Abstract
Glycosylphoshatidylinositol (GPI) anchors are remodeled during their transport to the cell surface. Newly synthesized proteins are transferred to a GPI anchor, consisting of diacylglycerol with conventional C16 and C18 fatty acids, whereas the lipid moiety in mature GPI-anchored proteins is exchanged to either diacylglycerol containing a C26:0 fatty acid in the sn-2 position or ceramide in Saccharomyces cerevisiae. Here, we report on PER1, a gene encoding a protein that is required for the GPI remodeling pathway. We found that GPI-anchored proteins could not associate with the detergent-resistant membranes in per1Δ cells. In addition, the mutant cells had a defect in the lipid remodeling from normal phosphatidylinositol (PI) to a C26 fatty acid–containing PI in the GPI anchor. In vitro analysis showed that PER1 is required for the production of lyso-GPI, suggesting that Per1p possesses or regulates the GPI-phospholipase A2 activity. We also found that human PERLD1 is a functional homologue of PER1. Our results demonstrate for the first time that PER1 encodes an evolutionary conserved component of the GPI anchor remodeling pathway, highlighting the close connection between the lipid remodeling of GPI and raft association of GPI-anchored proteins.
INTRODUCTION
A number of proteins are attached to the cell surface via glycosylphosphatidylinositol (GPI) anchors. Modification by GPI is a conserved posttranslational modification in yeast, protozoans, plants, and animals (Ferguson, 1999; McConville and Menon, 2000; Gillmor et al., 2005; Pittet and Conzelmann, 2006). More than 150 plasma membrane proteins are known to be anchored by GPI in mammalian cells. GPI-anchored proteins are functionally diverse and important for signal transduction, cell–cell interaction, cell adhesion, and host defense (Kinoshita et al., 1995). In the genome of Saccharomyces cerevisiae, more than 60 genes are predicted to encode GPI-anchored proteins. These proteins play important roles in cell wall biogenesis and assembly (Kapteyn et al., 1999).
The biosynthesis and attachment of GPI is carried out on the membrane of the endoplasmic reticulum (ER). The many genes involved in the GPI biosynthetic pathway have been well characterized in mammalian and yeast cells (Kinoshita and Inoue, 2000; Eisenhaber et al., 2003; Pittet and Conzelmann, 2006). The GPI anchor is synthesized in the ER by the stepwise addition of sugars, an acyl chain, and ethanolamine-phosphates to phosphatidylinositol (PI). When a complete GPI proprotein has been synthesized, the GPI transamidase complex removes the C-terminal GPI attachment signal peptide and links the thus generated new C-terminal end to the ethanolamine-phosphate of the complete GPI precursor lipid (Kinoshita and Inoue, 2000; Eisenhaber et al., 2003). After the attachment of GPI to the protein, the acyl group on the inositol residue of the GPI anchor is eliminated by Bst1p, a process required for the quality control of GPI-anchored proteins and their efficient transport from the ER to the Golgi (Tanaka et al., 2004; Fujita et al., 2006).
GPI-anchored proteins are transported from the ER to the Golgi apparatus via vesicles that are distinct from those containing other proteins such as the general amino acid permease Gap1p and pro-alpha factors in yeast (Muniz et al., 2001; Mayor and Riezman, 2004; Watanabe and Riezman, 2004). GPI-anchored proteins are also selectively transported to the apical surface in mammalian polarized cells (Schuck and Simons, 2006). The sorting in the secretory pathway seems to correlate with their association with microdomains, generally called lipid rafts (Brown and Rose, 1992; Mayor and Riezman, 2004). The lipid rafts are assumed to be sphingolipid- and sterol-rich membrane and can be biochemically isolated as a detergent-resistant membrane (DRM) fraction (Brown and Rose, 1992; Simons and Ikonen, 1997). There is not a good single definition of lipid rafts, but particular lipids form specific membrane structures that function as a platform for intracellular signaling and are required for the selective transport of proteins (Simons and Ikonen, 1997; Simons and Toomre, 2000; Mayor and Riezman, 2004). Although GPI-anchored proteins are one of the major components of lipid rafts, little is known about how GPI-anchored proteins are incorporated into and associate with them. In yeast, GPI-anchored proteins associate with the DRM fraction in the ER (Bagnat et al., 2000), whereas, in mammalian cells, the process takes place in the Golgi complex (Simons and Ikonen, 1997; Brown and London, 1998). This might be due to a difference in the compartment in which GPI lipid remodeling occurs.
In both yeast and mammalian cells, the lipid moieties of GPI are modified after transfer to proteins (Conzelmann et al., 1992; Tashima et al., 2006). In mammalian cells, this occurs during the transport of GPI anchors to the cell surface, but the precise modifications they undergo remain unclear. PGAP2, which is involved in lipid remodeling of GPI, is mainly found in the Golgi (Tashima et al., 2006), suggesting that the Golgi is the site of remodeling. On the other hand, in yeast, the lipid moieties of GPI-anchored proteins are exchanged mainly in the ER and somewhat in the Golgi (Sipos et al., 1997).
GPI is synthesized from conventional phosphatidylinositols (PIs), which contain an unsaturated fatty acid chain at the sn-2 position, whereas mature GPI-anchored proteins have diacylglycerol with a long-chain fatty acid in sn-2 or ceramides (Sipos et al., 1997). The molecular mechanisms of this pathway, however, are not well understood. A recent report showed that Gup1p is required for the addition of C26 fatty acids to the sn-2 position of GPI-anchored proteins and that the lyso-forms of GPI-anchors accumulate in gup1Δ cells (Bosson et al., 2006). It is also reported that GPI-anchored proteins harboring lyso-GPI are transported to the plasma membrane in PGAP2-deficient mammalian cells (Tashima et al., 2006). These reports suggest that an enzyme with phospholipase A2 (PLA2)-like activity removes an acyl-chain at the sn-2 position of GPI anchors during the remodeling of GPI in both yeast and mammals.
In this study, we found that yeast PER1, which was originally isolated as a gene involved in the unfolded protein response (Ng et al., 2000), is required for the GPI-PLA2 activity involved in the lipid remodeling of GPI-anchored proteins. Human PERLD1 fully complemented per1Δ phenotypes, indicating that a similar pathway exists in human. Our results further suggest that lipid remodeling of the GPI anchor is required for both the efficient transport of GPI-anchored proteins from the ER to the Golgi and their association with lipid rafts. Per1p and Gup1p are key molecules in these processes, demonstrating that long-chain fatty acids are necessary for the proper processing of GPI-anchored proteins.
MATERIALS AND METHODS
Media and Strains
YPAD and synthetic complete media are described elsewhere (Sherman, 1991). SDCA contains 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI), 2% glucose, 0.5% casamino acids, and 0.004% adenine sulfate. The yeast strains used in this study were gup1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gup1Δ::KanMX4; EUROSCARF), per1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 per1Δ::KanMX4; EUROSCARF), gup1Δ per1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gup1Δ::his5+ per1Δ::KanMX4; this study), YJY1 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; laboratory strain of the S288C background), and gpi7Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gpi7Δ::KanMX4; EUROSCARF). Gene disruption in yeast was performed using a one-step method as described previously (Longtine et al., 1998).
Plasmids
Construction of PER1, PER1-HA, and PER1-mRFP Plasmids.
The promoter and coding sequences of PER1 were cloned by amplification of genomic DNA using the primers PER1F (5′-AAAAACTAGTTGGAACATTGCACAAAGG-3′) and PER1R-NheI (5′-AAAAAAGCTTTTAGCTAGCGTACAATTGTCTATTACCCCAA-3′). The amplified fragment was digested with SpeI and HindIII and then purified. The purified fragment was ligated into pRS316T (CEN, URA3), which contains the GPI7 terminator region inserted into the multiple cloning site of XhoI/KpnI-digested pRS316 (Sikorski and Hieter, 1989; Fujita et al., 2004) to generate pMF917 (PER1, CEN, URA3). Three copies of the HA epitope were amplified and inserted into the NheI site of pMF917 to generate pMF918 (PER1-HA, CEN, URA3). The DNA fragment containing monomeric red fluorescent protein (mRFP; kindly provided by Dr. Roger Tsien, University of California, San Diego, La Jolla, CA) was also amplified and inserted into the NheI site of pMF917 to generate pMF921 (PER1-mRFP, CEN, URA3).
