Abstract
The Ire1p transmembrane receptor kinase/endonuclease transduces the unfolded protein response (UPR) from the endoplasmic reticulum (ER) to the nucleus in Saccharomyces cerevisiae. In this study, we analyzed the capacity of a highly basic sequence in the linker region of Ire1p to function as a nuclear localization sequence (NLS) both in vivo and in vitro. This 18-residue sequence is capable of targeting green fluorescent protein to the nucleus of yeast cells in a process requiring proteins involved in the Ran GTPase cycle that facilitates nuclear import. Mutagenic analysis and importin binding studies demonstrate that the Ire1p linker region contains overlapping potential NLSs: at least one classical NLS (within sequences 642KKKRKR647 and/or 653KKGR656) that is recognized by yeast importin α (Kap60p) and a novel βNLS (646KRGSRGGKKGRK657) that is recognized by several yeast importin β homologues. Kinetic binding data suggest that binding to importin β proteins would predominate in vivo. The UPR, and in particular ER stress-induced HAC1 mRNA splicing, is inhibited by point mutations in the Ire1p NLS that inhibit nuclear localization and also requires functional RanGAP and Ran GEF proteins. The NLS-dependent nuclear localization of Ire1p would thus seem to be central to its role in UPR signaling.
INTRODUCTION
Within the lumen of the endoplasmic reticulum (ER), a variety of resident ER proteins assist newly translocated nascent polypeptides to fold into their correct tertiary and quaternary structures (Stevens and Argon, 1999). These resident proteins include molecular chaperones that recognize and stabilize partially folded intermediates during polypeptide folding and assembly, as well as enzymes that catalyze rate-determining steps in folding, such as protein disulfide isomerase and peptidyl prolyl isomerases. Under normal growth conditions these chaperones and folding catalysts are synthesized constitutively and abundantly. However, their rates of synthesis can be increased significantly by the accumulation of mutant proteins in the ER or by a variety of stress conditions whose common denominator is thought to be the accumulation in the ER of unfolded polypeptides (Kozutsumi et al., 1988; Mori et al., 1992). This “unfolded protein response” (UPR) operates in yeast and higher eukaryotes to regulate the levels of ER chaperones and protein folding catalysts (for review, see Ma and Hendershot, 2001; Patil and Walter, 2001; Kaufman, 2002; Ron, 2002). Microarray analysis of yeast cells demonstrated that the UPR also activates genes encoding a variety of other proteins involved in diverse processes such as translocation, glycosylation and degradation of secretory proteins, lipid/inositol metabolism, cell wall biogenesis, vesicle trafficking/transport, and vacuolar protein sorting (Travers et al., 2000). Conserved elements (UPREs) are present in the promoter regions of many UPR-regulated yeast genes (Mori et al., 1992, 1998; Patil et al., 2004). Thus, an intracellular sensing system monitors events in the lumen of the yeast ER and transduces signals across the ER membrane and into the nucleus to activate the transcription of UPRE-controlled genes.
In Saccharomyces cerevisiae, the ER-to-nucleus (ERN) signal transduction pathway contains two unique components, the Ire1p/Ern1p transmembrane protein and the bZIP Hac1p transcription factor that binds UPREs. These components are not essential for vegetative growth, but they are absolutely necessary for survival under conditions that cause UPR stress (Cox et al., 1993; Mori et al., 1993). Ire1p contains in one molecule three of the essential components of the UPR pathway: the lumenal sensor, the mechanism for transducing the signal across the ER membrane, and the mechanism of transcriptional activation of UPRE-controlled genes. The glycosylated N-terminal portion of Ire1p is located in the ER lumen, and, apparently through binding to uncomplexed BiP (Kohno et al., 1993; Okamura et al., 2000), senses the load of misfolded proteins within the ER. The C-terminal half of Ire1p carries an essential protein kinase domain (Mori et al., 1993), which is activated by receptor dimerization (Shamu and Walter, 1996; Welihinda and Kaufman, 1996), as well as a C-terminal domain that functions as an RNA endonuclease after activation by UPR stress (Sidrauski and Walter, 1997). This endoribonuclease, together with Rlg1p (previously identified as a tRNA ligase) (Sidrauski et al., 1996), is required for the splicing of the mRNA that encodes Hac1p (Cox and Walter, 1996; Mori et al., 1996). Additional participants in the UPR response in yeast include the Ada2p and Ada5p subunits of the Gcn5 transcriptional coactivator complex, which interact with sequences within the C-terminal half of Ire1p (Welihinda et al., 2000), and the Gcn4p transcriptional activator, which cooperates with Hac1p to transactivate UPRE-containing genes (Patil et al., 2004).
The ERN signaling pathway outlined above presents a topological problem because it suggests that the C-terminal domain of Ire1p functions in the nucleus, where the Rlg1p ligase (Clark and Abelson, 1987) and the components of the Gcn5 complex are located. Thus, if Ire1p functions as an intact transmembrane protein (Shamu and Walter, 1996; see data below), it must be localized to the inner nuclear membrane, as has been demonstrated for the murine Ire1α homologue of yeast Ire1p (Lee et al., 2002). However, Ire1p, which contains an N-terminal hydrophobic signal sequence for targeting to the ER (Mori et al., 1993), must be synthesized on membrane-bound ribosomes that are present on the rough ER, which includes the outer nuclear membrane, but that are not present on the inner nuclear membrane. The inner and outer nuclear membranes are separated by nuclear pores that control the flow of macromolecules into and out of the nucleus (Rout et al., 2000). The signal-dependent trafficking of proteins and RNA species through these pores mediated by importins α and β is now well documented (Görlich and Kutay, 1999; Strom and Weis, 2001; Poon and Jans, 2005), but very little is known about translocation of membrane proteins between outer and inner nuclear membranes. Here, we demonstrate that a highly basic sequence within the linker region of Ire1p can mediate nuclear localization of green fluorescent protein and is recognized by both importin α and multiple importin β proteins. Mutations within this sequence that prevent the accumulation of Ire1p in the nucleus result in impaired processing of HAC1 mRNA and inhibition of the UPR. This work provides the first evidence that Ire1p interacts with the nuclear import machinery and suggests that nuclear localization of Ire1p is essential for UPR signaling.
MATERIALS AND METHODS
Strains and Plasmids
Descriptions and sources of the strains and plasmids used in this study are listed in Table 1. Yeast cells were grown in YP medium containing glucose (YPD) or synthetic complete media lacking appropriate amino acids (Kaiser et al., 1994). Strains containing a temperature-sensitive mutation were grown at the permissive temperature (23°C) before incubation under nonpermissive conditions (37°C). All plasmid manipulations were carried out using standard protocols (Sambrook et al., 1989). The sequences of oligonucleotides used in this study are available upon request. DNA sequence analysis was used to confirm the accuracy of introduced mutations.
Table 1.
List of yeast strains and plasmids
| Strain/plasmid | Genotype/description | Reference |
|---|---|---|
| KMY1005 | MATα leu2-3,112 ura3-52 his3Δ200 trp1Δ901 lys2-801 | 1 |
| KMY2005 | MATα leu2-3,112 ura3-52 his3Δ200 trp1Δ901 lys2-801 sec53-6 | 1 |
| KMY1115 | KMY1005 ern1Δ::TRP1 UPRE-lacZ::URA3 | 2 |
| KMY2115 | KMY2005 ern1Δ::TRP1 UPRE-lacZ::URA3 | 2 |
| PSY580 | MATa ura3-52 leu2Δ1 trp1Δ63 | 3 |
| PSY688 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 ade2Δ::hisGsrp1-31 | 4 |
| PSY713 | MATα ura3-52 leu2Δ1 trp1Δ63prp20-1 | 5 |
| PSY868 | MATα ura3-52 leu2Δ1 ade2Δ::hisGrna1-1 | 5 |
| PSY869 | MATα ura3-52 leu2Δ1 ade2Δ::hisG ade3rna1-1 (2μ-ADE3-RNA1) | 5 |
| PSY961 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200gsp1::HIS3(CEN-LEU2-gsp1-1) | 6 |
| PSY967 | MATα ura3-52 leu2Δ1 his3Δ200kap123Δ::HIS3 | 7 |
| PSY1042 | MATa ura3-52 trp1Δ63his3Δ200pse1-1 kap123Δ::HIS3 | 7 |
| PSY1103 | MATa ura3-52 leu2Δ1 trp1Δ63rsl1-4 | P. A. Silver |
| PSY1199 | MATα ura3-52 leu2Δ1 his3Δ200 ade2Δ::hisG ade8Δ100::KANRnmd5Δ::HIS3 | 8 |
| PSY1200 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 lys2sxm1Δ::HIS3 | 6 |
| PSY1201 | MATa ura3-52 leu2Δ1 trp1Δ63pse1-1 | 6 |
| PSY1664 | MATα kap104::URA3::HIS3 | 9 |
| PSY1267 | PSY580 with integrated KAP108-GFP replacing KAP108 (SXM1) | 10 |
| PSY1783 | PSY580 with integrated KAP114-GFP replacing KAP114 | 11 |
| PSY1786 | PSY580 with integrated KAP95-GFP replacing KAP95 (RSL1) | 10 |
| PSY2023 | PSY580 with integrated KAP104-GFP replacing KAP104 | P. A. Silver |
| PSY2024 | PSY580 with integrated KAP111-GFP replacing KAP111 (MTR10) | P. A. Silver |
| PSY2067 | PSY580 with integrated KAP123-GFP replacing KAP123 (YRB4) | P. A. Silver |
| PSY2079 | PSY580 with integrated KAP121-GFP replacing KAP121 (PSE1) | 10 |
| RG11907 | MATa ura3 leu2 his3 lys2ire1Δ::kanMX | Research Genetics (Huntsville, AL) |
| pERN1-EM | 2μ-based yeast vector, LEU2, ERN1 (IRE1) under ERN1 promoter | 12 |
| pERN1-BC | CEN6-ARS1-based yeast vector, LEU2, ERN1 under KAR2 promoter | 12 |
| pGFP-N-FUS | CEN6-ARS1-based yeast vector, URA3, GFP under MET promoter | 13 |
The references cited are as follows: 1.Mori et al. (1996);
