Abstract
Chromosome segregation during mitosis requires chromosomes to undergo bipolar attachment on spindle microtubules (MTs) and subsequent silencing of the spindle checkpoint. Here, we describe the identification and characterisation of a novel spindle and kinetochore (KT)-associated complex that is required for timely anaphase onset. The complex comprises at least two proteins, termed Ska1 (Spindle and KT Associated 1) and Ska2. Ska1 associates with KTs following MT attachment during prometaphase. Ska1 and Ska2 interact with each other and Ska1 is required for Ska2 stability in vivo. Depletion of either Ska1 or Ska2 by small interfering RNA results in the loss of both proteins from the KT. The absence of Ska proteins does not disrupt overall KT structure, but KT fibres show an increased cold-sensitivity. Most strikingly, Ska-depleted cells undergo a prolonged checkpoint-dependent delay in a metaphase-like state. This delay is characterised by the recruitment of Mad2 protein to a few KTs and the occasional loss of individual chromosomes from the metaphase plate. These data suggest that the Ska1/2 complex plays a critical role in the maintenance of the metaphase plate and/or spindle checkpoint silencing.
Keywords: kinetochore, Mad2, metaphase, microtubules, spindle checkpoint
Introduction
Upon entry into mitosis, the microtubule (MT) network is rearranged to form the mitotic spindle, which then brings about the segregation of sister chromatids. Central to this process is the proper attachment of spindle MTs to kinetochores (KTs), proteinaceous structures assembled on centromeric chromatin (Cleveland et al, 2003; Maiato et al, 2004). KT–MT interactions are important for both chromosome congression during prometaphase and subsequent chromosome segregation during anaphase. In addition, they regulate the activity of the spindle assembly checkpoint, a surveillance mechanism that monitors full MT attachment to KTs and/or the tension that develops in-between sister chromatids in response to bipolar attachment (Musacchio and Hardwick, 2002; Cleveland et al, 2003). The spindle checkpoint is silenced only after all KTs have undergone bipolar attachment and thus contributes to ensure the synchronous and error-free segregation of chromosomes.
As revealed by electron tomography, mammalian KTs display a trilaminar structure, comprising an inner KT plate directly adjacent to (and inclusive of) the centromeric heterochromatin and an outer plate with its associated corona (McEwen et al, 1998). In mammals, the outer KT plate provides about 20–30 end-on MT attachment sites (Rieder, 1982). Recent studies indicate that a multitude of proteins cooperate to bring about and regulate the highly dynamic KT–MT interactions that are required for chromosome movement during mitosis (Biggins and Walczak, 2003; Cleveland et al, 2003; McAinsh et al, 2003; Maiato et al, 2004; Rieder, 2005). Prominent among the proteins implicated in MT capture at KTs are the Hec1/Ndc80 complex (Ciferri et al, 2005; DeLuca et al, 2005; Emanuele et al, 2005), MT-dependent motors (Cleveland et al, 2003) and the MT plus end proteins (+TIPs) CLIP-170, CLASP and EB1 (Maiato et al, 2004). Moreover, several Ran-regulated proteins (e.g. RanGAP1, RanBP2, HURP) (Joseph et al, 2004; Sillje et al, 2006; Wong and Fang, 2006) have been implicated in the stabilisation of KT-associated MT bundles (KT-fibres).
A single unattached KT is sufficient to prevent anaphase onset by maintaining the spindle checkpoint in an active state (Rieder et al, 1994). The exact functioning of the spindle checkpoint is not yet understood, but a number of evolutionarily conserved spindle checkpoint proteins have been characterised. One prevailing model holds that the spindle checkpoint proteins Mad2 and/or BubR1 sequester Cdc20, an activating factor of the E3 ubiquitin-ligase known as anaphase promoting complex/cyclosome (APC/C). Inhibition of the APC/C then prevents the polyubiquitylation and hence degradation of key mitotic regulatory proteins, including cyclin B and securin (Peters, 2002). Upon bipolar attachment of all KTs to MTs the spindle checkpoint is silenced, followed by chromosome segregation and exit from mitosis (Musacchio and Hardwick, 2002).
To obtain further insight into the processes that regulate mitotic chromosome congression and segregation, we recently conducted a proteomic survey of the human mitotic spindle (Sauer et al, 2005). This resulted in the identification of 151 known spindle proteins, including centrosome and KT-associated proteins, as well as 154 previously uncharacterised components. Here, we have examined C18Orf24 (now termed Spindle and KT Associated 1 (Ska1)) and show that this novel protein indeed localises to both the mitotic spindle and KTs. Ska1 binds another novel spindle and KT-associated protein, FAM33A (now termed Ska2), with which it forms a KT-associated protein complex. Depletion of the Ska complex results in a prolonged mitotic delay even though most chromosomes are aligned in a near-perfect metaphase plate. The most straightforward interpretation of this unusual phenotype is that Ska-depleted cells are unable to maintain stable KT–MT interactions and concomitantly fail to satisfy the spindle assembly checkpoint.
Results
Identification of Ska1 at spindle MTs and outer KTs
C18Orf24 was previously identified as a putative spindle component in a mass spectrometry-based spindle inventory (Sauer et al, 2005). Inspection of the primary sequence of this 30 kDa protein revealed no known structural motifs, except for a predicted N-terminal coiled-coil domain. To determine if C18Orf24 constitutes a genuine spindle component, we transiently expressed the myc-tagged protein in HeLa S3 cells and analysed its localisation by indirect immunofluorescence (IF) microscopy. Co-staining with α-tubulin showed that the myc-tagged protein partly co-localised with spindle MTs in mitotic cells (Figure 1A). In addition, it localised to bright dots on chromosomes that were identified as KTs/centromeres by co-staining with CREST serum (Figure 1A). This localisation prompted us to designate C18Orf24 as Ska1. Database searches indicated the existence of putative Ska1 homologues in other vertebrates, as well as in Caenorhabditis elegans (NP_492739), Anopheles gambiae (mosquito) (EAL39257), Arabidopsis thaliana (NP_191625) and Oryza sativa (rice) (XP_478114). In contrast, no obvious homologues could be detected in yeast.
Figure 1.