Construction of Mutated per1-HA Plasmids.
We substituted S19, H102, K104, S118, S122, S173, H177, D315, and H326 with alanine using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and pMF918 as the template, generating pMF931 (per1-HA-S19A, CEN, URA3), pMF932 (per1-HA-H102A, CEN, URA3), pMF933 (per1-HA-K104A, CEN, URA3), pMF934 (per1-HA-S118A, CEN, URA3), pMF935 (per1-HA-S122A, CEN, URA3), pMF936 (per1-HA-S173A, CEN, URA3), pMF937 (per1-HA-H177A, CEN, URA3), pMF939 (per1-HA-D315A, CEN, URA3), and pMF940 (per1-HA-H326A, CEN, URA3), respectively.
Construction of PERLD1 Plasmid.
The open reading frame of the PERLD1 gene was amplified from the plasmid of the NIH mammalian gene collection clone (No. 3855206) using the primers PERLD1-F (5′-AAAAGAATTCATGGCCGGCCTGGCGGCG-3′) and PERLD1-R (5′-AAAAGTCGACTCAGTCCAGCTTGAACTTGTCC-3′). The amplified fragment was digested with EcoRI and SalI and then purified. The purified fragment was ligated into EcoRI/SalI-digested YEp352GAPII (Abe et al., 2003) to generate pMF942 (pTDH3-PERLD1, 2μ, URA3).
Construction of GFP-CWP2 Plasmid.
Green fluorescent protein (GFP)-tagged CWP2 plasmid was constructed from YEp51-ssGFP-GPI, which contains GFP-fused CWP2 under control of the GAL10 promoter (kindly provided by Drs. Kappei Tsukahara and Koji Sagane, Esai Co., Tokyo, Japan). We changed the promoter to the CWP2 promoter and subcloned the GFP-CWP2 fragment into pRS316 to generate pMF500 (GFP-CWP2, CEN, URA3).
Construction of mRFP-GAS1 and FLAG-GAS1 Plasmid.
Using pMF600 (Fujita et al., 2006) as a template, MluI and NdeI sites were introduced 69 base pairs downstream from the start codon of GAS1, and the fragment was subcloned into pRS315 (Sikorski and Hieter, 1989) to generate pMF605. The DNA fragment containing mRFP was amplified and inserted into the MluI-NdeI site of pMF605 to generate pMF923 (mRFP-GAS1, CEN, LEU2). Three copies of the FLAG epitope were amplified, inserted into the MluI-NdeI site of pMF605 to generate pMF924 (FLAG-GAS1, CEN, LEU2), and confirmed by sequencing.
Immunoblotting
Samples were denatured with SDS-sample buffer for 1 h at 4°C or for 10 min at 37°C for membrane proteins or 5 min at 95°C for soluble proteins. Protein samples (5 μl) were then separated by SDS-PAGE and electrophoretically transferred to a PVDF membrane. Gas1p was detected with anti-Gas1 peptide polyclonal antibody (1:2000; kindly provided by Dr. Katsura Hata, Eisai Co., Tokyo, Japan), and followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2000; Cell Signaling Technology, Danvers, MA). Per1-HA was detected with anti-HA mAb 16B12 (1:2000; Babco, Richmond, CA), followed by HRP-conjugated goat anti-mouse IgG (1:2000; Cell Signaling Technology). Dpm1p was detected with anti-Dpm1p mAb (1:2000; Invitrogen, Carlsbad, CA), followed by HRP-conjugated anti-mouse IgG (1:2000). Och1p was detected with anti-Och1p polyclonal antibody (1:2000; Nakayama et al., 1992), followed by HRP-conjugated anti-rabbit IgG (1:2000). Pgk1p was detected with anti-Pgk1p mAb (1:10,000; Invitrogen), followed by HRP-conjugated anti-mouse IgG (1:10,000). Prc1p was detected with anti-CPY mAb (1:5000; Invitrogen), followed by HRP-conjugated anti-mouse IgG (1:5000). Pho8p was detected with anti-Pho8p mAb (1:1000; Invitrogen), followed by HRP-conjugated anti-mouse IgG (1:1000). FLAG-tagged Gas1p (Flag-Gas1p) was detected with anti-FLAG mAb M2 (1:10,000; Sigma-Aldrich, St. Louis, MO), followed by HRP-conjugated goat anti-mouse IgG (1:10,000). Immunoreactive bands were visualized by chemiluminescence with ECL-plus reagents (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Subcellular Fractionation
Subcellular fractionation was performed as described previously with some modifications (Nishikawa et al., 1990). Cells were grown to 107 cells/ml in YPAD medium at 30°C and converted to spheroplasts as described previously (Nishikawa and Nakano, 1991). Spheroplasts (5 × 109) were suspended in 10 ml of ice-cold lysis buffer (0.3 M sorbitol, 0.1 M KCl, 50 mM Tris-HCl, pH 7.5, 1 mM EGTA, and 1 mM phenylmethylsulfonyl fluoride). The suspensions were vortexed for 10 s with glass beads in chilled Corex tubes and rested for 10 s on ice. The vortex was repeated 10 times. The lysates were subjected to the following centrifugation steps (all at 4°C): 300 × g for 5 min, 13,000 × g for 15 min, and 100,000 × g for 60 min. Aliquots from the 13,000 × g pellet and the 100,000 × g pellet and supernatant fractions were analyzed by SDS-PAGE and immunoblotting for Per1-HA.
Fluorescence Microscopy
To image Per1-mRFP, mRFP-Gas1, and GFP-Cwp2 proteins, cells grown to the exponential phase were collected and washed with synthetic complete medium. Fluorescence images were obtained using a BX50 fluorescence microscope (Olympus, Tokyo, Japan) and photographed with a microMax cooled CCD camera (Princeton Instruments, Trenton, NJ).
Pulse-Chase Experiments for Gas1p and CPY Maturation
Radiolabeling and immunoprecipitation to measure Gas1p and CPY maturation were performed as described previously (Sutterlin et al., 1997). Samples were separated by SDS-PAGE and analyzed using a Molecular Imager FX (Bio-Rad, Hercules, CA).
3H-Inositol Labeling of Lipids
Lipids from 3H-inostiol–labeled cells were extracted as described previously (Guillas et al., 2000), desalted by butanol/water partitioning, and separated by TLC using solvent 1 (10:10:3 CHCl3/CH3OH/H2O) for analysis of GPI intermediates or solvent 2 (55:45:5 CHCl3/CH3OH/0.25% KCl) for analysis of sphingolipids.
Isolation of DRMs
Cells were grown at 30°C in YPAD to the exponential phase, and 3 × 108 cells were collected. DRMs for the Gas1p analysis were isolated as described previously with a slight modification (Bagnat et al., 2000; Okamoto et al., 2006). After incubation with 1% Triton X-100 for 30 min on ice, the lysates were subjected to Optiprep density gradient floatation by centrifugation for 2 h at 40,000 rpm in a SW55Ti rotor (Beckman, Fullerton, CA). After centrifugation, six fractions of equal volume were collected starting from the top. Each fraction was mixed with sample buffer and subjected to SDS-PAGE and immunoblotting.
Isolation of the Lipid Moieties of GPI Anchors
3H-inositol–labeled PI moieties were prepared from GPI-anchored proteins as described previously (Sipos et al., 1997; Guillas et al., 2000). Lipids were analyzed by TLC on silica gel 60 plates using solvent 3 (55:45:10 CHCl3/CH3OH/0.25% KCl).