12. Mori et al. (1993);
13. Niedenthal et al. (1996). Note that ern1Δ = ire1Δ.
Mutations in ERN1/IRE1 encoding alterations in the classical nuclear localization signal (cNLS) were generated by oligonucleotide-mediated site-directed mutagenesis (Kunkel, 1985). Restriction fragments encompassing the mutated sequences were inserted into both the pERN1EM and pERN1BC plasmids, replacing the corresponding wild-type sequence. Plasmids capable of expressing GFP–Ire1p fusion proteins under the control of the MET25 promoter were constructed as follows: Nucleotides encoding the C-terminal region of Ire1p (residues 556–1115) were amplified by polymerase chain reaction (PCR) by using oligonucleotides VS1 and VS2 as primers and pERN1BC3 as the template. The primers also added terminal SpeI and XhoI restriction sites that were used to clone the amplified fragments between the SpeI and XhoI sites of pGFP-N-FUS to generate pGFIC. Plasmid pGFIC(K646A,R647A) was created from pGFIC using the complementary mutagenic oligonucleotides VS3 and VS4 and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmid pISGFFVITMC was created from pGFIC by inserting an XbaI–SpeI fragment from pC4-FV1E (Ariad Pharmaceuticals, Cambridge, MA) that encodes a variant FKBP (Spencer et al., 1993; Clackson et al., 1998) between the XbaI and SpeI sites of pGFIC, then inserting a double-stranded oligonucleotide (VS5/VS6) encoding the Ire1p transmembrane domain (residues 525–555) into the SpeI site of the resulting construct (pGFFVIC) to generate pGFFVITMC, and finally by inserting another double-stranded oligonucleotide (VS7/VS8) encoding the first 26 residues of Ire1p, which includes the ER targeting sequence, into the EcoR1 site of pGFFVITMC to generate pISGFFVITMC. DNA sequence analysis was used to confirm the correct insertion and orientation of the introduced nucleotide sequences at each step. The same procedures were used to generate pISGFFKITMC(K646A, R647A) from pGFIC(K646A, R647A). Nucleotides encoding wild-type or mutant versions of the linker region of Ire1p (residues 556–672) were amplified by PCR by using oligonucleotides VS1 and VS9 as primers and pERN1BC3 or pERN1BC(K644T,R645T) as templates. The primers also added terminal SpeI and XhoI restriction sites that were used to clone the amplified fragments into pGFP-N-FUS to give rise to pGFIL and pGFIL(K644T,R645T), respectively. To create the pGFIB series of plasmids, pairs of complementary oligonucleotides (VS10–VS19), which form duplexes encoding either wild-type or mutant versions of the 19 residue NLS (residues 642–660) flanked by SpeI and XhoI restriction sites, were phosphorylated using T4 polynucleotidyl kinase. Each complementary pair was then allowed to anneal in an Eppendorf thermocycler by using a program with a temperature drop of 5°C every 5 min starting from 95°C and ending at 23°C. The annealed duplex DNAs were purified on 15% nondenaturing polyacrylamide gels and cloned between the SpeI and XhoI sites of the pGFP-N-FUS vector. To generate pLG316, the KAP123 gene was amplified by PCR. The introduction of two flanking SmaI restriction sites within the oligonucleotide primers (LG1 and LG2) enabled in-frame subcloning of the KAP123 open reading frame (ORF) into the SmaI restriction site at the 3′ end of the glutathione S-transferase (GST) gene in pGEX-2TK.
Peptides
ENLS peptides (see sequences in Figures 4A and 9) were synthesized by continuous flow solid-phase synthesis by using fluorenylmethoxycarbonyl- or tert-butyloxycarbonyl-protected amino acids, purified using a model 1100 liquid chromatograph (Hewlett Packard, Palo Alto, CA) with a Hypersyl ODS micropreparative reverse phase-high performance liquid chromatography column (Hewlett Packard) and analyzed by matrix-assisted laser desorption ionization/time of flight mass spectrometry (Kratos Analytical, Manchester, Lancashire, United Kingdom) and amino acid analysis. The importin β1-recognized peptide representing parathryoid hormone related protein (PTHrP) residues 67–94 (YLTQETNKVETYKEQPLKTPGKKKKGKP) has been described previously (Lam et al., 1999).
Figure 4.
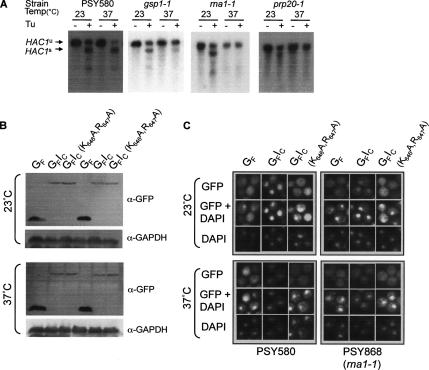
Components of the Ran cycle are required for HAC1 mRNA splicing and for import of Ire1p NLS-containing proteins. (A) ER stress-induced HAC1 mRNA splicing was assayed in wild-type yeast (strain PSY580) and strains carrying ts mutations in GSP1 (RanGTPase), RNA1 (RanGAP), or PRP20 (RanGEF). Cultures were grown at 23°C to an OD600 of 0.6 and then split into two aliquots, one of which was shifted to 37°C. After incubation for 4 h, tunicamycin was added to both cultures to a final concentration of 5 μg/ml, and a sample was taken immediately (−Tu) and snap-frozen before incubation was continued at 23 or 37°C. One hour later, a second sample (+Tu) was taken from each culture. Total RNA was extracted from each sample as described by Ausubel et al. (1995). After electrophoresis of glyoxal-denatured RNA through a 1.4% agarose gel containing 10 mM iodoacetic acid, the RNA was blotted to NYTRAN Plus, probed with randomly primed 32P-labeled HAC1 PCR products, and autoradiographed. The relative migration of the various HAC1 mRNA species is indicated on the right of the panel (u, unspliced; s, spliced). (B) PSY580 or PSY868 (rna1-1) yeast cells expressing GFP or GFP-Ire1C fusion proteins were grown at 23°C to an OD600 of 0.6 and then split into two aliquots, one of which was shifted to 37°C. After incubation for 4 h, protein extracts were prepared, resolved by SDS-PAGE, and analyzed by immunoblotting by using anti-GFP and anti-GAPDH antibodies. (C) Single colonies of PSY580 or PSY868 (rna1-1) yeast cells expressing GFP or GFP-Ire1C fusion proteins were grown at 23°C to an OD600 of 0.6 in SC-URA-MET medium and then split into two aliquots. One aliquot remained at 23°C, whereas the second aliquot was shifted to 37°C. After incubation for 4 h, the cells were examined by fluorescence microscopy.
Figure 9.
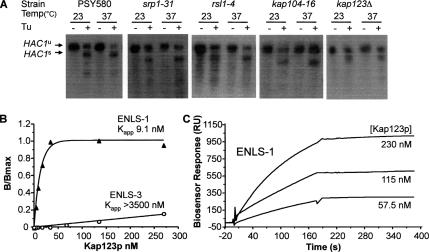
Redundancy of importin binding to the Ire1p NLS. (A) ER stress-induced HAC1 mRNA splicing was assayed in wild-type yeast (strain PSY580) and strains carrying ts mutations in SRP1 (Kap60p), RSL1 (Kap95p/karyopherin β1), KAP104 (Kap104p/karyopherin β2), and KAP123 (Kap123p/karyopherin β4) as described in the legend to Figure 4A. The relative migration of the spliced (s) and unspliced (u) HAC1 mRNA species is indicated on the right of the panel. (B and C) ENLS-1 and ENLS-3 peptides were tested for recognition by yeast importin Kap123p (β4) by using an ELISA-based binding assay (as described in the legend to Figure 6) (B) or a biosensor (as described in the legend to Figure 8) (C).
Expression of GST Fusion Proteins
Expression and purification of murine (m-PTAC58/Rch1 and PTAC97) or yeast (Kap60p or Kap95p) importin α and β subunits fused to the GST protein were performed as described previously (Lam et al., 1999). Human Ran was also expressed as a GST fusion protein (Hu and Jans, 1999) and then GST-free Ran was prepared by thrombin cleavage and loaded with nucleotides as described by Chi et al. (1996). All purified proteins were dialyzed against storage buffer (20 mM HEPES, pH 7.3, 100 mM KOAc, and 2 mM dithiothreitol [DTT]) and kept frozen at −80°C until use.
Enzyme-linked Immunosorbent Assay (ELISA)-based Importin Binding Assay
Binding of GST-importin fusion proteins to Ire1p peptides was quantitated using an ELISA-based assay (Hu and Jans, 1999). Where inhibition studies were performed, importin and Ran proteins were added simultaneously to the immobilized peptide. The amount of bound importin was determined using an antibody directed against the GST moiety of the importin fusion protein (goat anti-GST antibody; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Biosensor-based Importin Binding Assay
Biosensor analyses were performed using an optical biosensor (BIAcore 2000; BIAcore, Uppsala, Sweden) (Nice and Catimel, 1999). To obtain a defined orientation of peptide on the sensor surface and achieve optimum binding presentation for the importin proteins, the ENLS-1 peptide was thiol conjugated via the N-terminal cysteine on the sensor surface (Catimel et al., 1997). Binding data were generated by passing varying concentrations of GST–importin fusion proteins over the immobilized peptide.
Binding of Importin β-Green Fluorescent Protein (GFP) Fusion Proteins to Resin-linked NLS Peptides
ENLS-4 peptides (see Figure 9 legend for sequence) were linked via their N-terminal cysteine residues to thiopropyl-Sepharose 6B as described by the manufacturer (Pharmacia, Uppsala, Sweden). Aliquots of peptide-linked resin (25 μl of packed resin suspended in 50 μl of 0.1 M Tris-HCl, pH 7.0, 1 M NaCl, and 1 mM EDTA [EQ buffer]) were incubated for 1 h on ice with 50-μl aliquots of yeast cell extracts containing importin β-GFP fusion proteins, prepared as described by Seedorf et al. (1999). The unbound fraction was separated from resin by centrifugation in a Microfuge, and the resin was then washed three times with EQ buffer containing 0.5% Triton X-100 before bound importin-GFP proteins were eluted from the resin with 0.1 M Tris-HCl, pH 8.3, 1 M NaCl, 1 mM EDTA, and 20 mM DTT. In some experiments, lysates were incubated in the presence of 1 mM guanosine 5′-O-(3-thio)triphosphate (GTPγS) or GDP (Sigma-Aldrich, St. Louis, MO) before the peptide resin was added.
Cell Extracts and Immunoblotting
Protein extracts from yeast cells were made using EZ buffer [60 mM Tris-HCl, pH 6.8, 10% (vol/vol) glycerol, 2% (wt/vol) SDS, and 5% (vol/vol) β-mercaptoethanol] and quantitated using the Bradford protein assay kit (Bio-Rad, Hercules, CA). Volumes of extract containing equal amounts of total protein (∼150–200 μg) were boiled in 5X SDS sample buffer for 10 min and then loaded onto 8 or 12% acrylamide gels for PAGE. For immunoblot analysis, proteins were transferred onto nitrocellulose (0.45-μm Protran; Whatman Schleicher and Schuell, Dassel, Germany) via wet transfer in the Mini Trans-Blot Cell (Bio-Rad) and blocked with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20. Detection of Ire1p was performed using a polyclonal antibody directed against residues 32–259 of Ire1p/Ern1p (Mori et al., 1993), and GFP fusion proteins were detected using polyclonal anti-GFP antibodies (Seedorf et al., 1999). Subsequently, the membranes were probed with monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (a gift from Trevor Lithgow, University of Melbourne, Melbourne, Australia) as a control for protein loading. The proteins were detected using ECL reagents (Pierce Chemical, Rockford, IL, or Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions.
