Abstract
Retroviral restriction factor TRIM5α exhibits a high degree of sequence variation among primate species. It has been proposed that this diversity is the cumulative result of ancient, lineage-specific episodes of positive selection. Here, we describe the contribution of within-species variation to the evolution of TRIM5α. Sampling within two geographically distinct Old World monkey species revealed extensive polymorphism, including individual polymorphisms that predate speciation (shared polymorphism). In some instances, alleles were more closely related to orthologues of other species than to one another. Both silent and nonsynonymous changes clustered in two domains. Functional assays revealed consequences of polymorphism, including differential restriction of a small panel of retroviruses by very similar alleles. Together, these features indicate that the primate TRIM5α locus has evolved under balancing selection. Except for the MHC there are few, if any, examples of long-term balancing selection in primates. Our results suggest a complex evolutionary scenario, in which fixation of lineage-specific adaptations is superimposed on a subset of critical polymorphisms that predate speciation events and have been maintained by balancing selection for millions of years.
Keywords: HIV, retrovirus, restriction
TRIM5α (tri-partite motif protein 5, α isoform) was identified as the source of a major block to infection of Old World monkey (OWM) cells with HIV strain 1 (HIV-1) (1, 2). Human TRIM5α does not restrict HIV-1, a fact that is likely to have contributed to the extent of the AIDS pandemic. In the wake of these findings, TRIM5α orthologues were cloned from a wide variety of primate species, including Old and New World primates, and tested for retroviral restriction (3–5). Collectively, primate TRIM5α proteins can restrict a variety of retroviruses, and some orthologues have been shown to restrict multiple, unrelated retroviruses (reviewed in refs. 6–8). Different species display unique patterns of TRIM5α-mediated restriction. For example, rhesus TRIM5α restricts both HIV-1 and N-tropic murine leukemia virus (MLV-N), whereas human TRIM5α restricts MLV-N but not HIV-1 or simian immunodeficiency virus (SIV) (2, 9–11).
Several lines of evidence suggest that specificity of TRIM5α-mediated restriction is governed largely by the C-terminal SPRY/B30.2 domain (3, 12–15). Based on sequence similarity to known protein motifs, three other domains can be identified, including a RING domain, a B-box2, and a coiled-coil (CC) (reviewed in refs. 7 and 16). The contributions of the RING and B-box domains to retroviral restriction have not been defined, although both domains are important for function (14, 15, 17). The CC domain is involved in self-association of the protein and, along with the SPRY domain, contributes to viral specificity (14, 15, 18, 19). Evidence from several groups suggests that direct interaction of TRIM5α with the retroviral capsid is required for restriction (3, 20–25).
Previous analyses revealed considerable interspecies variability in primate TRIM5α sequence and provided evidence for positive selection operating on selected regions of the protein (3–5). To look for variation within species (polymorphism), we analyzed TRIM5α coding sequences from multiple individuals within two geographically and phylogenetically distinct species of OWMs, rhesus macaques (Macaca mulatta) and sooty mangabeys (Cercocebus atys). Rhesus macaques encompass several subspecies found throughout the Asian subcontinent (26). These animals are resistant to infection with HIV-1 but are highly susceptible to infection with certain strains of SIV and have therefore become an invaluable model for AIDS vaccine research (27, 28). Sooty mangabeys are found exclusively in Western Africa, where they are endemically infected with SIVsmm (29, 30). Despite robust levels of viral replication, infection does not result in any apparent signs of immunodeficiency or disease (31–34); for this reason, sooty mangabeys have also generated considerable interest as subjects of HIV/AIDS research.
Results
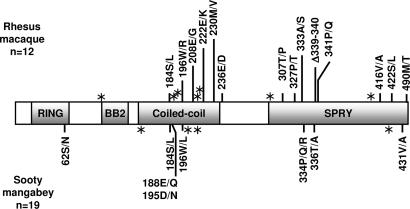
To identify sites of intraspecific polymorphism in rhesus macaques, multiple full-length TRIM5α cDNAs were derived from 12 unrelated animals and sequenced. Sequence alignments revealed an in-frame insertion/deletion polymorphism (positions 1015–1020, amino acids 339–340) and 20 common SNPs for which the minor allele was present in at least three individuals (Fig. 1 and Table 1). Interestingly, indel 1015–1020 and all nonsynonymous SNPs (nsSNPs) clustered exclusively in the CC and the SPRY domains. Unexpectedly, most synonymous polymorphisms also clustered in these two regions (Fig. 1). Genomic DNA samples from these and 14 additional animals were used to confirm the presence of each polymorphism (data not shown). At least three of these nsSNPs (amino acids 184, 196, and 307) were also found in two rhesus cell lines (9). Based on the distribution of 21 polymorphisms, rhesus macaque TRIM5α coding sequences were grouped into six alleles (Table 1). The amino acid sequence of allele 1 (Macaca mulatta allele-1, or Mamu-1) was identical to the published rhesus TRIM5α sequence (1), whereas the sequence of allele 6 (Mamu-6) was the most divergent [see supporting information (SI) Fig. 5].