Purification of Flag-Gas1p and Separation Using an Octyl-FF Column
Cells were grown in YPAD medium, and 8 × 109 cells were collected, washed, and resuspended in TNE buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail; Roche, Basel, Switzerland). The cell suspension was broken, and the debris was removed. The lysate was centrifuged at 10,000 × g for 20 min, and the pellets were resuspended in 900 μl TNE buffer, mixed with 100 μl 10× NP-40 (final 1%), and mixed with end-over-end rotation for 1 h at 4°C. The suspension was centrifuged at 10,000 × g for 20 min to remove insoluble membranes. The supernatant was mixed with 100 μl anti-FLAG beads (Sigma-Aldrich) and then incubated for 3 h at 4°C. The beads were then collected and washed, and Flag-Gas1p was eluted twice with 100 μl TNE buffer containing 0.1% NP-40 and 0.5 mg/ml 3× FLAG peptide (Sigma). A 10-μl aliquot of the Flag-Gas1p elute was added to 500 μl buffer C (0.1 M ammonium acetate, 5% 1-propanol, and 0.03% NP-40), loaded onto an Octyl-FF column (GE Healthcare), and separated using an AKTA prime protein purification system (GE Healthcare) at a flow rate of 0.5 ml/min and a gradient of 5–100% 1-propanol. Fractions of 1 ml were collected, dried, subjected to SDS-PAGE, and analyzed by immunoblotting using an anti-FLAG antibody, followed by HRP-conjugated anti-mouse IgG antibody.
PLA2 Treatment of Flag-Gas1p from per1Δ Cells
Flag-Gas1p purified from per1Δ cells was suspended in 500 μl buffer D (100 mM Tris-HCl, pH 7.5, 10 mM CaCl2, and 0.1% NP-40). After addition of bee venom PLA2 (Sigma) or buffer, the reaction mixture was incubated overnight at 37°C. The reaction was stopped by adding NaN3/NaF solution (final 10 mM). Anti-FLAG beads (20 μl) were added, and after a 3-h incubation at 4°C, the beads were collected by centrifugation and washed with TNE buffer containing 0.1% NP-40. Flag-Gas1p was eluted twice with 100 μl TNE buffer containing 0.1% NP-40 and 0.5 mg/ml 3× FLAG peptide (Sigma). The resulting repurified Flag-Gas1p was suspended in 400 μl buffer C, loaded onto an Octyl-FF, and analyzed by immunoblotting.
In Vitro Analysis of GPI-PLA2 Activity
Cells (2 × 108) were washed in TM buffer (100 mM Tris-HCl, pH 7.5, and 10 mM MgCl2), resuspended in 200 μl TM buffer, and broken with glass beads. Insoluble components were removed from the cell lysate, and the supernatant was centrifuged at 10,000 × g for 20 min. The pellet containing microsomes was resuspended in 100 μl TM buffer and stored at −80°C until use. Reaction mixtures containing 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM CaCl2, 1 mM dithiothreitol, 5 mM ATP, 30 μl Flag-Gas1p from per1Δ cells, and 50-μl microsome suspension was incubated on ice for 10 min. The mixture was incubated at 37°C for 30 min with end-over-end mixing. The reaction was stopped by the addition of NaN3/NaF solution (final 10 mM), after which 700 μl TNE buffer and 100 μl 10% NP-40 were added. The mixture was incubated for 1 h at 4°C with end-over-end mixing, centrifuged at 10,000 × g for 15 min at 4°C to remove the insoluble membranes, and mixed with 20 μl anti-FLAG beads. After incubation for 3 h at 4°C, the beads were collected by centrifugation and washed, and Flag-Gas1p was eluted twice with 100 μl TNE buffer containing 0.1% NP-40 and 0.5 mg/ml 3× FLAG peptide (Sigma). The resulting repurified Flag-Gas1p was suspended in 400 μl buffer C, loaded onto an Octyl-FF column, and analyzed by immunoblotting.
RESULTS
Per1p Is a Conserved Membrane Protein Localized in the ER
We previously found that inositol deacylation of the GPI anchor by Bst1p is required for both the efficient transport and the quality control of GPI-anchored proteins (Tanaka et al., 2004; Fujita et al., 2006). To elucidate the molecular mechanisms more precisely, we attempted to find novel genes related to BST1. Schuldiner et al. (2005) published comprehensive genetic-interaction maps, called E-MAPs (epistatic miniarray profiles). From the E-MAP analysis of the early secretory pathway, two uncharacterized genes, GUP1 and PER1, were found to genetically interact with BST1. Recently, it was reported that GUP1 encodes the O-acyltransferase involved in lipid remodeling of GPI-anchored proteins (Bosson et al., 2006). The function of PER1, however, has not been established.
PER1 was originally found as COS16, a mutational suppressor of the cdc1 mutant (Paidhungat and Garrett, 1998). Genetic analysis suggests that Cos16p participates in Mn2+ homeostasis in the vacuole; however, global localization analysis of GFP-tagged proteins showed that Per1p is localized in the ER (Tong et al., 2004). PER1 was also isolated by genetic screening as a gene involved in the unfolded protein response and protein folding (Ng et al., 2000). Mutation in PER1 showed a synthetic negative phenotype with IRE1, which is a sensor of the unfolded protein response in the ER lumen. MCD4, GPI10, BST1, and ERI1, all of which are required for the biosynthesis or modification of GPI, were also isolated from the synthetic negative screening with IRE1 (Ng et al., 2000; Ng, 2005), suggesting that PER1 is related to GPI modification. Although these genetic analyses indicate that PER1 is linked to several phenomena, the molecular function of Per1p has remained unknown.
We first characterized the phenotypes of per1 mutant cells. The per1Δ cells showed calcofluor white (CFW) sensitivity and temperature sensitivity at 37°C (Figure 1A), suggesting a defect in cell wall integrity. We next investigated the cellular localization of Per1p. The PER1 gene is predicted to encode a 357-amino acid membrane protein. Kyte-Doolittle hydropathy analysis and SOSUI suggest that Per1p has seven transmembrane domains. In addition, cellular fractionation studies suggest that Per1p is localized in the vacuole, and microscopic analysis of the GFP-fused protein indicates that it is present in the ER (Paidhungat and Garrett, 1998; Tong et al., 2004). In the current studies, we constructed C-terminally HA- and mRFP-tagged versions of Per1p to more precisely determine its subcellular localization. Both Per1-HA and Per1-mRFP rescued the CFW and temperature sensitivity of per1Δ cells, indicating that the tagged proteins were fully functional (Figure 1A). Microscopic analysis suggested that Per1-mRFP is mainly present in an intracellular compartment, probably the ER (Figure 1B). We further performed subcellular fractionation to examine the intracellular location in more detail. We detected Per1-HA as a 35-kDa protein mainly in the low-speed (13,000 × g) pellet (Figure 1C). This fraction typically contains large, dense membranes such as the ER and plasma membrane. The distribution was nearly identical to that of Dpm1p, an ER marker protein (Figure 1C). Och1p, a marker of Golgi-localized proteins, was founded in the high-speed (100,000 × g) pellet, and Pgk1p, a marker of cytosolic proteins, was present in the high-speed supernatant (Figure 1C). We also constructed FLAG-tagged versions of Per1p and examined its subcellular localization by fractionation. Like Per1-HA, Per1-FLAG was present mainly in the low-speed pellet (unpublished data). Collectively, these data suggest that Per1p resides in the ER.
Figure 1.
Per1p is mainly localized in the ER. (A) Tagged Per1 proteins are functional and complement the CFW sensitivity and the temperature sensitivity of per1Δ cells. Isogenic per1Δ cells carrying the pRS316T (vector), pMF917 (PER1), pMF918 (PER1-HA), and pMF921 (PER1-mRFP) were spotted on YPAD or YPAD plates containing 7.5 μg/ml CFW after making a 5× serially diluted solution and then incubating for 2 d at 30 or 37°C. (B) Localization of Per1-mRFP. per1Δ cells carrying pMF921 were visualized by fluorescence microscopy. DIC, differential interference contrast. (C) Fractionation of Per1-HA. Exponentially growing per1Δ cells carrying pMF918 were converted to spheroplasts, homogenized, and subjected to the following series of centrifugations: 300 × g for 5 min, 13,000 × g for 15 min, and 100,000 × g for 60 min. Aliquots were taken from the 13,000 × g pellet (P13) and the 100,000 × g pellet (P100) and supernatant (S100) and then analyzed by immunoblotting.