β-Galactosidase Assay
β-Galactosidase activity was assayed by the crude extract method of Kaiser et al. (1994). Protein concentrations were determined using the DC Protein Assay kit (Bio-Rad).
HAC1 mRNA Splicing
Total cellular RNA from intact yeast cells was isolated as described by Ausubel et al. (1995). Total RNA (5.4 μg) was denatured using glyoxal and separated by electrophoresis on a 1.4% agarose gel. Separated RNA species were transferred onto nitrocellulose membrane (Nylon N+; GE Healthcare) and probed with random primer 32P-labeled HAC1 PCR products. Autoradiography was performed either at room temperature or −70°C. Experimental details are reported in the figure legends.
Yeast Fluorescence Microscopy
Yeast cells expressing GFP fusion proteins were prepared for visualization as follows: log-phase cultures (OD600 = 0.6–0.8) were grown at 30°C in synthetic medium in the absence of methionine and uracil to induce expression from the MET25 promoter of pGFP-N-FUS vector. When the expressed fusion proteins contained the FKBP-based dimerization sequence (FV) dimerization domain (Spencer et al., 1993; Clackson et al., 1998), the cell-permeant dimerizer AP20187 (Ariad Pharmaceuticals) was added to the medium at a final concentration of 2 μM, and incubation was continued for 2 h. Cells were fixed by adding 1/10 volume of a 37% stock solution of formaldehyde directly to the medium and incubating for at least 30 min. Harvested cells were washed twice with 0.1 M potassium phosphate, pH 7.5, followed by two washes with 1X phosphate-buffered saline (PBS), and then resuspended in PBS (200 μl per 10 ml of culture). Twenty-microliter aliquots of cell suspensions were smeared evenly on cover slips coated with poly-l-lysine and allowed to dry. The coverslips were then inverted onto a 10-μl drop of Mowiol containing 5 ng/μl 4,6-diamidino-2-phenylindole (DAPI) on a glass slide. Fluorescence microscopy was carried out using either an Axioplan 2 imaging system (Carl Zeiss, Jena, Germany) and picture analysis using the Axioplan 3.0 software (Carl Zeiss), or an Olympus IX70 inverted microscope with picture analysis using Openlab software (Improvision, Lexington, MA). When desirable, fluorescence images were deconvoluted using Volocity software 3.7.0. Data sets for two channels were captured using GFP and DAPI filters. Exposure for the green channel was for 300 ms, whereas that for the blue DAPI channel was for 150 ms. Iterative deconvolution microscopy was performed on Z-stacks of 61 images with a step size of 0.2 μm. Significance was determined using 99.5% confidence limit or 25 iterations.
RESULTS
The Linker Region of Ire1p Contains a Nuclear Localization Sequence
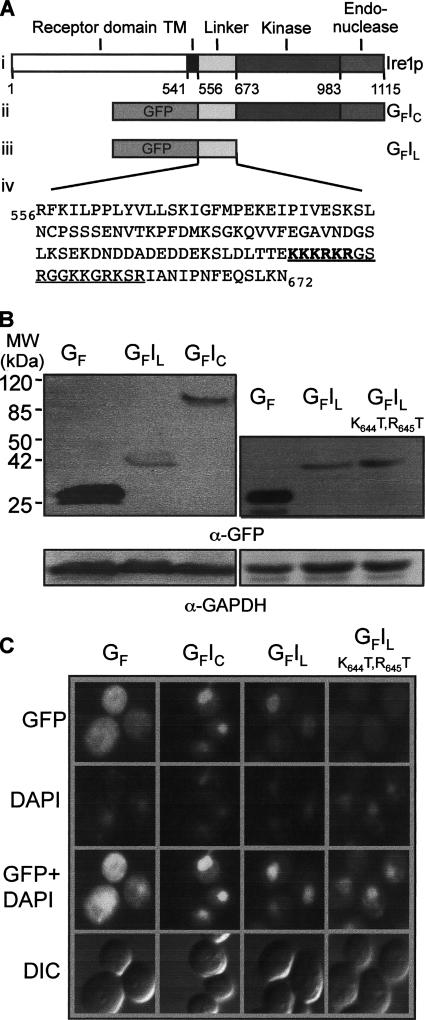
The kinase and endonuclease domains of Ire1p do not contain any clusters of basic residues that might act as NLSs. However, as noted previously (Mori et al., 1993) and shown in Figure 1A, the 117-residue linker region (residues 556–672) contains a highly basic sequence (residues 642–659, underlined) that includes a stretch of six residues (642KKKRKR647) that closely resemble the paradigmatic cNLS of SV40 large T antigen (127KKKRKV132; Dingwall and Laskey, 1991), as well as six additional lysine and arginine residues just downstream from this sequence.
Figure 1.
Sequences in the Ire1p linker region promote nuclear localization of GFP. (A) Ire1p is a 1115-amino acid transmembrane protein that contains a classic N-terminal hydrophobic signal sequence for ER localization. The various domains of the protein are indicated in i. GFP-fusion proteins containing the C-terminal domain (GFIC) or the linker sequence (GFIL) of Ire1p are shown in ii and iii, respectively. Shown in iv is the sequence of the linker domain, which contains a stretch of basic residues (in bold) that resembles the cNLS (PKKKRKV) of SV40 large T-antigen. This sequence is part of an 18-residue sequence (underlined) that is very rich in basic residues. (B) Equal amounts of protein extracts prepared from yeast cells expressing GFP or GFP-Ire1 fusion proteins were resolved by SDS-PAGE and analyzed by immunoblotting using anti-GFP and anti-GAPDH antibodies. (C) Single colonies of RG11907 yeast cells freshly transformed with plasmids encoding GFP or GFP–Ire1 fusion proteins were grown to an OD600 of 0.6 in SC-URA-MET medium before examination of the cells by fluorescence microscopy.
To determine whether the Ire1p linker sequence contains a functional NLS that could direct the nuclear localization of the GFP reporter, we constructed various plasmids expressing GFP–Ire1p fusion proteins, under the control of the MET25 promoter. The GFP moiety was fused either to the entire C-terminal domain (GFIC; Figure 1A, ii) or to the linker region (GFIL; Figure 1A, iii). The parental plasmid that expresses unfused GFP was used as the negative control. The pGFP, pGFIC, or pGFIL plasmids were introduced into RG11907 yeast cells, and the expression levels of GFP and GFP fusion proteins were analyzed by immunoblotting with an anti-GFP antibody (Figure 1B), whereas their intracellular localizations were analyzed by fluorescence microscopy (Figure 1C). Cells expressing unfused GFP displayed an even distribution of fluorescence (Figure 1C, first column, top), with the exception of a dark patch that does not colocalize with the nucleus (shown by DAPI staining, middle), but probably corresponds to the vacuole. The attachment of the entire C-terminal domain or just the linker sequence caused accumulation of GFP in the nucleus (Figure 1C, second and third columns). We concluded that the C-terminal domain of Ire1p contains a functional NLS that is likely to involve basic amino acids located between residues 642 and 659.
The potential cNLS in the basic sequence contains three overlapping versions of the cNLS consensus motif (K.R/K.x.R/K, Fontes et al., 2000). To determine whether one or more of these motifs is essential for nuclear targeting, a mutant version of pGFP-Ire1L was constructed in which threonine residues were substituted for K644 and R645 in the encoded fusion protein. These two amino acid substitutions interrupt all three versions of the cNLS motif and correspond to mutations that individually abolish or diminish the activity of the T-ag cNLS (Kalderon et al., 1984). Cells expressing the GFP-Ire1L(K644T,R645T) fusion protein (Figure 1B, see immunoblot in right-hand panel) showed greatly reduced nuclear fluorescence, which occurred at a level only slightly higher than that in the surrounding cytoplasm (Figure 1C, right-hand column). We concluded that residues K644 and R645 are important but not absolutely essential for nuclear accumulation of the fusion protein.
Mutation of the Potential cNLS in Ire1p Inhibits the UPR
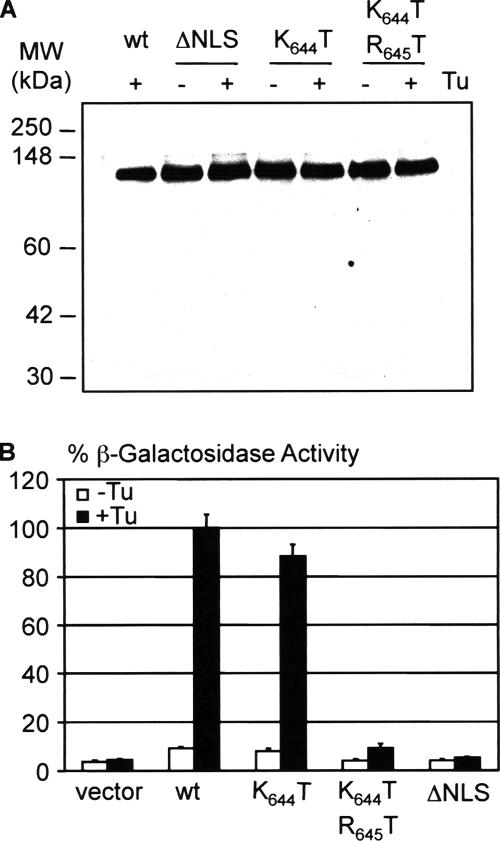
To test whether the overlapping cNLS motifs in the Ire1p linker region are required for UPR signaling, site-directed mutagenesis of the cDNA encoding full-length Ire1p was used to substitute threonine residues for K644, or for both K644 and R645, or to delete all six basic residues. The single K644T substitution leaves intact only the first of the three versions of the cNLS motif, whereas the deletion mutant, like the double substitution mutant, lacks all three versions. The wild-type and mutated Ire1 proteins were expressed in ire1Δ (ern1Δ) yeast cells by using single copy (CEN) or multicopy (2μ) vectors, and ER stress was imposed by treatment of cultures with tunicamycin, a drug that blocks protein glycosylation and causes the accumulation of unfolded proteins in the ER. Immunoblotting of protein extracts of unstressed and stressed cells demonstrated that the mutant proteins were expressed at levels equivalent to that of wild-type Ire1p (Figure 2A). No cleavage of the Ire1 polypeptides could be discerned after stress treatment, confirming previous data (Shamu and Walter, 1996) that UPR signaling does not involve proteolytic release of the kinase and endonuclease domains.
Figure 2.
