Abstract
Sir protein spreading along chromosomes and silencing in Saccharomyces cerevisiae requires the NAD+-dependent histone deacetylase activity of Sir2p. We tested whether this requirement could be bypassed at the HM loci and telomeres in cells containing a stably expressed, but catalytically inactive mutant of Sir2p, sir2-345p, plus histone mutants that mimic the hypoacetylated state normally created by Sir2p. Sir protein spreading was rescued in sir2-345 mutants expressing histones in which key lysine residues in their N-termini had been mutated to arginine. Mating in these mutants was also partially restored upon overexpression of Sir3p. Together, these results indicate that histone hypoacetylation is sufficient for Sir protein spreading in the absence of production of 2′-O-acetyl-ADP ribose by sir2p and Sir2p's enzymatic function for silencing can be bypassed in a subset of cells in a given population. These results also provide genetic evidence for the existence of additional critical substrates of Sir2p for silencing in vivo.
INTRODUCTION
Epigenetic processes play critical roles in biology by ensuring stable patterns of gene expression during normal growth and differentiation. Cells tightly control both the timing and location of formation of epigenetically regulated chromatin because the faithful inheritance of such structures will dictate transcriptional events at individual loci in future generations. Silencing represents an epigenetic process critical for maintaining stable states of gene expression and chromatin integrity in Saccharomyces cerevisiae. In yeast, the Sir proteins mediate silencing of mating-type genes at HML, HMR and of genes flanking telomeres, maintain telomere integrity, and regulate gene expression and genome stability at the rDNA locus (Rusché et al., 2003).
The formation of silent chromatin in S. cerevisiae occurs through multiple, genetically separable steps that have been best described at HMR. Silencing first requires the recruitment of Sir proteins to a site flanking the mating-type genes at HMR named the E silencer. A second, weaker silencer flanking HMR, HMR-I, does not recruit Sir proteins to HMR, but rather likely stabilizes silent chromatin once it has formed (Brand et al., 1985; McNally and Rine, 1991; Rivier et al., 1999; Rusché et al., 2002). During recruitment, Sir proteins interact with proteins bound to HMR-E, including the origin recognition complex (ORC), Rap1p, and Abf1p. Once Sir1p, Sir2p, Sir3p, and Sir4p become localized to HMR-E, additional Sir proteins are recruited through Sir–Sir interactions and then Sir2p, Sir3p, and Sir4p spread along the chromosome (Hoppe et al., 2002; Rusché et al., 2002; Bose et al., 2004; Rudner et al., 2005). Sir protein spreading from telomere ends occurs in a similar manner (Hoppe et al., 2002; Luo et al., 2002; Rudner et al., 2005).
This propagation of Sir proteins along the chromosome and silencing requires the NAD+-dependent histone deacetylase activity of Sir2p (Hoppe et al., 2002; Luo et al., 2002; Rusché et al., 2002). Sir2 homologues are prevalent across phyla, participate in processes ranging from aging to apoptosis to epigenetic gene regulation and share a common catalytic strategy. In the NAD+-dependent deacetylation reaction catalyzed by Sir2p, a glycosidic bond in NAD+ is broken as NAD+ is converted into 2′-O-acetyl-ADP ribose and nicotinamide each time an acetyl group is removed from a lysine residue. This deacetylation reaction has at least three potential purposes in silencing. The first purpose is to modify histones H3 and H4 to create high-affinity Sir protein-binding sites on nucleosomes to enable spreading and silencing (Braunstein et al., 1996; Grunstein, 1998; Moazed, 2001; Hoppe et al., 2002; Luo et al., 2002; Rusché et al., 2002). In support of this model, Sir3p and Sir4p bind to the N-terminal tails of histones H3 and H4, and Sir3p preferentially binds deacetylated tails of H4 in vitro (Hecht et al., 1995; Carmen et al., 2002), and Sir protein spreading and silencing is lost in catalytically inactive sir2 mutants that cannot deacetylate histones (Hoppe et al., 2002; Luo et al., 2002; Rusché et al., 2002).
The second purpose may be to generate a byproduct of the reaction, 2′-O-acetyl-ADP ribose, for use in forming silent chromatin. Consistent with this function, 2′-O-acetyl-ADP-ribose induces a conformational and stoichiometric change in Sir2/3/4 complexes in vitro (Liou et al., 2005) and sir2 mutants that cannot make 2′-O-acetyl-ADP ribose are defective in both Sir protein spreading and silencing (Hoppe et al., 2002; Luo et al., 2002; Rusché et al., 2002). The third purpose of this deacetylation reaction may be to generate energy to be used for either Sir protein spreading or to drive a conformational change needed to produce silent chromatin. The amount of free energy released during the hydrolysis of the glycosidic bond between nicotinamide and ribose in NAD+ is comparable to the free energy released during the hydrolysis of ATP (Rowen and Kornberg, 1951; Zatman et al., 1953). Thus, a significant amount of free energy is released during the formation of silent chromatin over large regions of chromosomal DNA that could be coupled to a conformational transition necessary for silencing.
Sir2p's demonstrated substrates in vitro include lysines 9 and 14 on histone H3 plus lysines 5, 8, and 12, and 16 on histone H4 (Imai et al., 2000; Tanny and Moazed, 2001; Borra et al., 2004; Tanny et al., 2004) and these sites are hypoacetylated in silent chromatin in vivo as well (Braunstein et al., 1996; Suka et al., 2001; Hoppe et al., 2002; Rusché et al., 2002; Kirchmaier and Rine, 2006). However, on histone H4, only lysine 16, but not lysines 5, 8, and 12 are critical for silencing (Johnson et al., 1990; Park and Szostak, 1990) and for Sir3p or Sir2/4p association in vitro (Liou et al., 2005). The major, evolutionarily conserved, histone acetyltransferase that makes substrates for Sir2p by targeting H4 K16 in cells is Sas2p (Kimura et al., 2002; Sutton et al., 2003). Sas2p also acetylates H3 K14 in vitro and tethering Sas2p to the chromosome can create a barrier that blocks the spread of silent chromatin (Donze and Kamakaka, 2001; Sutton et al., 2003). Thus, multiple lysine residues within the N-terminal tails of histones H3 and H4 can be deacetylated by Sir2p, but the influence of individual residues on the formation and stability of silent chromatin varies.
The sir2p mutant, sir2-345p lacks deacetylase activity and is defective in silencing (Imai et al., 2000). sir2-345p contains an Asn-to-Ala substitution at residue 345 within the active site. This evolutionarily conserved Asn residue has been proposed to participate in catalysis by positioning and activating a structurally conserved water molecule during the deacetylation reaction (Min et al., 2001; Chang et al., 2002; Zhao et al., 2003, 2004). Although sir2-345p is stably expressed, interacts with Sir3p and Sir4p, and is recruited to silencers, sir2-345p cannot deacetylate histones, does not support spreading of Sir2/3/4p across HMR in otherwise wild-type cells, and does not support silencing (Imai et al., 2000; Min et al., 2001; Rusché et al., 2002; Kirchmaier and Rine, 2006; see also Hoppe et al., 2002). Because enhanced histone acetylation, or loss of deacetylation, coincides with a loss of both silencing and Sir protein spreading in sir2-345 mutants, the defect in silencing correlates with an inability of these mutants to generate high-affinity binding sites for Sir3p and Sir4p on the N-terminal tails of histones H3 and H4 and an inability to generate 2′-O-acetyl-ADP ribose and release free energy (Rusché et al., 2002; Kirchmaier and Rine, 2006). Together, these observations motivated us to test if the requirement for Sir2p's catalytic activity might be bypassed if we created conditions in the cell that would ensure the deacetylated status of histones. Surprisingly, we identified conditions in which both Sir protein spreading and silencing could be partially restored in the absence of Sir2p's enzymatic activity.