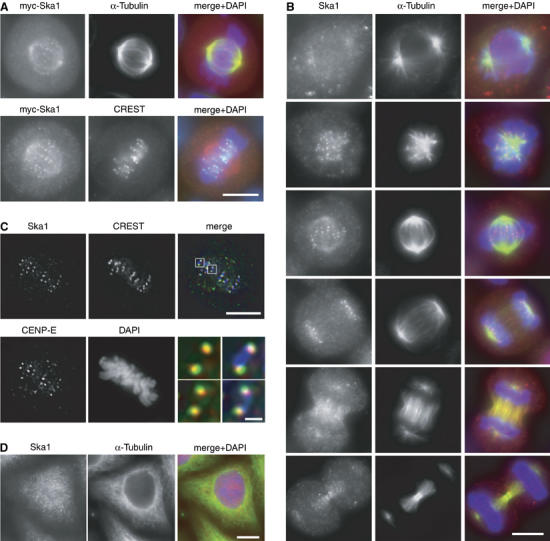
Mitotic spindle and outer KT localisation of Ska1. (A) HeLa S3 cells were transfected with a myc-tagged Ska1 construct and then fixed with PTEMF. Cells were stained with anti-myc 9E10 antibody (red), DAPI (DNA, blue) and with either anti-α-tubulin antibody (green) (upper panel) or CREST serum (green) (bottom panel). (B) HeLa S3 cells were fixed with PTEMF and then stained with anti-Ska1 antibody (red), anti-α-tubulin antibody (green) and DAPI (DNA, blue). (C) Same as in (B) except that cells were stained with anti-Ska1 antibody, anti-CENP-E antibody, CREST serum and DAPI followed by imaging with a Deltavision microscope. Merged pictures are single, deconvolved focal planes and show CREST (blue), CENP-E (green) and Ska1 (red). Right panels in the bottom row are magnifications of the above marked areas with scale bar indicating 1 μm. (D) An interphase cell is shown which was fixed with paraformaldehyde, permeabilised with Triton X–100 and stained as in (B). Scale bars=10 μm.
To investigate the localisation of endogenous Ska1, a polyclonal rabbit antibody was raised. This antibody detected a prominent band of the expected size in Western blots performed on whole HeLa S3 cell lysates (Supplementary Figure 1). It also stained both spindle and KT structures in mitotic cells (Figure 1B), confirming the localisation of the myc-Ska1 protein (Figure 1A). Weak spindle staining, particularly in the region of the forming poles, could already be observed in early prophase and persisted until mid-anaphase. Upon furrow ingression in late anaphase, Ska1 then localised diffusely to the central spindle and later to midbody structures. KT staining appeared during prometaphase, was most prominent from late prometaphase through mid-anaphase and then vanished in telophase. Co-staining of mitotic cells with anti-Ska1 antibodies and reagents detecting either the centromeres (CREST serum) or the outer KTs (anti-CENP-E) revealed that Ska1 partly co-localised with CENP-E, adjacent to the CREST staining (Figure 1C). Ska1 also co-localised with the outer KT protein Hec1 (A Hanisch and HHW Silljé, unpublished data), confirming that this protein is concentrated at the outer KTs. During interphase, anti-Ska1 antibodies produced a diffuse staining of both the cytoplasm and the nucleus, but no association with either MTs or centromeres could be observed (Figure 1D). Taken together, these results show that Ska1 is a novel mitotic spindle and KT-associated protein.
Requirements for Ska1 localisation to KTs
As Ska1 staining at KTs increased during prometaphase (Figure 1B), we asked whether this localisation might depend on KT–MT interactions. When cells were treated with the MT-depolymerising drugs nocodazole or colchicine, Ska1 failed to accumulate at KTs (Figure 2A; A Hanisch and HHW Silljé, unpublished data). Similarly, addition of nocodazole to cells already arrested in mitosis (by noscapine treatment (Zhou et al, 2002)) resulted in the loss of Ska1 from KTs, concomitant with the loss of MTs (Supplementary Figure 2A). Conversely, when cells were released from a nocodazole arrest, the KT localisation of Ska1 was restored in parallel with reformation of the spindle (Supplementary Figure 2B). Addition of the MT-stabilising drug taxol did not impair Ska1 localisation to KTs (Figure 2A), suggesting that the presence of MTs, rather than their dynamic properties was important for Ska1 localisation.
Figure 2.
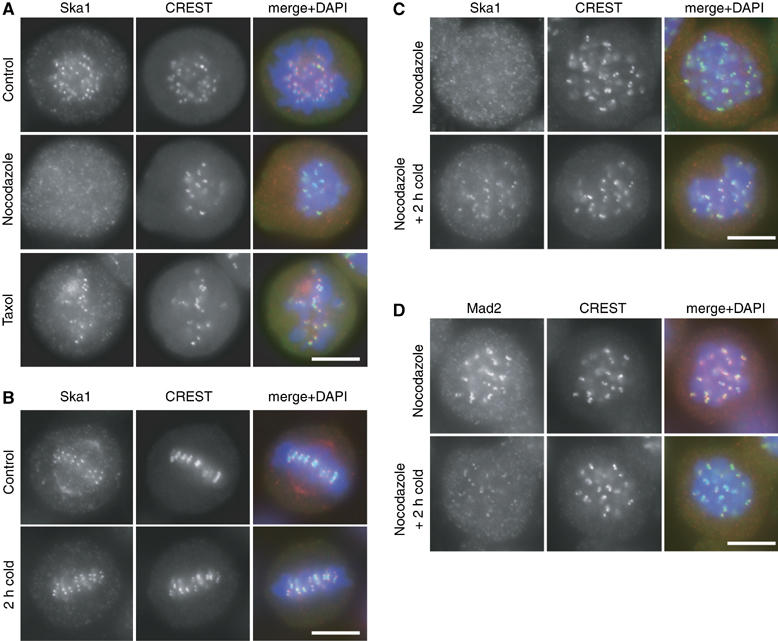
Ska1 KT localisation requires KT–MT attachment at physiological temperatures. (A) HeLa S3 cells were either left untreated as a control or treated for 14 h with nocodazole and taxol, respectively. After PTEMF fixation, cells were stained with anti-Ska1 antibody (red), CREST serum (green) and DAPI (DNA, blue). (B) HeLa S3 cells were fixed with PTEMF either directly (control) or after incubation in ice-cold medium for another 2 h. Cells were stained with anti-Ska1 antibody (red), CREST serum (green) and DAPI (DNA, blue). (C) HeLa S3 cells were treated with nocodazole for 14 h and then either fixed directly or incubated in ice-cold medium for another 2 h in the presence of nocodazole. Cells were fixed and stained as in (B). (D) HeLa S3 cells were treated as in (C) but stained with anti-Mad2 antibody (red), CREST serum (green) and DAPI (DNA, blue). Scale bars=10 μm.
Taken together, the above data suggested that KT–MT interactions regulate the accumulation and maintenance of Ska1 at KTs. In analogy to other proteins whose KT association had previously been shown to depend on MTs (Joseph et al, 2004), it was tempting to conclude that MTs are required for transporting Ska1 to KTs. Surprisingly, however, cold-induced depolymerisation of MTs did not result in the loss of Ska1 from KTs (Figure 2B), in striking contrast to the drug-induced effects described above. To rule out the persistence of short MTs in these experiments, cells were pre-treated with nocodazole and then incubated on ice in the continued presence of nocodazole. Under these conditions, Ska1 was initially lost from KTs in response to nocodazole, consistent with the data shown above, but KT staining was subsequently recovered upon incubation in the cold, even in the continued presence of nocodazole (Figure 2C; Supplementary Figure 3A). Interestingly, cold treatment of nocodazole-exposed cells did not restore KT localisation to RanGAP1 and RanBP2 (Supplementary Figure 3B; A Hanisch and HHW Silljé, unpublished data), two proteins that are known to depend on MTs for transport to the KT (Joseph et al, 2004). These data indicate that the dependency of Ska1 localisation on MT attachment does not simply reflect a requirement for MT-mediated transport. Instead, we conclude that the attachment of MTs to KTs either generates docking sites for the recruitment of Ska proteins or prevents their release, and that MT attachment can be mimicked by exposure to low temperature.