UPR signaling in cells expressing Ire1 proteins with mutant cNLS. (A) Wild-type and mutant Ire1p/Ern1p proteins were expressed in KMY2115 ern1Δ yeast cells from multicopy (2μ) pERN1-EM vectors (Mori et al., 1993). Mid-log cultures were incubated for 3 h at 23°C in the presence or absence of tunicamycin (Tu; 5 μg/ml final). Proteins were then extracted, and equal amounts of total proteins (measured by Bradford DC assay; Bio-Rad) were resolved by SDS-PAGE and analyzed by immunoblotting by using the anti-N1 Ire1p-specific primary antibody. (B) KMY1115 ern1Δ yeast cells containing wild-type or mutant Ire1 proteins expressed from single copy (CEN) pERN1-BC vectors (Mori et al., 1993) were grown to mid-logarithmic phase and then incubated at 30°C for 3 h in the presence (closed bars) or absence (open bars) of Tu (5 μg/ml final). Protein extracts were prepared and assayed for β-galactosidase activity by using a UPRE-controlled lacZ reporter gene (Mori et al., 1992). The results are presented as mean ± SD, based on duplicate determinations with three independent transformants and are normalized to 100% of the activity obtained with wild-type Ire1p.
The UPR response in cells expressing either the wild-type or mutated Ire1 proteins was measured by analysis of β-galactosidase activity in cells containing a UPRE-controlled lacZ reporter gene (Mori et al., 1993). As shown in Figure 2B, the single K644T substitution had little or no effect on signaling by Ire1p, but substitution of both K644 and R645 by threonine caused a significant (90%) decrease in the UPR response. This phenotype is consistent with the significant reduction observed in the nuclear targeting activity of the GFP-Ire1L protein containing the same amino acid substitutions (see above; Figure 1C). ERN signaling was essentially absent in cells expressing the ΔNLS mutant, suggesting either that one or more of the other basic residues in the putative cNLS are required in addition to residues K644 and R645 for maximal UPR signaling or that deletion of all six residues abolishes signaling as the result of misfolding of the mutant protein. However, the data are not compatible with some or all of residues 642–647 constituting a cNLS that is essential for the function of Ire1p because the double substitution mutant still retains measurable signaling activity (∼10%), despite lacking a cNLS motif. We therefore considered the possibility that the functional NLS involves additional residues within the extended basic sequence (residues 642–659) of the Ire1 linker (Figure 1A).
The Ire1p NLS Is an Extended Basic Sequence
Plasmids were constructed that encode GFP fused to Ire1p residues 642–659 (GFP-Ire1B), or mutant versions of this basic sequence in which lysine and arginine residues that lie downstream of R645 were substituted, either singly or in pairs, by alanine (Figure 3A). K642 and K643 were not targeted because parallel in vitro peptide binding studies described below had demonstrated that these residues were not required for binding to importin proteins. The pGFP-Ire1B plasmids were introduced into RG11907 yeast cells, and the expression levels of the fusion proteins were analyzed by immunoblotting with an anti-GFP antibody (Figure 3B). The intracellular localization of GFP was again analyzed by confocal microscopy (Figure 3C).
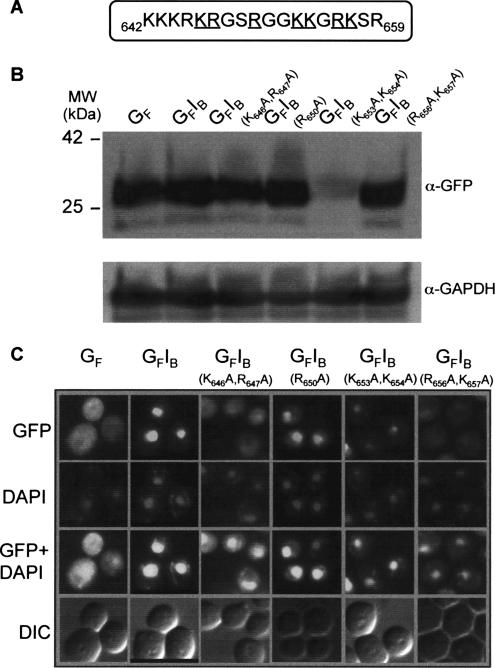
Figure 3.
Point mutations of basic residues in the Ire1p linker sequence inhibit nuclear localization. (A) Residues mutated within the Ire1p linker sequence in GFP-Ire1B fusion proteins are underlined. (B) Equal amounts of protein extracts prepared from yeast cells expressing GFP or GFP-Ire1B fusion proteins were resolved by SDS-PAGE and analyzed by immunoblotting using anti-GFP and anti-GAPDH antibodies. (C) Single colonies of RG11907 yeast cells freshly transformed with plasmids encoding GFP or GFP-Ire1B fusion proteins were grown to an OD600 of 0.6 in SC-URA-MET medium before examination of the cells by fluorescence microscopy.
Cells expressing GFP fused to the wild-type Ire1p basic sequence displayed bright nuclear fluorescence (Figure 3C, second column), confirming that this sequence functions as an efficient NLS. All of the substitution mutations caused reduced accumulation of the fusion proteins in the nucleus, although the extent of the reduction varied. The K646A,R647A and R656A,K657A double mutations almost completely prevented nuclear accumulation (Figure 3C, third and sixth columns), whereas the K653A,K654A double mutant, which consistently accumulated to lower levels than the other fusion proteins (Figure 3B), displayed both cytoplasmic and nuclear localization (Figure 3C, fifth column). The R650A mutant, although still predominantly localized in the nucleus, displayed significantly more cytoplasmic fluorescence than the construct containing the wild-type basic sequence (Figure 3C, fourth column). Together, these data define a minimal NLS consisting of 13 residues (R645–K657).
Components of the Ran Cycle Are Required for HAC1 mRNA Splicing and for Import of Ire1p NLS-containing Proteins
To determine whether components of the nuclear localization apparatus are essential for the early events of UPR signaling, we tested whether mutations in proteins involved in the Ran cycle required for the orientation of importin-mediated nuclear transport (reviewed by Görlich and Kutay, 1999) would affect ER stress-induced HAC1 mRNA splicing. We assayed HAC1 splicing rather than the induction of UPRE-controlled genes because the overall UPR signaling pathway requires two additional nuclear transport steps (export of HAC1 mRNA and import of the translated Hac1p transcription factor) that also might be affected in these mutants. Significant levels of stress-induced HAC1 mRNA splicing were observed at 23 and 37°C in wild-type yeast cells (strain PSY580; Figure 4A, first panel). Similar levels of splicing occurred at the permissive temperature of 23°C in cells carrying temperature-sensitive (ts) mutations in the genes encoding RanGTPase itself (gsp1Δ expressing gsp-1 strain PSY961), RanGAP (rna1-1 strain PSY868), and RanGEF (prp20-1 strain PSY713). However, after incubation at the nonpermissive temperature of 37°C, HAC1 mRNA splicing was significantly reduced in gsp-1 cells and completely inhibited in rna1-1 and prp20-1 cells.
To test whether the nuclear targeting activity of the Ire1p NLS is dependent on a functional Ran cycle, the pGFP, pGFP-Ire1C, and pGFP-Ire1C(K646A,R647A) plasmids were introduced into PSY580 and PSY868 (rna1-1) cells. The expression levels (Figure 4B) and intracellular localizations (Figure 4C) of GFP and the GFP fusion proteins were analyzed after growth at 23 or 37°C. The growth temperature had little or no effect on the expression levels of the GFP proteins (Figure 4B), and it did not alter the cellular localization of these proteins in the parental PSY580 cells. However, in rna1-1 cells the GFIC fusion protein was predominantly localized to the nucleus after incubation at 23°C, but it was redistributed to the cytoplasm after incubation at 37°C. By contrast, the largely cytoplasmic localizations of unfused GFP and the GFIC(K646A,R647A) fusion protein were not affected by the change in temperature. Essentially identical results were obtained when these experiments were repeated in PSY713 (prp20-1) cells (data not shown). Together, these results indicate that a functional Ran cycle is required for the nuclear targeting activity of the Ire1p NLS and for HAC1 mRNA splicing by Ire1p.
Membrane-anchored Ire1p Sequences Are Targeted to the Nuclear Membrane
Ire1p is normally present in yeast cells at such low levels (Mori et al., 1993) that the wild-type protein cannot be visualized by immunocytochemistry. A perinuclear and peripheral ER localization pattern has been reported for an Ire1p–GFP fusion protein expressed under the control of the constitutive TEF promoter (Kals et al., 2005). However, it is possible that this localization does not reflect the normal situation, because evidence from studies on the transport of membrane proteins targeted to the inner nuclear membrane suggests that this fusion protein could not be imported into the nucleus because the addition of GFP at the C terminus of Ire1p increases the size of the C-terminal domain beyond that (∼67 kDa) compatible with transport through the lateral channels of nuclear pore complexes (Holmer and Worman, 2001; Wu et al., 2002).
Membrane-anchored forms of Ire1p in which the majority of the N-terminal receptor domain of Ire1p is replaced by bZIP dimerization sequences activate UPR signaling (Liu et al., 2000), indicating that the Ire1p C-terminal domains of such constructs are targeted to the intracellular location(s) required for splicing of HAC1 mRNA. We designed a plasmid capable of expressing a membrane-anchored form of a GFP–Ire1 fusion protein in which the N-terminal receptor domain is replaced by GFP and an Fv domain, which facilitates dimerization upon addition of the cell permeant organic molecule AP20187 (Spencer et al., 1993; Clackson et al., 1998). In this construct, which was based upon the pGFP-N-FUS based vector, DNA sequences encoding the Ire1p ER targeting sequence, GFP and Fv were fused upstream of the IRE1 sequence encoding the transmembrane, linker, kinase, and endoribonuclease domains (Figure 5A). A parallel construct contained the K646A,R647A substitutions within the NLS in the linker sequence. These pISGFFVITMC and pISGFFVITMC(K646A,R647A) plasmids were introduced into RG11907 cells, and after addition of the AP20187 dimerization agent, the expression levels of the fusion proteins were measured by immunoblotting (Figure 5B), whereas their intracellular localization was analyzed by confocal microscopy (Figure 5C). Confocal images made under our standard conditions displayed faint perinuclear fluorescence in cells expressing the GFFVITMC fusion protein (Figure 5C, first column). This pattern was not apparent in cells expressing the GFFVITMC(K646A,R647A) protein (Figure 5C, second column). Deconvolution of images of cells expressing GFFVITMC (Figure 5C, third column) revealed a distinct pattern of perinuclear fluorescence that is very similar to that observed for a variety of GFP-fused nuclear pore and inner nuclear membrane proteins localized to the nuclear periphery of yeast cells (Huh et al., 2003). The fluorescence pattern for GFFVITMC particularly resembles that reported by Murthi and Hopper (2005) for Trm1-II, which normally resides as a peripherally associated protein of the yeast inner nuclear membrane. No reticular or cortical fluorescence characteristic of ER localization (Huh et al., 2003) was observed. Deconvolution of images of cells expressing GFFVITMC(K646A,R647A) (Figure 5C, fourth column) confirmed the absence of perinuclear fluorescence, indicating that the NLS mutations prevent targeting of the fusion protein to the nuclear membrane. Instead, a reticular pattern is observed. Although the “classic” ER pattern includes brighter perinuclear and cortical fluorescence, the localization data presented for a large collection of ER proteins by Huh et al. (2003) shows that patterns of generalized reticular fluorescence consistent with relatively even distribution of the fusion protein throughout the membrane system of the ER are not unusual, particularly for less abundant proteins. We therefore conclude that the membrane-anchored form of Ire1p is targeted to the inner nuclear membrane by an NLS-dependent process.
