Abstract
Preventing polyspermy during animal fertilization relies on modifications to the egg's extracellular matrix. On fertilization in sea urchins, the contents of cortical granules are secreted and rapidly assemble into the egg's extracellular vitelline layer, forming the fertilization envelope, a proteinaceous structure that protects the zygote from subsequent sperm. Here, we document rendezvin, a gene whose transcript is differentially spliced to yield proteins destined for either cortical granules or the vitelline layer. These distinctly trafficked variants reunite after cortical granule secretion at fertilization. Together, they help coordinate assembly of the functional fertilization envelope, whose proteome is now defined in full.
INTRODUCTION
Fusion of an egg and a sperm induces a series of cell surface alterations that hinder the penetration of additional sperm. These changes include both a transient membrane depolarization (the “fast block”) and modification of the extracellular matrix (the “slow” or “physical block”; for review, see Shapiro et al., 1989; Wessel et al., 2001; Wong and Wessel, 2006). In sea urchins, extracellular matrix modifications include assembly of a fertilization envelope (FE) seconds after gamete fusion, thereby establishing a physical barrier that protects the zygote. This new structure is a heterogeneous assembly of glycoproteins from both the vitelline layer (VL), a thin extracellular glycoprotein matrix attached to the egg surface, and the contents of nearly 15,000 cortical granules (CGs), secretory vesicles docked at the egg plasma membrane. Constituents of both egg components are synthesized simultaneously during oogenesis, but they accumulate within separate subcellular compartments (Anderson, 1968). After fertilization, CG contents are secreted and a fraction of these proteins selectively associates with the VL to form the FE.
Most of the FE mass originates from the CGs as freely soluble proteins that rapidly weave into the VL (Villacorta-Moeller and Carroll, 1982; Wong and Wessel, 2004). Three of these CG proteins contain tandem, low-density lipoprotein receptor type A (LDLrA) motifs: 1) proteoliaisin (>25 LDLrA repeats; Somers et al., 1989; Wong and Wessel, 2004), 2) SFE1 (22 LDLrAs; Wessel et al., 2000), and 3) SFE9 (15 LDLrAs; Wessel, 1995). Such enrichment in LDLrA repeats may facilitate the interaction of these structural proteins with the VL, as documented for the LDLrA-rich amino terminus of proteoliaisin (Somers and Shapiro, 1991). Defining these interactions is limited, however, because two major CG proteins remain unidentified and knowledge of VL constituents is sparse (Gache et al., 1983; Niman et al., 1984; Correa and Carroll, 1997).
We now identify the final, major CG-derived structural proteins and a VL constituent that together participate in FE construction. These proteins originate from one gene, and they are expressed specifically in oocytes via differential splicing of the transcript followed by distinct trafficking of the proteins to the CGs or the VL (Figure 5). We refer to this gene as rendezvin (RDZ) based on the reunification of these sibling proteins in the FE after fertilization. We also solve the FE proteome, and find that the self-associating RDZ proteins are central to the assembly of this classic physical block to polyspermy.
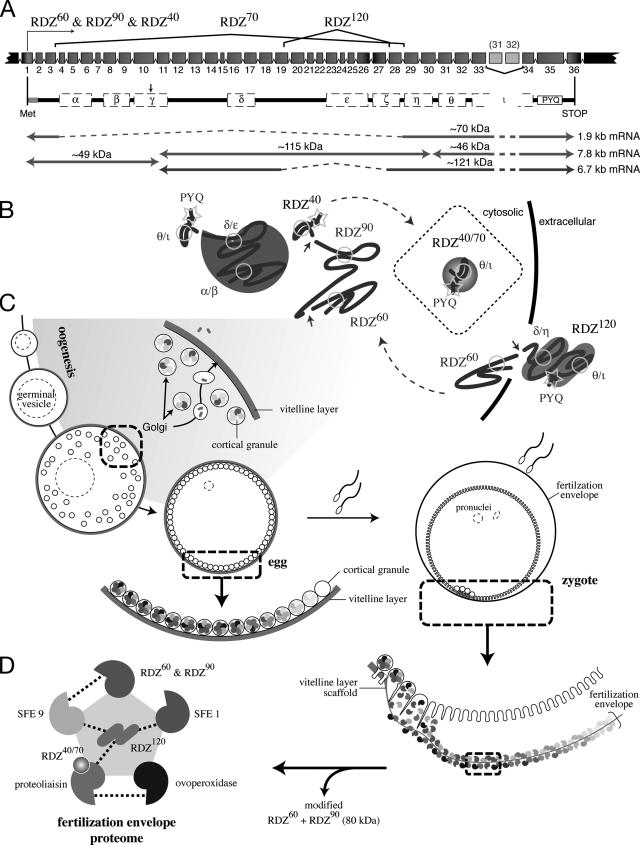
Figure 5.
Model of rendezvin protein expression and the fertilization envelope proteome. (A) Schematic of the S. purpuratus rendezvin exons, colored to match the predicted protein encoded, and resultant transcripts following posttranscriptional processing. The duplication of exons 31 (185 base pairs) and 32 (191 base pairs) is not retained in the most stable rdz transcript (8.2 kb versus stable 7.8 kb; our unpublished data). Spliced regions are shown with the corresponding isoform nomenclature used. Predicted primary structure, after removal of exons (dashed lines) and proteolysis (arrow; see text), are shown below with their predicted protein mass. The approximate transcript lengths that correspond to each isoform are listed on the right. Note the amino-terminal isoform (RDZ60; ∼49 kDa) is shared by two transcripts due to proteolysis. (B) Sketches of possible secondary structures for each RDZ component predicted by splice variants and immunoblot detection, including the interacting CUBs, are shown. (C) Diagram of oogenesis and fertilization, including a model for RDZ translation and protein trafficking within the oocyte (shaded region). Includes details of the egg and zygotic cortex undergoing exocytosis. (D) One binding network of interacting structural protein partners within the fertilization envelope is shown, following the color representation of Figure 4F.
MATERIALS AND METHODS
Animals
Adult Strongylocentrotus purpuratus and Lytechinus variegatus gametes were obtained and handled as described previously (Wong and Wessel, 2004).
Cloning and Sequencing
Both the 60- and 90-kDa bands separated by SDS-PAGE of S. purpuratus FEs were N-terminally microsequenced (Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL) and degenerate primers for the 90-kDa protein microsequence were used to screen an S. purpuratus ovary cDNA library, yielding a single amplicon. For L. variegatus sequence, we used a clone identified in our expression-tagged sequence collection (our unpublished data). Both the S. purpuratus and L. variegatus clones were used to generate 32P probes by using the RadPrime kit (Invitrogen, Carlsbad, CA) for hybridization screening λZAP cDNA libraries (Stratagene, La Jolla, CA) made from S. purpuratus or L. variegatus ovary poly-A+ RNA (Wong and Wessel, 2004). Alternatively, pan-VL antiserum (our unpublished data) was used in immunoscreens of the S. purpuratus λZAP cDNA library as described previously (Wong and Wessel, 2004). Resultant nucleotide sequence data were assembled and analyzed using Sequencher (Gene Codes, Grand Rapids, MI). Motif predictions were made using the SMART database (smart.embl-heidelberg.de/). All nucleotide and amino acid positions are reported from A = 1 of the predicted ATG initiation codon or methionine.
RNA Analysis
RNA probes, representing S. purpuratus sequence from fragment 193–652 (RDZCG probe) or 4678–5119 (RDZVL probe), were generated using the Megascript kit (Ambion, Austin, TX) in the presence of digoxigenin-UTP (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions. RNA samples were isolated by standard methods (Bruskin et al., 1981). Oocyte-enriched poly-A+ RNA was isolated using a poly(dT) column (Invitrogen). RNA gel blot and hybridization was performed as described previously (Bruskin et al., 1981). After hybridization to 10 ng/ml digoxigenin-labeled antisense probe, blots were treated with 2 μg/ml RNase A, washed in 1× SSC (150 mM NaCl and 15 mM sodium citrate), and probed for digoxigenin in maleate buffer (100 mM maleic acid, 185 mM NaCl, and 0.5% Tween 20, pH 8.0; Engler-Blum et al., 1993). Probes were detected using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium as described previously (McGadey, 1970).