MATERIALS AND METHODS
Plasmids and Strains
Yeast strains were generated by standard genetic techniques including genetic crosses, homologous recombination, one step gene replacement, and plasmid shuffling (Stearns et al., 1990; Wach et al., 1994; Adams et al., 1997; Goldstein and McCusker, 1999). Parental strain genotypes are described in Table 1, and plasmids used in this study are described in Table 2. The plasmid AK923 expressing histone mutants H3 K9,14R and H4 K16R was generated by site directed mutagenesis of pMP72 (Kelly et al., 2000) according to the Quick Change Site-Directed Mutagenesis Kit protocol (Stratagene, La Jolla, CA) using oligonucleotides oALK593 and oALK594 (Supplementary Table 2).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| JRY2726 | MATa his4 | P. Schatz |
| JRY2728 | MATα his4 | P. Schatz |
| W303 | MATa or α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | R. Rothstein |
| AKY1968 | W303 MATα hht1-hhf1Δ::LEU2 hht2- hhf2Δ::HIS3 plus PK189 | This studya |
| AKY944 | W303 MATa hht1-hhf1Δ::LEU2 hht2-hhf2Δ::HIS3 plus PK189 | This studya |
| AKY1101 | W303 MATα LEU2::sir2-345 sir2Δ::KanMX hht1-hhf1Δ::LEU2 hht2- hhf2Δ::HIS3 plus PK189 | This studya |
| AKY1103 | W303 MATa LEU2::sir2-345 sir2Δ::KanMX hht1-hhf1Δ::LEU2 hht2- hhf2Δ::HIS3 plus PK189 | This studya |
| AKY2765 | AKY1968 sir4Δ::NatMX | This studya |
| AKY2763 | AKY1101 sir4Δ::NatMX | This studya |
a Parental strains used in this study. See Table 2 for description of plasmids that were introduced into AKY1968, AKY944, AKY1101, AKY1103, AKY2765, and AKY2763 for experiments described in text.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| PK189 | HHT2 HHF2 ARS/CEN/URA3 | P. Kaufman |
| pwz-414-F13 | HHT2 HHF2 ARS/CEN/TRP1 | Zhang et al. (1998) |
| pMP3 | HHT2 HHF2 ARS/CEN/TRP1 | Kelly et al. (2000) |
| pMP72 | H3 K9,14R H4 ARS/CEN/TRP1 | Kelly et al. (2000) |
| AK923 | H3 K9,14R H4 K16R ARS/CEN/TRP1 | This study |
| pJR104 | SIR3 in YEp24 | Kimmerly and Rine (1987) |
| YEp24 | Vector; 2 μm/URA3 | Botstein et al. (1979) |
| pJR69 | SIR2 in YCp50 | J. Rine |
| YCp50 | Vector; ARS/CEN/URA3 | C. Mann, Ma et al. (1987) |
| pRS/345 | sir2-345 in pRS305 | Imai et al. (2000) |
| pFA6::kanMX4 | Wach et al. (1994) | |
| pFA6::natMX4 | Goldstein and McCusker (1999) |
Mating Assays
Patch and quantitative mating assays were conducted using two independent yeast strains for each genotype and were performed as described previously and as outlined in Table 3 and Figure 4 (van Leeuwen and Gottschling, 2002). Briefly, the mating efficiencies of each strain relative to wild type were determined using the following formula: (colonies on YM plate with indicated tester strain/colonies on YM plate with supplements)indicated strain/(colonies on YM plate with indicated tester strain/colonies on YM plate with supplements)SIR2 H3 H4, for each experiment.
Table 3.
Sir protein spreading in histone H3 K9,14R H4 K16R mutants does not restore silencing at HML and HMR in sir2-345 cells
| SIR2 | HHT2 HHF2 | Relative efficiency of matinga |
|
|---|---|---|---|
| MATα | MATa | ||
| SIR2 | H3/H4 | 1 | 1 |
| SIR2 | H3 K9,14R/H4 K16R | 0.27 ± 0.035 | 0.77 ± 0.22 |
| sir2-345 | H3/H4 | <1 × 10−5 (1 × 10−6, 1 × 10−6, 4 × 10−6) | <1 × 10−5 (1 × 10−6, 5.5 × 10−6, 0) |
| sir2-345 | H3 K9,14R/H4 K16R | <1 × 10−5 (0, 2 × 10−6, 1 × 10−6) | <1 × 10−5 (1 × 10−6, 1.1 × 10−5, 2 × 10−6) |
a The efficiency of mating of MATa SIR2 or MATα SIR2 plus wild-type histones H3 and H4 to the indicated tester strains JRY2728 (MATα) or JRY2726 (MATa), respectively, was determined relative to their plating efficiency (for MATa SIR2, 42 ± 8.6%, n = 3; for MATα SIR2, 100 ± 21%, n = 3) and was set to 1. The mating efficiencies of each strain relative to SIR2 cells expressing wild-type H3 and H4 was determined as indicated in Materials and Methods. Data reflects the average ± SD of three independent experiments, and data from individual experiments are shown in parentheses for sir2-345 strains.
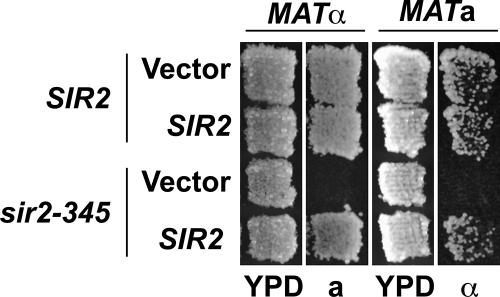
Figure 4.
SIR2 rescues silencing in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants at HMR and HML. sir2-345 was integrated at LEU2 and expressed from its endogenous promoter. SIR2 was expressed from its endogenous promoter on an ARS/CEN plasmid. Patch mating analysis. Haploid MATa or MATα cells were plated on rich media, YPD, grown 1 d at 30°C, and tested for silencing at HMR or HML, respectively, by replica plating to a (JRY2726) or α (JRY2728) mating-type tester lawns on minimal medium and incubating for 2 d at 30°C. Growth of diploid indicates that HML or HMR was silenced.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiments were performed using two independent yeast strains in three independent replicates of each experiment, unless noted in the figure legend, and were analyzed by real time PCR on an ABI Prism 7000 as described previously (Kirchmaier and Rine, 2006). Oligonucleotides are described in Supplementary Table 2. Statistical analyses were performed using the Wilcoxon rank sum test with MSTAT v.2.6. (http://mcardle.oncology.wisc.edu/mstat).
