Abstract
Proteins synthesized in the cytosol either remain there or are localized to a specific membrane and subsequently translocated to another cellular compartment. These extracytosolic proteins have to cross, or be inserted into, a phospholipid bilayer—a process governed by membrane-bound protein transporters designed to recognize and receive appropriate polypeptides and thread them through the membrane. One such translocation complex, SecY/Sec61, is found in every cell, in either the plasma membrane of bacteria and archaea or the endoplasmic reticulum membrane of eukaryotes. Recent structural findings, combined with previous genetic and biochemical studies, have helped to describe how the passage of proteins through the membrane might occur, but several points of uncertainty remain.
Keywords: protein translocation, channel, structure, conformational changes
Introduction
Membranes act as hydrophobic physical barriers that define the cell and compartmentalize biological reactions. Proteins need to be transported across these barriers to be correctly localized. To facilitate this process, several protein transport machines consisting of specific membrane proteins and soluble partners have evolved.
The transport of polymers, such as proteins, obviously involves complicated machines that can handle large and chemically diverse substrates. Moreover, specific substrate proteins have to be selected from the cytosol and appropriately sent through or into the membrane without significant leakage of small molecules, such as water or ions across the membrane. The elucidation of this remarkable mechanism has been the focus of a generation of researchers.
The secretory pathway
The current level of understanding of the secretory pathway has been attained through the classical application of genetic, biochemical and crystallographic technologies. More than 30 years ago, it was discovered that some proteins are synthesized as larger precursors, which mature after proteolytic modification (Milstein et al, 1972). This precursor was postulated to include a signal sequence to direct the proteins to the secretory pathway (Blobel & Dobberstein, 1975). Later research focused on the identification and characterization of the components of the secretory pathway. A genetic analysis of secretion in Escherichia coli identified the key components involved in protein translocation: SecA, SecY and SecE (Schatz & Beckwith, 1990). Some of these experiments used mutated and non-functional signal sequences to screen for second site suppressors. These genetic interactions were used to identify protein localization (prl) mutants found on genes important for protein translocation (Bedouelle et al, 1980; Derman et al, 1993; Emr et al, 1981; Schatz et al, 1991). The analysis of these mutations has not only helped to identify and characterize components of the translocation apparatus but also, more recently, has been important in the interpretation of the structure of the translocation pore.
A homologue of SecY in E. coli was identified as Sec61α in yeast and mammals (Deshaies & Schekman, 1987; Görlich et al, 1992). The respective subunits were found to form the core of a membrane-bound complex able to support protein translocation. Identification of ion conductance and an analysis of fluorescent probes present on translocating polypeptides showed that the Sec61 complex provides an aqueous environment for polypeptide translocation (Crowley et al, 1993, 1994; Simon & Blobel, 1991; Wirth et al, 2003). Seminal experiments reconstituted the transport of unfolded substrate polypeptide from purified components, from both E. coli and mammals (Arkowitz et al, 1993; Brundage et al, 1990; Görlich & Rapoport, 1993; Matlack et al, 1998). These studies found that the membrane-bound SecY and Sec61 complexes conduct polypeptides through or into the cytosolic or endoplasmic reticulum membrane, respectively, whilst various other factors use energy to push or pull the translocating polypeptide.
Probably all cells translocate proteins co-translationally; the ribosome–nascent chain complex is relayed to the membrane by the signal recognition particle, which guides the ribosome to engage with the translocon (Fig 1A; reviewed in Pool, 2005). Alternative post-translational pathways also exist. One mechanism that has been extensively studied in yeast involves a large assembly—including the Sec61 complex—bound to the endoplasmic reticulum membrane and a luminal heat-shock protein 70 (Hsp70) homologue called BiP, which uses ATP to pull the polypeptide through the membrane in a ratchet-like fashion (Matlack et al, 1999; Misselwitz et al, 1999; Panzner et al, 1995; Rapoport et al, 1999). Bacteria also have a post-translational pathway acting on the SecY complex at the cytosolic membrane. An ATPase—SecA—engages with a substrate polypeptide and the SecY complex to enable protein export, driven by ATP hydrolysis and membrane potential (Fig 1A; Economou & Wickner, 1994; Lill et al, 1989; Schiebel et al, 1991). A cytosolic chaperone—SecB—is also involved in this targeting pathway (Hartl et al, 1990).
