to wild type (A), the absolute values expressed as nanomoles/108 cells (B), and the difference from wild type for tyrosine, isoleucine, leucine, valine, methionine, and phenylalanine for the wild-type, atg22Δ (I), avt3Δ avt4Δ (II), and avt3Δ avt4Δ atg22Δ (III) strains (C). Tyrosine, isoleucine, and leucine accumulated in the mutants, compared with the controls valine, methionine, and phenylalanine. The table lists the vacuolar amino acid levels shown in the graph in B.
We were not able to examine levels of glutamine or asparagine, two other substrates of the Avt3/Avt4 effluxers because the concentrations of these amino acids could not be determined separately from glutamate and aspartate, respectively, by the methodology used; however, we continued the analysis by monitoring the levels of leucine and isoleucine, two putative substrates, as well as three control hydrophobic amino acids: valine, methionine, and phenylalanine. None of these amino acids displayed as great a -fold increase in accumulation in the mutant strains as seen with tyrosine (Figure 5A); however, we also examined the absolute values and the change in concentration in each amino acid (Figure 5, B and C). For example, the level of leucine in the atg22Δ avt3Δ avt4Δ triple mutant increased to 20.07 nmol/108 cells, compared with 15.58 nmol for the wild-type strain, representing a difference of 4.49 nmol, or a 1.3-fold increase. A similar change was seen in the isoleucine levels. In contrast, the control amino acids showed either a minimal increase or a decrease in concentration (Figure 5, B and C). The values for leucine and isoleucine showed a much lower -fold increase than tyrosine; however, this is partly due to the relatively low level of tyrosine accumulation in the vacuoles of wild-type cells. For tyrosine and leucine, the triple mutant demonstrated a greater accumulation of predicted substrate amino acids than seen with either the atg22Δ single mutant, or the avt3Δ avt4Δ double mutant. Taken together, these results suggested that Atg22 is a vacuolar effluxer for leucine, isoleucine, and tyrosine, sharing these same substrates with Avt3 and Avt4.
Putative Amino Acid Permeases Mediate the Connection between Autophagy and Maintenance of Protein Synthesis under Amino Acid Starvation Conditions
Recently, Onodera and Ohsumi (2005) reported an interesting new phenotype for autophagy-defective mutants. They found that bulk protein synthesis was substantially reduced under nitrogen starvation conditions in mutants, such as atg7Δ, atg1Δ and pep4Δ cells, compared with the wild type. They also demonstrated that the synthesis of certain proteins, such as the vacuolar proteinases Ape1 and Prc1 (carboxypeptidase Y), which are up-regulated at the protein expression level upon nitrogen starvation, was dramatically abrogated in the atg7Δ strain. Additionally, the total intracellular amino acid pool was reduced in atg7Δ cells. They interpreted these results to indicate that free amino acids pools generated during autophagy were a limiting factor for protein synthesis under starvation conditions. Here, we were interested in identifying the mechanistic connection between autophagy and maintenance of amino acid levels and hence protein synthesis. Based on the data we have already shown, we hypothesized that in a strain auxotrophic for a particular amino acid, a defect in efflux from the vacuole lumen for this amino acid would cause an intracellular shortage, which would result in a decline in the ability of the cell to maintain general protein synthesis. Because Atg22 may function in part as a leucine effluxer on the vacuolar membrane and our strain is auxotrophic for leucine, we decided to test the protein synthesis capacity in the atg22Δ mutant in the presence and absence of leucine.