In situ RNA hybridizations were completed using the probes generated for RNA blot analysis (see above), according to previously published protocols (Ransick et al., 1993; Wong et al., 2004). All samples were imaged by Nomarski light microscopy using an Orca-ER charge-coupled device (CCD) camera (Hammamatsu, Bridgewater, NJ) on an AxioPlan upright microscope (Carl Zeiss, Thornwood, NY) controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA).
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Primers used are listed in Supplemental Table 3. Single-tube RT-PCR reactions were conducted using the Access RT-PCR kit (Promega, Madison, WI) against 1 μg of total ovary RNA. Nested, alternative-splicing analysis was conducted in two rounds. Each first-round RT-PCR was processed through 1 h at 48°C, 95°C for 5 min, and then 45 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 4 min. One microliter of each RT-PCR reaction was then used in a standard PCR reaction with the appropriate inner nested primer pairs by using parameters 30 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 3 min. Multiplex RT-PCR reactions were run as described previously (Wong et al., 2004). For all reactions, one-fifth of the reaction was separated on a 2% agarose gel, stained with ethidium bromide, and imaged on a Typhoon fluorescent scanner using proprietary software (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Biochemical Fractionation of Eggs
Eggs were dejellied in ASW acidified with HCl (pH 5.2) (S. purpuratus) or washed in calcium-free seawater containing 10 mM EDTA (L. variegatus), followed by extensive reequilibration to artificial seawater (ASW), pH 8.0. These eggs were then used to isolate cell surface complex and fertilization envelope. Cell surface complex was obtained by methods published previously (Detering et al., 1977; Kinsey, 1986), with the addition of Complete Mini EDTA-free protease inhibitor cocktail (1 tablet per 10 ml of ASW; Roche Diagnostics) to the buffer. Soft fertilization envelopes (SFEs) were isolated from zygotes fertilized in the presence of the 1 mM 3-aminotriazole (Sigma-Aldrich, St. Louis, MO) as described previously (Wong and Wessel, 2004). Both samples were then solubilized in 1× SDS sample buffer.
Production of Polyclonal Antisera
Antiserum was raised in rabbits against bacterially overexpressed, recombinant fragments subcloned downstream of a glutathione S-transferase coding tag found in either a pGEX expression vector (GE Healthcare) or a 6x-histidine tag found in the pQE expression vectors (QIAGEN, Valencia, CA). Four independent regions of rendezvin were used to generate antiserum: anti-α was generated against residues 31–294 (SVES…FEEL), anti-γ against residues 355–600 (MDRE…SNWF), anti-δ against residues 530-1146 (LNQD…YKRQ), and anti-θ against residues 1363–1816 (CEYN…PNEP). All immunogen were expressed in Escherichia coli BL21 cells, column purified by nickel-column chromatography, and prepared and injected as described previously (Wong and Wessel, 2004).
Polyclonal Antibody Analysis
PAGE was accomplished in precast 4–20% polyacrylamide Tris-glycine gels (Life-Therapeutics, Frenchs Forest, New South Wales, Australia), unless otherwise noted. Immunoblot analysis was performed as described previously (Wong and Wessel, 2004). Immunofluorescence localization was performed on paraffin-embedded, Bouin's fixed tissue and on whole mount cells, embryos, or cortical lawns (prepared as described in Crabb and Jackson, 1985) fixed with 4% paraformaldehyde and then permeabilized with methanol. Antibody probing was performed as described previously for single (Wong and Wessel, 2004) and double labeling (Wessel et al., 1984). L. variegatus eggs and oocytes were stratified in a 16% final sucrose step gradient at 5000 × g for 10 min, washed in ASW, fixed with 10% formalin, and cleared with methanol before staining. Samples were imaged using either an LSM410 laser confocal microscope (Carl Zeiss Corporation, Thornwood, NY) driven by Renaissance software (Microcosm, Columbia, MD) or a TCS SP2 AOBS confocal scanning microscope (Leica Microsystems, Bannockburn, IL).
Electron microscopic analysis was performed on ovary sections with oocytes or on eggs fixed, processed, and immunoprobed as described previously (Wessel et al., 2000). Gold (90-nm) sections were examined and digitally recorded as described previously (Wong and Wessel, 2004). Colloidal gold particles were scored by location in the cortical granule or vitelline layer (see colorized domains in Figure 2G). Distribution of the particles is reported as percentage of total gold particles counted, representing fields from at least five different eggs.
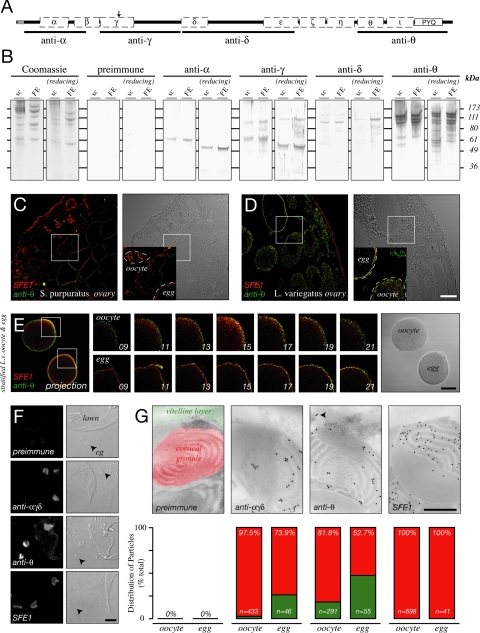
Figure 2.
Rendezvin variants localize to distinct organelles. (A) Schematic diagram of the S. purpuratus rendezvin open reading frame indicating regions used to generate specifically named antisera. Diagram includes the structural motifs of orthologues (Supplemental Figure 1). Note polyclonal anti-RDZ or anti-SFE1 sera, for a cortical granule protein (Wessel et al., 2000), were used as a control in labeling and transmission electron micrograph localization. (B) Comparison of contents of S. purpuratus surface complex (“sc”; 25 μg per lane) and FE (5 μg), stained for total protein (Coomassie), or probed with each serum. Samples were separated under nonreducing (left) and reducing (right) SDS-PAGE. Pattern from each antiserum (see A) compared with a mix of their respective preimmune serum. (C and D) Single confocal images of ovary sections from S. purpuratus (C) and L. variegatus (D) probed with anti-SFE1 (red) and anti-θ (green). Fluorescence signal is indicated on the left panel with respective differential interference contrast (DIC) images found in the right panels. Details (insets) are shown, with boxes indicating their origin: oocyte (oo) and eggs (egg) are outlined. Bar, 50 μm. (E) Permeabilized, stratified L. variegatus oocyte (oocyte) and egg (egg) double-stained for SFE1 (red) and anti-θ (green). Two-dimensional projection of optical sections (images 13–19) through an equatorial region of each cell is shown (left) with corresponding differential interference contrast image (right). Details of centrifugal poles per cell are shown as independent optical sections (middle panels). Z-distance between each optical section is 2.5 μm. Bar, 50 μm. (F) Single antibody staining of egg cortical lawns. Confocal fluorescence (2 airy units, left) and DIC (right) images are shown. Free cortical granules (cg) are indicated (arrowhead). Note membrane on lawns that have folded over and are exposed. Bar, 5 μm. (G) Transmission electron micrographs of immunogold localizations on S. purpuratus oocytes probed with anti-αγδ, anti-θ, or anti-SFE1. Quantitation of colloidal gold particle distribution, as percentage of total particles counted over 15–20 sections of oocytes or eggs. Percentiles refer to the cortical granule group; number of particles scored per treatment is indicated (n). Preimmune serum for anti-θ is shown, with colorization to identify the organelles analyzed in particle scoring. Arrowhead emphasizes particles in the vitelline layer. Bar, 500 nm.