Per1p Is required for the Efficient Transport of GPI-anchored Proteins
E-Map analysis predicted that PER1 may be involved in O-linked glycan synthesis or GPI biosynthesis (Schuldiner et al., 2005). Therefore, we next investigated whether PER1 encodes a gene related to GPI biosynthesis or transport. For this purpose, we examined the effect of gup1 and per1 deletion on the status of Gas1p, one of the most abundant GPI-anchored proteins in yeast. The ER form of Gas1p (105 kDa) is transported to the Golgi, where it is elongated with N- and O-glycans to form the Golgi-modified form of Gas1p (125 kDa; Popolo and Vai, 1999). Immunoblotting revealed that, although the Golgi form predominates in wild-type cells, only low levels of the Golgi and ER forms were detected in gup1Δ and per1Δ cells (Figure 2A). Immunoblotting for Dpm1p and carboxypeptidase Y (CPY) indicated nearly the same amounts of matured CPY in wild-type, gup1Δ, and per1Δ cells (Figure 2A). We further examined the maturation of Gas1p and CPY by pulse-chase analysis. The maturation of Gas1p was rapid in wild-type cells, with a half-time of <15 min, whereas its maturation was delayed in gup1Δ cells, as observed previously (Figure 2B; Bosson et al., 2006). In per1Δ cells, maturation of Gas1p was significantly delayed, with a half-time of ∼30 min (Figure 2B). In contrast, CPY was normally transported and processed with wild-type kinetics in gup1Δ and per1Δ cells (Figure 2B), indicating that PER1 is specifically required for the maturation of GPI-anchored proteins.
Figure 2.
Transport of GPI-anchored proteins is delayed in per1Δ cells. (A) The amount of Gas1p is significantly reduced in gup1Δ and per1Δ cells. Wild-type (WT), gup1Δ, and per1Δ cells were grown overnight to midlog phase. Equal cell numbers were lysed, and total protein extracts were analyzed by immunoblotting using anti-Gas1p, anti-Dpm1p, and anti-CPY. ER, the ER (105 kDa) form of Gas1p; Golgi, the Golgi (125 kDa) form of Gas1p; p1, precursor (ER) form of CPY; p2, precursor (Golgi) form of CPY; mature, mature (vacuolar) form of CPY. (B) ER-to-Golgi transport of Gas1p is specifically delayed in gup1Δ and per1Δ cells. WT, gup1Δ, and per1Δ cells were labeled with [35S]methionine and cysteine and then chased for the indicated periods of time. Gas1p and CPY were immunoprecipitated from cell lysates and separated by SDS-PAGE.
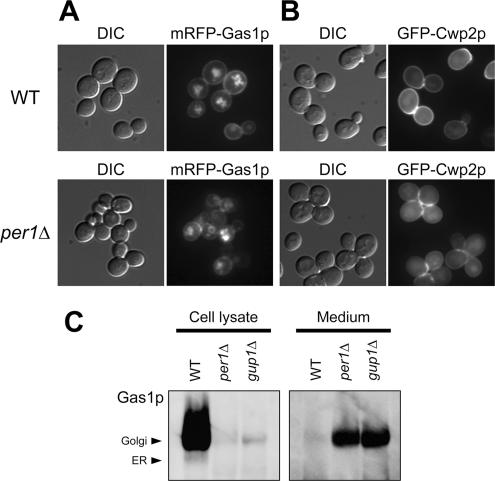
Next, we analyzed the cellular localization of GPI-anchored proteins in wild-type and per1Δ cells. We selected and observed two types of GPI-anchored proteins, mRFP-fused Gas1p, which is localized at the plasma membrane (Fujita et al., 2006), and GFP-fused Cwp2p, which is localized in the cell wall (Ram et al., 1998). As observed previously (Fujita et al., 2006), mRFP-Gas1p was present at the plasma membrane in wild-type cells (Figure 3A, top panel). In per1Δ cells, mRFP-Gas1p was localized at the plasma membrane, but the level of fluorescence was weaker than that in wild-type cells (Figure 3A, bottom panel). Although we also observed mRFP-Gas1p in the intracellular compartments both in WT and per1Δ cells, mRFP-Gas1p was particularly seen in the ER in per1Δ cells (Figure 3A, bottom panel). GFP-Cwp2p was clearly localized at the cell surface in wild-type cells (Figure 3B, top panel), whereas in per1Δ cells, it was localized at the cell surface, but the fluorescence intensity was much lower than in wild-type cells (Figure 3B, bottom panel). The per1Δ cells also aggregated, which is a phenotype frequently observed in cells with defects in the cell wall. Immunoblotting revealed only a small amount of matured Gas1p in per1Δ and gup1Δ cells (Figure 2A). In gup1Δ cells, a significant amount of Gas1p is lost from the plasma membrane into the culture medium (Bosson et al., 2006). We found here that a similar amount of Gas1p was released into the medium in per1Δ cells as in gup1Δ cells (Figure 3C). These results suggest that GPI-anchored proteins are not correctly transported and localized at the cell surface in per1Δ cells.
Figure 3.
GPI-anchored proteins are not correctly transported to the cell surface in per1Δ cells. Wild-type (WT) and per1Δ cells carrying pMF923 (mRFP-GAS1) (A) or WT and per1Δ cells carrying pMF500 (GFP-CWP2) (B) were visualized by fluorescence microscopy. DIC, differential interference contrast. (C) Gas1p is secreted into the medium in per1Δ cells. Cells (1 × 108) were grown to the exponential phase and harvested. Secreted proteins were precipitated from the medium using trichloroacetic acid (final 10%). Cell lysates and secreted proteins were analyzed by immunoblotting with an anti-FLAG antibody.
PER1 Is Not Involved in the Biosynthesis of the GPI Anchor or Ceramide
The delay in transport of GPI-anchored proteins from the ER to the Golgi can be caused at least by three defects. First, a failure in GPI anchor biosynthesis or GPI attachment to proteins would delay the exit of GPI-anchored proteins from the ER. Second, the delay could be caused by disruption of proteins specifically required for the ER-to-Golgi transport of GPI-anchored proteins. For instance, defects in ceramide synthesis are known to specifically affect ER-to-Golgi transport of GPI-anchored proteins (Watanabe et al., 2002). Third, disruption of proteins involved in remodeling of GPI after its attachment to proteins in the ER could delay the exit of GPI-anchored proteins from the ER. For example, BST1 and GUP1 encode proteins that function after GPI attachment and are required for the efficient transport of GPI-anchored proteins from the ER (Tanaka et al., 2004; Bosson et al., 2006).
To distinguish which of these defects accounts for the delay in the transport of GPI-anchored proteins in per1Δ cells, we first examined the accumulation of GPI intermediates and sphingolipids. Lipids from 3H-inositol–labeled cells were separated by TLC. Wild-type and gup1Δ cells did not accumulate GPI intermediates, whereas gpi7Δ cells accumulated a lipid intermediate (M4 in Figure 4A; Benachour et al., 1999; Fujita et al., 2004). In addition, intermediates were not accumulated in per1Δ or gup1Δ per1Δ cells, suggesting that PER1 is not involved in the biosynthesis of the GPI anchor or its attachment to protein (Figure 4A). Because sphingolipids are required for the stable membrane association and efficient transport of GPI-anchored proteins (Watanabe et al., 2002), we also analyzed sphingolipid biosynthesis. Lipid extracts from per1Δ cells and gup1Δ per1Δ cells showed identical TLC patterns as those from gup1Δ and gpi7Δ cells and contained normal amounts of mannose inositolphosphorylceramide and mannose diinositolphosphorylceramide, indicating that ceramide and sphingolipid synthesis is normal in per1Δ cells (Figure 4B). These results suggest that the delay in the transport of GPI-anchored proteins in per1Δ cells is not due to changes in GPI or ceramide biosynthesis.