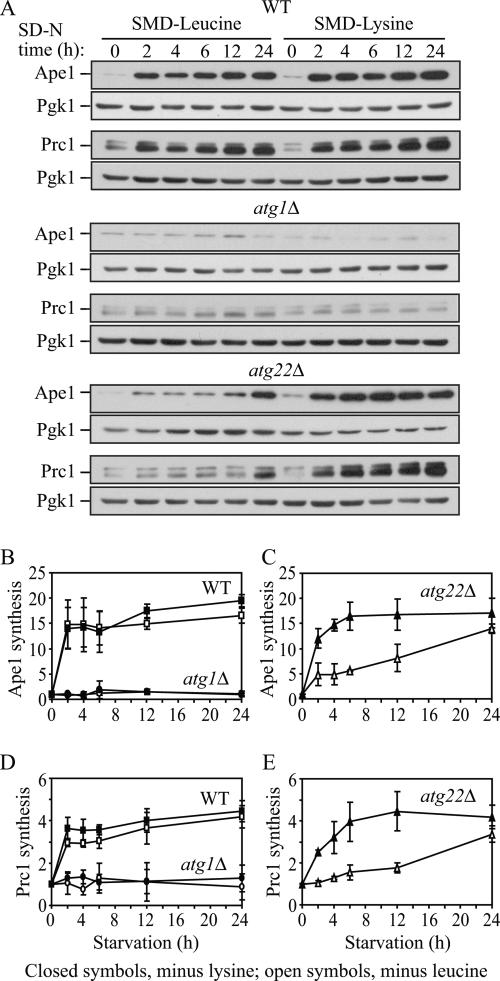
We first verified that the absence of a single amino acid would induce autophagy by using the GFP-Atg8 processing assay. The atg22Δ strain (his3 leu2 lys2 ura3) was grown in SMD lacking either leucine or lysine. Either condition induced autophagy at levels similar to those seen with nitrogen starvation based on processing of GFP-Atg8 (our unpublished data). Next, we carried out a protein synthesis assay. Cells were grown in SMD and then shifted to SMD-leucine or SMD-lysine, conditions that we had demonstrated would induce autophagy. To examine the effect on protein synthesis, we took advantage of the starvation-induced increase in Ape1 and Prc1 synthesis. Pgk1 did not show a substantial change in protein levels under these conditions and served as a loading control. Using immunoblot analysis, we quantified the protein levels of Ape1 and Prc1 in wild-type, atg1Δ, and atg22Δ cells. Under both SMD-leucine and SMD-lysine conditions, there was clear protein synthesis in wild-type cells, with up to a 19-fold increase in the Ape1 level and up to a 4.5-fold increase in the level of Prc1 (Figure 6, A, B, and D). In contrast, synthesis of Ape1 and Prc1 was essentially blocked in the atg1Δ cells under both SMD-leucine and SMD-lysine conditions, in agreement with the previous study by Onodera and Ohsumi (2005). In atg22Δ cells, in SMD-leucine both Ape1 and Prc1 synthesis were minimally elevated even after 12-h starvation and gradually increased by 14- and 3.4-fold, respectively, only after prolonged (24-h) starvation (Figure 6, A, C, and E). In contrast, in SMD-lysine Ape1 and Prc1 synthesis seemed similar to wild type and increased by 17- and 4.2-fold, respectively, with a substantial increase as early as 2 h. Thus, in the atg22Δ cells, a reduced level of Ape1 and Prc1 synthesis was only observed under SMD-leucine conditions, supporting our conclusion that Atg22 functions in part as a vacuolar leucine effluxer. Furthermore, this result supported our hypothesis that under amino acid deprivation conditions, permeases mediate the connection between autophagy and the maintenance of amino acid levels in the cytosol, and hence protein synthesis levels, through the efflux of amino acids resulting from autophagy.
Figure 6.
Protein synthesis dependent upon autophagic amino acids was partially defective in atg22Δ mutant cells. (A) The wild-type (SEY6210), atg1Δ (WHY1), and atg22Δ (ZFY6) cells were grown in SMD and then shifted to SMD-leucine or SMD-lysine to induce autophagy. At the indicated incubation time, aliquots were removed and cell lysates (1.0 OD600 unit of cells) were analyzed by immunoblot with antiserum against Ape1, Prc1, and Pgk1. Pgk1 was used as a loading control. (B–E) Quantification of immunoblots from SMD-leucine (open symbols) or SMD-lysine (closed symbols) in the wild-type (square), atg1Δ (circle), and atg22Δ (triangle) cells. Band intensities were quantified using NIH Image 1.62 (by Wayne Rasband, National Institutes of Health, Bethesda. MD). The data are the average of three independent experiments and the error bars indicate the SD.