Live Antibody Inhibition Assays
Effects of antibodies on FE assembly were assessed on S. purpuratus eggs dejellied in ASW acidified with HCl, pH 5.2, followed by extensive washing in ASW, pH 8.0. Eggs or zygotes were then preincubated for 1 h on ice with dilutions of whole sera, protein A-purified IgGs, or Fabs from antisera against RDZ or SFE1. The treated cells were then tested accordingly. For FEnotyping, resuspensions of eggs plus antisera (1:10 dilution) were transferred to an equal volume of 20 μg/ml A23187 in ASW. After 10 min at room temperature, eggs were viewed by bright-field and darkfield microscopy to document changes in FE birefringence. An aliquot of these activated eggs was fixed in 1% glutaraldehyde, 4% paraformaldehyde in calcium-free seawater, pH 8.0 (CFSW), for transmission electron microscopy. These fixed eggs were dehydrated and embedded in Spurrs as described previously (Spurr, 1969; Wessel et al., 1989). Gold (90-nm) sections were stained in 2% uranyl acetate and 0.002 g/ml lead citrate and then examined and digitally recorded as described previously (Wong and Wessel, 2004).
In Vivo Protein Competition Assays.
Biotinylated proteins of the FE (biotin-FE) were prepared as described previously (Wong and Wessel, 2004). Rhodaminated proteins of the FE (Rh-FE) were reacted with 2 μg/ml rhodamine B isothiocyanate (Sigma-Aldrich, St. Louis, MO) and then dialyzed against CFSW as described previously (Ettensohn and McClay, 1988). Denatured Rh-FE was prepared by exposure to 1 mM dithiothreitol and incubated at 95°C for 5 min, followed by extensive dialysis against CFSW. Both biotin-FE and Rh-FE samples were bound to antibody-preincubated eggs and/or zygotes. For biotin-FE, eggs and zygotes were incubated in a 1:10 dilution of biotinylated-FE for 2 h on ice, followed by ASW washes. Biotin was detected using Cy3-conjugated Extravidin (Sigma-Aldrich). Samples were then fixed in fresh 4% paraformaldehyde in ASW overnight at 4°C before visualization on a TCS SP2 AOBS confocal scanning microscope (Leica Microsystems). Equatorial fluorescence intensity at the egg surface or in the FE was measured using MetaMorph software (Molecular Devices). For Rh-FE, eggs were incubated in up to 10 μg/ml Rh-SFE for 5 min on ice and then washed rapidly in ASW, gently collecting the cells by hand centrifugation. Samples were fixed in fresh 1% paraformaldehyde in CFSW overnight at 4°C before visualization with an Orca-ER CCD camera (Hammamatsu) on an AxioPlan microscope fitted with tetramethylrhodamine B isothiocyanate dichroics (Carl Zeiss) and controlled by MetaMorph software (Molecular Devices). Epifluorescence intensity of the egg was measured using MetaMorph software (Molecular Devices).
Statistical significance was assessed using Student's two-tailed t test. Evaluation was made after applying the Bonferroni correction for the number of treatments compared.
V5-Recombinant Protein Expression in Drosophila Schneider S2 Cells
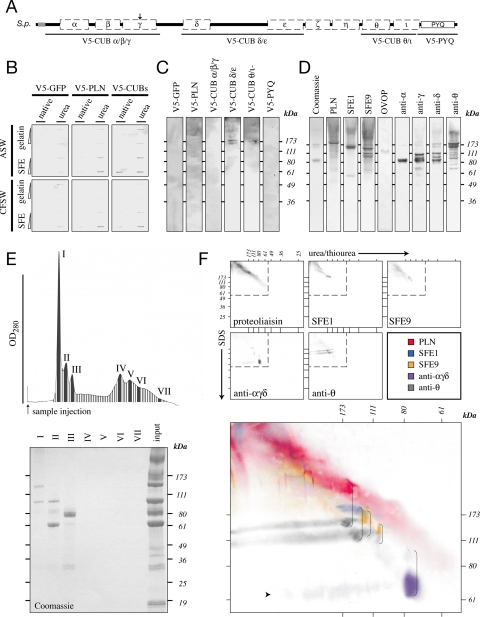
Regions of RDZ enriched for complement C1r/C1s, Uegf and bone morphogenic protein-1 (CUBs) or the proline/tyrosine/glutamate (PYQ) repeats were independently subcloned, in frame, into the fusion vector pcoHyg/MT, which contains sequence from pCoHygro and pMT/BiP/V5 insect expression vectors (Invitrogen; see Supplemental Figure 4). The regions of RDZ subcloned into pcoHyg/MT include V5-CUBα/β/γ, spanning amino acids 41–505 (RWAV…YRSI); V5-CUBδ/ε, spanning amino acids 691-1110 (GANI…SKTS); V5-CUBθ/ι, spanning amino acids 1363–1708 (CEYN…NVHA); and V5-PYQ, spanning 1708–1834 (SNVH…PAPY). As a control, the amino-terminal region of S. purpuratus proteoliaisin (Wong and Wessel, 2004) was also subcloned into pcoHyg/MT, spanning residues 17–365 (CSGS…LTQP).
Three micrograms of each recombinant vector was transfected into Drosophila Schneider S2 cells using a 1:5 ratio of DNA (micrograms)-to-Superfect (microliters) (QIAGEN) according to manufacturer's instructions. Recombinant cells exhibiting plasmid integration were selected by culturing with 300 μg/ml hygromycin B in Drosophila Serum Free Media supplemented with 20 mM l-glutamine, 100 mg/ml penicillin/streptomycin, and 10% heat-inactivated fetal calf serum (Invitrogen). Expression of recombinant V5-RDZ domains was induced in confluent populations by using 500 mM CuSO4. Protein was harvested from their conditioned media 7 d after induction, dialyzed against 150 mM NaCl, 8 mM CaCl2, 10 mM Tris, pH 8.0, and purified by nickel-affinity chromatography. Eluates enriched for the recombinant proteins were concentrated and equilibrated in 150 mM NaCl, 10 mM Tris, pH 8.0, by using Amicon Ultra-50 columns (Millipore, Billerica, MA). Masses of V5 protein used per experiment were normalized to anti-V5 monoclonal antibody-reactive equivalents, relative to the signal produced by the CUBα/β/γ construct on a standard immunoblot for the V5 epitope by using 1 μg of total protein per construct (data not shown). In general, the following dilutions were used: green fluorescent protein (GFP), 1:20; proteoliaisin (PLN), 1:5; CUBα/β/γ, no dilution; CUBδ/ε, 1:5; CUBθ/ι, 1:10; and PYQ, no dilution.
Protein–Protein Interaction Assays
Multiple methods for testing protein interactions among S. purpuratus FEs were used.
Urea-PAGE.
Isolated FEs were solubilized in a loading buffer containing 8 M urea, 2 M thiourea, and 20% sucrose for 10 min at room temperature. The proteins were separated in a 10% Tris borate-EDTA-urea gel (Invitrogen), blotted to nitrocellulose according to manufacturer's instructions, and briefly rinsed with distilled water. Blots were used for overlay assays or immunodetection (see above).
Overlays.
Isolated FEs were blotted in slots onto nitrocellulose in buffer containing 50 mM EDTA or 8 M urea/2 M thiourea, or separated by urea-PAGE (see above). All blots were briefly rinsed in distilled water to remove urea and then blocked in 10% fetal bovine serum-CFSW or ASW. These blots were exposed to appropriate V5-tagged protein for 16 h. Slot blots were incubated with 1 μg of V5-reactive equivalents of total protein (0.25 μg of each RDZ construct versus 1 μg of GFP or PLN), whereas urea-PAGE blots were incubated with 10 μg of V5-reactive protein (one-half the mass for paired constructs). After two washes with appropriate seawater, interacting proteins were fixed by incubating with 0.1% glutaraldehyde in seawater. Glutaraldehyde was quenched with a 10-min wash in 1 mg/ml glycine in ASW, and then the blots were probed for the V5 epitope by using commercial anti-V5 monoclonal antibodies (Invitrogen) as described above.
Gel Filtration.