RNA Analyses
Total RNA was isolated from logarithmically growing cells as described previously (Schmitt et al., 1990), and 4 μg total RNA was incubated with 1 U DNase I (Sigma, St. Louis, MO) to degrade genomic DNA. cDNAs were then synthesized from 2 μg DNaseI-treated RNA using random hexamer primers and 200 U M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) in the presence of 40 U RNaseOUT (Invitrogen). cDNAs were diluted 1:75 in water, and 1:150 of the diluted samples were used per reaction when analyzed by real-time PCR using Sybr Green PCR master mix (Applied Biosystems, Foster City, CA), according to the manufacturer's protocols. Real-time PCR was performed using oligonucleotides listed in Supplementary Table 2 and was analyzed on an ABI Prism 7000. Quantification was performed using the comparative CT method according to the manufacturer's instructions (User Bulletin 2, ABI Prism 7700 Sequence Detection System, Columbia, MD). Analysis was conducted with two independent yeast strains for each genotype and represents the averages of three independent experiments. For each experiment, data represented the average of three PCRs for each transcript. Statistical analyses were performed using the Wilcoxon rank sum test with MSTAT v.2.6.
RESULTS
Histone Mutants Rescue Sir Protein Spreading at HM Loci in Catalytically Inactive sir2 Cells
The key catalytic role of Sir2p in silencing is widely thought to be to deacetylate lysines within the N-terminal tails of histones H3 and H4. Acetylation of lysine residues neutralizes their positive charge, whereas deacetylation by Sir2p restores their charge and creates high-affinity Sir protein-binding sites on histones H3 and H4 to permit Sir protein spreading (Hoppe et al., 2002; Luo et al., 2002; Rusché et al., 2002). To test if this is the only critical catalytic role of Sir2p, we generated SIR2 and sir2-345 yeast expressing either wild-type histones H3 and H4 or mutant histones H3 and H4 in which key substrates of Sir2p, including lysines 9 and 14 of H3 and lysine 16 of H4, were mutated to arginine. Mutating lysine residues to arginine prevents their acetylation and retains their positive charge and thus mimics the hypoacetylated state. We asked what steps in forming silent chromatin could be restored in these strains by monitoring Sir protein spreading at the HM loci by chromatin immunoprecipitation (ChIP) and quantitative real-time PCR (Figure 1, A and B, and Table 4) and monitoring silencing at HML and HMR by reverse transcription, RT-PCR (Figure 2), and quantitative mating assays (Table 3).
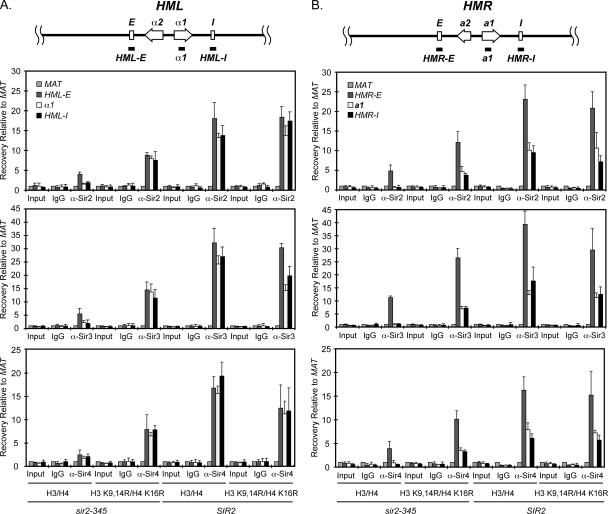
Figure 1.
Sir protein spreading is restored at HML and HMR in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants. (A) Sir protein association at HML. (B) Sir protein association at HMR. Sir protein association in SIR2 or sir2-345 yeast lacking chromosomal copies of the histone H3 and H4 genes and containing wild-type or mutant H3 and H4 expressed from an ARS/CEN plasmid was monitored by chromatin immunoprecipitation, ChIP. ChIPs using IgG, anti-Sir2p, Sir3p, and Sir4p antibodies were measured by quantitative real time PCR using primers that amplified HML-E, HMLα1, HML-I, HMR-E, HMRa1, HMR-I, or MAT (see Supplementary Table 2). The regions amplified at HML and HMR are noted at top of the figure (not to scale). Efficiency of co-precipitation of each locus is expressed relative to MAT and was calculated as: Locus IP/MAT IP = 2(MAT CT − Locus CT). Data are expressed as average ± SD; n = 3.
Table 4.
Sir proteins spread efficiently at HMR in histone H3 K9,14R H4 K16R mutants
| SIR2 | HHT2 HHF2 | Sir protein ChIP | Percent efficiency of Sir bindinga |
|
|---|---|---|---|---|
| a1/E | I/E | |||
| SIR2 | H3/H4 | Sir2p | 45 ± 14 | 41 ± 4.0 |
| SIR2 | H3 K9,14R/H4 K16R | Sir2p | 56 ± 34 | 37 ± 16 |
| sir2-345 | H3/H4 | Sir2p | 15 ± 6.2b | 13 ± 5.6b |
| sir2-345 | H3 K9,14R/H4 K16R | Sir2p | 41 ± 18 | 32 ± 4.5 |
| SIR2 | H3/H4 | Sir3p | 32 ± 5.8 | 46 ± 17 |
| SIR2 | H3 K9,14R/H4 K16R | Sir3p | 39 ± 8.3 | 44 ± 14 |
| sir2-345 | H3/H4 | Sir3p | 7.7 ± 2.6b | 11 ± 10b |
| sir2-345 | H3 K9,14R/H4 K16R | Sir3p | 27 ± 4.8 | 28 ± 5.2 |
| SIR2 | H3/H4 | Sir4p | 49 ± 8.1 | 38 ± 2.6 |
| SIR2 | H3 K9,14R/H4 K16R | Sir4p | 50 ± 13 | 39 ± 8.7 |
| sir2-345 | H3/H4 | Sir4p | 25 ± 17b | 15 ± 3.8b |
| sir2-345 | H3 K9,14R/H4 K16R | Sir4p | 35 ± 2.8 | 33 ± 3.3 |
a The efficiency of association of each Sir protein at a1 or HMR-I relative to HMR-E silencer for each experiment in Figure 1B was determined using the following formula: (2(MAT CT − Locus CT))/(2(MAT CT − E silencer CT)) × 100, and the average ± SD (n = 3) is shown. The association of each Sir protein at HMR-E in each yeast strain has been set to 100%.
b Sir2/3/4 protein association at HMRa1 and HMR-I are at background levels (see Figure 1B). For ChIP of Sir2p, Sir3p, and Sir4p, the background signal at MAT relative to HMR-E in sir2-345 cells expressing wild-type H3/H4 is MAT/HMR-E = 22 ± 5.7, 8.9 ± 0.44, and 28 ± 9.1%, respectively (n = 3).
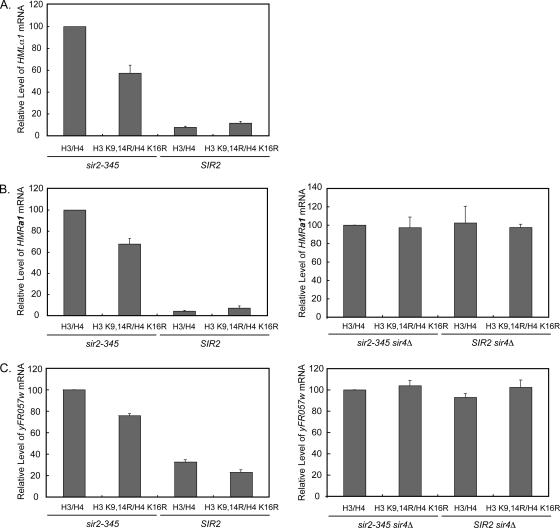
Figure 2.