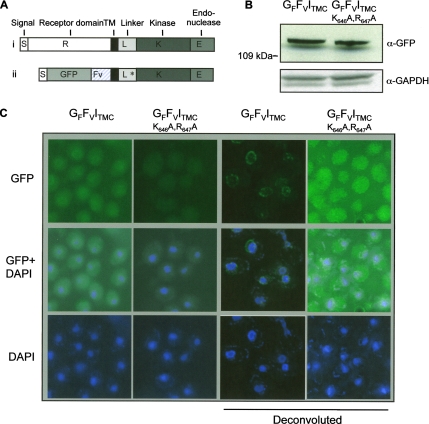
Figure 5.
Membrane anchored Ire1p sequences are targeted to the nuclear membrane. (A) Schematic diagrams of i) wild-type Ire1p and ii) a membrane-anchored form of a GFP–Ire1 fusion protein in which the N-terminal luminal domain of Ire1p is replaced by GFP and Fv. The various domains of the protein are indicated as follows: S, Ire1p ER targeting sequence; R, N-terminal receptor domain of Ire1p; TM, Ire1p transmembrane domain; L, K, E, linker, kinase, and endoribonuclease domains of Ire1p; GFP, green fluorescent protein; and Fv, variant FKBP domain. The asterisk in the linker domain of the GFFVITMC fusion protein indicates the position of amino acid substitutions in the NLS of the GFFVITMC(K646A,R647A) mutant. (B) RG11907 cells expressing GFFVITMC or GFFVITMC(K646A,R647A) fusion proteins were grown at 30°C to an OD600 of 0.6 in SC-URA-MET medium and then treated for 2 h with 2 μM AP20187 dimerizer before cell extracts were prepared. Equal amounts of total protein (∼200 μg) were resolved by SDS-PAGE and analyzed by immunoblotting using anti-GFP and anti-GAPDH antibodies. (C) Single colonies of RG11907 yeast cells freshly transformed with plasmids encoding GFFVITMC or GFFVITMC(K646A,R647A) were grown to an OD600 of 0.6 in SC-URA-MET medium. The cultures were then treated for 2 h with 2 μM AP20187 dimerizer before the cells were examined by fluorescence microscopy. Iterative deconvolution microscopy was performed using Volocity restoration software 3.7.0, and confidence limit was set to 99.5% or 25 cycles of iteration.
The Basic Sequence in the Ire1p Linker Domain Is Recognized with High Affinity by Importin β
To characterize the Ire1p NLS by using in vitro techniques, we synthesized a peptide containing the cNLS sequence and the adjacent basic residues for use in an ELISA-based importin binding assay (Hu and Jans, 1999). During synthesis of this peptide (ENLS-1, residues 642–660 of Ire1p), we also obtained two N-terminally truncated early termination peptides (ENLS-2; residues 646–660, and ENLS-3; residues 648–660). The three peptides (see sequences in Figure 6A) were purified and tested for recognition by the yeast importins Kap60p/Srp1p (the sole importin α in S. cerevisiae) and Kap95p/Rsl1p (importin β1), which in vivo form the importin α/β heterodimer involved in nuclear import of cNLS-containing substrates (see Figure 6B for a representative experiment and Table 2 for pooled data).
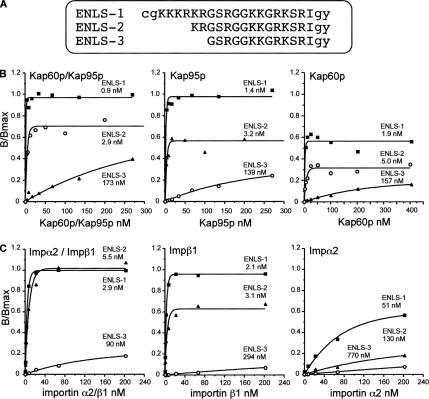
Figure 6.
Binding of yeast and murine importins to synthetic Ire1p NLS peptides as determined using an ELISA-based binding assay. (A) Sequences of synthetic Ire1p NLS peptides. Lowercase letters represent residues additional to the Ire1p linker sequence (uppercase letters). The three peptides were tested for recognition by yeast importins (Kap60p/α and Kap95p/β1) (B) or murine importins (Imp α2 and β1) (C). A standard ELISA-based binding assay was performed (see Materials and Methods). The KD values are shown on each graph below the peptide name. Results shown are from a single typical experiment, performed in triplicate, with pooled data shown in Table 2.
Table 2.
Binding parameters for the interaction of importins with synthetic Ire1p NLS peptides
| Yeast importin binding parameter |
||||||
|---|---|---|---|---|---|---|
| Kap60p/Kap95p | Kap95p | Kap60p | ||||
| A | ||||||
| Peptide | KD (nM) | Bmax (% α/β) | KD (nM) | Bmax (% α/β) | KD (nM) | Bmax (% α/β) |
| ENLS-1 cgKKKRKRG…Igy | 1.7 ± 0.3 (5) | 100 | 1.6 ± 0.1 (4) | 92 ± 4 | 2.2 ± 0.5 (4) | 66 ± 4 |
| ENLS-2 KRG…Igy | 2.5 ± 0.6 | 79 ± 3.2 | 3.2 ± 1.1 | 66 ± 5 | 5.0 ± 1.1 | 36 ± 2 |
| ENLS-3 G…Igy | 83 ± 47 (2) | 41 ± 13 | 113 ± 26 (2) | 30 ± 4 | 126 ± 40 (3) | 17 ± 6 |
| Mouse importin binding parameter |
||||||
|---|---|---|---|---|---|---|
| Impα2/β1 | Impβ1 | Impα2 | ||||
| B | ||||||
| Peptide | KD (nM) | Bmax (% α/β) | KD (nM) | Bmax (% α/β) | KD (nM) | Bmax (% α/β) |
| ENLS-1 cgKKKRKRG…Igy | 1.7 ± 0.6 (3) | 100 | 1.9 ± 0.3 (2) | 96 ± 0.3 | 31 ± 20 (2) | 81 ± 26 |
| ENLS-2 KRG…Igy | 2.7 ± 1.4 (3) | 101 ± 8.9 | 3.3 ± 0.2 (2) | 83 ± 21 | 82 ± 47 (2) | 48 ± 21 |
| ENLS-3 G…Igy | 45 ± 23 (3) | 28.2 ± 6.1 | 234 ± 60 (2) | 27 ± 8 | 558 ± 211 (2) | 27 ± 15 |
| ENLS-1 binding parameter |
||||||
|---|---|---|---|---|---|---|
| ka × 10−4 (M−1 s−1) | χ2 | kd × 105 (s−1) | χ2 | KD (nM) | ||
| C | ||||||
| Kap60p | 2.0 | 0.045 | 25 | 0.049 | 12.5 | |
| Kap95p | 5.4 | 0.039 | 2.8 | 0.122 | 0.52 | |
| Impα2 | — | — | — | |||
| Impβ1 | 28.3 | 0.025 | 330 | 0.043 | 11.8 | |
| Kap123p | 4.0 | 0.052 | 4.35 | 0.058 | 1.1 | |
Importin binding parameters were determined using an ELISA-based binding assay as described in Materials and Methods from experimental data fitted as shown in Figure 6. The results for the apparent dissociation constants (KD, representing the concentration of importin at which the level of binding is half-maximal) and the maximal level of importin bound (Bmax, normalized relative to that obtained for ENLS-1 when both α and β importins are added) are shown as the means ± SE (n in parentheses), where n is not indicated, the SE is determined from the curve fit. The apparent association (ka) and dissociation rate constants (kd) derived from the biosensor analysis of the interaction between immobilized ENLS-1 peptide and importins α2, β1, Kap60p, Kap95p, and Kap123p (Figure 8) were calculated from regions of the sensorgrams where 1:1 Langmurien interactions seemed to be operative. The apparent dissociation constant KD is the concentration of importin at which the level of binding is half-maximal. The binding of importin α2 was too low to perform kinetic analysis. The accuracy of the fit between experimental data and fitted curves (χ2) was estimated by chi-square analysis (Catimel et al., 1997; Nice and Catimel, 1999).
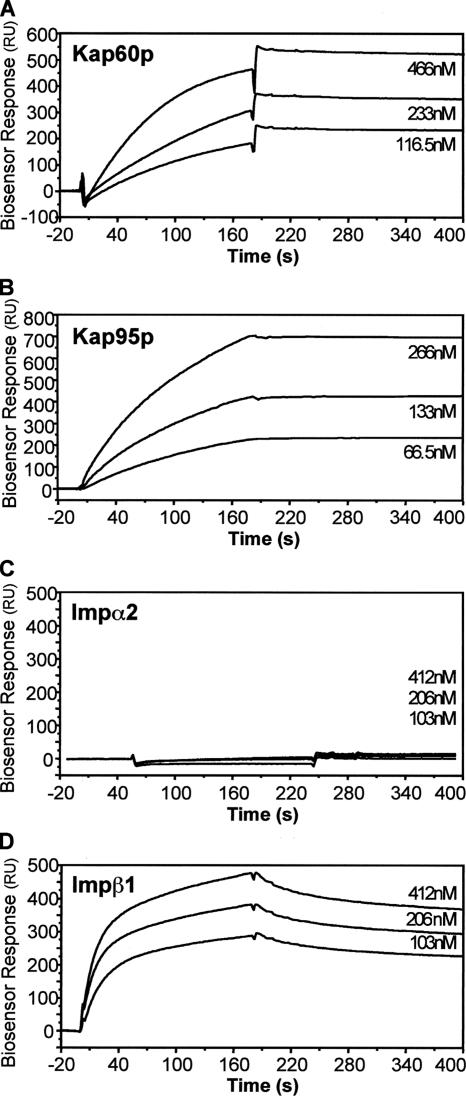
The Kap60p/Kap95p heterodimer bound ENLS-1 with very high affinity (1.7 nM). Unexpectedly, Kap95p and Kap60p alone bound ENLS-1 with affinities (1.6 and 2.2 nM, respectively) very similar to that of the heterodimer, although Kap60p exhibited somewhat lower maximal binding. These data imply that ENLS-1 contains recognition sites for both importin α and importin β1. ENLS-2 was also bound by the heterodimer and by the two individual importins with similarly high affinities (albeit in each case with reduced Bmax), indicating that the first four amino acids of the putative cNLS (642KKKR645, absent in ENLS-2) are dispensable for in vitro binding by the importins and suggesting that importin α may recognize a second minimal cNLS motif (653KKGR656) present toward the C terminus of both peptides. ENLS-3 was recognized with greatly reduced affinity by both importins, suggesting that the last two basic amino acids of the putative cNLS (residues K646 and R647) are part of the recognition site for Kap95p and that these residues may be required (together with 653KKGR656) to form a bipartite cNLS for Kap60p binding.