Fertilization envelope proteins were separated using a HiPrep 16/60 Sephacryl column (GE Healthcare) in the presence of 1 M NaCl, 1 M urea, and 50 mM EDTA. Two-milliliter fractions were collected simultaneously with a constant read-out of the optical absorbance at 280 nm. Protein in fractions of interest were concentrated by chloroform/methanol precipitation (Wessel and Flugge, 1984) and then solubilized for SDS-PAGE. Protein content in the gel was visualized with Coomassie blue stain.
Two-Dimensional PAGE.
Two micrograms of total FE was separated first by urea-PAGE (see above). Individual lanes were excised and equilibrated in Tris-glycine electrophoresis buffer containing SDS (2.5 mM Tris, 20 mM glycine, and 0.1% SDS, pH 8.0) before separation in a 4–20% Tris-glycine gel (Invitrogen). Gels were blotted and probed with antisera against PLN (Somers and Shapiro, 1989), SFE1 (Wessel et al., 2000), SFE9 (Wessel, 1995), and RDZ as described above.
RESULTS
Rendezvin Identification
Rendezvin (rdz) cDNA was assembled from independent screens for FE proteins in two sea urchin species. Amino-terminal microsequences of the 60- and 90-kDa CG-derived S. purpuratus FE proteins were merged with a single contig from an independent screen for VL-derived FE proteins (Supplemental Figure 1). The rdz gene consists of 36 unique exons (∼5900 base pairs), excluding the duplications of exons 31 (185 base pairs) and 32 (191 base pairs) located between exons 33 and 34 (Supplemental Figure 1A; Song et al., 2006).
The deduced protein sequence contains CUB domains and a repetitive PYQ-rich motif at the carboxy terminus that is reminiscent of repeats found in extracellular matrix proteins (Lyons et al., 1993; Wong and Wessel, 2004). The S. purpuratus open reading frame is 76.3% identical its orthologue in Lytechinus variegatus (Supplemental Figure 1B), a species that diverged from S. purpuratus more than 50 million years ago (Smith, 1988; Smith et al., 1992). Other FE proteins are shared between these two species, but these non-CUB–containing orthologues exhibit sequence divergence not observed in RDZ (Wong and Wessel, 2004). The high percentage of sequence identity between rdz orthologues implies that the products of this gene may be key structural elements of the FE.
Alternative mRNA Splicing in Oocytes
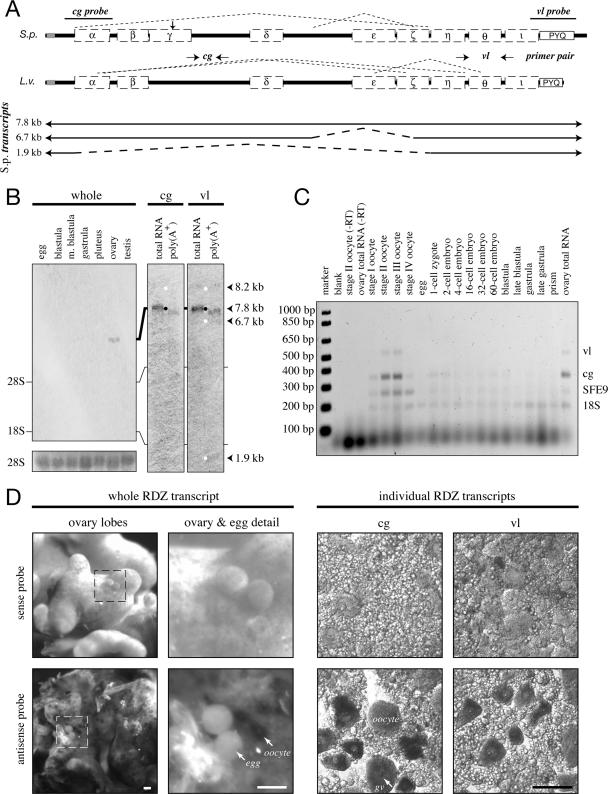
Rendezvin mRNAs are exclusive to oocytes in both S. purpuratus (Figure 1D) and L. variegatus (Figure 1C). This gene is represented in these cells by three alternatively spliced transcripts showing variable abundance (Figure 1B). The sequence of these alternatively spliced sequences were identified by RT-PCR in both S. purpuratus and L. variegatus (Supplemental Figure 2; sequences not shown), suggesting this RNA-based mechanism of functional diversification is conserved for rdz.
Figure 1.
Rendezvin transcripts are exclusive to oocytes. (A, top) Schematic identifying splicing organization predicted by RT-PCR fragments in Figure 2 (dashed lines), probe locations (S. purpuratus), and primer pairs (L. variegatus) relative to the predicted open reading frames, including motifs (see Supplemental Figure 1). cg, cortical granule; vl, vitelline layer. (A, bottom) Diagram of S. purpuratus coding strand RNA identified by sequenced RT-PCR products, listed with likely size of each transcript based on RNA blot pattern. (B) RNA blots probing S. purpuratus total RNA (10 μg) from different developmental stages and gonads, or S. purpuratus ovary total RNA (2.5 μg) and oocyte-enriched poly-A+ RNA (1 μg). The developmental blot (left) was probed with an equal mixture of antisense RNA probes. Duplicate RNA blots (right) were probed with either rdz probe and then overexposed to identify all major transcripts (white dots). Approximate size of each identified transcript is also shown, calculated based on migration distance (our unpublished data). rRNA bands are indicated. Ethidium bromide staining for the 28S rRNA bands is shown as loading controls for the developmental blot. (C) Ethidium bromide image of multiplex reverse transcriptase PCR amplifications from two-cell/embryo equivalents of whole L. variegatus cells or embryos, or 250 ng of ovary total RNA. Primer pairs for the vitelline layer or cortical granule transcript of rendezvin, the cortical granule protein SFE9, and the 18S rRNA (Supplemental Table 3) were used to amplify specific products from total lysates of each stage indicated. Oocyte staging was based on the relative nuclear-to-cytoplasmic ratio, as described previously (Wong et al., 2004). Controls include no template (“blank”) and reactions without reverse transcriptase (“-RT”). (D) In situ hybridizations of rendezvin on S. purpuratus ovary and eggs. Left set shows intact ovary lobes and ovulated eggs, including a magnified image. Right set shows tissue smears of ovary hybridized with either probe (see A). Both sense (top images) and antisense (bottom images) are shown. Oocytes (oo), their germinal vesicles (gv), and eggs are labeled. Bars, 100 μm.
Rendezvin Proteins Are Differentially Targeted within the Oocyte and Egg
We generated four independent antibodies against select regions of RDZ to assess where the different epitopes of the alternatively spliced isoforms reside. These antibodies are named according to the most amino-terminal CUB domain of the immunogen, for example, anti-α was generated against CUBs α-β (Figure 2A).
Each antibody detects RDZ protein in biochemical preparations of the egg cortex and the FE of S. purpuratus (Figure 2B) and L. variegatus (data not shown), consistent with the sequence identity between these orthologues. Different regions of RDZ segregate in the S. purpuratus egg: CUBs α-ε (detected by a mix of anti-α, -γ, and -δ; or collectively [anti-αγδ]) accumulate in CGs as 60- (RDZ60) and 90-kDa (RDZ90) isoforms, whereas the epitope CUBθ to PYQ [anti-θ] is found in both CGs (as RDZ40 and RDZ70; data not shown) and the VL (as RDZ120). We also observe an 80-kDa mass with anti-αγδ that occurs only in biochemical collections of CG contents (data not shown). This mass is absent from the egg cortex (Figure 2B) and does not remain with the zygote after fertilization (data not shown). Based on these observations, we suggest that the 80-kDa mass is a cross-linked form of RDZ60 and/or RDZ90.