Effects of Sir protein spreading on transcription from HM loci and yFR057w at telomere VIR. Transcription of HMLα1 (A), HMRa1 (B), or yFR057w mRNA (C) in the indicated strains was monitored by quantitative real-time PCR. mRNA levels from each locus relative to an internal control, SCR1, was determined for each cell line and then expressed relative to that observed in sir2-345 cells containing wild-type histones H3 and H4, which has been set to 100%. Data were calculated as follows; 2[(locus CT−SCR1 CT) sir2-345 H3 H4 and SIR4 or sir4Δ − (locus CT − SCR1 CT)a] × 100, where a is the indicated genotype. Data are expressed as average ± SD; n = 3. The regions amplified to monitor α1, a1, and yFR057w mRNA are noted at top of Figures 1 and 3 (not to scale). In sir2-345 cells expressing H3 K9,14R H4 K16R mutants, a1, α1, and yFR057w mRNA levels were significantly reduced to 69 ± 4.6, 57 ± 7.4, and 76 ± 1.8%, respectively, of the levels observed in sir2-345 cells expressing wild-type histones; p = 0.018 for each comparison.
Our analysis at HML in SIR2 cells indicated Sir2, Sir3, and Sir4 proteins associated efficiently at HML-E and spread throughout HMLα1 and HML-I in cells expressing either wild-type histones H3/H4 or the H3 K9,14R H4 K16R mutants (Figure 1A) and HMLα1 was silenced (Figure 2A and Table 3). Thus, the deacetylation reaction by Sir2p at these histone residues was not necessary for Sir protein spreading or silencing at HML. These data and the data described below also imply that, if localized production of 2′-O-acetyl-ADP ribose or energy release by Sir2p was needed for spreading or silencing, additional substrates of Sir2p beyond those already known to be critical for silencing must exist in vivo (see below and Discussion).
We then examined Sir protein association at HML in sir2-345 cells. In sir2-345 cells containing wild-type histones, Sir proteins loaded primarily at HML-E and were weakly associated throughout HML (Figure 1A) and silencing was lost (Figure 2A and Table 3). In contrast, efficient Sir protein association across the HML locus in sir2-345 cells was restored in histone H3 K9,14R H4 K16R mutants. Although the overall levels of Sir proteins at HML were reduced in sir2-345 cells expressing the histone H3 K9,14R H4 K16R mutants, the pattern of Sir protein association at α1 or HML-I relative to HML-E was generally similar to that observed for SIR2 cells containing wild-type or mutant histones (Figure 1A and Supplementary Table 1). Thus, mimicking deacetylated histone tails was sufficient to restore Sir protein spreading throughout the HML locus in the absence of the catalytic activity of Sir2p and therefore the release of free energy and the localized production of 2′-O-acetyl-ADP-ribose by Sir2p.
We also analyzed Sir protein association at HMR and found Sir2, Sir3, and Sir4 proteins associated throughout HMR in SIR2 cells expressing either wild-type histones H3 and H4 or the H3 K9,14R H4 K16R mutants (Figure 1B and Supplementary Table 1) and HMRa1 was efficiently silenced in both strains (Figure 2B and Table 3). Therefore, like at HML, the deacetylation reaction by Sir2p at these key histone residues was not necessary for Sir protein spreading or silencing.
In sir2-345 mutants expressing wild-type H3 and H4, the Sir proteins were recruited to HMR-E, but the Sir proteins did not detectably spread throughout the HMR locus (Figure 1B) and silencing was lost (Figure 2B and Table 3) as we had observed previously (Rusché et al., 2002; Kirchmaier and Rine, 2006). In contrast, Sir protein association throughout HMR was restored in sir2-345 cells expressing the histone H3 K9,14R H4 K16R mutants, albeit the overall levels of Sir2p, Sir3p, and Sir4p at HMR were reduced relative to SIR2 cells, as had been observed at HML (Figure 1B and Supplementary Table 1). We next asked whether this pattern of reduced Sir protein association at HMR in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants occurred throughout HMR or reflected primarily a defect in Sir protein spreading from HMR-E to the rest of the HMR locus. Such spreading defects would be observed as a reduction in Sir protein association at a1 or HMR-I relative to HMR-E, whereas a general defect in association throughout the HMR locus would indicate that, once recruited to HMR, the Sir proteins spread efficiently throughout HMR. We compared the level of Sir2, Sir3, and Sir4 proteins at a1 or HMR-I relative to HMR-E in each strain from Figure 1B (Table 4). The efficiency of spreading of Sir2p and Sir4p from HMR-E to a1 and HMR-I was similar in sir2-345 and SIR2 cells expressing histone H3 K9,14R H4 K16R mutants. In contrast, the level of Sir3p was slightly reduced at a1 and HMR-I in sir2-345 cells relative to SIR2 cells (p = 0.025 and p = 0.063, respectively, n = 3; Table 4, see also Supplementary Table 1). In general, these results are consistent with an overall reduction in localization of Sir proteins to HMR in sir2-345 cells expressing hypoacetylated histone mutants, but efficient Sir protein spreading having occurred whenever Sir proteins were recruited to the HMR locus. Thus, the hypoacetylated histone mutants restored Sir protein association throughout the HMR locus in sir2-345 cells.
Sir Protein Spreading Does Not Stably Restore Silencing
We next tested whether Sir protein spreading led to silencing by monitoring transcription of α1 from HML and a1 from HMR (Figure 2, A and B). Transcription of α1 and a1 mRNA was silenced in SIR2 cells expressing either wild-type or mutant histones, and transcription of both α1 and a1 mRNA was restored in sir2-345 cells expressing wild-type histones, as anticipated. In sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants, the HM loci were derepressed relative to SIR2 cells expressing either wild-type or mutant histones, indicating a defect in silencing. However, in sir2-345 strains, both α1 and a1 mRNA levels were also significantly reduced in cells expressing histone H3 K9,14R H4 K16R mutants relative to those expressing wild-type histones (p = 0.018, n = 3 for each comparison).
To determine whether this reduction in transcription was due to Sir protein spreading or to the histone mutations themselves adversely affecting transcription, we monitored a1 mRNA from HMR in cells containing either wild-type histones or the hypoacetylated histone mutants but lacking SIR4. Sir2p and Sir3p are no longer recruited to HMR in sir4Δ cells (Rusché et al., 2002), allowing us to test specifically the effect of histone mutants on transcription. Transcription of a1 mRNA in sir2-345 sir4Δ and SIR2 sir4Δ cells expressing histone H3 K9,14R H4 K16R mutants was restored to the levels observed in cells expressing wild-type histones (Figure 2B). These results indicated that Sir protein spreading, but not the histone H3 K9,14R H4 K16R mutants interfered with transcription.
To further explore the influence of Sir protein spreading on silencing, we performed quantitative mating assays with these strains (Table 3). The ability of yeast to mate is dependent both on their ability to respond to mating pheromones of the opposite mating-type and on their ability to produce their own mating-type pheromone. Exposure to mating-type pheromones from the opposite mating-type will induce yeast to arrest in G1 to mate. The N terminal tail of histone H4 influences mating-type pheromone production. Cells lacking residues 4–19 of histone H4 are defective in producing both α factor and a factor (Kayne et al., 1988), raising the possibility that the histone mutants alone could prevent pheromone production and thereby block mating. However, because SIR2 cells expressing the histone H3 K9,14R H4 K16R mutants readily mated (Table 3), our data indicate that pheromones could be produced in SIR2 cells expressing histone mutants.