ELISA was then used to analyze the recognition of the ENLS peptides by murine importin α2/Rch1 and its binding partner importin β1 (see Figure 6C for a representative experiment and Table 2 for pooled data). ENLS-1 and ENLS-2 were both recognized by the importin α2/β1 heterodimer or by importin β1 with very high affinities similar to those exhibited by the yeast importins. Again the ENLS-3 peptide bound with low affinity, indicating that residues K646 and R647 are part of the minimal βNLS sequence recognized by both yeast Kap95p and murine importin β1, i.e., 646KRGSRGGKKGRKSRI660. The ENLS-1 peptide was bound only weakly by importin α2, indicating that the putative cNLS, 642KKKRKR647, does not represent a high-affinity binding site for importin α2, despite its close similarity to the T antigen cNLS (see above). Importin α2 is also unable to bind the alternative cNLS, present within ENLS-2, that is recognized by Kap60p. Thus, the yeast and murine importin α proteins differ significantly in their ability to recognize the Ire1p NLS.
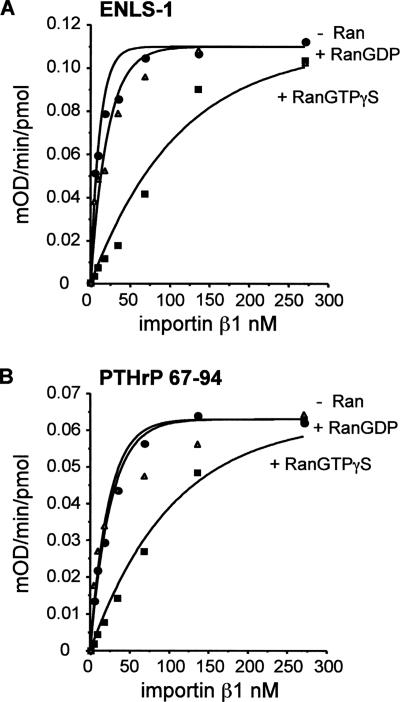
Binding of NLS-bearing cargo to importin β proteins can be reversed by RanGTP but not by RanGDP (for review, see Görlich and Kutay, 1999). We therefore investigated the effect of Ran on the interaction of the Ire1p NLS with importin β1. As shown in Figure 7, RanGTPγS significantly decreased the binding of ENLS-1 (or a control NLS peptide, PTHrP 67–94) to a GST–importin β1 fusion protein, whereas RanGDP consistently had little or no effect on peptide binding.
Figure 7.
Effect of Ran proteins on the binding of importin β1 to ENLS-1 peptide. Purified recombinant human RanGTPγS and RanGDP proteins (final concentrations 215 nM) were tested for their ability to inhibit the binding of ENLS-1 or PTHrP (67–94) peptides by murine importin β1 by using an ELISA-based binding assay as described in Materials and Methods. The sequence of ENLS-1 is shown in Figure 4A and that of PTHrP (67–94) in Materials and Methods. The KD values derived from data in the absence of added nucleotides were 2.1 ± 0.58 nM for ENLS-1 and 7.6 ± 3.1 nM for PTHrP (67–94).
Kinetic Analysis of Binding of Yeast and Murine Importins to the Ire1p NLS
We used a BIAcore biosensor to characterize the kinetic parameters of binding of the yeast and murine importins to the Ire1p NLS(s). ENLS-1 peptides were linked to the biosensor chip via their N-terminal cysteine residues, and binding data generated by passing increasing concentrations of the GST-importin fusion proteins over the immobilized peptides. As shown by ELISA, yeast Kap60p and Kap95p and murine importin β1 all showed significant binding, whereas murine importin α2 bound ENLS-1 very poorly, if at all (Figure 8 and Table 2). The affinity of Kap95p for the NLS peptide (KD = 0.52 nM) was 20- to 25-fold greater than that of Kap60p (12.5 nM) or murine importin β1 (11.8 nM). The association rates of the two yeast importins were comparable (ka = 5.4 and 2.0 × 104 M−1 s−1). Murine importin β1 bound more rapidly (28.3 × 104 M−1 s−1) but also dissociated at an extremely fast rate (330 × 10−5 s−1). Kap95p displayed the most stable binding, with an apparent dissociation rate (kd = 2.8 × 10−5 s−1), significantly lower than that displayed by Kap60p (25 × 10−5 s−1). These data suggest that within yeast cells, binding of Ire1p to Kap95p should be favored over binding to Kap60p and that importin β-dependent nuclear import should predominate in vivo.
Figure 8.
Biosensor analysis of the interaction between immobilized ENLS-1 peptide and importins Kap60p, Kap95p, α2, and β1. Varying concentrations (shown on the right of each panel) of yeast importins Kap60p/α and Kap95p/β1 (A and B) and murine importins α2 and β1 (C and D) were injected over immobilized ENLS-1 peptide (see Materials and Methods). The sensorgrams shown have been corrected for the corresponding signal obtained when the sample was passed over a blank derivatized control channel.
Redundancy of Importin Binding to the Ire1p NLS
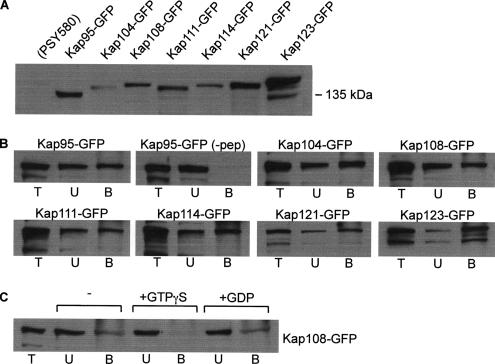
To determine whether Kap60p or Kap95p is essential for the function of Ire1p in UPR signaling in vivo in yeast cells, we tested the capacity of yeast cell mutants that are conditionally deficient in these importins to support ER stress-mediated HAC1 mRNA splicing. We observed little or no difference in the extent of stress-induced HAC1 mRNA splicing at permissive or nonpermissive temperatures in wild-type yeast cells (strain PSY580) and cells carrying ts mutations in Kap60p (srp1-31 strain PSY688) or Kap95p (rsl1-4 strain PSY1103) (Figure 9A, 1-3), even though these strains display defects in nuclear import in vivo at nonpermissive temperatures (Loeb et al., 1995; Koepp et al., 1996). We also analyzed HAC1 mRNA splicing in a number of other strains bearing ts mutations or deletions in the genes encoding several other importin β family members involved in nuclear import in yeast (Kap104p, Kap108p/Sxm1p, Kap121p/Pse1p, Kap123p/Yrb4p, or Nmd5p, see Table 1 for strain descriptions and references). Again, no defects in splicing were observed (Figure 9A; data not shown). The results described previously (Figure 4) for cells carrying ts mutations in the genes encoding RanGTPase, RanGAP, and RanGEF indicated that nuclear targeting by the Ire1p NLS involves Ran cycle-dependent, importin-mediated nuclear transport, suggesting that the lack of a requirement for any individual importin protein is due to redundant binding of two or more importins to Ire1p. Because our binding data suggested that Kap60p would play a minor role in vivo, we surmised that more than one importin β family member might recognize the Ire1p NLS. To test this hypothesis, we first analyzed the binding of the ENLS-1 peptide to a GST–Kap123 fusion protein using both the ELISA assay and the BIAcore biosensor. We observed (Figure 9, B and C) that the Kap123 protein exhibited high affinity for the Ire1p NLS, with binding and kinetic constants very similar to those previously observed for Kap95p (Table 2). To determine whether the Ire1p NLS is recognized by additional members of the importin β family, protein extracts were prepared from seven yeast strains each expressing a different importin β family member that is fused at its C terminus to a bright derivative of GFP (Morehouse et al., 1999; Seedorf et al., 1999). Their individual levels of expression were verified by immunoblotting by using the anti-GFP antibody (Figure 10A). The data shown in Figure 10B demonstrate that all seven of these importin β–GFP fusion proteins could be removed from the extracts by incubation with ENLS-4 peptide (see Figure 10 legend for sequence), which had been linked via its N-terminal cysteine residues to thiopropyl-Sepharose 6B. The fusion proteins did not bind to peptide-free resin (see Figure 10B, top row, second panel for Kap95p-GFP; data not shown for the other fusion proteins). Addition of nonhydrolysable GTPγS decreased the binding of the Kap108 fusion protein to the NLS peptide-linked resin, presumably by maintaining Ran present in the cell extract in its active form, whereas the addition of GDP had no effect (Figure 10C).
Figure 10.
The Ire1p NLS binds to multiple importin β–GFP fusion proteins. (A) Whole cell lysates prepared from PSY580 cells or seven derivative strains bearing GFP fusions to various importin β proteins were resolved by SDS-PAGE and immunoblotted by using α-GFP antibodies (Seedorf et al., 1999). (B) The lysates were incubated with ENLS-4 peptide (NH2-CGKRGSRGGKKGRKSRIGY-COOH) linked to thiol-Sepharose beads, and the bound fusion proteins were eluted with DTT. Equal proportions of the total lysates (T), the unbound fractions (U), and the bound and eluted fractions (B) were resolved by SDS-PAGE and immunoblotted using α-GFP antibodies. (C) The ENLS-4/Kap108p interaction is disrupted by a nonhydrolysable GTP analogue. Lysates were incubated in the presence or absence of 1 mM GTPγS or 1 mM GDP before binding as described in B.
A Consensus Sequence Recognized by Multiple Importin β Family Members?
The capacity of Ire1p to interact with multiple importin β proteins is reminiscent of the interaction of ribosomal proteins and histones with two or more importin β family members (Schlenstedt et al., 1997; Rout et al., 1997; Jäkel and Görlich, 1998; Claussen et al., 1999; Muhlhausser et al., 2001; Mosammaparast et al., 2002). Significantly, the combined NLS we have defined for Ire1p displays a high degree of similarity to the highly basic sequences present within the β importin binding (BIB) domains defined for the yeast L25 and human rpL23a ribosomal proteins and to the short BIB NLSs defined for Xenopus and human rpL5 and for human histones H3 and H4 (Table 3A). The NLS of Ire1p also resembles closely the NLSs defined for eight additional nonribosomal proteins known to translocate into the nucleus in an importin β-specific manner (Table 3B). Alignment of all these sequences yielded a “βNLS consensus” sequence (Table 3C). When we interrogated the mammalian and yeast protein sequence databases with this consensus sequence, a very significant proportion of the proteins identified were additional ribosomal subunits (see Table 3D). Although many of the other sequences identified as containing the consensus were hypothetical ORFs, the set included a number of nonribosomal proteins confirmed to have a nuclear localization (Table 3E) as well as a large variety of other proteins that function in the nucleus, including transcription factors, polymerases, and mRNA processing enzymes (data not shown).