Immunolocalization of RDZ in oocytes and eggs recapitulates the accumulation of its constituents in the CG or VL (Figure 2, C–G). Consistent with the life cycle of CG proteins (Wessel et al., 2001), the amino-terminal–containing variants of RDZ [anti-αγδ] are in vesicles found throughout the oocytes, but they are limited to the egg cortex after maturation. The PYQ-containing carboxy terminus of RDZ [anti-θ], however, is detected in both CGs and the VL (Figure 2, F–G). Two features of rdz expression are particularly relevant to understanding the localization of the PYQ-containing fragment (also see Discussion): First, the 120-kDa S. purpuratus protein (RDZ120; [anti-θ]) was originally identified at the egg surface by 125I-radiolabeling (Glabe and Vacquier, 1977), biotinylation (data not shown; Haley and Wessel, 2004), and by partial removal of VL components from the egg surface by using reducing agents (data not shown; Epel et al., 1970; Carroll et al., 1977). Second, a minor population of PYQ-containing protein is also stored in CGs (Figure 2, F and G) and released only upon CG exocytosis (data not shown). These RDZ40 and RDZ70 isoforms [anti-θ] correspond to the coding regions of the 7.8-kb 3′ end (a result of posttranslational proteolysis from RDZ90) and 1.9-kb rdz transcripts, respectively (Figure 5).
Assembly of Fertilization Envelope Constituents Requires the Carboxy Terminus of Rendezvin
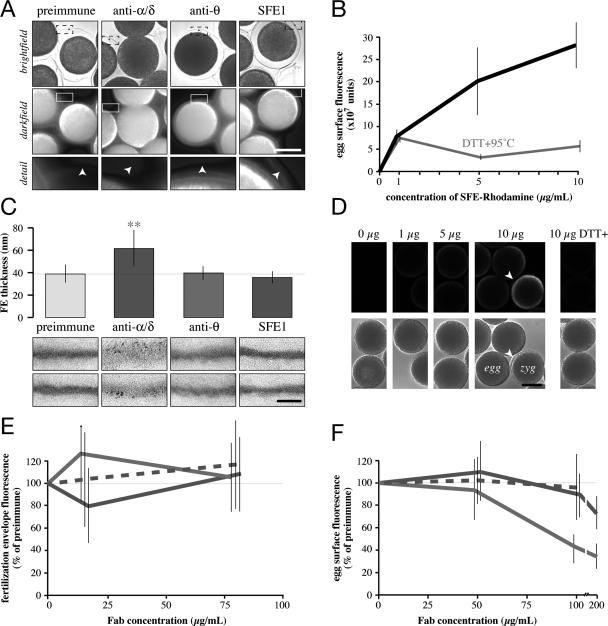
Based on the sequence conservation and their identical localization in S. purpuratus and L. variegatus eggs, we hypothesized that the individual RDZ components play similar, structural roles during FE assembly. We tested such a function by using our panel of antibodies against the different RDZ fragments in the species with more abundant VL and FE mass, S. purpuratus (Figure 3). Sterically blocking RDZ60 and RDZ90 [anti-αγδ] interferes with FE assembly and structure in vivo (Figure 4, A and C). Anti-RDZ competition assays using labeled populations of predominantly CG-derived proteins (Figure 3, B and D; also Wong and Wessel, 2004) reveal that Fabs against RDZ [anti-αγδ or -θ] or SFE1 do not significantly affect in vivo assembly of soluble CG proteins onto preexisting FEs, suggesting the FE is replete with binding sites (Figure 3E). Incorporation of the same CG proteins within the egg VL, however, is coordinated by the PYQ-containing isoform RDZ120 because anti-θ dose-dependently inhibits binding to the egg surface (Figure 3F).
Figure 3.
Rendezvin components play distinct and essential roles in the fertilization envelope. (A) Analysis of FE birefringence, comparing bright-field (top) and darkfield (middle; composite of thresholded pseudocolor images emphasizing the intensity of light derived from the FE overlaying grey scale darkfield image) images of eggs activated by ionophore in the presence of antisera (1:20 dilution). Darkfield details of FEs (bottom) from one segment of the larger image (box) are shown, with arrowheads indicating the FE as seen by bright-field. Bar, 50 μm. (C) Quantitation of the thickness of respective FEs formed in the presence of 1:20 dilutions of antisera, with representative electron micrographs of each FE (bottom). Vertical bars indicate SD. **p < 10−9 among all treatments. Bar, 500 nm. (E) Competition of soluble, biotinylated S. purpuratus FE proteins to intact FEs in the presence of increasing concentrations of mixed Fabs. The x-axis is shown logarithmically. Vertical lines indicate SD. Color coordinated to C. (B) Titration of native (black) or denatured (gray) S. purpuratus rhodamine-FE binding to eggs (see Materials and Methods). Vertical lines indicate SD. (D) Representative confocal images showing dose-dependent binding of rhodamine-FE to the egg surface. Arrowhead identifies a partial FE showing higher binding intensity, consistent with results from E. Bar, 50 μm. (F) Competition of rhodamine-FE binding to the egg by purified Fabs. Color coordinated to C. Vertical lines indicate SD.
Figure 4.
Rendezvin is integral to fertilization envelope assembly. (A) Schematic diagram identifying regions of S. purpuratus rendezvin used to generate V5-recombinant protein. The amino-terminal sequence of S. purpuratus PLN, previously reported to interact with other FE proteins (Somers and Shapiro, 1991), was also used. (B and C) V5-recombinants were tested for binding to immobilized FEs (see Materials and Methods). (B) One and 5 μg of urea-denatured gelatin or FE was probed with a total of 5 μg of V5-recombinant protein in seawater containing (ASW) or lacking (CFSW) free calcium. (C) Proteins separated by urea-PAGE were probed with a total of 10 μg of V5-reactive equivalents of individual or paired V5-recombinants in CFSW. (D) One microgram of FE was separated into subcomplexes by urea-PAGE and immunoblotted for individual proteins or domains of RDZ to identify coassociating proteins. (E) UV absorption profile and eluting fractions of 10 μg of total FE separated by gel filtration identify seven major protein peaks (top). Only the first three peaks contain significant protein as detected by SDS-PAGE and Coomassie staining (bottom). (F) Two micrograms of FE were separated by two-dimensional urea-SDS-PAGE and immunoblotted for the major structural proteins found within the fertilization envelope (top, gray). Details superimposing all the immunoblots (bottom, color) indicate structural proteins that interact endogenously in a pairwise manner (brackets). The faint streak of a 70-kDa protein is indicated (arrowhead).
Binding Partners of Rendezvin and the Fertilization Envelope Proteome
Cloning rdz completed the identification of all the major FE constituents. Thus, we could now define the relationship of the major proteins within the FE proteome with respect to the most conserved element, RDZ. We first generated carboxy terminal, V5-epitope-tagged recombinant proteins encoding specific domains from S. purpuratus RDZ (Figure 4A). As a control, we included the amino-terminal region of proteoliaisin, the region that binds both ovoperoxidase, and the VL (Somers and Shapiro, 1991). These recombinants were expressed in Drosophila Schneider S2 cells by using an inducible secretion system (see Materials and Methods; Supplemental Figure 4) to favor normal processing of the CUBs (Bork and Beckmann, 1993).
Recombinant RDZ proteins bind to solubilized FE proteins in a concentration-dependent manner, independent of calcium (Figure 4B). Fusions V5-CUBs δ/ε and θ/ι even associate with 170-kDa FE subcomplexes separated by urea/thiourea-PAGE (Figure 4C). The larger subcomplex contains PLN- and PYQ-containing RDZ isoforms [anti-θ], whereas the smaller subcomplex contains SFE1, SFE9, and a minor amount of RDZ90 [anti-δ] (Figure 4D). The population of proteins in these subcomplexes is consistent with coimmunoprecipitation results by using the recombinant CUBs that identify SFE9 in association with V5-CUBδ/ε and SFE1 with V5-CUBα/β/γ + δ/ε and with PYQ (our unpublished data). Together, these data suggest that RDZ can bind all the major structural FE proteins.