Figure 4.
The biosynthesis of GPI and ceramide synthesis is normal in per1Δ cells. Cells (2 × 108) were labeled with [3H]inositol (50 μCi) for 2 h at 30°C. Lipids were extracted from wild-type (WT), gup1Δ, per1Δ, gup1Δ per1Δ, and gpi7Δ cells, desalted, and analyzed by TLC using (A) solvent 1 (10:10:3 CHCl3/CH3OH/H2O) or (B) solvent 2 (55:45:5 CHCl3/CH3OH/0.25% KCl). PI, phosphatidylinositol; M(IP)2C, mannose diinositolphosphorylceramide; M4, GPI intermediate accumulated in gpi7Δ cells; IPCs, inositolphosphorylceramides; MIPC, mannose inositolphosphorylceramide; lyso-PI, lyso form of PI.
PER1 Is Required for the Raft Association of GPI-anchored Proteins
GPI-anchored proteins are one of the components of lipid rafts at the plasma membrane in eukaryotes. Because these compartments are generated in the ER in yeast (Bagnat et al., 2000), we next addressed whether GPI-anchored proteins are sorted to the DRMs in per1Δ cells. Wild-type, gup1Δ, and per1Δ cells were disrupted and extracted with 1% Triton X-100 at 4°C. The extracts were then fractionated by centrifugation on an Optiprep density gradient. In wild-type cells, Gas1p was located predominantly in fraction 2, which corresponded to DRMs (Figure 5). Gas1p derived from gup1Δ cells was entirely located in the detergent-soluble fractions as previously reported (Figure 5; Bosson et al., 2006), indicating that a change in the lipid moiety of the GPI-anchored proteins alters the properties of the lipid rafts. In per1Δ cells, there was a dramatic decrease in the amount of Gas1p associated with DRMs (Figure 5). The plasma membrane proton ATPase Pma1p, a known DRM-associated protein, was primarily located in the DRMs in wild-type cells (Figure 5). Pma1p was also mostly associated with the DRMs in gup1Δ and per1Δ cells, indicating that raft formation itself is normal (Figure 5). The alkaline phosphatase Pho8p, a type II membrane protein at the vacuole, was used as a marker of detergent-soluble fractions. The fractionation patterns of Pho8p were almost same in these cells (Figure 5). These results indicate that Per1p is required for the association of GPI-anchored proteins with lipid rafts and that it might be involved in the remodeling of their lipids.
Figure 5.
GPI-anchored proteins are not associated with DRMs in per1Δ cells. Cells were grown to the exponential phase in YPAD medium. Wild-type (WT), gup1Δ, per1Δ cells were disrupted with glass beads, extracted with Triton X-100, and subjected to Optiprep density gradient centrifugation. Six fractions were collected and analyzed by immunoblotting with antibodies against Gas1p, Pma1p, and Pho8p. Pma1p was used as a marker for DRM-associated proteins, and Pho8p was used as a marker for non-DRM proteins.
The per1Δ Cells Have a Defect in Lipid Remodeling of the GPI Anchor
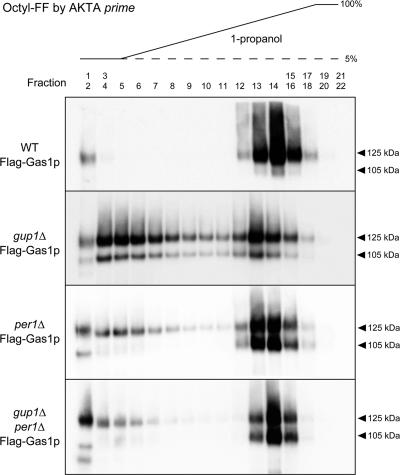
The fractionation of DRMs suggested that the lipid moiety of GPI is somehow abnormal in per1Δ cells. To examine this further, we constructed Flag-Gas1p, in which a 3× FLAG-tag was inserted in Gas1p after the cleavable N-terminal signal sequence. Immunoblotting revealed that Flag-Gas1p was normally matured in wild-type cells. We analyzed the hydrophobicity of the proteins by fractionation on octyl-Sepharose using elution with a 1-propanol gradient. Flag-Gas1p purified from wild-type cells eluted in fractions 13–16 (Figure 6, WT), whereas Flag-Gas1p purified from gup1Δ cells eluted in fractions 3–7 and 13–14 (Figure 6, gup1Δ). In gup1Δ cells, it is reported that most GPI-anchored proteins harbor a lyso-form lipid (Bosson et al., 2006). We therefore suspected that the lipid moiety of the earlier fractions of Flag-Gas1p in the gup1Δ cells is lyso-GPI. We further found that Flag-Gas1p purified from per1Δ cells (Figure 6, per1Δ) showed a different chromatographic profile than Flag-Gas1p from wild-type and gup1Δ cells. Both the ER and Golgi forms of Flag-Gas1p mainly eluted in fractions 13–16, but only the Golgi form eluted in the slightly earlier fractions (fractions 3–7). These results are discussed further in the Discussion. Flag-Gas1p purified from gup1Δ per1Δ double-mutant cells eluted at similar fractions as that purified from per1Δ cells (Figure 6, gup1Δ per1Δ), strongly supporting the idea that Per1p functions before Gup1p, which is a C26-fatty acid acyltransferase for lyso-GPI.
Figure 6.
Per1p acts before the Gup1p function. Cells producing Flag-Gas1p were grown to the exponential phase in YPAD medium, and Flag-Gas1p was purified from the cell lysate with anti-FLAG beads and eluted with 3× FLAG-peptide. Flag-Gas1p purified from wild-type (WT), gup1Δ, per1Δ, and gup1Δ per1Δ cells was separated by chromatography on an Octyl-FF column and eluted with a 5–100% gradient of 1-propanol. Fractions were collected, dried, separated by SDS-PAGE, and analyzed by immunoblotting with an anti-FLAG antibody.
In the lipid remodeling pathway for GPI-anchored proteins in yeast, it is thought that an unknown lipase first removes the acyl in sn-2 and then Gup1p adds the C26:0 fatty acid to generate pG1, which can be further remodeled from a diacylglycerol-type to ceramide-type lipids (Bosson et al., 2006; Pittet and Conzelmann, 2006). To investigate the lipid structure of GPI-anchored proteins in per1Δ cells, we next analyzed the PI moiety obtained from GPI-anchored proteins by well-established analytical methods on TLC (Sipos et al., 1997; Guillas et al., 2000). Cells were labeled with [3H]inositol, lysed, and delipidated completely. The glycoproteins were then concentrated by concanavalin A-Sepharose and then digested with pronase. The PI moieties were quantitatively liberated from the GPI-anchored peptides by nitrous acid deamination, separated by TLC, and detected by autoradiography (Guillas et al., 2000). In wild-type cells, we detected at least three lipid moieties (Figure 7, lane 3) in similar ratios as reported previously (Sipos et al., 1997; Benachour et al., 1999): pG1, a phosphatidylinositol with a long-chain fatty acid at the sn-2 position; inositolphosphorylceramide (IPC)/B, a IPC consisting of phytosphingosine and a C26:0 fatty acid; and IPC/C, a IPC consisting phytosphingosine and a hydroxylated C26:0 fatty acid. In contrast, gup1Δ cells accumulated lyso-PI moieties instead of pG1 and IPCs (Figure 7, lane 4). PI moieties from GPI-anchored proteins in gpi7Δ cells were also analyzed as a control because ceramide remodeling in the Golgi was significantly reduced in this mutant (Benachour et al., 1999). As reported previously, the amount of IPC/C, which is remodeled in the Golgi, was significantly reduced, and the relative amount of pG1 was increased in gpi7Δ cells (Figure 7, lane 7; Benachour et al., 1999). The per1Δ cells mainly accumulated a normal PI containing short-chain fatty acids at sn-2 (Figure 7, lane 5). We did not detect other inositol-containing lipid moieties, pG1, or lyso-PI, but we detected a low amount of IPC/C. The gup1Δ per1Δ double-mutant cells accumulated PI but not lyso-PI (Figure 7, lane 6), supporting the idea that Per1p acts before the C26-fatty acid acylation at sn-2 by Gup1p. In addition, we found that IPC/C is incorporated into the GPI-anchored proteins (Figure 7, lane 6). These results strongly suggest that Per1p is involved in the lipid remodeling of the GPI anchor and is required for the formation of lyso-GPI, which is used by Gup1p to produce long-chain fatty acid–containing GPI anchors.