In contrast to Ska1, the KT localisation of the spindle checkpoint protein Mad2 is lost upon MT attachment (Waters et al, 1998), but can readily be restored by nocodazole treatment (Figure 2D). Remarkably, however, the association of Mad2 with KTs was strongly reduced when nocodazole-treated cells were incubated in the cold (Figure 2D). Thus, the KT localisation of Mad2 appears to depend on conditions that are exactly opposite to those required for Ska1 recruitment (compare Figures 2C and D). The possible implications of these results are considered in the Discussion.
To determine which proteins might be required for KT localisation of Ska1, several KT-associated proteins were depleted by small interfering RNA (siRNA). Of all the proteins tested, only depletion of the outer KT protein Hec1 clearly reduced the concentration of Ska1 at KTs (Figure 3A). In contrast, depletion of Bub1, Aurora-B, CENP-E, CENP-F, RanBP2, RanGAP1, CLIP170 and EB1 did not significantly affect Ska1 localisation to KTs (A Hanisch and HHW Silljé, unpublished data). Cold treatment of Hec1-depleted cells did not restore Ska1 to the KT, although it did so in control-treated cells (Figure 3B), arguing for a structural effect of Hec1 depletion on the Ska1 docking site(s) rather than an indirect effect through impaired KT–MT interaction (Ciferri et al, 2005; DeLuca et al, 2005). We have explored the possibility that Hec1 might recruit Ska1 to the KT via a direct interaction, but failed to detect Hec1 in Ska1 immunoprecipitates (Supplementary Figure 4). Similarly, none of several KT and centromere proteins analysed could be found in Ska1 immunoprecipitates (Supplementary Figure 4).
Figure 3.
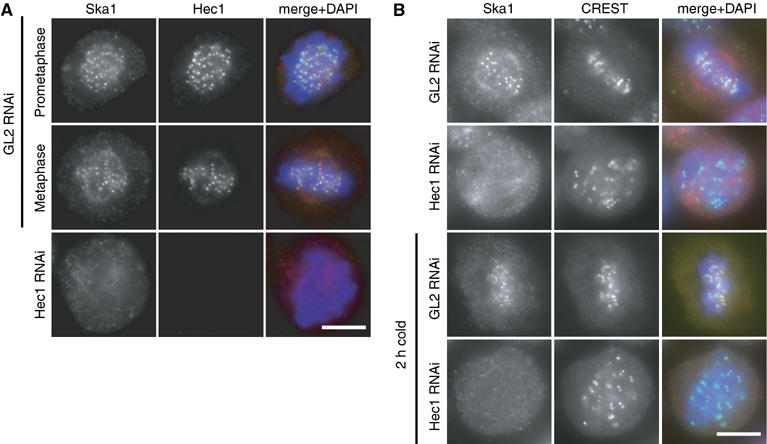
Ska1 KT localisation requires the presence of Hec1 even in the cold. (A) HeLa S3 cells were treated for 48 h with control (GL2) and Hec1-specific siRNAs, respectively, then fixed with PTEMF and stained with anti-Ska1 (red), anti-Hec1 (green) and DAPI (DNA, blue). (B) HeLa S3 cells were treated for 48 h with control (GL2) and Hec1-specific siRNAs, respectively, before half of the samples were incubated for 2 h in ice-cold medium. Cells were fixed as in (A) but stained with anti-Ska1 antibody (red), CREST serum (green) and DAPI (DNA, blue). Scale bars=10 μm.
Ska1 interacts with Ska2 (FAM33A)
As the above experiments had failed to reveal any binding partners of Ska1, we performed a yeast two-hybrid screen with full-length Ska1 as bait. This screen did not yield known KT proteins, but resulted in the repeated isolation of cDNAs coding for a 14 kDa protein (Figure 4A), which we now refer to as Ska2 (formerly this hypothetical protein had been designated FAM33A). Database searches revealed the existence of Ska2 homologues in several other vertebrate but not invertebrate species. Considering that Ska1 genes are present in both invertebrates and plants, we assume that the difficulty to detect Ska2 homologues outside of vertebrates most likely reflects its small size and the absence of known diagnostic motifs.
Figure 4.
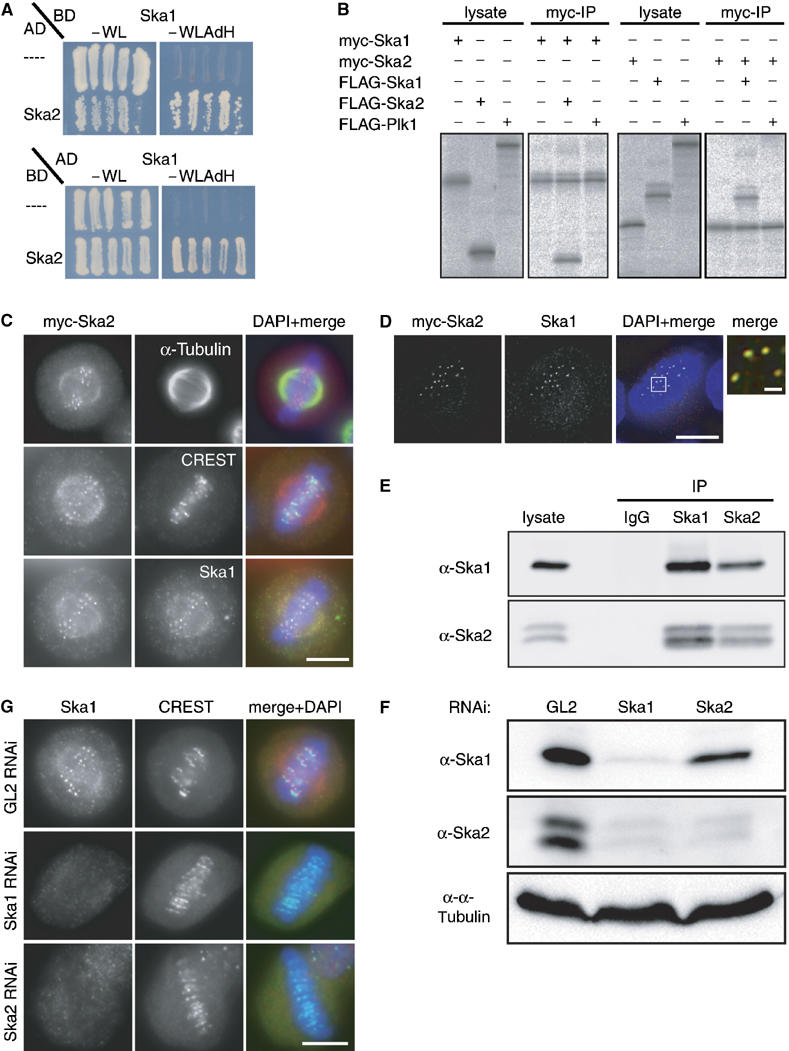
Ska1 interacts with another novel protein termed Ska2 (FAM33A). (A) Yeast two-hybrid interaction between Ska1 expressed as a binding domain (BD) fusion and Ska2 expressed as an activation domain (AD) fusion (top panel). In the bottom panel, Ska1 is expressed from the AD vector and Ska2 from the BD vector. As negative controls, the empty AD or BD (−) vectors were used. Interactions were reflected by growth on selective medium (−WLAdH, right panels). For control, growth on nonselective plates is also shown (−WL, left panels). (B) Myc- and FLAG-tagged versions of Ska1, Ska2 and Plk1 (negative control) were produced in different combinations by IVT in the presence of 35S-labelled methionine. Myc-tagged Ska1 (left panels) or Ska2 (right panels) were subsequently immunoprecipitated and IVT input and myc-immunoprecipitates were analysed by SDS–PAGE followed by autoradiography. (C) Myc-tagged Ska2 was transiently expressed in HeLa S3 cells for 48 h. After PTEMF fixation cells were stained with anti-myc 9E10 antibody (red), DAPI (DNA, blue) and with either anti α-tubulin antibody (upper panel), CREST serum (middle panel) or anti-Ska1 antibody (bottom panel) (all in green). (D) As in (C) after staining with anti-myc 9E10 antibody (red), anti-Ska1 antibody (green) and DAPI (DNA, blue), cells were imaged with a Deltavision microscope. Pictures are single, deconvolved focal planes. Right panel is a magnification of the marked area in the merged picture with scale bar indicating 1 μm. (E) Lysates were prepared from mitotic HeLa S3 cells. They were then used for immunoprecipitations with anti-Ska1 antibody, anti-Ska2 antibody and rabbit IgGs (negative control), respectively. Lysate and immune complexes were separated by SDS–PAGE and probed by Western blotting with anti-Ska1 and anti-Ska2 antibodies, as indicated. (F) HeLa S3 cells were treated for 48 h with control (GL2) and Ska1 and Ska2-specific siRNAs, respectively. Equal amounts of cell extracts were separated by SDS–PAGE and probed by Western blotting with anti-Ska1 and anti-Ska2 antibodies. Detection of α-tubulin was used as a loading control. (G) HeLa S3 cells were treated for 48 h with control (GL2), Ska1- and Ska2-specific siRNAs, respectively, then fixed with PTEMF and stained with anti-Ska1 antibody (red), CREST serum (green) and DAPI (DNA, blue). Scale bars=10 μm.