We next determined the specific interactions within the FE proteome. Separation of soluble FEs by gel filtration in the presence of urea and EDTA generated three high-molecular-weight peaks (I–III) and four broad peaks (IV–VII) (Figure 4E). When separated by SDS-PAGE and identified by mass, peak I contains SFE9 and the amino terminus of RDZ60 [CUBα-γ], peak II contains RDZ60 and RDZ90 [CUBα-γ and δ-ε], and peak III contains ovoperoxidase. Yet some FE proteins were missing, specifically, proteoliaisin and SFE1. Because the urea/thiourea separation maintained discrete subcomplexes that contained all the FE constituents (Figure 4D), we proceeded to use two-dimensional electrophoresis to resolve the urea/thiourea-insensitive complexes by SDS-PAGE (Figure 4F). This method identified coassociating proteins by their vertical alignment, revealing the following pairs: RDZ60 [anti-α]-RDZ90 [anti-δ]; SFE9-RDZ90 [anti-δ]; and SFE9, SFE1, and proteoliaisin with a 120- and 140-kDa PYQ-containing protein [anti-θ]. The pairing of RDZ60 [anti-α] and RDZ90 [anti-δ] is consistent with tandem mass spectroscopy identification of “LLVVTFHDFQLER” and “DSDVPNWADIGIR” peptides, respectively (our unpublished data), from the 80-kDa urea/thiourea–PAGE subcomplex (Figure 4D). The PYQ-containing [anti-θ] 120- and 140-kDa proteins are likely the two most abundant forms of the VL-derived RDZ120 (Figure 2B), the promiscuous isoform that pairs with all the major non-RDZ proteins derived from the CG. Surprisingly, there is no definitive binding partner for CG-derived RDZ70 [anti-θ]; our conclusion, based on the overlap of its horizontal migration pattern with other CG-derived proteins, is that RDZ70 and its sibling RDZ40 (Figures 2A and 5) associate with proteoliaisin (Figure 4F).
Most of the interactions identified within the FE are consistent with previous observations. For example, the association of proteoliaisin with the VL-component RDZ120 [anti-θ] is in agreement with previous experiments showing purified, endogenous proteoliaisin binds to the egg VL (Weidman et al., 1985). Also, RDZ90 [anti-δ] preferentially associates with a 60-kDa protein, presumed to be RDZ60 [anti-α], in an interaction that is reminiscent of a 69-Å Stokes radius complex shown to consist of a 108- and 59-kDa protein (Shapiro et al., 1989). Finally, the extensive interaction of RDZ120 [anti-θ] is consistent with the observation that the egg VL transitions into an insoluble FE core masked by the addition of all the CG content proteins (Carroll et al., 1986), presumably making it less accessible to antibodies (Figure 3E).
DISCUSSION
The first apparent effect of this union [between sperm and egg] is the almost sudden appearance of a perfectly transparent envelope that encircles the yolk [egg] at a certain distance, which is manifested by the appearance of a circular line…. I saw this envelope manifest when in contact with very small number of spermatozoids….
–A. Derbès, 1847 (translated by E. Briggs)
After 150 yr, the major structural proteins of this “circular line” and how they assemble to form an “envelope” have now been identified (Weidman et al., 1985; Wessel, 1995; Wessel et al., 2000; Wong and Wessel, 2004). Here, we document the final components of this FE, which originate from rdz. This gene encodes sibling proteins, derived from alternative splicing, that play an essential role during FE construction.
We propose the following model of differential rdz expression during oogenesis, and its ultimate fate in the zygote (Figure 5). First, the full-length rdz transcript is alternatively spliced into at least three forms, with the most abundant pool of mRNA encoding the parent of RDZ60, RDZ90, and RDZ40. Two significantly less-abundant transcripts are also created, encoding the VL-destined RDZ120 (6.7 kb) and RDZ70 (1.9 kb). During posttranslational processing, S. purpuratus RDZ60 is cleaved from its carboxy-terminal partners (RDZ90-RDZ40 or RDZ120) at serine protease-targeted lysine and arginine residues (…MIK|RST…; the homologous site has not been determined for L. variegatus), whereas another cleavage separates RDZ90 from RDZ40 between CUBs η and θ. Differential trafficking of each variant follows secretory paths destined for either the CG or the VL; we postulate that CUBε and/or protein-binding interactions within RDZ and with its other partners may contribute to the distinct sorting observed. After fertilization, these segregated siblings reunite within the FE, likely via heterologous CUB interactions.
Could the presence of CUB domains in RDZ account for its organizational role within the FE proteome? CUB modules are classically defined as 110 amino acids that contain four positionally conserved cysteines (Bork and Beckmann, 1993). They are present in many extracellular matrix proteins, receptors, and fertilization-related proteins such as the sea urchin vitelline post protein p160 (Haley and Wessel, 2004), the egg bindin receptor 1 (Kamei and Glabe, 2003), the embryonic matrix protein hyalin (Song et al., 2006), ascidian sperm acrosin (Kodama et al., 2001), and mammalian spermadhesins (Topfer-Petersen et al., 1998). Among the fertilization-specific proteins, two sperm-derived proteins have been shown to directly bind the extracellular matrices of their respective eggs, consistent with the involvement of CUBs in gamete binding in vivo (Dostalova et al., 1995; Calvete et al., 1996; Topfer-Petersen et al., 2000; Kodama et al., 2001). Crystal structures of the CUB domain suggest this 10 β-strand motif sandwiches a hydrophobic core between two sheets of five β-strands (Romero et al., 1997; Varela et al., 1997). Two quaternary structures are possible: the edges of two CUB monomers may join to form a functional carbohydrate binding pocket (Romero et al., 1997; Varela et al., 1997) or a monomer can share interact with other monomers via its edges, creating a plane of β-strands accessible from either side (Romero et al., 1997). In RDZ, intraprotein CUB associations, for example, between CUBα and -β; CUBδ and -ε or -η; and CUBθ and -ι, are likely used to establish surfaces where other FE proteins can aggregate (Figure 5D). We do not believe CUBγ participates in such intraprotein CUB–CUB associations because it is cleaved in S. purpuratus. The positional similarity of CUBγ cysteines to other CUBs (our unpublished data), however, suggests that this domain may still be involved in homologous CUB interactions, as exemplified by sea urchin fibropellin C/uEGF I, which lacks two conserved cysteines yet participates in apical extracellular matrix interactions (Bisgrove and Raff, 1993; Laitinen et al., 1999). Heterodimeric CUB–CUB interactions may also occur through associations with CUBs in other proteins. For example, RDZ120 could bind the CUB-rich p160 (Haley and Wessel, 2004). Such a pairing is consistent with the protease-dependent release of VL constituents such as RDZ120 from the egg (Carroll et al., 1977), which parallels the loss of the p160 ectodomain by CGSP1 (Haley and Wessel, 2004).
Although CUB domains and LDLrA repeats are common throughout the animal kingdom, where they function in protein–protein binding (Bork and Beckmann, 1993; Sugiyama et al., 2000; Topfer-Petersen et al., 2000; Kodama et al., 2001; Guo et al., 2004; Song et al., 2006), our results provide the first direct evidence that these two modules interact in a specific, high-affinity manner. How might the CUBs of RDZ120 organize a matrix enriched with LDLrAs (Wong et al., 2004; Wong and Wessel, 2004)Oogenesis: single If the lectin-like function of CUBs is used, then the proteins may be associating through carbohydrate–protein interactions. The limited reactivity of lectins to the VL (Correa and Carroll, 1997) and FE (Wong, unpublished data) is consistent with this model because RDZ CUBs would occupy and compete for all potential targets of these exogenous probes. However, the inability to block FE assembly with monosaccharides (Weidman and Shapiro, 1987) and the low percentage of carbohydrates within the VL by mass (Glabe and Vacquier, 1977) suggest otherwise. Instead, we hypothesize the predominant quaternary fold in RDZ is one that optimizes protein–protein interactions, thereby establishing an extensive network of binding surfaces.