Table 3.
Amino acid sequence alignment of the Ire1p NLS and the NLSs (defined or putative) of various other importin β-interacting and nuclear proteins
| Mammalian importin binding | Yeast importin binding | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ||||||||||||
| Protein(Ref.) | ||||||||||||
| NLS sequence (defined or proposed) | α | β1 | Tr | RBP5 | RBP7 | RBP9 | α | β1 | β2 | β3 | β4 | β* |
| y Ire1p | ||||||||||||
| KKKRKRGSRGGKKGRKSRI | − | ++ | + | +++ | ++ | ++ | +++ | ++ | ||||
| y L25(1) | ||||||||||||
| KKAVVKGTNGKKALKVRT | ++ | |||||||||||
| h rpL23a(2) | ||||||||||||
| KKKKIRTSPTFRRPKTLRL | +++ | +++ | ++ | +++ | +++ | +++ | +++ | |||||
| h rpS7(2) | ||||||||||||
| KPTRKSRTKNKQKRPRS | + | +++ | − | + | +++ | |||||||
| h rpL5(2,3) | ||||||||||||
| RRRREGKTDYYARKRLV | − | +++ | − | (+) | +++ | |||||||
| x rpL5(4) | ||||||||||||
| KNKAYFKRYQVKFRRRRE | ++ | ++ | ++ | ++ | ||||||||
| h or y H3(5,6) | ||||||||||||
| KQTARKSTGGKAPRKQL or GKAPRKQLASKAARKSA | + | +++ | +++ | + | + | + | +++ | |||||
| h or y H4(5,6) | ||||||||||||
| KGGKGLGKGGAKRHRKILR | + | + | ++ | + | + | + | + | |||||
| B | ||||||||||||
| y Gal4p(7) | ||||||||||||
| RLKKLKCSKEKPKCAKCLK | +++ | |||||||||||
| n AlcR(8) | ||||||||||||
| RSKAKGAAPRARTKKART | +++ | ++ | ||||||||||
| h,m,x NF-YA(9) | ||||||||||||
| KRRQARAKLEAEGKIPK and KERRKYLHESRHRHAMARKR | +++ | ++ | ++ | ++ | ||||||||
| h Smad3(10) | ||||||||||||
| CEKAVKSLVKKLKKTG | − | +++ | ||||||||||
| h IGFBP-3(11) | ||||||||||||
| KKKQCRPSKGRKRGFCWCV | + | +++ | ||||||||||
| h IGFBP-5(11) | ||||||||||||
| KRKQCKPSRGRKRGICWCV | + | +++ | ||||||||||
| h TCPTP(12) | ||||||||||||
| KVQQQMKQRLNENERKRKR | − | +++ | ||||||||||
| h PTHrP(13) | ||||||||||||
| KVETYKEQPLKTPGKKKKGK | − | +++ | + | +++ | ||||||||
| C | ||||||||||||
| CONSENSUS | ||||||||||||
| Z2-4.X2.Z.X4-5.Z2 | ||||||||||||
| D | ||||||||||||
| h rpL7a | ||||||||||||
| KKAAGKGDVPTKRPPVLR | ||||||||||||
| h rpL27a | ||||||||||||
| KKRKKKSYTTPKKNKHKR | ||||||||||||
| h rpL41 | ||||||||||||
| KKRMRRLKRKRRKMRQR | ||||||||||||
| h rpS24 | ||||||||||||
| KKKTSRKQRKERKNRMKK | ||||||||||||
| y rpL6p-like | ||||||||||||
| KKKREEQIKKKRSNKNKFV | ||||||||||||
| Notes | Location | |||||||||||
| E | ||||||||||||
| y Mrh1p(14) | ||||||||||||
| KKKKSKKSKKSKKSKKSE | Membrane protein related to Hsp30 | Plasma membrane and nuclear envelope | ||||||||||
| y Sen1p(15) | ||||||||||||
| KKEKKKSKADDKKKNNKK | RNA helicase | Nuclear matrix | ||||||||||
| h CARSCYP(16) | ||||||||||||
| KKRKKKHRKNSRKHKKEK | Cyclophilin PPIase | Nuclear matrix, colocalizes with mRNA | ||||||||||
| r CARSCYP(16) | ||||||||||||
| KKKHRKNSRKHKKEKKKR | Cyclophilin PPIase | Splicing factors | ||||||||||
The 19-residue combined Ire1p NLS is shown with the 13-residue minimal importin β-binding sequence highlighted in bold. A consensus motif (third box, Z denotes K or R) was defined on the basis of sequence similarities (bolded residues) between the Ire1p NLS and sequences within either ribosomal and histone proteins (first box) or other proteins (second box) previously reported to interact with one or more importin β proteins. Yeast importin proteins are designated α (Kap60p), β1 (Kap95p), β2 (Kap104p), β3 (Kap121p), β4 (Kap123p), and β* (Kaps 108p, 111p, and 114p). The fourth and fifth boxes contain examples of ribosomal and nonribosomal nuclear proteins identified by interrogation of mammalian and yeast protein sequence databases as containing an exact or very close fit to the consensus sequence. The consensus sequence demands only five positions that should be basic residues, but the sequences identified frequently matched the Ire1p NLS at additional positions (in particular a pair of basic residues aligning with K653 and K654 of Ire1p), providing confidence that we have not randomly extracted basic sequences. y, yeast; h, human; m, mouse; n, Aspergillus nidulans; r, rat; and x, Xenopus.
The references cited are 1. Schlenstedt et al. (1997);
3. Rosorius et al. (2000);
5. Muhlhauser et al. (2001);
9. Kahle et al. (2005). The two sequences lie within the portion of the conserved C-terminal sequence of NF-YA that contains the ncNLS;
10. Xiao et al. (2000);
11. Schedlich et al. (2000). IGFBP denotes insulin-like growth factor-binding protein;
12. Tiganis et al. (1997). TCPTP denotes T-cell protein tyrosine phosphatase;
13. Lam et al. (1999). PTHrP denotes parathyroid hormone-related protein;
14. J. R. Aris, annotation of MRH1/YDR033W in the Stanford Saccharomyces Genome Database;
15. Ursic et al. (1995). The consensus sequence identified lies within the 231 amino acid sequence reported to contain the Sen1p NLS.
16. Bourquin et al. (1997). Human and rat CAR-SCYP are members of the cyclophilin family of peptidylprolyl isomerases. The motif identified is located outside the cyclophilin domain, in a region not shared with cyclophilin family members that function in other cellular locations.
DISCUSSION
The starting point for this work was a puzzling topological issue associated with Ire1p, the receptor that senses the load of unfolded proteins in the ER and transduces the signal across the ER membrane. The IRE1/ERN1 gene encodes the first type I transmembrane receptor kinase characterized in yeast (Mori et al., 1993; Cox et al., 1993) and the first such receptor in eukaryotic cells known to signal across an internal membrane. The C-terminal portion of Ire1p carries a UPR stress-activated endoribonuclease domain that participates in splicing the mRNA precursor encoding the Hac1p transcription factor (Sidrauski and Walter, 1997). The question arose as to how the cytoplasmic domain of Ire1p could be targeted to the inner nuclear membrane, the location of the Rlg1p ligase that completes the unconventional splicing reaction (Clark and Abelson, 1987). In this study we analyzed the capacity of a highly basic sequence in the linker region of Ire1p to function as an NLS both in vivo and in vitro. We found that the 18 residue basic sequence, which includes a stretch of six residues that closely resembles a cNLS (Mori et al., 1993), is capable of targeting GFP to the nucleus of yeast cells in a process that requires proteins involved in the Ran GTPase cycle that facilitates nuclear import. The UPR, and in particular stress-induced HAC1 mRNA splicing, is inhibited by point mutations in the Ire1p NLS that inhibit nuclear localization and also require functional RanGAP and Ran GEF proteins.
Mutagenic analysis and importin binding studies demonstrated that the Ire1p linker region contains overlapping potential NLSs: at least one cNLS (within sequences 642KKKRKR647 and/or 653KKGR656) that can be recognized efficiently by yeast importin α (Kap60p), but only poorly by murine importin α2, and a novel βNLS (646KRGSRGGKKGRK657) that is recognized by several yeast β importins and by murine importin β1. In vivo in yeast, Ire1p thus has the capacity to interact with Kap60p or an array of different importin βs; hence, it can traffic to the nucleus either via the classical importin α/β heterodimer pathway or via various importin β-mediated, importin α-independent pathways. Clearly, our kinetic data suggest that binding to importin β proteins would predominate.
Jäkel and Görlich (1998) suggested that BIB domains contain an archetypal import signal, still present in many ribosomal proteins, which was originally the recognition motif for the evolutionary progenitor of present importin β-like import receptors. They further proposed that during diversification of these receptors in evolution, they acquired additional specialized binding sites such as that in importin β1 for the IBB domain of importin α or that in transportin for proteins displaying the M9 NLS. The crystal structure of importin β1 (residues 1–485) bound to the nonclassical NLS of PTHrP (Cingolani et al., 2002) supports this hypothesis by defining a “prototypical” cargo binding site that is distinct from the site that interacts with the importin α IBB domain. We think that the βNLS we have identified in Ire1p corresponds to an archetypal BIB motif: it is recognized by at least seven different importin β family members and displays significant sequence similarity to a portion of the BIB domains defined for the yeast L25 and human L23 ribosomal proteins. The ancient character of this motif seems consistent with other seemingly archaic features of the UPR signaling pathway, such as the use of a nonconventional splicing mechanism (Cox and Walter, 1996; Kawahara et al., 1997), which shares components of the tRNA maturation system, to mediate transcriptional control.