To confirm the interaction between Ska1 and Ska2, tagged versions of the two proteins were produced in vitro by coupled transcription–translation, in the presence of 35S-methionine and immunoprecipitations were performed. Myc-tagged Ska1 and Ska2 readily precipitated FLAG-Ska2 and FLAG-Ska1, respectively. In contrast, FLAG-tagged Polo-like kinase 1 (Plk1), used as a negative control, was not co-precipitated, attesting to the specificity of the observed interactions (Figure 4B). Next, we analysed the localisation of transiently expressed myc-tagged Ska2 in HeLa S3 cells (Figure 4C). Similar to Ska1, Ska2 showed faint spindle localisation as well as prominent KT localisation. Moreover, myc-Ska2 co-localised exactly with Ska1 (Figure 4C), as confirmed by high-resolution analysis of a single deconvolved Z-stack (Figure 4D). Together these experiments identify Ska2 as a second novel spindle and KT-associated protein and in vitro binding partner of Ska1.
An antibody raised against Ska2 recognised a single protein in interphase cells, but two forms in mitotically-arrested cells (Supplementary Figure 5A). Both bands were sensitive to siRNA-mediated depletion (Figure 4F), indicating that they represent variant or modified forms of Ska2. To investigate the interaction of Ska1 and Ska2 in vivo, co-immunoprecipitation experiments were performed on lysates prepared from HeLa S3 cells that had been synchronised in M phase by nocodazole treatment, followed by a 40 min release to allow for spindle reformation (Figure 4E). Whereas neither Ska1 nor Ska2 was precipitated by control IgGs, both forms of Ska2 could readily be detected in anti-Ska1 precipitates and, conversely, anti-Ska2 antibodies brought down Ska1 (Figure 4E), demonstrating the existence of a specific complex in vivo. To analyse whether the stability and localisation of Ska1 and/or Ska2 depends on complex formation, we depleted each protein individually, using specific siRNA oligonucleotides. As shown by Western blotting, Ska1 was efficiently depleted within 48 h of siRNA treatment (Figure 4F). Interestingly, this resulted in the concomitant disappearance of both forms of Ska2, indicating that Ska2 proteins require Ska1 for stability. Ska2 could also be depleted efficiently, but in this case, the level of Ska1 was not significantly affected (Figure 4F). We also examined the influence of complex formation on the localisation of Ska proteins. When either Ska1 or Ska2 was depleted by siRNA, Ska1 was lost from the spindle and KTs (Figure 4G), indicating that complex formation between the two proteins is required for correct localisation of Ska1. The converse experiment could not be carried out, because the available anti-Ska2 antibody does not perform well in IF experiments. However, as the bulk of Ska2 is degraded upon Ska1 depletion (see above), one would predict that Ska2 is lost from spindle and KT structures when Ska1 is absent. Taken together, these results establish that Ska2 requires Ska1 for stability, whereas Ska1 requires Ska2 for correct localisation.
The cell cycle expression of Ska1 and Ska2 was explored by Western blot analysis. Both proteins were present at similar levels in asynchronous, G1/S phase (aphidicolin-treated) and mitotic (nocodazole/taxol-treated) cells (Supplementary Figure 5A). To explore whether the slower migrating form of Ska2 present in mitotically arrested cells reflects phosphorylation, its sensitivity to phosphatase treatment was examined. No change in Ska2 migration could be observed in response to alkaline phosphatase, type 1 phosphatase or lambda phosphatase, although BubR1 and Cdc27, two mitotic phosphoproteins known to undergo phosphorylation-induced mobility changes (Li et al, 1999; Kraft et al, 2003), readily responded to these treatments (Supplementary Figure 5B; A Hanisch and HHW Silljé, unpublished data). Thus, Ska2 is either phosphorylated on a site that is particularly resistant to phosphatase or, alternatively, subjected to another type of modification. Whatever its molecular identity, the variant Ska2 persisted for more than 2 h after release of nocodazole-arrested cells, long after cyclin B was degraded (Supplementary Figure 5C). These results show that both Ska1 and Ska2 proteins are expressed at near constant levels throughout the cell cycle, but that an as yet unidentified variant of Ska2 is present throughout mitosis.