The pairings of VL-derived RDZ120 domains with proteoliaisin, SFE1, and SFE9 and of RDZ90 with SFE9 and RDZ60 are likely occurring via ionic interactions. This model is consistent with the reported functional requirement of acidic residues within the LDLrA repeats and their ligands (Esser et al., 1988; Russell et al., 1989; Rong et al., 1998; Andersen et al., 2000). The affinity of RDZ120 for LDLrA-containing structural proteins and the exclusive association of RDZ60 with RDZ90 suggest that CUB domains are quite selective. Such specificity toward a binding partner is likely affected by both the CUB domain's and its ligand's primary sequence. Indeed, such a mechanism complements how LDLrAs maintain their differential binding affinities for various partners (Esser et al., 1988; Russell et al., 1989). In CUB–LDLrA interactions within the fertilization envelope, for example, CUB domains could provide selective pressure to maintain, and even embellish, the use of LDLrAs in proteins with which they associate (Song et al., 2006). In general, the more high-affinity interacting domains a protein retains, the more freedom a structurally unrelated domain in that same protein has to diverge without losing affinity for its original binding partner. Thus, minor alterations to an otherwise conserved protein foundation can greatly expand the function of an extracellular matrix, possibly resulting in diversification and specialization of the tissues that rely on these evolving matrices. Some cases where this can be seen in action include protein superfamilies such as laminin, fibronectin, and collagen, whose members are used throughout the animal kingdom in matrices as diverse as the basal lamina to tendons and ligaments. Back at the level of the single cell, the paradoxical relationship between conservation of function and sequence radiation is best exemplified by the constituents of the physical block to polyspermy (Wong and Wessel, 2006), especially the one first documented by Derbès as the egg's “envelope.”
Supplementary Material
Abbreviations used:
- CG
cortical granule
- FE
fertilization envelope
- RDZ
rendezvin
- VL
vitelline layer.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-07-0634) on September 27, 2006.
REFERENCES
- Andersen O. M., Christensen L. L., Christensen P. A., Sorensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., Thogersen H. C. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP). A conserved acidic residue in the complement-type repeats is important for recognition of RAP. J. Biol. Chem. 2000;275:21017–21024. doi: 10.1074/jbc.M000507200. [DOI] [PubMed] [Google Scholar]
- Anderson E. Oocyte differentiation in the sea urchin, Arbacia punctulata, with particular reference to the origin of cortical granules and their participation in the cortical reaction. J. Cell Biol. 1968;37:514–539. doi: 10.1083/jcb.37.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove B. W., Raff R. A. The SpEGF III gene encodes a member of the fibropellins: EGF repeat-containing proteins that form the apical lamina of the sea urchin embryo. Dev. Biol. 1993;157:526–538. doi: 10.1006/dbio.1993.1155. [DOI] [PubMed] [Google Scholar]
- Bork P., Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Bruskin A. M., Tyner A. L., Wells D. E., Showman R. M., Klein W. H. Accumulation in embryogenesis of five mRNAs enriched in the ectoderm of the sea urchin pluteus. Dev. Biol. 1981;87:308–318. doi: 10.1016/0012-1606(81)90154-8. [DOI] [PubMed] [Google Scholar]
- Calvete J. J., Carrera E., Sanz L., Topfer-Petersen E. Boar spermadhesins AQN-1 and AQN- 3, oligosaccharide and zona pellucida binding characteristics. Biol. Chem. 1996;377:521–527. doi: 10.1515/bchm3.1996.377.7-8.521. [DOI] [PubMed] [Google Scholar]
- Carroll D. J., Acevedo-Duncan M., Justice R. W., Santiago L. Structure, assembly and function of the surface envelope (fertilization envelope) from eggs of the sea urchins, Strongylocentrotus purpuratus. Adv. Exp. Med. Biol. 1986;207:261–291. doi: 10.1007/978-1-4613-2255-9_16. [DOI] [PubMed] [Google Scholar]
- Carroll E. J., Jr, Byrd E. W., Epel D. A novel procedure for obtaining denuded sea urchin eggs and observations on the role of the vitelline layer in sperm reception and egg activation. Exp. Cell Res. 1977;108:365–374. doi: 10.1016/s0014-4827(77)80043-8. [DOI] [PubMed] [Google Scholar]
- Correa L. M., Carroll E. J., Jr Characterization of the vitelline envelope of the sea urchin Strongylocentrotus purpuratus. Dev. Growth Differ. 1997;39:69–85. doi: 10.1046/j.1440-169x.1997.00008.x. [DOI] [PubMed] [Google Scholar]
- Crabb J. H., Jackson R. C. In vitro reconstitution of exocytosis from plasma membrane and isolated secretory vesicles. J. Cell Biol. 1985;101:2263–2273. doi: 10.1083/jcb.101.6.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detering N. K., Decker G. L., Schmell E. D., Lennarz W. J. Isolation and characterization of plasma membrane-associated cortical granules from sea urchin eggs. J. Cell Biol. 1977;75:899–914. doi: 10.1083/jcb.75.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostalova Z., Calvete J. J., Sanz L., Topfer-Petersen E. Boar spermadhesin AWN-1. Oligosaccharide and zona pellucida binding characteristics. Eur. J. Biochem. 1995;230:329–336. doi: 10.1111/j.1432-1033.1995.tb20567.x. [DOI] [PubMed] [Google Scholar]
- Engler-Blum G., Meier M., Frank J., Muller G. A. Reduction of background problems in nonradioactive northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- Epel D., Weaver A. M., Mazia D. Methods for removal of the vitelline membrane of sea urchin eggs. I. Use of dithiothreitol (Cleland Reagent) Exp. Cell Res. 1970;61:64–68. doi: 10.1016/0014-4827(70)90257-0. [DOI] [PubMed] [Google Scholar]
- Esser V., Limbird L. E., Brown M. S., Goldstein J. L., Russell D. W. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 1988;263:13282–13290. [PubMed] [Google Scholar]
- Ettensohn C. A., McClay D. R. Cell lineage conversion in the sea urchin embryo. Dev. Biol. 1988;125:396–409. doi: 10.1016/0012-1606(88)90220-5. [DOI] [PubMed] [Google Scholar]
- Gache C., Niman H. L., Vacquier V. D. Monoclonal antibodies to the sea urchin egg vitelline layer inhibit fertilization by blocking sperm adhesion. Exp. Cell Res. 1983;147:75–84. doi: 10.1016/0014-4827(83)90272-0. [DOI] [PubMed] [Google Scholar]
- Glabe C. G., Vacquier V. D. Isolation and characterization of the vitelline layer of sea urchin eggs. J. Cell Biol. 1977;75:410–421. doi: 10.1083/jcb.75.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Yu X., Rihani K., Wang Q. Y., Rong L. The role of a conserved acidic residue in calcium-dependent protein folding for a low density lipoprotein (LDL)-A module: implications in structure and function for the LDL receptor superfamily. J. Biol. Chem. 2004;279:16629–16637. doi: 10.1074/jbc.M400157200. [DOI] [PubMed] [Google Scholar]
- Haley S. A., Wessel G. M. Proteolytic cleavage of the cell surface protein p160 is required for detachment of the fertilization envelope in the sea urchin. Dev. Biol. 2004;272:191–202. doi: 10.1016/j.ydbio.2004.03.043. [DOI] [PubMed] [Google Scholar]
- Kamei N., Glabe C. G. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey W. H. Purification and properties of the egg plasma membrane. In: Schroeder T. E., editor. Methods in Cell Biology. vol. 27. Orlando, FL: Academic Press; 1986. pp. 139–152. [DOI] [PubMed] [Google Scholar]