Fig. 1.
Common polymorphisms in OWM TRIM5α coding sequences cluster in the CC and SPRY domains. A diagram depicting the 497-aa TRIM5α protein and its functional domains is shown. Locations of frequent, nonsynonymous polymorphisms are shown above and below the TRIM5α schematic. Sites of common, synonymous polymorphisms are indicated by asterisks.
Table 1.
Common alleles of TRIM5α from rhesus macaques
| DNA | 273 | 551 | 558 | 567 | 586 | 623 | 645 | 651 | 664 | 688 | 708 | 843 | 919 | 979 | 997 | 1015–1020 | 1022/1016 | 1221/1215 | 1247/1241 | 1265/1259 | 1469/1463 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | 91 | 184 | 186 | 189 | 196 | 208 | 215 | 217 | 222 | 230 | 236 | 281 | 307 | 327 | 333 | 339–340 | 341/339 | 405/407 | 416/414 | 422/420 | 490/488 |
| Rhesus | G | S | D | Q | W | E | S | T | E | M | E | P | T | P | A | TF | P | G | V | S | M |
| Chimp | G | L | D | Q | W | E | S | T | E | V | D | P | P | P | A | ΔΔ | P | G | A | F | M |
| Human | G | L | D | Q | W | E | S | T | E | L | D | P | P | P | A | ΔΔ | Q | G | A | F | M |
| Mamu-1 | G | S | D | Q | W | E | S | T | E | M | E | P | T | P | A | TF | P | G | V | S | M |
| Mamu-2 | G | S | D | Q | W | E | S | T | E | M | E | P | T | P | A | TF | P | G | A | S | M |
| Mamu-3 | G | S | D | Q | W | E | S | T | E | M | D | P | P | P | A | TF | P | G | V | S | M |
| Mamu-4 | G | S | D | Q | W | E | S | T | E | M | E | P | P | P | S | ΔΔ | Q | G | V | L | M |
| Mamu-5 | G | L | D | Q | R | E | S | T | K | V | D | P | P | T | S | ΔΔ | Q | G | V | L | M |
| Mamu-6 | G | L | D | Q | R | G | S | T | K | V | D | P | P | T | S | ΔΔ | Q | G | V | L | T |
The amino acid sequence of each polymorphic site is compared with published sequences from rhesus macaque (AA558505.1), human, and chimpanzee.
We typed 54 additional animals by high-resolution DNA melting analysis of the region surrounding indel 1015–1020 (SI Fig. 6). Thirty-four of 54 (63%) were heterozygous in this region. In a subset of these animals, the presence of indel 1015–20 was confirmed by polyacrylamide-gel electrophoresis and BanI digestion (SI Fig. 6E). Because the C-terminal half of the CC domain harbored many of the observed polymorphisms, the same animals were surveyed by HR-DNA melting analysis of a region spanning nt 508–750; Twenty-eight of 54 animals (52%) were heterozygous in this region (SI Fig. 6). Thus, polymorphism in these two short segments is a common property of the rhesus TRIM5 locus.
We next surveyed 19 sooty mangabeys and identified 12 polymorphisms that occurred at high frequency (the minor variant was present in three or more individuals). The distribution of polymorphisms among the sooty mangabey sequences permitted preliminary classification of four alleles (Table 2). The clustering of polymorphisms was strikingly similar to rhesus macaques, with synonymous and nsSNPs localized predominantly in the CC and SPRY domains (Fig. 1).
Table 2.
TRIM5α alleles from sooty mangabeys
| DNA | 185 | 402 | 551 | 562 | 587 | 604 | 645 | 989 | 1001 | 1028 | 1292 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | 62 | 134 | 184 | 188 | 196 | 205 | 215 | 330 | 334 | 343 | 431 |
| Rhesus | S | E | S/L | E | W/R | L | S | M | P | L | V |
| Chimp | S | E | L | E | W | L | S | I | R | F | V |
| Human | S | E | L | E | W | L | S | I | R | F | V |
| Gorilla | S | E | L | E | W | L | R | I | Q | F | V |
| Ceat-1 | S | E | S | E | W | L | S | M | R | H | A |
| Ceat-2 | S | E | L | Q | L | L | S | M | Q | H | V |
| Ceat-3 | N | E | S | E | W | L | S | M | R | H | A |
| Ceat-4 | S | E | S | E | W | L | S | T | P | L | V |
The amino acid sequence for each polymorphic site is compared with published sequences from rhesus macaque, human, chimpanzee, and gorilla.
Several studies report that residue 334 of TRIM5α and adjacent residues influence target-specificity (14, 15). Among known primate TRIM5α orthologues, this residue is a proline, a glutamine, or an arginine (3–5). Our analysis revealed the presence of a 334Q/R polymorphism in sooty mangabeys. The 334R was present in at least one copy in 9 of 19 sooty mangabeys, and 9 individuals had at least one copy of the 334Q allele. Surprisingly, a third variant (334P) was also identified in one animal. Repeated cloning and sequencing of TRIM5α from this animal, by both RT-PCR of cellular RNA and by PCR of genomic DNA, has identified only the 334P variant, suggesting that this individual may be homozygous (334P/334P).