Mating of MATa cells to the opposite mating-type in G1 phase is also dependent on silencing of HMLα, and mating of MATα cells is likewise dependent on silencing at HMRa. The histone H3 K9,14R H4 K16R mutants supported silencing at both HMR and HML in SIR2 cells, albeit silencing at HML was less efficient in SIR2 cells expressing hypoacetylated histone mutants relative to wild-type histones (p = 0.025, n = 3). And, as expected, sir2-345 cells expressing wild-type histones did not mate (Table 3). Likewise, both MATa and MATα sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants were five orders of magnitude less efficient at mating than SIR2 cells expressing either wild-type or mutant histones, despite Sir protein association at HMRa1 and HMLα1 being restored to ∼45–62% and 42–97%, respectively, of the levels observed for SIR2 cells (Figure 1 and Supplementary Table 1) and transcription from the HM loci being significantly reduced (Figure 2, A and B). Together, these results indicate that although Sir protein spreading significantly diminished transcription from the HM loci, mating was not restored.
Histone Mutants Rescue Sir Protein Spreading at Telomere VIR in Catalytically Inactive sir2 Cells
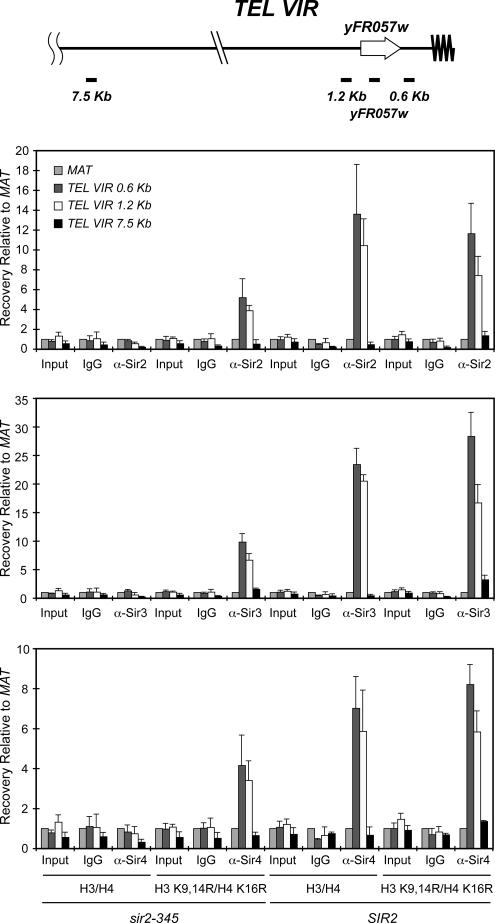
Like HM silencing, Sir-mediated silencing of genes near telomeres is position-dependent, but not gene-specific. Telomere position effect requires Sir2p, Sir3p, and Sir4p, and the Sir proteins are recruited to telomeres through interactions with telomere-associated proteins, including Rap1p (Luo et al., 2002). In SIR2 cells, Sir proteins spread along the chromosome from the telomere ends, whereas Sir protein spreading is lost in catalytically defective sir2 mutants (Hoppe et al., 2002; Luo et al., 2002). To determine whether Sir protein spreading at telomeres could be restored if histones H3 and H4 were hypoacetylated, we examined Sir protein spreading at telomere VIR by ChIP (Figure 3). In SIR2 cells expressing either wild-type or hypoacetylated histone mutants, Sir proteins spread to at least 1.2 kb from the end of telomere VIR, whereas in sir2-345 cells containing wild-type histones, Sir spreading was not detected. In contrast, Sir proteins spread to the 1.2-kb region in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants, albeit less efficiently than in SIR2 cells (Figure 3). These results indicate that, like at the HM loci, the histone H3 K9,14R H4 K16R mutants rescued Sir protein spreading in sir2-345 cells at telomere VIR.
Figure 3.
Sir proteins spread at telomere VIR in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants. Sir protein association at sites flanking telomere VIR was monitored by ChIP using IgG, and anti-Sir2p, Sir3p, and Sir4p antibodies were measured by quantitative real-time PCR using primers that amplified DNA 0.5, 1.2, and 7.5 kb from the end of telomere VIR (see Supplementary Table 2). The regions amplified at telomere VIR are noted at top of the figure (not to scale). Efficiency of co-precipitation of each locus was determined as described in Figure 1; n = 3.
We then determined the effect of Sir protein spreading on transcription near the telomere by monitoring expression of yFR057w, which lies from 1.1 to 0.65 kb from the end of telomere VIR (Figure 3). yFR057w was silenced in SIR2 cells expressing either wild-type histones or H3 K9,14R H4 K16R mutants (Figure 2C). In contrast, yFR057w was derepressed in sir2-345 strains relative to SIR2 cells expressing either wild-type or mutant histones. (For SIR2 cells expressing wild-type histones vs. sir2-345 cells expressing wild-type histones, p = 0.018; for SIR2 cells expressing wild-type histones vs. sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants, p = 0.025; for SIR2 cells expressing histone H3 K9,14R H4 K16R mutants vs. sir2-345 cells expressing wild-type histones, p = 0.018; and for SIR2 cells expressing histone H3 K9,14R H4 K16R mutants vs. sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants, p = 0.025. n = 3 for each comparison). And, as was seen for both HMRa1 and HMLα1 mRNA, transcription of yFR057w was slightly reduced upon restoring Sir protein spreading in sir2-345 cells with histone H3 K9,14R H4 K16R mutants relative to sir2-345 cells containing wild-type histones (p = 0.018, n = 3). This reduction was Sir-dependent, not histone mutant-dependent (Figure 2C). Together, these results imply that hypoacetylated histones were sufficient for Sir protein spreading at HML, HMR and the region flanking telomere VIR, but Sir protein spreading was not sufficient for silencing to be maintained.
Overexpression of SIR3 Restores Silencing at HM Loci in sir2-345 Cells in which Sir Protein Spreading Had Been Rescued
The above analyses indicated that Sir protein spreading could be restored in sir2-345 cells by providing histones that mimicked the hypoacetylated state, but the level of Sir proteins at HML, HMR and the region flanking telomere VIR were reduced compared with SIR2 cells. These findings suggested that the amount of Sir proteins present in sir2-345 cells was insufficient to maintain silencing. As the histone mutants potentially created high-affinity binding sites for Sir proteins throughout the genome, we considered whether Sir proteins could be relocalizing in sir2-345 cells and whether this might limit the amount of Sir proteins available to associate with the HM loci. However, in SIR2 cells expressing the same hypoacetylated histone mutants, the same potential binding sites would exist throughout the genome yet silencing still occurred (Figure 2, Table 3). And, we did not detect elevated levels of our control locus MAT, coprecipitating with any of the Sir proteins in sir2 or SIR2 strains with mutant relative to wild-type histones (data not shown). Thus, although Sir relocalization could potentially contribute to the observed defects in silencing, a dilution effect alone was not likely the sole cause of the silencing defect in sir2-345 cells expressing hypoacetylated histones.
Therefore, we first asked whether sir2-345 cells expressing hypoacetylated histone mutants could support silencing under appropriate conditions. To do so, we introduced a wild-type copy of SIR2 expressed from its own promoter on an ARS/CEN plasmid into sir2-345 cells containing the histone H3 K9,14R H4 K16R mutants (Figure 4). Silencing in these yeast was rescued, indicating that sir2-345p did not prohibit silencing at HMR or HML under these conditions and that, to enable silencing, Sir2p likely deacetylated a key substrate that sir2-345p could not.