Ska1 and Ska2 are required for proper mitotic progression
To examine the functional consequence of depleting the Ska complex, HeLa S3 cells were depleted of Ska proteins and then examined by IF microscopy. First, we asked whether the absence of the Ska complex disrupts KT structure. None of the KT- and centromere-associated proteins analysed, including Bub1, BubR1, Mad1, Hec1, Aurora-B, CENP-E, CENP-F, RanBP2, RanGAP1, CLIP170 and EB1, was significantly affected in its localisation upon depletion of Ska proteins (A Hanisch and HHW Silljé, unpublished data), indicating that the Ska complex is not required for overall KT structure. However, when cells were examined 48 h after siRNA-mediated depletion of either Ska1 or Ska2, a strong increase in mitotic index could be observed (Figure 5A). Analysis of the resulting mitotic cells by 4′,6-diamidino-2-phenylindole (DAPI) staining revealed that most of them showed a metaphase-like appearance (Figure 5B), with apparently normal mitotic spindles (Figure 6A, left column). Whereas many disturbances of mitotic progression result primarily in an increased prometaphase population (Biggins and Walczak, 2003), we were surprised to find that 60% of all pre-anaphase cells showed chromosomes aligned in a near-perfect metaphase plate. This strongly indicates that depletion of the Ska complex did not primarily impair chromosome congression, but instead caused cells to arrest or delay in a metaphase-like state. To investigate this unusual mitotic phenotype in more detail, time-lapse video microscopy was performed on HeLa S3 cells that stably express a histone H2B-GFP fusion protein (Figure 5C; Supplementary Movies 1, 2 and 3). These experiments confirmed that mitotic progression was strongly delayed in Ska-depleted cells and that most of the delay occurred in a metaphase-like state. Eventually, however, cells were able to progress through mitosis (Figure 5C; Supplementary Movies 2 and 3).
Figure 5.
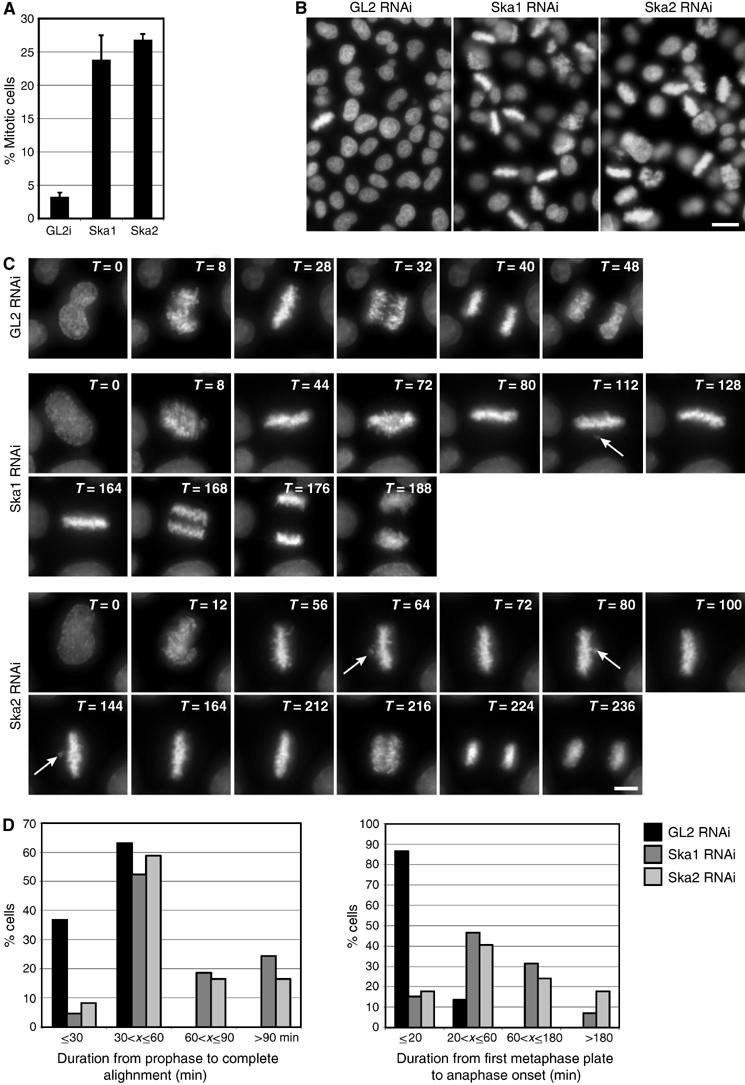
Depletion of Ska1 and Ska2 results in a metaphase delay. (A) HeLa S3 cells were treated for 48 h with control (GL2), or Ska1- and Ska2-specific siRNAs, respectively. The mitotic indexes were determined by light microscopy. (B) The same cells as in (A) were fixed and stained with DAPI (DNA). Scale bar=20 μm. (C) Live-cell imaging of H2B-GFP expressing HeLa S3 cells. Selected images show H2B-GFP stained chromosomes of Hela S3 cells progressing through mitosis. Cells were treated with GL2 (control), or Ska1- and Ska2-specific siRNAs, respectively, for 30 h before filming. T=0 was defined as the time point at which chromosome condensation became evident (prophase). Time points are indicated in minutes. Arrows point to chromosomes that have transiently moved out of the metaphase plate. Scale bar=10 μm. (D) The durations of different periods from prophase to anaphase onset were calculated from time-lapse movies (C) of at least 37 control (GL2), and at least 76 Ska1 and Ska2 siRNA-treated cells. T=0 was defined as in (C). Complete alignment was defined as the duration from T=0 to the first time point at which a perfect metaphase plate was observed and anaphase onset was calculated from the first frame at which chromosome segregation was visible. Histograms show the percentages of mitotic cells that had progressed within the indicated time frames from prophase to the first complete metaphase plate (left panel) and from first metaphase plate to anaphase onset (right panel).
Figure 6.
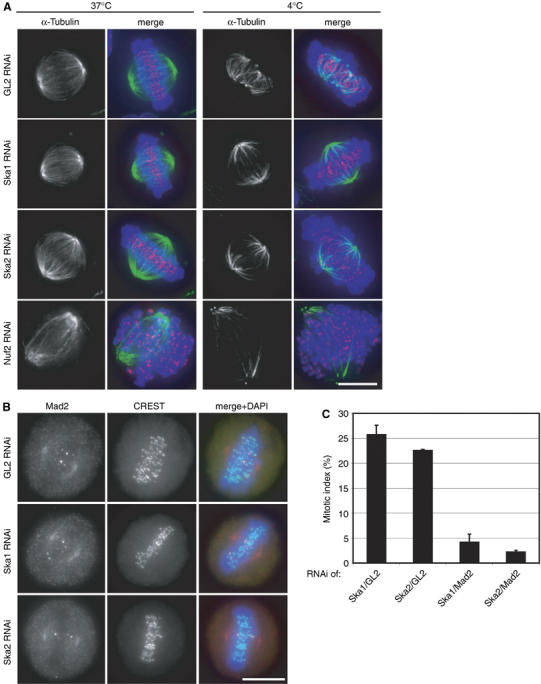
Characterisation of Ska-depletion phenotype. (A) HeLa S3 cells were treated for 48 h with control (GL2), Ska1-, Ska2- and Nuf2-specific siRNAs, respectively. They were fixed directly (left column, 37°C) or after incubation for 10 min at 4°C (right column, 4°C) before staining with anti-α-tubulin antibody (green), CREST serum (red) and DAPI (DNA, blue). (B) HeLa S3 cells were treated for 48 h with control (GL2), or Ska1- and Ska2-specific siRNAs, respectively, then fixed with PTEMF and stained with anti-Mad2 antibody (red), CREST serum (green) and DAPI (DNA, blue). Scale bars=10 μm. (C) Quantification of the mitotic indices (300 cells each) after 48 h of Ska1 and Ska2 siRNA together with control GL2 siRNA or after simultaneous depletion of Mad2.