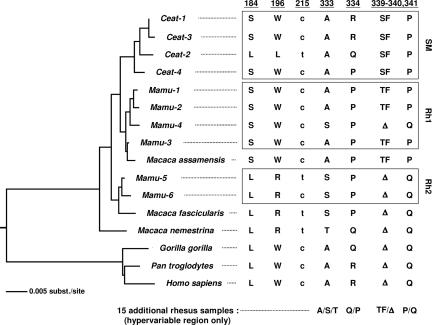
The maintenance of polymorphism over very long evolutionary periods is unlikely unless some form of balancing selection acts to counter the processes of purifying selection and random genetic drift (35, 36). Nonetheless, we identified polymorphisms at seven sites in OWM TRIM5α that predate speciation. Three of these polymorphisms clustered within a 94-nt stretch of exon 4 (184, 196, and 215) and four within a 25-nt stretch of exon 8 (333, 334, 339–340, and 341) (Fig. 2). Three polymorphisms (184S/L, 215c/t, and 334P/Q/R) are unambiguous cases of shared polymorphism (i.e., both alleles were present at high frequency in two or more lineages, indicating that the polymorphism arose in a common ancestor). Among these, the 334P/Q/R polymorphism is especially intriguing, because the presence of the Q and R states in both the OWM and hominid lineages suggests that both alleles (at position 334) have been maintained for >23 million years (37). The closely linked 184S/L (nonsynonymous) and 215c/t (synonymous) polymorphisms are shared by rhesus macaques and sooty mangabeys and therefore appeared no less than 8–10 million years ago (38). The 215c/t change is presumably neutral, and may have persisted because of tight linkage with nearby selected sites (Fig. 1). At sites 196, 333, 339–340, and 341 polymorphism was detected in only one of the two species sampled; however, each of the alternative states was found in at least two other species (Fig. 2). Therefore, these polymorphisms also predate speciation, and for each site, at least one extant species continues to maintain both alleles. For example, residue 196 is polymorphic in rhesus macaques (196W/R), but the R variant is present in Macacca fascicularis and Macacca nemestrina, and the W variant is found in sooty mangabey, Macacca assamensis, and hominids (Fig. 2).
Fig. 2.
Phylogenetic analysis reveals unusual features of TRIM5α evolution in Old World primates. A neighbor-joining tree is depicted; a similar topology was obtained by using maximum parsimony. Tree was rooted by designating the human/chimpanzee/gorilla cluster as an outgroup. Alleles described in this study include Ceat-1 through Ceat-4 and Mamu-1 through Mamu-6. Additional sequences included Macaca assamensis (accession nos. AY899873.1–AY899879.1), Macaca fascicularis (AB210052), Macaca nemestrina (AY899887.1–AY899893.1), Homo sapiens (NM_033034), Gorilla gorilla (DQ298178), and Pan troglodytes (NM_001012650). The sequences at sites representing putative transspecies polymorphisms are shown.
To determine the evolutionary relationship between rhesus macaque and sooty mangabey TRIM5α alleles, all sequences were subjected to phylogenetic analysis. Sequences from additional Macaca species and hominids were included. Alignments were analyzed by neighbor-joining (Fig. 2) and maximum parsimony (MP) (data not shown), and trees were subsequently rooted by using the hominid cluster as an outgroup. There was strong bootstrap support for the sooty mangabey cluster (98% of 1,000 replicates) and for the hominid cluster (100%). Within Macaca sp., support was strong for the terminal nodes leading to the Mamu-1/Mamu-2 grouping (90%) and the Mamu-5/Mamu-6 grouping (82%). Resolution within Macaca was weaker (55–76%). MP analysis identified eight minimal trees of equal length (L = 175); all eight preserved the mangabey, hominid, and Mamu-1/Mamu-2 clusters. Alleles did not cluster by species; for example, Mamu-5 and Mamu-6 clustered with M. fascicularis, whereas Mamu-1–Mamu-4 clustered with M. assamensis and all four sooty mangabey alleles. This finding suggests that ancestral alleles diverged before speciation and have been evolving independently in different lineages. It is noteworthy that we identified multiple examples of ancient polymorphism in a limited sampling of only two species (n = 12 and n = 19). Thus, it is likely that additional sampling within these, and other, primate species will reveal more extensive shared polymorphism in the TRIM5 locus.
When maximum parsimony analysis was constrained to identify only trees that conform to established phylogenetic relationships between hominids, mangabeys, and macaques (39), the pattern at several sites required additional evolutionary assumptions (minimal tree length increased from 175 to 185 steps), including multiple instances of reversal and convergence (SI Fig. 7). Published estimates put the substitution rate for primate nuclear DNA sequences in the range of 10−8 to 10−9 substitutions per site per year (for example, see ref. 40). Therefore, the probability of an identical polymorphism occurring more than once since the divergence of Old World primates (<25 million years) is very low. More importantly, to explain the pattern of linked changes depicted in Fig. 2 as the result of convergent/parallel evolution would require not just one, but multiple, identical substitutions to occur in similar or identical combinations more than once in a relatively short evolutionary time span. This is highly unlikely, given the low substitution rate, even in the presence of positive selection. Although unusual, the more probable explanation is long-term persistence of multiple alleles because of some form of balancing selection.