Vacuolar Release of Amino Acids during Starvation Is Essential for Survival
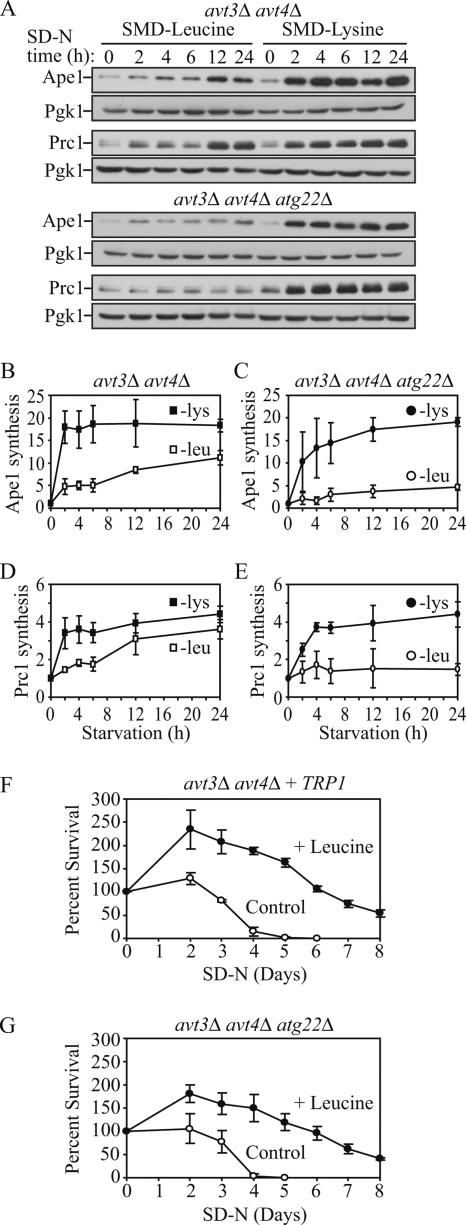
Having established that permeases mediate autophagy and its physiological role in terms of maintenance of protein synthesis under conditions of amino acid starvation, we decided to investigate whether they are also needed for cell viability, examining the Atg22 and Avt3/Avt4 proteins. Based on our data, the atg22Δ cells gradually lost viability through 12 d of starvation (Figure 4A), and Ape1 and Prc1 protein synthesis in the atg22Δ cells was severely but not completely blocked in SMD-leucine (Figure 6). Thus, we hypothesized that Avt3/Avt4 could partially compensate for the defect seen in the absence of Atg22. To address this possibility, we examined an avt3Δ avt4Δ strain auxotrophic for both leucine and lysine. Ape1 and Prc1 synthesis were compared under either SMD-leucine or SMD-lysine conditions (Figure 7, A, B, and D). Pgk1 again served as a loading control. In the avt3Δ avt4Δ cells, Ape1 and Prc1 synthesis was delayed and showed a clear increase only after 12-h starvation in SMD-leucine, whereas there was normal synthesis in SMD-lysine, in support of a role for Avt3/Avt4 as leucine permeases. Accordingly, we hypothesized that the absence of Atg22 along with Avt3/Avt4 might completely block Ape1 and Prc1 synthesis. Thus, we further tested protein synthesis in the atg22Δ avt3Δ avt4Δ strain. Ape1 and Prc1 synthesis was hardly detected even after 24-h starvation in SMD-leucine in the triple deletion strain, but it was relatively unaffected in SMD-lysine conditions (Figure 7, A, C, and E). Together, these results supported the premise that Atg22, Avt3, and Avt4 are redundant with regard to their function as leucine effluxers on the vacuolar membrane.
Figure 7.