Quantification of the above data showed that control (GL2)-depleted cells proceeded from prophase to anaphase onset in 47±9.7 min. In Ska1- and Ska2-depleted cells this period was highly variable, but on average took about four times longer (179±141 and 174±120 min, respectively). Most remarkably, most Ska1- and Ska2-depleted cells (57 and 67%, respectively), as well as all (GL2- treated) controls, completed alignment of chromosomes in a metaphase plate within 60 min after onset of chromosome condensation (Figure 5D, left histogram). This indicates that although a defect in chromosome congression contributed to the lengthening of mitotic progression, it did not constitute the primary reason for the strong delay seen in most Ska-depleted cells. Instead, the clearest difference between control cells and Ska-depleted cells was that almost all control cells initiated anaphase within 20 min of metaphase plate formation (average of 14 min), whereas Ska1- and Ska2-depleted cells spent highly variable amounts of time at this metaphase-like state, requiring an average of 76 and 100 min, respectively, for progressing to anaphase onset (Figure 5D, right histogram). Moreover, about 60% of the delayed Ska-depleted cells were unable to keep all chromosomes fully aligned in a metaphase plate, so that, occasionally, individual chromosomes moved out of the plate and back in (Figure 5C; Supplementary Movies 2 and 3).
Role of Ska complex in stabilisation of KT–MT interaction and checkpoint silencing
To further characterise the molecular mechanism underlying the Ska depletion phenotype, Ska-depleted cells were challenged by short exposure to low temperature, a procedure known to test the stability of KT-fibres (Rieder, 1981). When compared to control cells and Nuf2-depleted cells – which are known to be impaired in KT–MT attachment (DeLuca et al, 2002) – the KT-fibres of Ska-depleted cells showed an intermediate cold-sensitivity (Figure 6A). This result suggests that KT-fibres, although functional, were weakened, in keeping with the observation that a majority of Ska-depleted cells was able to accomplish near-normal chromosome congression (Figure 5D, left histogram), but then failed to maintain a metaphase plate (Figure 5C; Supplementary Movies 2 and 3). One straightforward interpretation of these results is that a transient impairment of K-fibre stability caused the occasional loss of KT–MT interaction, allowing individual chromosomes to escape from the metaphase plate. Consistent with this view, most of the metaphase plates present in Ska-depleted cells contained at least one or two Mad2-positive KTs (85% for Ska1-depleted, 87% for Ska2-depleted metaphase cells, s.d. ⩽2) (Figure 6B). As the KT association of Mad2 usually (albeit not invariably) (Martin-Lluesma et al, 2002) correlates with an active spindle checkpoint, this observation indicates that Ska-depleted cells failed to turn off the spindle checkpoint. Indeed, the mitotic delay induced by Ska depletion could readily be overcome by co-depletion of Mad2, demonstrating its dependency on the checkpoint (Figure 6C). Furthermore, although interkinetochore distances were normal among the aligned chromosomes in Ska1- and Ska2-depleted cells (1.60±0.3 and 1.63±0.3 μm, respectively, as compared to 1.59±0.3 μm in control cells), the rare ‘escaper' chromosomes showed interkinetochore distances similar to those seen in nocodazole-arrested cells (0.83±0.2 μm), suggesting a reduction in tension only at the rare chromosomes that had been lost from the metaphase plate. Taken together, these results lead us to propose that the Ska complex is required primarily for the maintenance (rather than the establishment) of a metaphase plate and/or the silencing of the spindle checkpoint.
Discussion
In this study, we describe the characterisation of Ska1, a novel spindle and KT-associated protein originally identified as a candidate spindle component by mass spectrometry (Sauer et al, 2005). Furthermore, we identify a second novel protein, termed Ska2 that interacts with Ska1 both in vitro and in vivo. Complex formation between the two proteins is required for Ska2 stability as well as Ska protein localisation to spindle MTs and KTs. During prometaphase, the Ska complex associates with KTs only after MT attachment, suggesting that this complex is not required for KT–MT interactions per se. Depletion studies by siRNA showed that the absence of the Ska complex did not overtly affect general KT structure. Furthermore, although increased cold-sensitivity suggests a weakening of KT-fibres, initial chromosome congression was only modestly impaired. Instead, depletion of the Ska complex caused cells to spend prolonged periods of time in a metaphase-like state characterised by occasional loss of individual chromosomes and a persistent activation of the spindle checkpoint. Taken together, our data indicate that the Ska complex is required for the maintenance of chromosomes in a fully aligned metaphase plate and for checkpoint silencing.
Requirements for KT localisation of Ska proteins
At physiological temperature, Ska1 localisation to KTs required KT–MT attachments, but at low temperature the protein associated with KTs in the absence of MTs. Other proteins, including RanGAP1 and RanBP2, have previously been shown to localise to KTs in an MT-dependent manner and this is thought to reflect MT-dependent transport (Joseph et al, 2004). However, the fact that Ska1 showed a strong concentration at KTs of cold-treated cells, even though MTs were absent, argues that Ska1 does not depend on MTs for its transport to KTs. Instead, the data suggest that Ska1 binding to KTs depends on docking sites that, at physiological temperature, are created by MT attachment, but those can also be generated when cells are incubated in the cold, in the absence of MTs. One possibility is that docking sites are generated by a shift in the balance of opposing enzymatic activities (e.g., a disturbance of kinase/phosphatase equilibria). Alternatively, it is possible that Ska proteins turn over constitutively at KTs and that their release requires an energy-dependent enzymatic activity, which is blocked by MT attachment (at physiological temperature) or cold treatment.
Regardless of the molecular nature of the Ska docking sites, it is striking that Ska proteins and Mad2 display opposite requirements for KT localisation under all conditions examined. At physiological temperature, Ska docking sites are generated in response to MTs attachment, whereas, concomitantly, Mad2 is lost from KTs. Conversely, nocodazole treatment abolishes Ska localisation to KTs but causes the recruitment of Mad2, and finally, cold treatment restores Ska docking sites even in the absence of MTs, while inducing the loss of Mad2. Considering the striking correlation between the creation of Ska binding sites and the loss of Mad2 from KTs, the question arises of whether the recruitment of the Ska complex directly contributes to the release of Mad2 from KTs. However, the observation that Mad2 was absent from the KTs of most aligned chromosomes in Ska-depleted cells argues against a direct, active role of the Ska complex in Mad2 displacement. Instead, a particular structural change produced at KTs upon MT attachment (or cold treatment in the absence of MTs) may cause both Ska recruitment and Mad2 displacement.
Does the Ska complex functionally resemble the yeast Dash/Dam1 complex?