An unusual feature of these sequences is the absence of silent changes outside of the polymorphic clusters (Fig. 1). A prediction of balancing selection operating over long evolutionary periods is that selectively neutral changes closely linked to selected sites may also be preserved (“hitchhiking”) (35), but with increasing distance from the region under balancing selection, neutral substitutions will be uncoupled from selected sites by recombination and will be increasingly subject to loss or fixation by random genetic drift (41, 42). Thus, the observed pattern is consistent with a locus that has been under balancing selection over a long evolutionary time span.
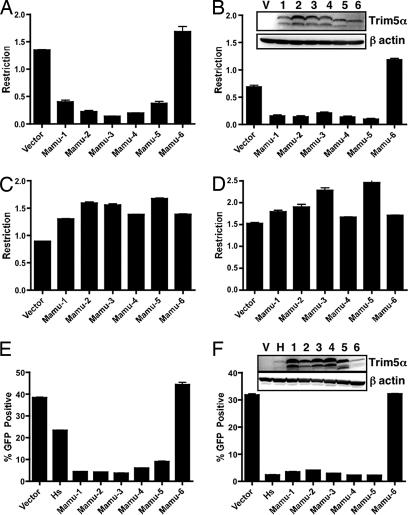
The expectation of sequences evolving under balancing selection is functional differentiation of allelic variants. To look for evidence of functional diversification, we assessed the consequences of TRIM5α polymorphism on viral restriction. We used two independent assays to assess viral restriction: a two-color FACS-based assay where HA-tagged rhesus TRIM5α alleles were transiently expressed (43, 44) and an infectivity assay using cell lines stably expressing untagged versions of each allele. Single-cycle lentiviruses (HIV and SIV), a γ-retrovirus (MLV-N) and a β-retrovirus [Mason–Pfizer monkey virus (MPMV)] were used as targets of restriction. Both assays demonstrated that Mamu-1, -2, -3, -4, and -5 efficiently restricted HIV-1 and MLV-N, whereas Mamu-6 did not restrict either retrovirus (Fig. 3 and SI figs 8 and 9). The failure of Mamu-6 to inhibit HIV-1 and MLV-N infection was not due to gross differences in protein expression in either assay, because Mamu-6 expression was comparable to restriction-competent alleles tested in parallel (Fig. 3 B and F Inset). Although we cannot dismiss differences in expression and/or stability of TRIM5α alleles as contributors to differences in restriction, we believe that levels of expression are not solely responsible for the observed difference in restriction by Mamu-6 in these in vitro assays. Interestingly, although alleles Mamu 1–5 differed at several residues in the CC and SPRY domains, all were comparable in their antiretroviral activity. However, Mamu-6, which differs from Mamu-5 by only 2 aa, one in the CC and one in the SPRY domain, lacked antiretroviral activity against the viruses tested.
Fig. 3.
Rhesus macaque TRIM5α alleles differ in their ability to restrict retroviral infection. Retroviral restriction was assayed by using a transient two-color (A–D) and infection of stable cell lines (E–F). For the two-color assay, permissive cells expressing human and various rhesus TRIM5α alleles (Mamu-1 to Mamu-6) and coexpressing ECFP were challenged with single-cycle HIV-1 (A), MLV-N (B), SIVmac (C), or MPMV (D) viruses carrying an EGFP reporter. The ratio of infected, TRIM5α containing cells to infected, TRIM5α-null cells was determined by FACS analysis. As a negative control for restriction, cells expressing ECFP alone were also challenged. Stable cell lines expressing rhesus alleles Mamu-1 to Mamu-6 as well as human TRIM5α (Hs), were challenged with either HIV-1 (E) or MLV-N (F) expressing an EGFP reporter. The number of infected (GFP-positive) cells was determined by FACS analysis. Error bars represent three independent infections. Representative data from one of three independent experiments is shown. The level of TRIM5α protein in transduced or stable cells was determined by Western blotting using antibody against the HA tag (Inset in B) or TRIM5α (Inset in F). Levels of β-actin are also shown to demonstrate equal loading of cell lysate.