In vivo, overexpression of SIR3 can restore silencing in SIR2 cells with silencing defects (e.g., Santos-Rosa et al., 2004). In vitro, the amount of Sir3p interacting with Sir2/4p is elevated in the presence of a product of Sir2p's NAD+-dependent histone deacetylation reaction, 2′-O-acetyl-ADP ribose (Liou et al., 2005). Because the catalytically inactive sir2-345p is defective in generating 2′-O-acetyl-ADP ribose and Sir3p spreading was reduced relative to the other Sirs at HMR in sir2-345 cells (Table 4), it was possible that Sir3p was somehow limiting at HMR in sir2-345 cells containing hypoacetylated histone mutants, leading to the defect in silencing. We thus considered whether overexpression of Sir3p could restore silencing in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants. To test whether silencing could be rescued at the HM loci, we introduced a 2 μm plasmid containing SIR3 expressed from its endogenous promoter into sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants and performed quantitative mating assays (Table 5, Supplementary Figure 1, and data not shown). Overexpression of Sir3p enhanced the efficiency of mating in MATα sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants by ∼7000-fold relative to vector alone, or to 9% the efficiency of SIR2 cells expressing histone H3 K9,14R H4 K16R mutants. Mating in MATa sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants was also enhanced by SIR3 overexpression by over 130-fold compared with vector alone, or to 1% the efficiency of SIR2 cells expressing histone H3 K9,14R H4 K16R mutants (Table 5). Thus, elevated expression of Sir3p partially bypassed the requirement for the enzymatic activity of Sir2p for silencing.
Table 5.
SIR3 overexpression in histone H3 K9,14R H4 K16R mutants rescues silencing at HML and HMR in sir2-345 cells
| SIR2 | HHT2 HHF2 | SIR3 | Relative efficiency of matinga |
|
|---|---|---|---|---|
| MATα | MATa | |||
| SIR2 | H3/H4 | Vector | 1 | 1 |
| SIR2 | H3 K9,14R/H4 K16R | Vector | 0.13 ± 0.029 | 0.78 ± 0.21 |
| sir2-345 | H3/H4 | Vector | <1 × 10−5 (0, 1 × 10−5, 0, 0) | <1 × 10−5 (0, 0, 7 × 10−5) |
| sir2-345 | H3 K9,14R/H4 K16R | Vector | <1 × 10−5 | <1 × 10−5 |
| SIR2 | H3/H4 | SIR3 | 0.98 ± 0.34 | 0.90 ± 0.091 |
| SIR2 | H3 K9,14R/H4 K16R | SIR3 | 0.65 ± 0.16 | 0.91 ± 0.13 |
| sir2-345 | H3/H4 | SIR3 | <1 × 10−5 (1 × 10−5, 0, 0, 0) | <1 × 10−5 (0, 2 × 10−5, 1 × 10−5) |
| sir2-345 | H3 K9,14R/H4 K16R | SIR3 | 0.0013 ± 0.00051 | 0.068 ± 0.027 |
a The efficiency of mating of MATa SIR2 or MATα SIR2 plus wild-type histones H3 and H4 and vector to the indicated tester strains JRY2728 (MATα) or JRY2726 (MATa), respectively, was determined relative to their plating efficiency (for MATa SIR2, 87 ± 16%, n = 4; for MATα SIR2, 97 ± 6.3%, n = 3) and was set to 1. The mating efficiencies of each strain relative to SIR2 cells expressing wild-type H3 and H4 was determined as indicated in Materials and Methods. Data reflect the average ± SD of three or four independent experiments, and data from individual experiments are shown in parentheses when any mating events were observed.
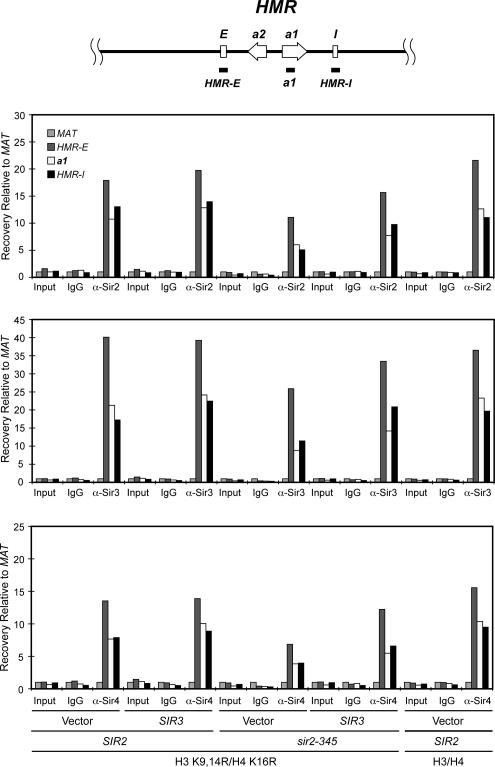
To determine directly the impact of overexpression of Sir3p on Sir protein association at HMR, we performed ChIP assays (Figure 5). Overexpression of Sir3 enhanced the association of Sir2p, Sir3p, and Sir4p throughout the HMR locus in sir2-345 mutants expressing histone H3 K9,14R H4 K16R mutants relative to cells containing vector alone, but did not restore Sir protein association to wild-type levels (Figure 5). Together, these results indicate that silencing can be restored, albeit inefficiently, in cells expressing catalytically inactive sir2p.
Figure 5.
SIR3 overexpression enhances Sir protein association throughout the HMR locus. Sir protein association in SIR2 or sir2-345 yeast containing wild-type or mutant H3K9,14R H4 K16R histones and overexpressing SIR3 was monitored by ChIP as in Figure 1. The regions amplified at HMR are noted at top of the figure (not to scale). The average of two independent experiments is shown.
DISCUSSION
The key observations reported here include the restoration of Sir protein spreading across HMR, HML and the region flanking telomere VIR in cells containing a catalytically inactive mutant of Sir2p and mutant histones H3 and H4 with key N-terminal residues that could not be acetylated. Significantly, these findings demonstrated the catalytic activity of Sir2p was not required for Sir protein complexes to spread efficiently across hypoacetylated chromatin. These observations also indicated that Sir protein spreading was insufficient to maintain silencing, but that mating could be partially restored upon overexpression of Sir3p. Moreover, these data led to the prediction that at least one additional key substrate of Sir2p exists that serves as a critical step in forming silent chromatin as silencing occurred in SIR2 cells expressing the hypoacetylated histone mutants.