- Kodama E., Baba T., Yokosawa H., Sawada H. cDNA cloning and functional analysis of ascidian sperm proacrosin. J. Biol. Chem. 2001;276:24594–24600. doi: 10.1074/jbc.M011370200. [DOI] [PubMed] [Google Scholar]
- Laitinen O. H., Airenne K. J., Marttila A. T., Kulik T., Porkka E., Bayer E. A., Wilchek M., Kulomaa M. S. Mutation of a critical tryptophan to lysine in avidin or streptavidin may explain why sea urchin fibropellin adopts an avidin-like domain. FEBS Lett. 1999;461:52–58. doi: 10.1016/s0014-5793(99)01423-4. [DOI] [PubMed] [Google Scholar]
- Lyons C. E., Payette K. L., Price J. L., Huang R. C. Expression and structural analysis of a teleost homolog of a mammalian zona pellucida gene. J. Biol. Chem. 1993;268:21351–21358. [PubMed] [Google Scholar]
- McGadey J. A tetrazolium method for non-specific alkaline phosphatase. Histochemie. 1970;23:180–184. doi: 10.1007/BF00305851. [DOI] [PubMed] [Google Scholar]
- Niman H. L., Hough-Evans B. R., Vacquier V. D., Britten R. J., Lerner R. A., Davidson E. H. Proteins of the sea urchin egg vitelline layer. Dev. Biol. 1984;102:390–401. doi: 10.1016/0012-1606(84)90203-3. [DOI] [PubMed] [Google Scholar]
- Ransick A., Ernst S., Britten R. J., Davidson E. H. Whole mount in situ hybridization shows Endo 16 to be a marker for the vegetal plate territory in sea urchin embryos. Mech. Dev. 1993;42:117–124. doi: 10.1016/0925-4773(93)90001-e. [DOI] [PubMed] [Google Scholar]
- Romero A., Romao M. J., Varela P. F., Kolln I., Dias J. M., Carvalho A. L., Sanz L., Topfer-Petersen E., Calvete J. J. The crystal structures of two spermadhesins reveal the CUB domain fold. Nat. Struct. Biol. 1997;4:783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- Rong L., Gendron K., Bates P. Conversion of a human low-density lipoprotein receptor ligand- binding repeat to a virus receptor: identification of residues important for ligand specificity. Proc. Natl. Acad. Sci. USA. 1998;95:8467–8472. doi: 10.1073/pnas.95.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Brown M. S., Goldstein J. L. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 1989;264:21682–21688. [PubMed] [Google Scholar]
- Shapiro B. M., Somers C. E., Weidman P. J. Extracellular remodeling during fertilization. In: Schatten H., Schatten G., editors. The Cell Biology of Fertilization. San Diego: Academic Press; 1989. [Google Scholar]
- Smith A. B. Phylogenetic relationship, divergence times, and rates of molecular evolution for camarodont sea urchins. Mol. Biol. Evol. 1988;5:345–365. [Google Scholar]
- Smith A. B., Lafay B., Christen R. Comparative variation of morphological and molecular evolution through geologic time: 28S ribosomal RNA versus morphology in echinoids. Philos Trans. R. Soc. Lond. B Biol. Sci. 1992;338:365–382. doi: 10.1098/rstb.1992.0155. [DOI] [PubMed] [Google Scholar]
- Somers C. E., Battaglia D. E., Shapiro B. M. Localization and developmental fate of ovoperoxidase and proteoliaisin, two proteins involved in fertilization envelope assembly. Dev. Biol. 1989;131:226–235. doi: 10.1016/s0012-1606(89)80054-5. [DOI] [PubMed] [Google Scholar]
- Somers C. E., Shapiro B. M. The heme environment of ovoperoxidase as determined by optical spectroscopy. J. Biol. Chem. 1989;264:17231–17235. [PubMed] [Google Scholar]
- Somers C. E., Shapiro B. M. Functional domains of proteoliaisin, the adhesive protein that orchestrates fertilization envelope assembly. J. Biol. Chem. 1991;266:16870–16875. [PubMed] [Google Scholar]
- Song J. L., Wong J. L., Wessel G. M. Oogenesis: single cell development and differentiation. Dev. Biol. 2007 doi: 10.1016/j.ydbio.2006.07.041. (in press) [DOI] [PubMed] [Google Scholar]
- Spurr A. A low viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., et al. A novel low-density lipoprotein receptor-related protein mediating cellular uptake of apolipoprotein E-enriched beta-VLDL in vitro. Biochemistry. 2000;39:15817–15825. doi: 10.1021/bi001583s. [DOI] [PubMed] [Google Scholar]
- Topfer-Petersen E., Petrounkina A. M., Ekhlasi-Hundrieser M. Oocyte-sperm interactions. Anim. Reprod. Sci. 2000;60–61:653–662. doi: 10.1016/s0378-4320(00)00128-7. [DOI] [PubMed] [Google Scholar]
- Topfer-Petersen E., Romero A., Varela P. F., Ekhlasi-Hundrieser M., Dostalova Z., Sanz L., Calvete J. J. Spermadhesins: a new protein family. Facts, hypotheses and perspectives. Andrologia. 1998;30:217–224. doi: 10.1111/j.1439-0272.1998.tb01163.x. [DOI] [PubMed] [Google Scholar]
- Varela P. F., Romero A., Sanz L., Romao M. J., Topfer-Petersen E., Calvete J. J. The 2.4 A resolution crystal structure of boar seminal plasma PSP-I/PSP-II: a zona pellucida-binding glycoprotein heterodimer of the spermadhesin family built by a CUB domain architecture. J. Mol. Biol. 1997;274:635–649. doi: 10.1006/jmbi.1997.1424. [DOI] [PubMed] [Google Scholar]
- Villacorta-Moeller M. N., Carroll E. J., Jr Sea urchin embryo fertilization envelope: immunological evidence that soluble envelope proteins are derived from cortical granule secretions. Dev. Biol. 1982;94:415–424. doi: 10.1016/0012-1606(82)90358-x. [DOI] [PubMed] [Google Scholar]
- Weidman P. J., Kay E. S., Shapiro B. M. Assembly of the sea urchin fertilization membrane: isolation of proteoliaisin, a calcium-dependent ovoperoxidase binding protein. J. Cell Biol. 1985;100:938–946. doi: 10.1083/jcb.100.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman P. J., Shapiro B. M. Regulation of extracellular matrix assembly: in vitro reconstitution of a partial fertilization envelope from isolated components. J. Cell Biol. 1987;105:561–567. doi: 10.1083/jcb.105.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D., Flugge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wessel G. M. A protein of the sea urchin cortical granules is targeted to the fertilization envelope and contains an LDL-receptor-like motif. Dev. Biol. 1995;167:388–397. doi: 10.1006/dbio.1995.1033. [DOI] [PubMed] [Google Scholar]
- Wessel G. M., Brooks J. M., Green E., Haley S., Voronina E., Wong J., Zaydfudim V., Conner S. The biology of cortical granules. Int. Rev. Cytol. 2001;209:117–206. doi: 10.1016/s0074-7696(01)09012-x. [DOI] [PubMed] [Google Scholar]
- Wessel G. M., Conner S., Laidlaw M., Harrison J., LaFleur G. J., Jr SFE1, a constituent of the fertilization envelope in the sea urchin is made by oocytes and contains low-density lipoprotein-receptor-like repeats. Biol. Reprod. 2000;63:1706–1712. doi: 10.1095/biolreprod63.6.1706. [DOI] [PubMed] [Google Scholar]
- Wessel G. M., Goldberg L., Lennarz W. J., Klein W. H. Gastrulation in the sea urchin is accompanied by the accumulation of an endoderm-specific mRNA. Dev. Biol. 1989;136:526–536. doi: 10.1016/0012-1606(89)90278-9. [DOI] [PubMed] [Google Scholar]
- Wessel G. M., Marchase R. B., McClay D. R. Ontogeny of the basal lamina in the sea urchin embryo. Dev. Biol. 1984;103:235–245. doi: 10.1016/0012-1606(84)90025-3. [DOI] [PubMed] [Google Scholar]
- Wong J. L., Créton R., Wessel G. M. The oxidative burst at fertilization is dependent upon activation of the dual oxidase Udx1. Dev. Cell. 2004;7:801–814. doi: 10.1016/j.devcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Wong J. L., Wessel G. M. Major components of a sea urchin block to polyspermy are structurally and functionally conserved. Evol. Dev. 2004;6:134–153. doi: 10.1111/j.1525-142X.2004.04019.x. [DOI] [PubMed] [Google Scholar]
- Wong J. L., Wessel G. M. Defending the zygote: search for the ancestral animal block to polyspermy. Curr. Top Dev. Biol. 2006;72:1–151. doi: 10.1016/S0070-2153(05)72001-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.