Protein synthesis and survival dependent on autophagic amino acids was reduced in avt3Δ avt4Δ mutant cells, and the defect was exacerbated by the atg22Δ mutation. (A) avt3Δ avt4Δ (ZFY20) and avt3Δ avt4Δ atg22Δ (ZFY22) cells were grown and analyzed for up-regulated protein synthesis as described in the legend to Figure 6. (B–E) Quantification of immunoblots from SMD-leucine (open symbols) or SMD-lysine (closed symbols) in the avt3Δ avt4Δ (square), or avt3Δ avt4Δ atg22Δ (circle) cells. Band intensities were quantified as described in the legend to Figure 6. The loss of leucine effluxers blocked synthesis of Ape1 and Prc1 during leucine depletion. The avt3Δ avt4Δ strain harboring pRS414 (TRP1) (F) and avt3Δ avt4Δ atg22Δ (G) cells were grown in SMD to mid-log phase and incubated in SD-N or SD-N containing 100 μg/ml leucine. At the indicated days, viability was determined as described in the legend to Figure 4. The error bars indicate the SD of three independent experiments. The reduced viability of the avt3Δ avt4Δ, and avt3Δ avt4Δ atg22Δ mutant cells was partially rescued by addition of leucine.
To bring our analysis full circle, we decided to examine viability in avt3Δ avt4Δ and atg22Δ avt3Δ avt4Δ cells. The avt3Δ avt4Δ strain was transformed with a plasmid carrying the TRP1 gene to balance the prototrophy of the atg22Δ::TRP1 deletion, leaving both strains auxotrophic for only leucine and lysine. The two strains were then grown in SMD to mid-log phase and incubated in SD-N, or SD-N containing leucine, and viability was measured as described previously. The avt3Δ avt4Δ culture lost viability between 5 and 6 d in SD-N (Figure 7F), whereas the atg22Δ avt3Δ avt4Δ cells displayed a slightly more severe phenotype, completely losing viability by day 4 (Figure 7G). Addition of leucine rescued the loss of viability for both mutants for several days, probably until this nutrient became limiting. These results combined with the result seen with atg22Δ cells shown in Figure 4, suggested that if the efflux of amino acids resulting from autophagy was blocked, the cytosolic shortage of these amino acids would cause a decreased level of general protein synthesis, and, ultimately, the cells would lose the ability to survive starvation of the corresponding amino acids.
DISCUSSION
The atg22Δ Mutant Is Not Defective in Autophagic Body Breakdown
One of the main roles of autophagy is to degrade cytoplasm and recycle the resulting macromolecules for reuse in the synthesis of essential components during nutrient stress. Accordingly, autophagic bodies that are delivered into the vacuole lumen must be disintegrated and broken down (Klionsky, 2005). The topic of vesicle breakdown is important, because the vacuole is the terminal destination for many cellular delivery pathways; however, little is known about the process of lipid recycling within this organelle. For example, Atg15 is the only putative lipase so far associated with this organelle. In addition, it is not even known how the vacuole membrane is itself protected from degradation. This complication extends to all heterotypic fusion events. That is, the vesicle membrane that fuses with the vacuole must be rapidly removed or protected to prevent its degradation while part of the vacuole-limiting membrane and the subsequent loss of vacuolar integrity, which could be deleterious to the cell. Finally, with regard to autophagy, a difference in the susceptibility to lysis of Cvt versus autophagic bodies as suggested by Suriapranata et al. (2000) might have provided insight into the origin of the sequestering vesicle membrane, a topic of considerable debate.
As we have demonstrated in this article, in contrast to the previously reported data, Atg22 is not directly involved in intravacuolar vesicle lysis. The steps of autophagy up to and including autophagic body breakdown are essentially normal in the atg22Δ mutant, although there is a kinetic delay in breakdown (Figures 1 and 2). Our data disagree with the conclusion from the previous work (Suriapranata et al., 2000) that the primary role of Atg22 is in the breakdown of autophagic, but not Cvt, bodies. Thus, no data at present suggest a substantial difference between the membranes used for autophagy and the Cvt pathway, nor do they indicate a fundamental difference between autophagic and Cvt bodies with regard to the mechanism of degradation. This finding is supported by recent studies indicating that the origin of the membrane for the Cvt pathway and autophagy is probably the same (Reggiori et al., 2004).