Figure 7.
Lipid remodeling of the GPI anchor is abnormal in per1Δ cells. Cells (2 × 108) were labeled with [3H]inositol (50 μCi) for 2 h at 30°C. GPI-anchored proteins of wild-type (WT), gup1Δ, per1Δ, gup1Δ per1Δ, and gpi7Δ cells were concentrated from the delipidated proteins, after which lipid moieties from GPI anchors were released with nitrous acid and analyzed by TLC using solvent 3 (55:45:10 CHCl3/CH3OH/0.25% KCl). Lipids extracted from WT cells were also included as a control (WT Free lipid; note that lane 1 was scanned at a lower sensitivity than lane 2). pG1, phosphatidylinositol with a C26:0 fatty acid in sn-2 position; PI, phosphatidylinositol; IPC/B, inositolphosphorylceramide consisting of phytosphingosine and a C26:0 fatty acid; IPC/C, inositolphosphorylceramide consisting of phytosphingosine and a hydroxylated C26:0 fatty acid; lyso-PI, lyso form of PI.
Per1p Is Required for the GPI-PLA2 Activity
Our results show that Per1p is involved in the formation of lyso-GPI after the attachment of GPI to proteins in the ER. We next investigated whether Per1p is required for the GPI-PLA2 activity by performing an in vitro assay, followed by fractionation of Flag-Gas1p. First, Flag-Gas1p purified from per1Δ cells (per1Δ Flag-Gas1p) was incubated with buffer or commercially available PLA2 and then fractionated by chromatography on octyl-Sepharose with a 1-propanol gradient. The elution profile for per1Δ Flag-Gas1p incubated with buffer was similar to that of untreated Flag-Gas1p from per1Δ cells (Figure 6, per1Δ, and Figure 8A, +buffer). In contrast, in the elution profile for per1Δ Flag-Gas1p incubated with PLA2, the bands in fractions 13–16 were shifted to fractions 3–8, so that the elution pattern was similar to that of Flag-Gas1p purified from gup1Δ cells (Figure 6, gup1Δ, and Figure 8A, +PLA2), indicating that the lipid moiety in the earlier fractions from Flag-Gas1p is lyso-GPI. Next, the per1Δ Flag-Gas1p was incubated for 30 min with microsomes prepared from wild-type, gup1Δ, or per1Δ cells, repurified, and analyzed by octyl-Sepharose chromatography. The elution profile for Flag-Gas1p incubated with per1Δ microsomes was the same as that for untreated Flag-Gas1p from per1Δ cells (Figure 8B, +per1Δ microsome for 30 min). In contrast, Flag-Gas1p incubated with wild-type microsomes (+WT microsome for 30 min) or gup1Δ microsome (+gup1Δ microsome for 30 min) was eluted much earlier (fractions 3–8), corresponding to Flag-Gas1p containing lyso-GPI (Figure 8B). When the reaction with microsomes was extended to more than 60 min, Flag-Gas1p was not detected in earlier fractions, probably because of degradation (unpublished data). These results suggest that Per1p is required for the GPI-PLA2 activity.
Figure 8.
PER1 is required for the formation of lyso-GPI. (A) PLA2 treatment of Flag-Gas1p from per1Δ cells. Flag-Gas1p purified from per1Δ cells was incubated overnight at 37°C with buffer only (+buffer) or PLA2 (+PLA2), collected, washed, and eluted with 3× FLAG peptide. The repurified Flag-Gas1p was separated by chromatography on an Octyl-FF column, and eluted fractions were analyzed by immunoblotting as described in Figure 6. (B) In vitro analysis of GPI-PLA2 activity. Flag-Gas1p purified from per1Δ cells was incubated at 37°C for 30 min with microsomal fractions from per1Δ, wild-type (WT), gup1Δ cells. The mixture was solubilized, and Flag-Gas1p was repurified and separated on an Octyl-FF column. Eluted fractions were analyzed by immunoblotting as described in Figure 6.
Human PERLD1 Is a Functional Homologue of Yeast PER1
A search of the databases for homologue(s) of yeast PER1 identified PERLD1 as a homologous gene in humans (Figure 9A). The predicted amino acid sequences of yeast PER1 and human PERLD1 shared 28% identity. To examine whether human PERLD1 is a functional homologue of PER1, we introduced a plasmid expressing PERLD1 cDNA into the per1Δ cells and examined their CFW and temperature sensitivities. We found that the PERLD1 gene restored both the CFW sensitivity and temperature sensitivity at 37°C to per1Δ cells (Figure 9B). We also found that the amount of mature Gas1p was restored to normal levels in per1Δ cells carrying PERLD1 (Figure 9C). These results clearly indicate that human PERLD1 is a functional homologue of yeast PER1.
Figure 9.
Human PERLD1 is a functional homologue of Per1p, and H177 and H326 are required for Per1p activity. (A) Alignment of the amino acid sequences of Per1p homologues. Amino acid sequences of Homo sapiens (GenBank Accession No. AAH10652), Mus musculus (Accession No. NP_001028709), Schizosaccharomyces pombe (Accession No. CAB90152), and S. cerevisiae (Accession No. NP_009973) proteins were aligned using ClustalW. (B) Spot test of CFW and temperature sensitivities in per1 mutant cells. Top panels, isogenic per1Δ cells carrying the pRS316T (vector), pMF917 (PER1), or pMF942 (Human PERLD1) were spotted on YPAD or YPAD plates containing 7.5 μg/ml CFW after making a 5× serially diluted solution and then incubated for 2 d at 30 or 37°C. Bottom panels, isogenic per1Δ cells carrying the pRS316T (vector), pMF918 (PER1-HA), pMF931 (S19A), pMF932 (H102A), pMF933 (K104A), pMF934 (S118A), pMF935 (S122A), pMF936 (S173A), pMF937 (H177A), pMF939 (D315A), or pMF940 (H326A) were spotted on YPAD or YPAD plates containing 7.5 μg/ml CFW and then incubated for 2 d at 30 or 37°C. (C) Human PERLD1 expression restores the amount of Gas1p to per1Δ cells. Isogenic per1Δ cells carrying pRS316T, pMF917, or pMF942 were grown overnight to midlog phase. Equal cell numbers were lysed, and total protein extracts were analyzed by immunoblotting using anti-Gas1p and anti-Dpm1p. (D) H177 and H326 are essential for Per1p activity. Isogenic per1Δ cells carrying the pRS316T, pMF918, pMF931, pMF932, pMF933, pMF934, pMF935, pMF936, pMF937, pMF939, or pMF940 were grown overnight to midlog phase. Equal cell numbers were lysed, and total protein extracts were analyzed by immunoblotting using anti-Gas1p, anti-HA, and anti-Dpm1p.