As indicated by comparative sequence analyses, homologues of Ska1 are present in the genomes of vertebrates and invertebrates, as well as plants. Ska2 genes could be identified with confidence only in vertebrates, but considering the functional interaction between Ska1 and Ska2, we presume that Ska2 genes are also present in invertebrates and plants, although they are impossible to detect by current search algorithms. Neither Ska1 nor Ska2 homologues could be identified in yeast, but again, this does not necessarily imply that functional homologues are truly absent. In recent years, it has become increasingly apparent that yeast and vertebrate KTs share many more components than had previously been appreciated (Meraldi et al, 2006). One notable exception to this conclusion concerns the yeast Dash/Dam1 complex, comprising 10 different small subunits for which no counterpart has yet been identified in mammals (Cheeseman et al, 2001; Miranda et al, 2005; Westermann et al, 2005). It is intriguing, therefore, that several of the properties of the Ska complex described here are reminiscent of results described for the yeast Dash/Dam1 complex. Specifically, the Dash/Dam1 complex of Saccharomyces cerevisiae has been shown to localise to spindle MTs and KTs (Hofmann et al, 1998; Cheeseman et al, 2001) and to require both an intact Ndc80 complex (the yeast homologue of the human Hec1 complex) and MTs for KT localisation (Janke et al, 2002). Moreover, this budding yeast complex is known to contribute to the stabilisation of KT–MT interactions (Cheeseman et al, 2001; Miranda et al, 2005). Interestingly, the Dash/Dam1 complex is essential for viability in Saccharomyces cerevisiae (Hofmann et al, 1998) but not in Schizosaccharomyces pombe (Sanchez-Perez et al, 2005). Instead, mutations in Schizosaccharomyces pombe Dash/Dam1 proteins result in a delayed anaphase onset and persistent spindle checkpoint activation (Sanchez-Perez et al, 2005), similar to the Ska1 and Ska2 depletion phenotype described here. Thus, in spite of the absence of obvious sequence similarity between Ska1, Ska2 and any of the Dash/Dam1 complex components, it is possible that the human Ska complex and the yeast Dash/Dam1 complex perform at least partially similar roles. One interesting question for the future is whether Ska proteins are able to form ring structures around MTs, as described for the Dash/Dam1 complex (Miranda et al, 2005; Westermann et al, 2005).
Ska complex is required for timely anaphase onset
Depletion of human Ska proteins did not affect the localisation of any other KT protein examined so far, arguing that the Ska complex is not required for overall KT structure. Moreover, the mitotic spindle appeared to form normally in Ska-depleted cells and although KT-fibres were weakened, chromosome congression was not fundamentally impaired in a majority of cells. However, it would be premature to exclude that a complete (genetic) knockout of Ska1 and Ska2 might reveal a more severe phenotype. The most striking consequence of siRNA-mediated depletion of Ska proteins was a delayed anaphase onset preceded by a prolonged metaphase-like state. This unusual phenotype was characterised by individual chromosomes occasionally moving out of (and back into) the metaphase plate, rare KTs staining positive for Mad2 and persistent spindle checkpoint activation. On the basis of these results, we propose that the Ska complex is required for stabilising KT–MT attachments and/or checkpoint silencing. In principle, the failure of Ska-depleted cells to silence the spindle checkpoint could be a consequence of an occasional destabilisation of KT–MT interactions at individual chromosomes. Alternatively, Ska-depleted cells could primarily be impaired in switching off the checkpoint, in which case the occasional loss of individual chromosomes from the metaphase plate could be a consequence of the extended metaphase delay. In particular, it is tempting to speculate that the Ska complex could contribute to functionally down regulate some of the KT-associated proteins that are responsible for relaying the inhibitory checkpoint signal (e.g. one or several of the checkpoint kinases). Thus, we anticipate that the discovery of the vertebrate Ska complex will prompt new lines of enquiry into the coupling of MT attachment to the KT and checkpoint silencing.
Materials and methods
Cloning procedures
For cloning of Ska1 (C18Orf24), a cDNA clone (IMAGp998H169724Q) was obtained from the ‘Deutsches Ressourcenzentrum für Genomforschung' (RZPD). This cDNA was cloned in-frame into a pcDNA3.1 vector (Invitrogen, Carlsbad, CA) encoding either an N-terminal 3xmyc tag or a FLAG tag. Ska2 (FAM33A) was cloned into the same vectors using a yeast two-hybrid clone as PCR template.
Antibody production
In order to produce Ska1- and Ska2-specific antibodies, the polyhistidine-tagged proteins were expressed in Escherichia coli from pET-28 vectors (EMD Biosciences, Madison, WI) and purified under denaturing conditions. New Zealand's white rabbits were used to produce antisera against these recombinant proteins (Charles River Laboratories, Romans, France). The sera were affinity purified using AminoLink Plus Immobilization Kit (Pierce Biotechnology, Rockford, IL) coated with the respective antigens according to the manufacturer's protocol.
Cell culture and synchronisation
HeLa S3 cells were grown at 37°C under 5% CO2 in DMEM (Invitrogen), supplemented with 10% FCS and penicillin–streptomycin (100 IU/ml and 100 μg/ml, respectively). Cell synchronisation and K-fibre stability assays were described previously (Sillje et al, 2006).
Transient transfections and siRNA
Plasmid transfections were performed using FUGENE6 reagent (Roche Diagnostics, Indianapolis, IN), according to the manufacturer's instructions. SiRNA duplexes were transfected using Oligofectamine (Invitrogen) as described elsewhere (Elbashir et al, 2001). The sequence of the siRNA duplex for targeting Ska1 was: 5′-CCC GCT TAA CCT ATA ATC AAA-3′ and for Ska2: 5′-AAG AAA TCA AGA CTA ATC ATC TT-3′ (Qiagen, Hilden, Germany). Similar results were obtained with two other siRNAs targeting different sequences in Ska1: 5′-CTG GAG ATT TGT GTC AAT AAT-3′ and Ska2: 5′-TTT CAC ATG CCA GAT TTA TGA-3′. Hec1, Mad2 and Nuf2 were depleted using established siRNAs targeting published sequences (DeLuca et al, 2002; Stucke et al, 2004). As a control, a duplex (GL2) targeting luciferase was used (Elbashir et al, 2001).
IF microscopy
Cells were grown on coverslips and fixed and permeabilised as described previously (Sillje et al, 2006). Primary antibodies used in this study were rabbit anti-Ska1 serum (1:5000), mouse mAb anti-myc (1:10, 9E10 tissue culture supernatant), mouse mAb anti-α-tubulin-FITC (1:1000, Santa-Cruz Biotechnology, Santa Cruz, CA), human CREST autoimmuneserum (1:5000, Immunovision, Springdale, AR), mouse mAb anti-Hec1 (1:1000, Abcam, Cambridge, UK), rabbit anti-Mad2 (1:1000, Bethyl, Montgomery, TX) and goat anti-RanGAP1 and anti-RanBP2 (1:1000, 1:500, respectively, gifts from Dr Frauke Melchior, University. of Göttingen, Germany). Primary antibodies were detected with Alexa-Fluor-488- and Alexa-Fluor-555-conjugated goat anti-mouse, anti-rabbit or anti-goat IgGs (1:1000, Molecular Probes, Eugene, OR), respectively. DNA was stained with DAPI (2 μg/ml).