Atg22 Is a Putative Amino Acid Effluxer on the Vacuolar Membrane
Biochemical analysis of Atg22 indicates that it is an integral membrane protein (Figure 3). Localization of Atg22-GFP suggests that this protein is localized on the vacuole-limiting membrane. These data fit well with the predictions based on its amino acid sequence that Atg22 functions as a permease. Analysis of amino acid concentrations suggested that Atg22 is an effluxer for tyrosine, but it also revealed the vacuolar accumulation of leucine in the atg22Δ mutant (Figure 5). We were unsuccessful in carrying out analyses of amino acid efflux by using vacuoles containing radiolabeled amino acids; thus, we do not have direct evidence for the function of Atg22 as a leucine effluxer. However, the Avt3/4 proteins have been characterized as permeases for leucine and other amino acids in addition to tyrosine. The defect in protein synthesis seen in the absence of leucine in the atg22Δ strain along with the viability assays suggested that leucine is also a substrate for Atg22 (Figures 4, 6, and 7).
Release of Amino Acid from the Vacuole Is Essential for Viability as the Last Stage of Autophagy
The finding that Ape1 synthesis in the atg22Δ cells was severely but not completely blocked in SMD-leucine (Figure 6) indicated there might be redundant leucine effluxers that could partially compensate for the defect. At present, only Avt3/Avt4 and Avt6 have been identified as vacuolar amino acid effluxers in S. cerevisiae, and leucine has been shown to be a possible substrate of Avt3 and Avt4 (Sato et al., 1984). Analysis of both Ape1 and Prc1 protein synthesis and cell viability in avt3Δ avt4Δ and atg22Δ avt3Δ avt4Δ strains (Figure 7) suggested that all three proteins are redundant leucine effluxers and part of the same family of permeases. In support of this finding, vacuolar amino acid analysis indicated an increase in leucine, isoleucine, and tyrosine levels in the double and triple mutants (Figure 5). We note that there is precedence for redundancy in vacuolar permeases; the Vba1, Vba2, and Vba3 proteins all seem to function in influx of basic amino acids (Shimazu et al., 2005). To examine the nature of this redundancy, we transformed the avt3Δ avt4Δ strain with a plasmid overexpressing the ATG22 gene under the control of the CUP1 promoter. Overexpression of Atg22, however, did not rescue the decrease in viability of the avt3Δ avt4Δ strain or rescue the defect in protein synthesis in medium lacking leucine (our unpublished data), suggesting that the functions of these permeases are not completely overlapping. Finally, these results verify the previously untested assumption that vacuolar amino acid efflux is essential for cell viability under starvation conditions.
Breakdown of Autophagic Bodies Is Kinetically Delayed in the atg22Δ Mutant
Although the steps of autophagy up to and including breakdown are essentially normal in the atg22Δ mutant, there is a partial accumulation of autophagic bodies. The present study, however, makes it clear that the atg22Δ mutant displays only a kinetic block in breakdown (Figure 2). One response of cells to long-term amino acid or nitrogen starvation is the up-regulation of certain genes, including those for some autophagy-related proteins (such as Atg8) and vacuolar proteinases (such as Pep4, Prb1, and Prc1) (Gasch et al., 2000). Thus, the role of Atg22 as a leucine effluxer provides an explanation for an indirect effect on autophagic body breakdown. The atg22Δ mutant will be limited for leucine in strains that are leu2 auxotrophs when incubated in starvation conditions. As a result, proteins that are normally up-regulated during starvation will not be synthesized. The proteinases Pep4, Prb1, and Prc1 are critical for breakdown of autophagic bodies, and they are normally substantially induced during starvation. The strain used by Suriapranata et al. (2000) as well as those used in the present study are leu2 mutants. Thus, we propose that the inability to synthesize adequate levels of Pep4, Prb1, and Prc1, which are normally up-regulated approximately eight-, nine-, and sevenfold under nitrogen starvation, respectively (Gasch et al., 2000; Figures 6 and 7), could account for the observed kinetic delay in autophagic body degradation.