Histone Hypocetylation Permits Sir Protein Spreading
Although Sir2p's deacetylase activity is not required for Sir2p or other Sir proteins to localize to silencers, Sir2p's catalytic activity is required for Sir protein spreading across HMR (Figures 1 and 6; Rusché et al., 2002; Kirchmaier and Rine, 2006) and in telomere regions (Figure 3) in cells with wild-type histones (Hoppe et al., 2002; Luo et al., 2002). Sir2p targets several acetylated lysine residues in the N-termini of histones H3 and H4 (Imai et al., 2000; Borra et al., 2004). However, the hypoacetylated state of only a subset of these residues is critical for silencing (Johnson et al., 1990; Park and Szostak, 1990 [see also Megee et al., 1990; Hecht et al., 1995]). We analyzed Sir protein spreading in histone mutants in which key lysine residues were mutated to mimic hypoacetylated states. We found that H3 K9,14R H4 K16R mutants rescued Sir protein spreading at the HM loci and telomere VIR (Figures 1 and 3). Although the overall level of Sir proteins at these loci was somewhat reduced in sir2-345 cells upon rescuing Sir protein spreading, at HMR, the efficiency of spreading from HMR-E to a1 or HMR-I was largely similar in SIR2 cells (Table 4). Our experiments did not distinguish whether a fraction of sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants in the analyzed population had recruited Sir proteins to the silencer and, in these cells, Sirs spread as efficiently as in SIR strains or whether all cells had fewer Sir proteins throughout the HM loci.
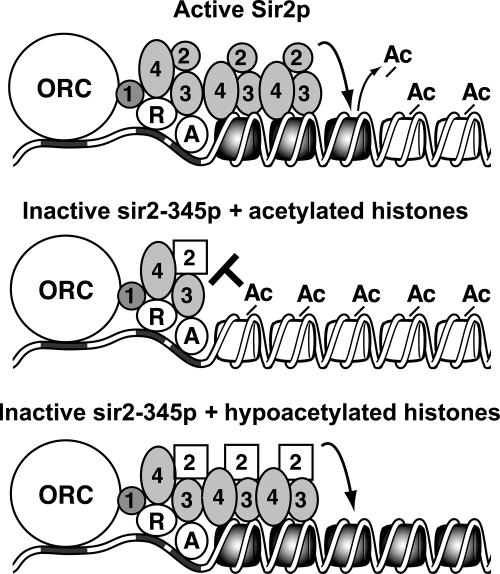
Figure 6.
Model for Sir protein spreading. Sir proteins are recruited to silencers through protein–protein interactions with ORC and Rap1p (R) and Abf1p (A). Sir2p then deacetylates lysine residues on N terminal tails of histones H3 and H4 of the adjacent nucleosome, enabling Sir3 and Sir4p to bind to H3 and H4. Repetitive deacetylation and binding cycles enable Sir proteins to spread along the chromosome (top). In cells expressing catalytically inactive sir2-345p, Sir proteins are recruited to the silencer but cannot spread along the chromosome because of acetylated lysine residues in the N-terminal tails of histones H3 and H4 (middle). Sir protein spreading is rescued in cells expressing sir2-345p and histone H3 and H4 mutants that mimic the hypoacetylated state (bottom). An additional unknown step(s) is required to form silent chromatin.
The Influence of Sir Protein Spreading on Silencing
When we monitored transcription as a direct readout of silencing, we observed a Sir protein-dependent reduction in a1 mRNA from HMR, α1 mRNA from HML, and mRNA from yFR057w in sir2-345 cells with hypoacetylated versus wild-type histones (Figure 2). This reduction in transcription correlated with the restoration of Sir protein spreading. This reduction was not, however, as efficient as that observed in SIR2 cells expressing either hypoacetylated or wild-type histones. In contrast, when we monitored mating as an indirect readout of silencing, SIR2 cells expressing these histone mutants readily mated, but mating was not restored in sir2-345 cells (Table 3) unless SIR3 was overexpressed (Table 5).
At least three possible mechanisms could account for how the observed levels of Sir protein spreading the sir2-345 cells expressing histone H3 K9,14R H4 K16 R mutants could lead to a Sir-dependent reduction in transcription from the HM loci. The first possibility was that two populations of cells were present in the cultures of sir2-345 cells expressing histone H3 K9,14R H4 K16 R mutants; one with Sirs spread throughout the HM loci, heritably silencing transcription, and one without Sir protein spreading and, therefore, expressing normal levels of transcripts from the HM loci. In this scenario, however, we would have expected to detect this former silenced population in the quantitative mating assays, which we did not (Table 3).
The second possibility was that in the sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants, Sir proteins had spread throughout the HM loci in all cells creating a structure that interfered with the probability of promoter access by RNA Polymerase II, as in SIR2 cells upon silent chromatin formation (Chen and Widom, 2005). Yet, unlike SIR2 cells, in the mutants, the silent chromatin that formed was somehow unstable and allowed continual, but reduced levels of transcription from the HM loci in all cells. This instability could be caused by a subtle structural defect of sir2-345p or by the continued acetylated status of a key target of Sir2p (see below). Sir relocalization throughout the genome due to the hypoacetylated histones could also contribute to this instability. However, Sir relocalization likely could not account for all of the silencing defect in sir2-345 cells because histone H3 K9,14R H4 K16R mutants supported both Sir protein spreading and silencing in SIR2 cells (Figures 1–5, Tables 3–5, and Supplementary Table 1). Regardless of the mechanism of the silencing defect, in this second scenario, none of the cells would have been expected to be able to mate, which was our observation (Table 3).
The third possibility was that silencing was efficiently established in sir2-345 cells expressing histone H3 K9,14R H4 K16R mutants but then was disrupted for any of the above reasons at a later point in the cell cycle, resulting in transcription from the HM loci in G1 phase. As the HM loci must be silenced in G1 phase for cells to mate, this scenario would have also led to the observed complete defect in mating. Future analyses should distinguish between these latter two possibilities.
Silencing and Targets of Sir2p
Together, our findings indicate that hypoacetylated histone tails are sufficient for Sir protein spreading (Figure 6) and a Sir-dependent reduction in transcription, but are not sufficient for stable silent chromatin. For silencing to be established, Sir2p may have additional critical substrate(s) in vivo. These substrates could theoretically be other acetylated lysine residues on histones or, alternatively, other components of silent chromatin or the transcription machinery. Key targets could either be components that must be “activated” by deacetylation to permit silencing or inhibitors that must be “inactivated” by deacetylation for silencing to occur. Consistent with the notion of additional targets, Sir2p has been reported to mono-ADP-ribosylate itself in vitro and this mono-ADP-ribosylation requires catalytically active Sir2p in vivo (Tanny et al., 1999), but the role of this modification is unknown. Also, co-expression of Sir2p restored silencing in sir2-345 cells in which Sir protein spreading had been rescued (Figure 4). In contrast, co-expression of a different catalytically defective mutant of SIR2, sir2 H364Y, has been reported to inhibit telomeric silencing in SIR2 cells, and, if greatly overexpressed, inhibit rDNA silencing as well (Tanny et al., 1999). Understanding these differences in sensitivity of each locus and of different sir2 mutants awaits further study.
Sir Protein Spreading during the Formation of Silent Chromatin
We have previously shown that Sir proteins are recruited to silencers, histones H3 and H4 are deacetylated, and Sir proteins can spread across HMR in G1 phase, before establishing silencing in S phase (Kirchmaier and Rine, 2006; see also Lau et al., 2002). These observations indicate that the presence of Sir proteins at HMR is insufficient to mediate silencing, and the S phase–regulated step reflects an event that occurs after recruitment (Lau et al., 2002; Kirchmaier and Rine, 2006). Here, we demonstrate a second condition in which Sir spreading can occur before silencing. And, recent observations indicate targeted acetyltransferases can disrupt silencing without preventing Sir association (Yu et al., 2006). Together, these results raise the possibility that Sir2p may have an additional, and as yet unrecognized, activity that is required to establish silent chromatin. Consistent with Sir2p having additional functions beyond enabling Sir protein spreading, temperature-sensitive alleles of SIR2 with mitotic-specific silencing defects have been reported (Matecic et al., 2006).