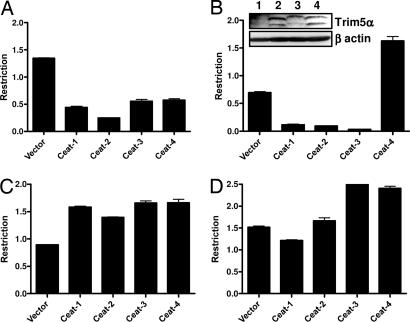
The restriction properties of the four sooty mangabey TRIM5α alleles were also assayed by using the two-color FACS assay (Fig. 4) and stable cell lines (SI Fig. 10). All four sooty alleles had moderate-to-weak restriction activity against HIV-1, and three (Ceat-1, -2, and -3) strongly inhibited MLV-N infection. Ceat-4, which differs from Ceat-2 at three sites in the SPRY domain, was unique in that it failed to restrict MLV-N. Ceat-1 protein levels were low in comparison with the other alleles in both transient and stable expression (Fig. 4B Inset and SI Fig. 10); however, levels of expression cannot account for the observed differences in restriction activity because cells expressing Ceat-1 still efficiently restrict MLV-N (Fig. 4B). None of the alleles from either species restricted SIV or MPMV (Figs. 3 and 4).
Fig. 4.
Restriction properties of sooty mangabey TRIM5α alleles. Cells expressing sooty mangabey TRIM5α alleles Ceat-1 to Ceat-4 were tested for their ability to inhibit infection by HIV-1 (A), MLV-N (B), SIVmac (C), or MPMV (D). Two-color restriction assays were carried out as described in Fig. 3. Representative data from one of three independent experiments is shown. Levels of HA-tagged TRIM5α protein in transduced cells is shown (Inset in B).
Discussion
We surveyed TRIM5α sequences from multiple individuals within two species of OWM to look for evidence of within-species variation. Despite clear geographic separation and an estimated divergence time of >8 million years ago, both species displayed a strikingly similar pattern of polymorphism. Notable features included the presence of multiple alleles and the coclustering of silent and amino acid replacement polymorphisms. Altogether, we identified 20 common SNPs and an insertion/deletion among just 12, unrelated rhesus macaques, and 12 common SNPs among 19 sooty mangabeys. By contrast, over a thousand human individuals have been surveyed, leading to the identification of less than a dozen SNPs in the human TRIM5α coding sequence (45–48).
Well documented examples of shared polymorphism are found in the incompatibility systems of plants and fungi and the MHC loci of many species (49–53). Other than genes within the MHC, there are few examples of shared polymorphism in primates. A recent survey of >6 × 106 base pairs of shared human and chimpanzee sequence for which SNP data were available in both species, identified only 11 instances of shared polymorphism outside the MHC, a number similar to that expected by chance (54). In contrast, we identified three shared SNPs in TRIM5α (of only 1494 bp) and four additional polymorphisms that clearly predate radiation of modern OWMs. Shared, ancient polymorphisms constitute compelling evidence for long-term balancing selection at the TRIM5 locus of Old World primates. Long-term balancing selection, involving the maintenance of multiple alleles over millions of years and through multiple speciation events, is thought to be rare, the best documented examples being self-incompatability systems of certain plants and fungi and loci of the MHC (41) and possibly the ABO blood group system (55). This is in contrast to classical examples of balancing selection, such as the β-globin gene cluster and bitter-taste receptors, which typically involve a small number of alleles confined to an extant species or a population within a species (56–59).
Previous reports described significant interspecies variability in TRIM5α (3–5, 13). However, in these studies each species was typically represented by a single, orthologous sequence. Thus, sites that appeared to be variable between species might actually represent sites of variation within species. Indeed, we found that at least four sites previously shown to contribute to interspecies variability were, in fact, polymorphic (327P/T, 334P/Q/R, 341P/Q, and indel339–340). The extent to which presumed interspecies variation can actually be attributed to polymorphism will require further sampling within multiple species.
Sawyer et al. (3) identified a 13-aa patch clearly evolving under positive selection in the SPRY domain of OWMs and hominids, and similar results have been reported by others (5, 13). We identified 10 nsSNPs in the SPRY domain, five of which mapped to this same stretch of 13 aa. Sawyer et al. proposed a model in which a history of virus–host interactions has selected for fixation of species-specific adaptations (“evolutionary arms-race” scenario) (3). Our finding that some sites of interspecies variability are in fact polymorphic doesn't conflict with this model but does suggest a more complex scenario in which lineage-specific amino acid replacements are superimposed over a subset of polymorphisms that predate speciation and have been maintained for millions of years by balancing selection.
Given published evidence that the SPRY domain is a specificity determinant (reviewed in refs. 6–8), the finding of polymorphism in this domain was not surprising. But what might explain the presence of balanced polymorphism in the CC domain? One possibility is that the CC is also involved in targeting, perhaps by interacting with viral targets directly or through as yet unidentified protein intermediates. Because the CC mediates multimerization, changes could also act at a distance to modify the spatial arrangement of SPRY domains, thus giving rise to new or altered specificities. An alternative and intriguing possibility is that polymorphism in the CC might have been selected secondarily to changes in the SPRY domain to minimize dominant-negative interference when different alleles are expressed in heterozygous individuals. Although speculative, support for this notion comes from previous reports that isoforms of TRIM5 and other TRIMs lacking functional SPRY domains can act as dominant-negative inhibitors of TRIM5α function (15, 18, 60, 61).