IF microscopy was performed using a Zeiss Axioplan II microscope (Zeiss, Jena, Germany) with Apochromat × 40 and × 63 oil immersion objectives, as described before (Sillje et al, 2006). For high-resolution images a Deltavision microscope (Applied Precision, Issaquah, WA) on an Olympus IX71 base, equipped with a PlanApo 60 × /1.40 oil immersion objective and a CoolSNAP HQ camera (Photometrics) was used for collecting 0.15 μm-distanced optical sections in the z-axis. Images at single focal planes (Figures 1C and 4D) were processed with a deconvolution algorithm, and optical sections were projected into one picture using Softworx software (Applied Precision). The same software was used to measure interkinetochore distances in images at single focal planes (n=20 cells/total 235 KTs for control, n=19 cells/total 189 KTs for Ska-depleted cells and n=16 cells/total 189 KTs for nocodazole-treated cells). Images were cropped in Adobe Photoshop 6.0, and then sized and placed in figures using Adobe Illustrator 10 (Adobe Systems, San Jose, CA).
Live-cell imaging
For live-cell imaging, an HeLa S3 cell line stably expressing histone H2B-GFP was used (Sillje et al, 2006). Cells were treated with siRNAs for 30 h, before changing the medium to CO2-independent medium and the culture dish was placed onto a heated sample stage within a heated chamber (37°C). Live-cell imaging was performed using the Plan Apo 40 × /0.95 objective on the above-described Deltavision microsope and Softworx software was used to collect and process data. Images were captured with 10% neutral density and 200 ms exposure times in 4 min intervals for 18 h with three stacks per field spaced 3 μm each.
Yeast two-hybrid analysis
A yeast two-hybrid screen was performed using a system described previously (James et al, 1996). Ska1 cDNA in the pFBT9 Gal4 DNA binding domain vector was used to screen a human HEK293 two-hybrid library (BD Clontech, Mountain View, CA). Clones able to activate both the Ade2 and His3 selection markers, specifically in the presence of the bait, were selected.
In vitro coupled transcription translation
The respective 3xmyc- and FLAG-tagged proteins were produced by in vitro coupled transcription translation (IVT) in the presence of 35S-methionine using the TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI). For immunoprecipitation these reactions were diluted in LS buffer (50 mM Tris pH 8.0, 100 mM NaCl, 0.1% NP40 and protease inhibitors) and incubated with anti-myc antibody (9E10) coated Protein G beads (Pierce Biotechnology, Rockford, IL) for 90 min at 4°C on a rotating wheel. After washing, samples were boiled in sample buffer and equal protein amounts of input and myc-precipitates were separated by SDS–PAGE and visualised by autoradiography.
Cell extracts, immunoprecipitation and Western blot analysis
Preparation of cell extracts and Western blot analysis were described previously (Hanisch et al, 2006). For immunoprecipation experiments Affi-Prep Protein A Support beads (Bio-Rad Laboratories, Hercules, CA) coated with either rabbit anti-Ska1, rabbit anti-Ska2 or rabbit IgGs as a control were used. For Western blot analysis, affinity purified rabbit anti-Ska1 (2.5 μg/ml), affinity purified rabbit anti-Ska2 (5 μg/ml), mAb anti-α-tubulin (1:1000, Sigma), mAb anti-Hec1 (1:1000, Abcam, Cambridge, UK), goat anti-RanGAP1, goat anti-RanBP2 (1:750, 1:6000, respectively, gift from Frauke Melchior), mAb anti-CLIP-170 (1:100, gift from Dr Franck Perez, Institut Curie, Paris, France), mAb anti-EB1 (1:1000, BD Transduction Laboratories), mAb anti-Plk1 (1:10, PL2, tissue culture supernatant), mAb anti-Cdc27 (1:250, BD Transduction Laboratories), mAb anti-BubR1 (1:10, 68–3–9, tissue culture supernatant), mAb anti-Bub1 (undiluted, 61–22–2, tissue culture supernatant), mAb anti-Aurora B (1:500, BD Transduction Laboratories), mAb anti-Mps1 (undiluted, 3-472-1, tissue culture supernatant), rabbit anti-MCAK (1:100, Cytoskeleton, Denver, CO), mAb anti-CENP-F (1:1000, BD Transduction Laboratories), goat anti-CENP-E (1:200, Santa Cruz), mAb anti-CENP-A (1:1000, MBL, Naka-ku Nagoya, Japan) and mAb anti-Cyclin B (1:1000, Upstate) were used and detected by ECL Supersignal (Pierce Biotechnology, Rockford, IL).
Phosphatase assay
HeLa S3 cells were arrested with either 1.6 μg/ml aphidicolin (G1/S-phase) or with 150 ng/ml nocodazole (M-phase) for 14 h. The corresponding cell lysates were either treated with Alkaline Phosphatase (Roche) or left untreated (in the presence of phosphatase inhibitors) for 1 h at 30°C. The phosphatase reaction was stopped by addition of sample buffer followed by boiling. Equal protein amounts were loaded and separated by SDS–PAGE, followed by Western blot analysis.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Figures and Legends
Acknowledgments
We thank A Wehner for technical assistance, K Hofmann for sequence analysis, F Melchior and F Perez for generously providing us with antibodies and all our colleagues in the department for helpful discussions. This study was supported by the Max Planck Society, the ‘Deutsche Forschungsgemeinschaft' (SFB646) and the ‘Fonds der Chemischen Industrie'.
References
- Biggins S, Walczak CE (2003) Captivating capture: how microtubules attach to kinetochores. Curr Biol 13: R449–R460 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001) Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, De LJ, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human ndc80–hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280: 29088–29095 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT (2005) Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 16: 4882–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Wehner A, Nigg EA, Sillje HH (2006) Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell 17: 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG (1998) Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol 143: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E (2002) Four new subunits of the Dam1–Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 21: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M (2004) The RanGAP1-RanBP2 complex is essential for microtubule–kinetochore interactions in vivo. Curr Biol 14: 611–617 [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM (2003) Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J 22: 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lan Z, Wu H, Wu S, Meadows J, Chen J, Zhu V, Dai W (1999) BUBR1 phosphorylation is regulated during mitotic checkpoint activation. Cell Growth Differ 10: 769–775 [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19: 519–539 [DOI] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL (1998) A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De WP, Sorger PK, Harrison SC (2005) The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG (2002) The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol 3: 731–741 [DOI] [PubMed] [Google Scholar]
- Peters JM (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9: 931–943 [DOI] [PubMed] [Google Scholar]
- Rieder CL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84: 145–158 [DOI] [PubMed] [Google Scholar]
- Rieder CL (1982) The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol 79: 1–58 [DOI] [PubMed] [Google Scholar]
- Rieder CL (2005) Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 114: 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G (1994) Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol 127: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB (2005) The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J 24: 2931–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH (2005) Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 4: 35–43 [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA (2006) HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742 [DOI] [PubMed] [Google Scholar]
- Stucke VM, Baumann C, Nigg EA (2004) Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma 113: 1–15 [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 141: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, vila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G (2005) Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Wong J, Fang G (2006) HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol 173: 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Panda D, Landen JW, Wilson L, Joshi HC (2002) Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint. J Biol Chem 277: 17200–17208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Figures and Legends