Although Per1p is required for GPI-PLA2 activity, we do not know whether Per1p is itself a GPI-PLA2 or a factor that regulates GPI-PLA2. The lipase motif of PLA2 is known to contain a putative catalytic serine (Wong and Schotz, 2002; Akoh et al., 2004), and other reports suggest that a histidine in the active site is essential for the activity of secreted PLA2 (Berg et al., 2001; Edwards et al., 2002). Although Per1p does not have a lipase-like motif, several serine and histidine residues are conserved among PER1 homologues. We tried to identify the amino acids that are essential for Per1p activity. On the basis of the amino acid alignment of PER1 homologues between budding yeast, fission yeast, mouse, and human, we substituted conserved residues S19, H102, K104, S118, S122, S173, H177, D315, and H326 residues with alanine (Figure 9A). The S19A, H102A, K104A, S118A, S122A, S173A, and D315A mutants of per1 complemented the CFW sensitivity and temperature sensitivity at 37°C in per1Δ cells (Figure 9B). These mutants also restored the amount of mature Gas1p (Figure 9D). In addition, the amounts of HA-tagged Per1-H177A and Per1-H326A were similar to that of wild-type Per1-HA and the other HA-tagged functional mutants; however, the per1-H177A and per1-H326A did not rescue the phenotypes found in per1Δ cells (Figure 9, B and D). Both H177 and H326 residues are found in the conserved regions, and several reports on the crystal structure of PLA2 have suggested that the active site histidine residue attacks the ester carbonyl at the sn-2 position of phospholipids (Berg et al., 2001; Edwards et al., 2002). Although it remains unclear whether Per1p itself possesses GPI-PLA2 activity, it is likely that the regions containing H177 and H326 are important for the function of Per1p.
DISCUSSION
In the current study, we showed that Per1p is involved in the lipid remodeling of GPI-anchored proteins. Our data suggest that Per1p is required for the GPI-PLA2 activity necessary for the formation of C26:0 fatty acid–containing GPI. We also found that lipid remodeling by Per1p is required for the efficient transport of GPI-anchored proteins from the ER to the Golgi and for their association with lipid rafts.
Lipid moieties of yeast GPI are dynamically remodeled in the ER (Figure 10; Sipos et al., 1997). First, after attachment of GPI to proteins, the acyl group on the inositol residue of GPI anchor is eliminated by Bst1p (Tanaka et al., 2004; Fujita et al., 2006). Next, the acyl portion in sn-2 of PI moiety is removed. PI prepared from GPI-anchored proteins in per1Δ cells contained only the conventional PI moiety, but not pG1 or lyso-PI. Our in vitro analysis suggests that Per1p is directly involved in the formation of lyso-PI from PI. Gup1p adds the C26:0 fatty acid in sn-2 to generate pG1 (Bosson et al., 2006). The lipids of many GPI-anchored proteins are then changed from diacylglycerol-type to ceramide-type in the ER (IPC/B) and Golgi (IPC/C; Reggiori et al., 1997; Sipos et al., 1997). In the gup1Δ per1Δ double-mutant cells, we detected significant amounts of IPC/C in addition to PI, indicating that a small number of conventional diacylglycerol-type of GPI-anchored proteins could be replaced by ceramide-type GPI-anchored proteins in the Golgi of these cells.
Figure 10.
Lipid remodeling pathway of GPI-anchored proteins in yeast. This model for the lipid remodeling pathway of GPI anchors in yeast based on previous reports (Sipos et al., 1997; Bosson et al., 2006). After attachment of GPI to proteins, the acyl group on the inositol of the GPI anchor is eliminated by Bst1p. The acyl portion at sn-2 of the PI moiety is removed to generate lyso-PI. Gup1p adds a C26:0 acyl chain at sn-2 of lyso-PI to generate pG1. The various lipids of GPI-anchored proteins are then changed from diacylglycerol-type to ceramide-type in the ER (IPC/B) and Golgi (IPC/C). A small number of conventional diacylglycerol-type GPI-anchored proteins can replace ceramide-type GPI-anchored proteins in the Golgi. Our current studies indicate that Per1p is required for the production of lyso-GPI during the remodeling of GPI-anchored proteins.
Gas1p could not associate with DRMs in per1Δ cells, strongly suggesting that lipid remodeling is required for the association of GPI-anchored proteins with lipid rafts. In yeast cells, phospholipids including PI usually contain an unsaturated fatty acid in the sn-2 position, and the major species of PI is sn-1-palmitoyl (C16:0)-sn-2-oleoyl (C18:1)-PI (Schneiter et al., 1999). The GPI intermediates are synthesized from these species (Sipos et al., 1997). In contrast, the PI moieties of mature GPI-anchored proteins are pG1 or IPCs containing very long saturated fatty acids (Fankhauser et al., 1993). Our results suggest that these remodeling reactions are required for the incorporation of GPI-anchored proteins into the DRMs. In yeast, GPI-anchored proteins are associated with DRMs from the ER, whereas incorporation of proteins into DRMs in mammalian cells takes place in the Golgi (Simons and Ikonen, 1997; Brown and London, 1998; Bagnat et al., 2000). Our results are in accordance with the fact that, in yeast, lipid remodeling is mainly carried out in the ER, whereas mammalian lipid remodeling mainly occurs in the Golgi.
The association of GPI-anchored proteins with lipid rafts is required for their tight anchoring to the plasma membrane as indicated by the dramatic release of Gas1p into the culture medium in gup1Δ cells (Bosson et al., 2006). We also found an equal release of Gas1p in per1Δ cells. In the current studies, the Golgi but not the ER form of Flag-Gas1p from per1Δ cells eluted in earlier fractions during chromatography on octyl-Sepharose. Usually, GPI-anchored proteins contain very long fatty acids and are tightly bound to the membrane. In the per1Δ cells, however, the GPI-anchored proteins could not associate with microdomains such as lipid rafts and are not stable at the Golgi or the plasma membrane. A number of enzymes possessing phospholipase B activity, which eliminates phospholipid acyl chains at both sn-1 and sn-2, are localized at the cell surface in yeast (Merkel et al., 2005). In the per1Δ cells, unremodeled GPI-anchored proteins might be recognized as substrates by phospholipase Bs. This could explain why the Golgi form of Flag-Gas1p eluted in the earlier fractions from the octyl-Sepharose column. Structural analysis of the GPI anchor in the earlier fractions would help determine the validity of this hypothesis, but the small amounts of GPI-anchored proteins in the per1Δ cells currently make such studies difficult.
The transport of Gas1p from the ER to the Golgi was substantially delayed in the per1Δ cells. Ceramide is required for the specific transport of GPI-anchored proteins from the ER to the Golgi (Watanabe et al., 2002). By participating in the formation of DRMs in the ER, ceramide may help drive the incorporation of GPI-anchored proteins into ER-derived vesicles. The transport could be delayed because the unremodeled GPI-anchored proteins are not tightly associated with ceramide-rich vesicles. Further analysis is needed to determine whether the unremodeled GPI-anchored proteins are transported from the ER to the Golgi in the GPI-dependent vesicles (Muniz et al., 2001; Mayor and Riezman, 2004; Watanabe and Riezman, 2004).
In the bloodstream form of African trypanosome Trypanosoma brucei, the fatty acids of GPI intermediates are replaced by myristic acid (C14:0) through sequential deacylation and reacylation reactions on sn-2 followed by sn-1 before the GPI is attached to the protein (Ferguson, 1999; Ferguson et al., 1999). We were unable to find PER1 homologues in the genome database of T. brucei, suggesting that other enzymes are required for the deacylation of GPI precursors. The lipid remodeling reactions of GPI anchors in yeast and mammals differ at several points from those in T. brucei; the remodeling reactions in T. brucei are carried out before the attachment of GPI to the protein, and myristic acids are incorporated not only at sn-2 but also at sn-1 in the GPI precursor, whereas, in yeast and human, the GPI lipid remodeling is carried out after the attachment of GPI to proteins, and only a fatty acid at the sn-2 position of the GPI anchor is exchanged (Ferguson et al., 1999; Pittet and Conzelmann, 2006). We found several genes involved in the acylation or deacylation of GPI anchors in the genome database of T. brucei, including homologues of GUP1 and PGAP1/BST1, but we did not find homologues of PIG-W/GWT1 and PGAP2, suggesting that several of the GPI modification reactions are carried out by distinct enzymes in yeast, mammals, and protozoans.