Even though the genetic distance between alleles in both species was low (<2%), we were able to functionally differentiate alleles based on an initial screening of four retroviruses. Although this pattern may be unique to the viral agents we chose to test, it potentially reflects selection to maintain functional diversity. It has been argued that diversity in the MHC is driven by heterozygote advantage, the notion that heterozygotes, who express twice as many alleles of each MHC locus than homozygotes, benefit by being able to recognize a broader range of epitopes (53, 62); perhaps a similar mechanism accounts for maintenance of polymorphism in OWM TRIM5α. One might even speculate that formation of TRIM5α heteromultimers in heterozygous individuals could lead to specificities not encoded in the amino acid sequences of the individual alleles. Although the nature of the agents driving selection is unknown, it is worth noting that a number of diverse retroviruses are common among Old World primates, including betaretroviruses (simian retrovirus, MPMV), deltaretroviruses (primate T lymphotropic viruses), gammaretroviruses (gibbon ape leukemia virus, baboon endogenous virus), lentiviruses (SIVs), and spumaretroviruses (simian foamy virus) (63).
Previous reports have shown that sequence variation in TRIM5α is responsible for the appearance of species-specific patterns of TRIM5α-mediated restriction (reviewed in refs. 6–8). Our findings of polymorphism in two divergent lineages of OWMs indicate that specificity varies not only between species but also within species. Thus, the notion of species-specificity may be oversimplified, particularly in the case of species where only a single TRIM5α allele has been tested. To date, extensive sampling within a species has been reported only for human (45–48).
The current release of the rhesus macaque genome sequence (www.ensembl.org/Macaca_mulatta/index.html) does not contain duplication(s) of TRIM5, and our data are consistent with a single, multiallelic locus. Nonetheless, we cannot rule out the possibility that expansion/contraction of TRIM5 copy number during the course of primate evolution has contributed to the maintenance of specific amino acid replacements (i.e., that some of the described differences could have been selectively maintained not as alleles of one locus, but as sequences of paralogous loci). Further population-level sampling within multiple branches of the primate order will be necessary to clarify the complex and interesting evolutionary history of the TRIM5 locus.
Closely related, naturally occurring alleles of TRIM5α also provide unique tools for the study of function. For example, identifying the polymorphisms responsible for differential restriction may identify previously overlooked residues of TRIM5α important for target-specificity, clarify additional contributions of the CC domain to function, and potentially aid in identification of cellular cofactors.
Materials and Methods
Cells.
HEK293T/17 and feline renal fibroblasts (CRFK) were obtained from American Type Culture Collection (Manassas, VA) and grown in DMEM/10% FBS. The viral packaging cell line GP2–293 expressing viral gag and pol proteins was obtained from Clontech (Mountain View, CA) and grown in DMEM/10% FBS.
Recombinant Viruses.
Recombinant retroviruses were produced by cotransfection of HEK293T/17 cells with appropriate plasmids by using the Transfectin Lipid Reagent (Bio-Rad, Hercules, CA). At 72 h after transfection, cell-free supernatant was collected and viral titer was determined by infection of CRFK cells and subsequent enumeration of GFP+ cells by FACS. Recombinant HIV-1 viruses were produced by cotransfection with pNL43ΔenvFL, pVSV-G (Clontech) and pLenti-GFP. SIVmac recombinant viruses were produced by cotransfection with pHDM.G, pFSΔPRΔINEGFP, and pGPFusion as described in ref. 64. Plasmids for production of SIVmac recombinant viruses were a gift of David Evans (New England Primate Research Center/Harvard Medical School). Production of N-tropic MLV (MLV-N) was carried out by cotransfection with pCIGN (gift of Jonathan Stoye, Medical Research Council, London, U.K.), pVSV-G, and pLXIN-EGFP. To generate a replication-defective, GFP expressing M-PMV vector, EGFP was PCR amplified from pEGFP-N1 (Clontech) by using pfu polymerase (Stratagene, La Jolla, CA) the 5′ primer GFP-F (5′-CTCTCCTCGAGATCGCTCTTTCCCTTGTTCACAGATATGGTGAGCAAGGGCGAG-3′) and the 3′ primer GFP-R (5′-GTAAGGCTAAGCTTACTTGTACAGCTC GTCCA-3′). The resulting 768-bp fragment was inserted into the plasmid pSARM-X (65) by using the restriction sites XhoI and BlpI. In the resulting vector, pSARM-EGFP, EGFP replaces the env gene and uses the endogenous splice-acceptor site of the env gene for its expression. Production of MPMV was carried out by cotransfection with pSARM-EGFP and pVSV-G.
Establishment of B Lymphoblastoid Cell Lines (BLCL).
Autologous BLCL were established as described (66). Briefly, B cells were transformed by incubating peripheral blood mononuclear cells with herpesvirus papio in the presence of 1 μg/ml cyclosporine. BLCL were propagated in RPMI medium 1640/20% FBS.
Sequencing of TRIM5α cDNA.