In addition to deacetylation, multiple other changes to histone modifications occur over time during the formation of silent chromatin (Katan-Khaykovich and Struhl, 2005). Because histone H3 K9,14R H4 K16R mutants support silencing in SIR2 cells (Figures 2 and 4, Tables 1 and 3), we anticipate that these mutations do not block silencing in sir2-345 cells by preventing such changes on histones from occurring. Dissection of which changes to histone modifications occur upon Sir protein spreading versus those needed for silencing should lead to insights into how silent chromatin is assembled.
Sir2, Free Energy Release, O-Acetyl-ADP Ribose, and Silencing
The inability of silencing to be restored upon rescuing Sir protein spreading may be related to the lack of free energy being released or of 2′-O-acetyl-ADP ribose being produced by sir2-345p. It is possible that sir2-345p cannot readily undergo a critical energy- or 2′-O-acetyl-ADP ribose–driven structural transition needed for silencing, but not for Sir protein spreading. 2′-O-acetyl-ADP ribose, induces conformational and stoichiometric changes in Sir2/3/4 complexes in vitro (Liou et al., 2005). Specifically, 2′-O-acetyl-ADP-ribose alters interactions between Sir3p and Sir2/Sir4p in which the Sir2/3/4p complex is converted to a form that contains more Sir3p relative to Sir2p and Sir4p. Such a change could be required in vivo for silencing. However, because Sir spreading occurs in sir2-345 mutants (Figures 1, 3, and 6), generation of 2′-O-acetyl-ADP ribose locally by Sir2p is not necessary for Sir protein spreading. This study does not, however, rule out 2′-O-acetyl-ADP ribose being supplied in trans by Sir2p paralogs, the Hst proteins. By overexpressing Sir3p in sir2-345 mutants in which Sir spreading was rescued, we may have bypassed the role of 2′-O-acetyl-ADP ribose in inducing such changes in vivo and thereby rescued silencing in some cells (Figure 6 and Table 5). Regardless, because Sir spreading and histone deacetylation, and therefore energy release and generation of 2′-O-acetyl-ADP ribose, occurs in G1 phase before establishing silencing (Kirchmaier and Rine, 2006; see also Lau et al., 2002), these roles of the catalytic activity of Sir2p alone are not sufficient to establish silencing.
Future
The results of this study have reframed the issues regarding the NAD+-dependent deacetylase function of Sir2p in epigenetic processes. One objective is to determine what additional substrate(s) of Sir2p are critical for silencing. Another objective is to determine whether the deacetylated state of that substrate is key for silencing or whether the deacetylation reaction itself and 2′-O-acetyl-ADP ribose are crucial for forming silent chromatin. Addressing these questions should enhance our understanding of how cells epigenetically regulate gene expression and the roles of Sir2p in epigenetic processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Briggs and Joe Ogas for helpful discussions and Scott Briggs, Paul Kaufman, Erin Osborn, and Jasper Rine and our reviewers for invaluable comments on this manuscript. We thank Leonard Guarente, Paul Kaufman, Mark Parthun, Jasper Rine, and Sharon Roth for reagents, strains, and plasmids. This work was supported by U.S. Department of Agriculture Hatch Grant IND053072 (A.L.K.) and Kimmel Scholar Award SKF-03-010 (A.L.K.). This project was also supported by an American Cancer Society Institutional Research Grant to the Purdue Cancer Center. This is article number 2006-18027 from the Purdue University Agricultural Experiment Station.
Abbreviations used:
- SIR
Silent Information Regulator
- ChIP
chromatin immunoprecipitation.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0669) on October 11, 2006.
REFERENCES
- Adams A., Gottschling D. E., Kaiser C. A., Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Borra M. T., Langer M. R., Slama J. T., Denu J. M. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- Bose M. E., McConnell K. H., Gardner-Aukema K. A., Muller U., Weinreich M., Keck J. L., Fox C. A. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 2004;24:774–786. doi: 10.1128/MCB.24.2.774-786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel R. E., Allis C. D., Turner B. M., Broach J. R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen A. A., Milne L., Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- Chang J. H., Kim H. C., Hwang K. Y., Lee J. W., Jackson S. P., Bell S. D., Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- Chen L., Widom J. Mechanism of transcriptional silencing in yeast. Cell. 2005;120:37–48. doi: 10.1016/j.cell.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Donze D., Kamakaka R. T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hoppe G., Tanny J., Rudner A., Gerber S., Danaie S., Gygi S., Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne P. S., Kim U.-J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Qin S., Gottschling D. E., Parthun M. R. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W. J., Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol. Cell. Biol. 1987;7:4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Kirchmaier A. L., Rine J. Cell-cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:852–862. doi: 10.1128/MCB.26.3.852-862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Blitzblau H., Bell S. P. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 2002;16:2935–2945. doi: 10.1101/gad.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Luo K., Vega-Palas M. A., Grunstein M. Rap1-Sir4 binding independent of other Sir, yku or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Matecic M., Martins-Taylor K., Hickman M., Tanny J., Moazed D., Holmes S. G. New alleles of SIR2 define cell cycle specific silencing functions. Genetics. 2006;173:1939–1950. doi: 10.1534/genetics.106.055491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F. J., Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P. C., Morgan B. A., Mittman B. A., Smith M. M. Genetic analysis of histone H4, essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Min J., Landry J., Sternglanz R., Xu R. M. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Park E.-C., Szostak J. W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier D. H., Ekena J. L., Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen J. W., Kornberg A. The phosphorolysis of nicotinamide riboside. J. Biol. Chem. 1951;193:497–507. [PubMed] [Google Scholar]
- Rudner A. D., Hall B. E., Ellenberger T., Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché L. N., Kirchmaier A. L., Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché L. N., Kirchmaier A. L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Bannister A. J., Dehe P. M., Geli V., Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Ma H., Botstein D. Manipulating yeast genome using plasmid vectors. Methods Enzymol. 1990;185:280–297. doi: 10.1016/0076-6879(90)85025-j. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Sutton A., Shia W. J., Band D., Kaufman P. D., Osada S., Workman J. L., Sternglanz R. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 2003;278:16887–16892. doi: 10.1074/jbc.M210709200. [DOI] [PubMed] [Google Scholar]
- Tanny J. C., Dowd G. J., Huang J., Hilz H., Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tanny J. C., Kirkpatrick D. S., Gerber S. A., Gygi S. P., Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gottschling D. E. Assays for gene silencing in yeast. Methods Enzymol. 2002;350:165–186. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yu Q., Sandmeier J., Xu H., Zou Y., Bi X. Mechanism of the long range anti-silencing function of targeted histone acetyltransferases in yeast. J. Biol. Chem. 2006;281:3980–3988. doi: 10.1074/jbc.M510140200. [DOI] [PubMed] [Google Scholar]
- Zatman L. J., Kaplan N. O., Colowick S. P. Inhibition of spleen diphosphopyridine nucleotidase by nicotinamide, an exchange reaction. J. Biol. Chem. 1953;200:197–212. [PubMed] [Google Scholar]
- Zhang W., Bone J. R., Edmondson D. G., Turner B. M., Roth S. Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Chai X., Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Zhao K., Harshaw R., Chai X., Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. USA. 2004;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.