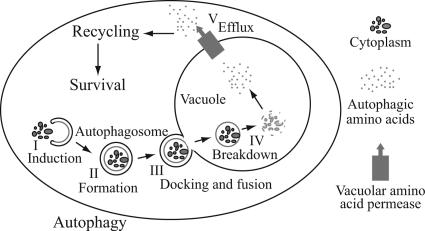
Recycling Model for Autophagy
When cells lack essential nutrients, autophagy is induced, which generates an internal nutrient pool to supply the missing components essential for survival (Kuma et al., 2004; Levine and Klionsky, 2004; Lum et al., 2005). However, there are no mechanistic data that specifically connect the breakdown process with subsequent cytosolic protein synthesis. In this study, using genetics and biochemical assays, we have provided this link by demonstrating that 1) Atg22 most likely functions as a leucine effluxer; and 2) vacuolar permeases mediate the connection between autophagy and its associated protein degradation, and maintenance of amino acid levels in the cytosol and hence protein synthesis levels. Thus, our study has allowed us to propose a “recycling” model that includes the efflux of macromolecules from the vacuole as the final step of autophagy (Figure 8).
Figure 8.
Model for autophagy including the final step of amino acid efflux. Autophagy is depicted as occurring in five discrete stages as shown. Additional steps including cargo recognition, vesicle nucleation, and retrieval of Atg proteins have been omitted for simplicity. See the text for a discussion of the role of efflux.
The vacuole/lysosome is a highly complex organelle that is characterized as having an acidic lumen and harboring a range of hydrolytic enzymes. These hydrolases are involved in the degradation of various substrates, and in concert with vacuolar permeases, they allow the homeostatic control of cytosolic nutrients used for anabolic and catabolic processes. Importantly, the vacuole is not a “dead-end” compartment, and this study is the first report with the efflux process being defined as the final step of autophagy.
ACKNOWLEDGMENTS
We thank members of the Klionsky laboratory for reading the manuscript and for helpful discussions. This work was supported by the National Institutes of Health Public Health Service Grant GM53396 (to D.J.K.).
Abbreviations used:
- Ape1
aminopeptidase I
- Cvt
cytoplasm-to-vacuole targeting
- Prc1
carboxypeptidase Y.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0479) on October 4, 2006.
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr., Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple U. D., Eskelinen E.-L., Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Epple U. D., Suriapranata I., Eskelinen E.-L., Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMbase–a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Huang W.-P., Scott S. V., Kim J., Klionsky D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kim J., Dalton V. M., Eggerton K. P., Scott S. V., Klionsky D. J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Klionsky D. J. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., Thompson C. B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Nelissen B., De Wachter R., Goffeau A. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997;21:113–134. doi: 10.1111/j.1574-6976.1997.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Ohki R., Murata M. bmr3, a third multidrug transporter gene of Bacillus subtilis. J. Bacteriol. 1997;179:1423–1427. doi: 10.1128/jb.179.4.1423-1427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y., Kitamoto K., Anraku Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 1988;170:2676–2682. doi: 10.1128/jb.170.6.2676-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera J., Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R., Konczal D., McIntire S. L. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 2001;276:23849–23857. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- Sato T., Ohsumi Y., Anraku Y. An arginine/histidine exchange transport system in vacuolar-membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 1984;259:11509–11511. [PubMed] [Google Scholar]
- Shimazu M., Sekito T., Akiyama K., Ohsumi Y., Kakinuma Y. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4851–4857. doi: 10.1074/jbc.M412617200. [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriapranata I., Epple U. D., Bernreuther D., Bredschneider M., Sovarasteanu K., Thumm M. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 2000;113:4025–4033. doi: 10.1242/jcs.113.22.4025. [DOI] [PubMed] [Google Scholar]
- Teter S. A., Eggerton K. P., Scott S. V., Kim J., Fischer A. M., Klionsky D. J. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]