Total RNA was isolated from EBV-immortalized B cell lines created from 12 rhesus macaques and 19 sooty mangabeys by using the Rneasy Mini kit; (Qiagen, Valencia, CA). Untagged, full-length TRIM5α cDNA clones were generated by RT-PCR with primers T5aNotIF (5′-GCGGCCGCATGGCTTCTGGAATC-3′) and T5aRIR (5′-CGCGAATTCTCAAGAGCTTGGTGAGC-3′) by using the SuperScript One-Step kit (Invitrogen). PCR products were cloned directly into the TOPO TA vector (Invitrogen). Three to 18 independent cDNA clones from individual rhesus macaques and one to five from individual sooty mangabeys were used for automated sequence analysis (Retrogen, San Diego, CA). To generate C-terminally tagged TRIM5a, alleles were amplified with primers T5aNotIF and T5aAgeIHA-R (5′-CCACCGGTGGCTCAAGCGTAGTCTGGGACGTCGTATGGGTAGCCGCCAGAGCTTGGTGAGCACAGAG-3′).
Phylogenetic Analyses.
Sequences were assembled by using the SeqMan application (DNAstar, Madison, WI), aligned by using the CLUSTAL W method and imported into PAUP (version 4.0b10; Sinauer Associates, Sunderland, MA) (67) as a single Nexus file. Unrooted trees were generated by using both Neighbor-Joining and Parsimony analysis settings. Distances were computed as both absolute differences and by using the Kimura 2-parameter model with nearly identical results. Gaps were treated as “missing data.” To root trees, hominid sequences were designated as a monophyletic outgroup.
Two-Color Restriction Assay.
The restriction assay was performed as described (43, 44). Specific details are provided in SI Supporting Materials and Methods.
Restriction in Stable Cell Lines.
Cell lines stably expressing were generated by transducing CRFK cells with VSV-pseudotyped virus made from empty pQCXIN vector (BD Biosciences, Franklin Lakes, NJ) or pQCXIN into which rhesus macaque and sooty mangabey TRIM5α alleles had been cloned at NotI and EcoRI sites. At 24 h after transduction, cells were selected with 1 mg/ml G418 and subsequently maintained in 0.5 mg/ml G418. For infection, 5 × 104cells per well were seeded into 12-well dishes, infected with retrovirus the following day, and prepared for FACS analysis as described above.
Immunoblotting.
CRFK cells were transduced as described above. At 5 days after transduction, cells were lysed in MPER lysis reagent (Pierce, Rockford, IL). Total protein was measured by using a BCA assay kit (Pierce) and equal amounts (100 μg) were separated by SDS/PAGE. Untagged TRIM5α was detected with rabbit polyclonal anti-TRIM5α(CT) antibody (ProSci, Poway, CA) and horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (Pierce). HA-tagged TRIM5α was detected with monoclonal anti-HA antibody (Covance, Berkeley, CA) and HRP-conjugated anti-mouse IgG secondary antibody (Pierce).
Supplementary Material
Acknowledgments
We thank J. Stoye, G. Towers, D. Evans, and N. Landau for critical reading of the manuscript and H. Malik, J. Stoye, and G. Towers for helpful advice.
Abbreviations
- CC
coiled coil
- CRFK
feline renal fibroblasts
- MLV-N
N-tropic murine leukemia virus
- MPMV
Mason–Pfizer monkey virus
- nsSNP
nonsynonymous SNP
- OWM
Old World monkey
- SIV
simian immunodeficiency virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF113914–EF113923).
This article contains supporting information online at www.pnas.org/cgi/content/full/0605838103/DC1.
References
- 1.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Yap MW, Nisole S, Lynch C, Stoye JP. Proc Natl Acad Sci USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawyer SL, Wu LI, Emerman M, Malik HS. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. J Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu HL, Wang YQ, Liao CH, Kuang YQ, Zheng YT, Su B. Gene. 2005;362:109–116. doi: 10.1016/j.gene.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 6.Goff SP. Mol Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Nisole S, Stoye JP, Saib A. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz PD. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 9.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Proc Natl Acad Sci USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keckesova Z, Ylinen LM, Towers GJ. Proc Natl Acad Sci USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stremlau M, Perron M, Welikala S, Sodroski J. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B, Gold B, O'Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yap MW, Nisole S, Stoye JP. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. J Biol Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 18.Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, Strack B, Si Z, Sodroski J. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebastian S, Luban J. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. J Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayah DM, Luban J. J Virol. 2004;78:12066–12070. doi: 10.1128/JVI.78.21.12066-12070.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisole S, Lynch C, Stoye JP, Yap MW. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens CM, Song B, Perron MJ, Yang PC, Stremlau M, Sodroski J. J Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napier JR, Napier PH. The Natural History of Primates. Cambridge, MA: MIT Press; 1985. [Google Scholar]