The remodeling pathway requiring Per1p is conserved among yeast and mammals. In this study, we uncovered the molecular mechanisms of lipid remodeling of the GPI anchor. Investigations of Per1p should clarify the roles of the very long saturated fatty acids of GPI-anchored proteins. Our data suggest that lipid remodeling of GPI-anchored proteins is essential for their association with specific lipid microdomains and for their efficient transport from the ER to the Golgi in yeast.
ACKNOWLEDGMENTS
We are grateful to Taroh Kinoshita and Yusuke Maeda for helpful discussions. We also thank Katsura Hata for providing the anti-Gas1 peptide polyclonal antibody; Kappei Tsukahara and Koji Sagane for providing plasmids; and Roger Tsien for providing the mRFP plasmid. Finally, we thank Michiyo Okamoto, Hiroto Hirayama, and Takuji Oka for stimulating discussions. M.F. was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science. This research was partly supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Abbreviations used:
- CFW
calcofluor white
- DRM
detergent-resistant membrane
- ER
endoplasmic reticulum
- GPI
glycosylphosphatidylinositol
- IPC
inositolphosphorylceramide
- IPC/B
IPC consisting of phytosphingosine and a C26:0 fatty acid
- IPC/C
IPC consisting of phytosphingosine and a hydroxylated C26:0 fatty acid
- mRFP
monomeric red fluorescent protein
- pG1
phosphatidylinositol with a C26:0 fatty acid in sn-2 position
- PI
phosphatidylinositol
- PLA2
phospholipase A2
- TLC
thin-layer chromatography.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0715) on October 4, 2006.
REFERENCES
- Abe H., Shimma Y., Jigami Y. In vitro oligosaccharide synthesis using intact yeast cells that display glycosyltransferases at the cell surface through cell wall-anchored protein Pir. Glycobiology. 2003;13:87–95. doi: 10.1093/glycob/cwg014. [DOI] [PubMed] [Google Scholar]
- Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachour A., Sipos G., Flury I., Reggiori F., Canivenc-Gansel E., Vionnet C., Conzelmann A., Benghezal M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 1999;274:15251–15261. doi: 10.1074/jbc.274.21.15251. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Gelb M. H., Tsai M. D., Jain M. K. Interfacial enzymology: the secreted phospholipase A(2)-paradigm. Chem. Rev. 2001;101:2613–2654. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell. 2006;17:2636–2645. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., London E. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Rose J. K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Conzelmann A., Puoti A., Lester R. L., Desponds C. Two different types of lipid moieties are present in glycophosphoinositol-anchored membrane proteins of Saccharomyces cerevisiae. EMBO J. 1992;11:457–466. doi: 10.1002/j.1460-2075.1992.tb05075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. H., Thompson D., Baker S. F., Wood S. P., Wilton D. C. The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism. Biochemistry. 2002;41:15468–15476. doi: 10.1021/bi020485z. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Maurer-Stroh S., Novatchkova M., Schneider G., Eisenhaber F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays. 2003;25:367–385. doi: 10.1002/bies.10254. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Homans S. W., Thomas-Oates J. E., McConville M. J., Desponds C., Conzelmann A., Ferguson M. A. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:26365–26374. [PubMed] [Google Scholar]
- Ferguson M. A. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 1999;112(Pt 17):2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A., Brimacombe J. S., Brown J. R., Crossman A., Dix A., Field R. A., Guther M. L., Milne K. G., Sharma D. K., Smith T. K. The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim. Biophys. Acta. 1999;1455:327–340. doi: 10.1016/s0925-4439(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-o T., Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 2006;17:834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Yoko-o T., Okamoto M., Jigami Y. GPI7 involved in glycosylphosphatidylinositol biosynthesis is essential for yeast cell separation. J. Biol. Chem. 2004;279:51869–51879. doi: 10.1074/jbc.M405232200. [DOI] [PubMed] [Google Scholar]
- Gillmor C. S., Lukowitz W., Brininstool G., Sedbrook J. C., Hamann T., Poindexter P., Somerville C. Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell. 2005;17:1128–1140. doi: 10.1105/tpc.105.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillas I., Pfefferli M., Conzelmann A. Analysis of ceramides present in glycosylphosphatidylinositol anchored proteins of Saccharomyces cerevisiae. Methods Enzymol. 2000;312:506–515. doi: 10.1016/s0076-6879(00)12935-0. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., Van Den Ende H., Klis F. M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta. 1999;1426:373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Inoue N., Takeda J. Defective glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria. Adv. Immunol. 1995;60:57–103. doi: 10.1016/s0065-2776(08)60584-2. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mayor S., Riezman H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- McConville M. J., Menon A. K. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol. 2000;17:1–16. doi: 10.1080/096876800294443. [DOI] [PubMed] [Google Scholar]
- Merkel O., Oskolkova O. V., Raab F., El-Toukhy R., Paltauf F. Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae. Biochem. J. 2005;387:489–496. doi: 10.1042/BJ20041272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M., Morsomme P., Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. T. Screening for mutants defective in secretory protein maturation and ER quality control. Methods. 2005;35:366–372. doi: 10.1016/j.ymeth.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Nakano A. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim. Biophys. Acta. 1991;1093:135–143. doi: 10.1016/0167-4889(91)90114-d. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Umemoto N., Ohsumi Y., Nakano A., Anraku Y. Biogenesis of vacuolar membrane glycoproteins of yeast Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:7440–7448. [PubMed] [Google Scholar]
- Okamoto M., Yoko-o T., Umemura M., Nakayama K., Jigami Y. Glycosylphosphatidylinositol-anchored proteins are required for the transport of detergent-resistant microdomain-associated membrane proteins Tat2p and Fur4p. J. Biol. Chem. 2006;281:4013–4023. doi: 10.1074/jbc.M504684200. [DOI] [PubMed] [Google Scholar]
- Paidhungat M., Garrett S. Cdc1 and the vacuole coordinately regulate Mn2+ homeostasis in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1787–1798. doi: 10.1093/genetics/148.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2006 doi: 10.1016/j.bbalip.2006.05.015. (in press) [DOI] [PubMed] [Google Scholar]
- Popolo L., Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- Ram A. F., Van den Ende H., Klis F. M. Green fluorescent protein-cell wall fusion proteins are covalently incorporated into the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1998;162:249–255. doi: 10.1111/j.1574-6968.1998.tb13006.x. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Canivenc-Gansel E., Conzelmann A. Lipid remodeling leads to the introduction and exchange of defined ceramides on GPI proteins in the ER and Golgi of Saccharomyces cerevisiae. EMBO J. 1997;16:3506–3518. doi: 10.1093/emboj/16.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Simons K. Controversy fuels trafficking of GPI-anchored proteins. J. Cell Biol. 2006;172:963–965. doi: 10.1083/jcb.200603015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Sipos G., Reggiori F., Vionnet C., Conzelmann A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997;16:3494–3505. doi: 10.1093/emboj/16.12.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Doering T. L., Schimmoller F., Schroder S., Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 1997;110(Pt 21):2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Maeda Y., Tashima Y., Kinoshita T. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., Maeda Y. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell. 2006;17:1410–1420. doi: 10.1091/mbc.E05-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Funato K., Venkataraman K., Futerman A. H., Riezman H. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J. Biol. Chem. 2002;277:49538–49544. doi: 10.1074/jbc.M206209200. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Riezman H. Differential ER exit in yeast and mammalian cells. Curr. Opin. Cell Biol. 2004;16:350–355. doi: 10.1016/j.ceb.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Wong H., Schotz M. C. The lipase gene family. J. Lipid Res. 2002;43:993–999. doi: 10.1194/jlr.r200007-jlr200. [DOI] [PubMed] [Google Scholar]