Ire1p, which contains an N-terminal hydrophobic signal sequence for targeting to the ER (Mori et al., 1993), must initially be inserted across the ER membrane with its N-terminal receptor domain located in the ER lumen, and its C-terminal linker, kinase, and endonuclease domains located in the cytoplasm. Because the outer nuclear membrane is contiguous with the rest of the ER membrane system of the cell, Ire1p could either be co- or posttranslationally inserted across the outer nuclear membrane or move to this location by diffusion from other portions of the ER. Two possible mechanisms could then be envisaged to transfer the C-terminal domains of the molecule into the nucleus. The first model, which has well-characterized precedents in the activation in mammalian cells of the SREBP receptor in response to low cholesterol (Brown and Goldstein, 1997) or of the ATF6 bZIP protein in response to ER stress (Haze et al., 1999; Yoshida et al., 2000), involves proteolytic cleavage of Ire1p at or near the cytoplasmic face of the membrane, followed by import of the released C-terminal domain into the nucleus. However, we and others have consistently failed to detect any stress-induced cleavage of Ire1p (Figure 2A; Shamu and Walter, 1996), so we support an alternative model, which involves movement of the intact Ire1p transmembrane protein around the periphery of the nuclear pore, such that the N-terminal receptor domain remains in the ER lumen, whereas the C-terminal domains are translocated into the nuclear matrix. Lateral diffusion of transmembrane proteins around the pore membrane from the ER membrane to the inner nuclear membrane of mammalian cells has been demonstrated for the lamin B receptor and emerin by using fluorescence recovery after photobleaching (for review, see Holmer and Worman, 2001). Studies such as these indicate that the nuclear pore does not pose a barrier to movement of transmembrane proteins between the outer and inner nuclear membranes, provided the nucleocytoplasmic domain is smaller than ∼67 kDa (Holmer and Worman, 2001). However, this diffusional process is very slow and although sequences promoting localization to the inner nuclear membrane have been mapped to the nucleocytoplasmic and transmembrane domains of these and other inner nuclear membrane proteins (Holmer and Worman, 2001; Wu et al., 2002), there is no evidence to indicate that the nuclear import apparatus actively facilitates the transport process. Instead, localization is apparently driven by trapping of the proteins in the inner nuclear membrane by their association with nucleoplasmic components such as lamins and/or heterochromatin, or by interaction between their transmembrane segments and those of other inner nuclear membrane proteins (Holmer and Worman, 2001; Wu et al., 2002). Our findings that Ire1p contains an essential importin β-binding NLS that targets membrane-anchored GFP–Ire1p to the inner nuclear membrane, together with the very recent report of importin-mediated inner nuclear membrane localization of the S. cerevisiae Heh1 and Heh2 proteins (King et al., 2006), document the interaction of integral membrane proteins with the nuclear import machinery. This interaction can apparently occur either via the importin α/importin β1 (karyopherin–α/karyopherin-β1) complex, reported by King et al. (2006) to be the sole route for the Heh1 and Heh2 proteins, or in the case of Ire1p, via a variety of different importin family members. The distinct importin-binding specificities of the Heh and Ire1 proteins probably reflect differences in their NLSs; thus, although a 14-residue NLS (124PKKKRKKRSSKANK137) identified in Heh2p displays significant sequence identity with the N-terminal “cNLS” region of the Ire1p NLS (642KKKRKRGSRGGKKGRK657), little identity is evident with the extended “βNLS” region (italicized).
The data reported in this article, as well as our unpublished data on the localization and splicing activity of various GFP–Fv–Ire1 fusion proteins (Vodala, S., and Gething, M.-J., unpublished data) demonstrate a strong correlation between the degree to which Ire1p is localized in the nucleus and its capacity to cleave the HAC1 mRNA precursor and signal the UPR. However, Ruegsegger et al. (2001) reported that splicing of HAC1 mRNA precursors that have accumulated on stalled polyribosomes can occur in the cytoplasm. To reconcile these apparently disparate findings, we suggest that the import of Ire1p into the nucleus is not required for processing of the preexisting pool of stalled polyribosome-associated HAC1 mRNA immediately upon induction of ER stress, but, as the UPR continues, is essential for splicing of newly synthesized HAC1 mRNA precursor in the nucleus. Nuclear import of Ire1p may also be necessary to facilitate its interaction with Ada5p, which is essential for HAC1 mRNA splicing (Welihinda et al., 2000).
ACKNOWLEDGMENTS
We thank Clive Slaughter and Brad Reik (University of Texas Southwestern Medical Center, Dallas, TX) for assistance with peptide synthesis and purification, Kazu Mori (Kyoto University, Kyoto, Japan) and Trevor Lithow (University of Melbourne) for gifts of plasmids and antibodies, and Judy Callaghan (University of Melbourne) for advice and assistance with fluorescence microscopy. This work was supported by a grant to M.-J.G. from the Australian Research Council.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0292) on October 11, 2006.
REFERENCES
- Aitchison J. D., Blobel G., Rout M. P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. 3rd ed. New York: John Wiley & Sons; 1995. Short Protocols in Molecular Biology; pp. 13-46–13-47. [Google Scholar]
- Bourquin J.-P., Stagljar I., Meier P., Moosmann P., Silke J., Baechi T., Georgiev O., Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Catimel B., Nerrie M., Lee F. T., Scott A. M., Ritter G., Welt S., Old L. J., Burgess A. W., Nice E. C. Kinetic analysis of the interaction between the monoclonal antibody A33 and its colonic epithelial antigen by use of an optical biosensor. A comparison of immobilisation strategies. J. Chromatogr. 1997;776:15–30. doi: 10.1016/s0021-9673(97)00087-3. [DOI] [PubMed] [Google Scholar]
- Chan C. K., Hubner S., Hu W., Jans D. A. Mutual exclusivity of DNA binding and nuclear localization signal recognition by the yeast transcription factor GAL 4, implications for nonviral DNA delivery. Gene Ther. 1998;5:1204–1212. doi: 10.1038/sj.gt.3300708. [DOI] [PubMed] [Google Scholar]
- Chi N. C., Adam E. J., Visser G. D., Adam S. A. RanBP1 stabilises the interaction of Ran with p97 nuclear protein import. J. Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Bednenko J., Gillespie M. T., Gerace L. Molecular basis for recognition of a nonclassical nuclear localization signal by importin β. Mol. Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- Clackson T., et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Abelson J. The subnuclear localization of tRNA ligase in yeast. J. Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M., Rudt F., Pieler T. Functional modules in ribosomal protein L5 for ribonucleoprotein complex formation and nucleocytoplasmic transport. J. Biol. Chem. 1999;274:33951–33958. doi: 10.1074/jbc.274.48.33951. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences–a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin β homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes M. R., Teh T., Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localisation sequences by mammalian importin-alpha. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Cell Biol. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmer L., Worman H. J. Inner nuclear membrane proteins: functions and targeting. Cell Mol. Life Sci. 2001;58:1741–1747. doi: 10.1007/PL00000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Jans D. A. Efficiency of importin α/β-mediated nuclear localisation sequence recognition and nuclear import. Differential role of NTF2. J. Biol. Chem. 1999;274:15820–15827. doi: 10.1074/jbc.274.22.15820. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jäkel S., Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle J., Baake M., Doenecke D., Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol. Cell Biol. 2005;25:5339–5354. doi: 10.1128/MCB.25.13.5339-5354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics–A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kals M., Natter K., Thallinger G. G., Trajanoski Z., Kohlwein S. D. YPL.db 2, the yeast protein localization database, version 2.0. Yeast. 2005;22:213–218. doi: 10.1002/yea.1204. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T., Yanagi H., Yura T., Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Lusk C. P., Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Koepp D. M., Wong D. H., Corbett A. H., Silver P. A. Dynamic localisation of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M.-J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.H.C., Briggs L. J., Hu W., Martin T. J., Gillespie M. T., Jans D. A. Importin β recognises parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin α. J. Biol. Chem. 1999;274:7391–7398. doi: 10.1074/jbc.274.11.7391. [DOI] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen S., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. Y., Schröder M., Kaufman R. J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Loeb J.D.J., Schlensted G., Pellman D., Kornitzer D., Silver P. A., Fink G. R. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Morehouse H., Buratowski R. M., Silver P. A., Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M.-J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2(BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signaling from the ER to the nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M.-J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K., Ogawa N., Kawahara T., Yanagi H., Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998;730:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 2002;277:862–868. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- Muhlhausser P., Muller E.-C., Otto A., Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A., Hopper A. K. Genome-wide screen for inner nuclear membrane targeting in Saccharomyces cerevisiae: Roles for N-Acylation and an integral membrane protein. Genetics. 2005;170:1553–1560. doi: 10.1534/genetics.105.043620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice E. C., Catimel B. Instrumental biosensors: new perspectives for the analysis of biomolecular interactions. Bioessays. 1999;21:339–352. doi: 10.1002/(SICI)1521-1878(199904)21:4<339::AID-BIES11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Nikolaev I., Cochet M. F., Felenbok B. Nuclear import of zinc binuclear cluster proteins proceeds through multiple, overlapping transport pathways. Eukaryot. Cell. 2003;2:209–221. doi: 10.1128/EC.2.2.209-221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R. K., Riles L., Johnston M., Hegemann J. H. Green fluorescent protein as a marker for gene expression and subcellular localisation in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Okamura K., Kimata Y., Higashio H., Tsuru A., Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Patil C. K., Li H., Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:1208–1223. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon I. K., Jans D. A. Regulation of nuclear transport: central role in development and transformation. Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Ron D. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout B. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout B. P., Blobel G., Aitchison J. D. A distinct nuclear pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Ruegsegger U., Leber J. H., Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schedlich L. J., Le Page S. L., Firth S. M., Briggs L. J., Jans D. A., Baxter R. C. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by importin β subunit. J. Biol. Chem. 2000;275:23462–23470. doi: 10.1074/jbc.M002208200. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G., Smirnova E., Deane R., Solsbacher J., Kutay U., Görlick D., Ponsting H., Bischoff F. R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M., Damelin M., Kahana J., Taura T., Silver P. A. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M., Silver P. A. Importin/karyoherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu C. E., Walter P. Oligomerisation and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1–20. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Cox J. S., Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Spencer D. M., Wandless T. J., Schreiber S. L., Crabtree G. R. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Stevens F. J., Argon Y. Protein folding in the ER. Semin. Cell Dev. Biol. 1999;10:443–454. doi: 10.1006/scdb.1999.0315. [DOI] [PubMed] [Google Scholar]
- Strom A. C., Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. Reviews3008, Epub 2001 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiganis T., Flint A. J., Adam S. A., Tonks N. K. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J. Biol. Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Ursic D., DeMarini D. J., Culbertson M. R. Inactivation of the yeast Sen1 protein affects the localisation of nucleolar proteins. Mol. Gen. Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- Welihinda A. A., Kaufman R. J. The unfolded protein response pathway in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Welihinda A. A., Tirasophon W., Kaufman R. J. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J. Biol. Chem. 2000;275:3377–3381. doi: 10.1074/jbc.275.5.3377. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wong D. H., Corbett A. H., Kent H. M., Stewart M., Silver P. A. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol. Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Lin F., Howard H. J. Intracellular trafficking of MAN1, an integral membrane protein of the inner nuclear envelope inner membrane. J. Cell Sci. 2002;115:1361–1372. doi: 10.1242/jcs.115.7.1361. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Mori K. ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Liu X., Lodish H. F. Importin β mediates nuclear translocation of Smad 3. J. Biol. Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]