- 27.Desrosiers RC. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 28.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. Nat Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Telfer P, Reed P, Zhang L, Getti A, Ho DD, Marx PA. J Med Primatol. 1995;24:108–115. doi: 10.1111/j.1600-0684.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 30.Fultz PN, Gordon TP, Anderson DC, McClure HM. AIDS. 1990;4:619–625. doi: 10.1097/00002030-199007000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti LA, Lewin SR, Zhang L, Gettie A, Luckay A, Martin LN, Skulsky E, Ho DD, Cheng-Mayer C, Marx PA. J Virol. 2000;74:1209–1223. doi: 10.1128/jvi.74.3.1209-1223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rey-Cuille MA, Berthier JL, Bomsel-Demontoy MC, Chaduc Y, Montagnier L, Hovanessian AG, Chakrabarti LA. J Virol. 1998;72:3872–3886. doi: 10.1128/jvi.72.5.3872-3886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvestri G, Sodora DL, Koup RA, Paiardini M, O'Neil SP, McClure HM, Staprans SI, Feinberg MB. Immunity. 2003;18:441–452. doi: 10.1016/s1074-7613(03)00060-8. [DOI] [PubMed] [Google Scholar]
- 34.Silvestri G. J Med Primatol. 2005;34:243–252. doi: 10.1111/j.1600-0684.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- 35.Takahata N, Nei M. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark AG. Proc Natl Acad Sci USA. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. J Hum Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Li QQ, Zhang YP. Biochem Genet. 2005;43:375–386. doi: 10.1007/s10528-005-6777-z. [DOI] [PubMed] [Google Scholar]
- 39.Purvis A. Philos Trans R Soc London B Biol Sci. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z, Jin L, Fu YX, Ramsay M, Jenkins T, Leskinen E, Pamilo P, Trexler M, Patthy L, Jorde LB, et al. Proc Natl Acad Sci USA. 2000;97:11354–11358. doi: 10.1073/pnas.200348197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlesworth D. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes AL. Immunogenetics. 2000;51:473–486. doi: 10.1007/s002510050646. [DOI] [PubMed] [Google Scholar]
- 43.Bishop KN, Bock M, Towers G, Stoye JP. J Virol. 2001;75:5182–5188. doi: 10.1128/JVI.75.11.5182-5188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bock M, Bishop KN, Towers G, Stoye JP. J Virol. 2000;74:7422–7430. doi: 10.1128/jvi.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speelmon EC, Livingston-Rosanoff D, Li SS, Vu Q, Bui J, Geraghty DE, Zhao LP, McElrath MJ. J Virol. 2006;80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS. Curr Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 47.Javanbakht H, An P, Gold B, Petersen DC, O'Huigin C, Nelson GW, O'Brien SJ, Kirk GD, Detels R, Buchbinder S, et al. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 48.Goldschmidt V, Bleiber G, May M, Martinez R, Ortiz M, Telenti A. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richman A. Mol Ecol. 2000;9:1953–1963. doi: 10.1046/j.1365-294x.2000.01125.x. [DOI] [PubMed] [Google Scholar]
- 50.Richman AD, Kohn JR. Trends Ecol Evol. 1996;11:497–502. doi: 10.1016/s0169-5347(96)10051-3. [DOI] [PubMed] [Google Scholar]
- 51.May G, Shaw F, Badrane H, Vekemans X. Proc Natl Acad Sci USA. 1999;96:9172–9177. doi: 10.1073/pnas.96.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes AL, Nei M. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 53.Hughes AL, Yeager M. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 54.Asthana S, Schmidt S, Sunyaev S. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Martinko JM, Vincek V, Klein D, Klein J. Immunogenetics. 1993;37:274–278. doi: 10.1007/BF00187453. [DOI] [PubMed] [Google Scholar]
- 56.Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, Vazquez M, Dunn DM, Meyerhof W, Weiss RB, Bamshad MJ. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 57.Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tadmouri GO, Garguier N, Demont J, Perrin P, Basak AN. Hum Biol. 2001;73:661–674. doi: 10.1353/hub.2001.0075. [DOI] [PubMed] [Google Scholar]
- 59.Currat M, Trabuchet G, Rees D, Perrin P, Harding RM, Clegg JB, Langaney A, Excoffier L. Am J Hum Genet. 2002;70:207–223. doi: 10.1086/338304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Li Y, Stremlau M, Yuan W, Song B, Perron M, Sodroski J. J Virol. 2006;80:6198–6206. doi: 10.1128/JVI.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berthoux L, Sebastian S, Sayah DM, Luban J. J Virol. 2005;79:7883–7888. doi: 10.1128/JVI.79.12.7883-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parham P, Ohta T. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 63.Medawar PB. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Plainview, NY: Cold Spring Harbor Lab Press; 1997. [PubMed] [Google Scholar]
- 64.Evans DT, Bricker JE, Desrosiers RC. J Virol. 2004;78:11715–11725. doi: 10.1128/JVI.78.21.11715-11725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song C, Hunter E. J Virol. 2003;77:7779–7785. doi: 10.1128/JVI.77.14.7779-7785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaur A, Grant RM, Means RE, McClure H, Feinberg M, Johnson RP. J Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swofford DL. Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0b10. Sunderland, MA: Sinauer Associates; 2003. paup*. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.