Figure 1.
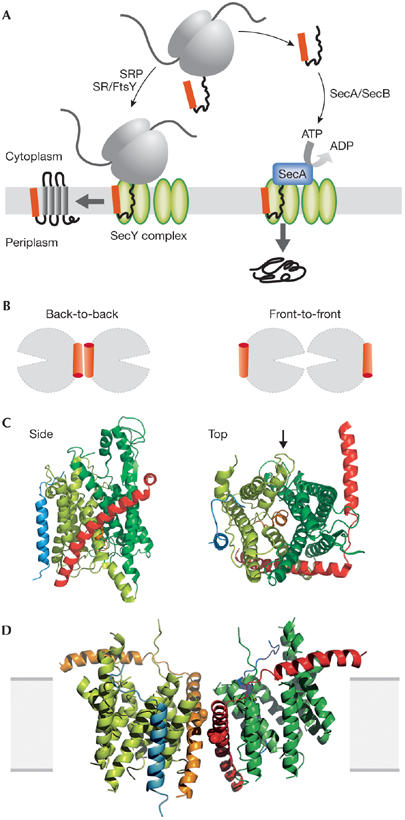
Protein translocation through the SecYEG complex. (A) Schematic overview of protein translocation and membrane protein insertion through the SecY complex in bacteria. Nascent chains having a signal sequence (red) emerge from the ribosomes (grey) and are localized to the SecY complex (green) co-translationally by the signal recognition particle (SRP) and FtsY/SRP receptor (SR; left). Alternatively, they can be presented post-translationally through SecA or SecB (right). (B) Schematic of two possible dimeric conformations of SecYEG, shown looking down onto the membrane. In the back-to-back form, the SecE transmembrane domains (orange cylinders) are juxtaposed and the lateral gates face outwards; the front-to-front configuration places the lateral gates close to one another. (C) Ribbon representation of the Methanococcus jannaschii SecYEβ structure, solved by X-ray crystallography (Protein Data Base file ; van den Berg et al, 2004): the side view is from within the plane of the membrane and the top view is from the cytoplasmic side. SecY is shown in green, with the two halves of the clam shell in light and dark shades. SecE and Secβ (SecG) are shown in red and blue, respectively. The plug helix of SecY is shown in orange and an arrow indicates the lateral gate. (D) Homology model of the dimeric, membrane-bound Escherichia coli SecYEG structure, built using coordinates of the M. jannaschii structure and then docked into the electron microscopy map from the membrane-bound two-dimensional crystals (only the membrane-bound domains were modelled; Bostina et al, 2005). SecY (green), E (red/orange) and G (blue) are shown in different shades for the two monomers. Residue Leu 106 in SecE, at which cysteine crosslinking between monomers can be achieved, is shown as spheres. The membrane is shown in grey.
The structures of the SecY complex (Breyton et al, 2002; Mitra et al, 2005; van den Berg et al, 2004), SecB (Xu et al, 2000), SecA (Hunt et al, 2002) and ribosomes (Ban et al, 2000; Wimberly et al, 2000) have all contributed tremendously to our understanding of these reactions. In the following sections, we focus primarily on the structures of the SecY complex and discuss what they reveal about protein translocation.
The SecY complex
The SecY (or Sec61) complex provides a flexible conduit for proteins to pass across or into the membrane. It is composed of three integral membrane proteins—SecY, E and G—which respectively contain 10, 3 and 2 transmembrane α-helices in E. coli (Douville et al, 1995). In archaea and eukaryotes, the equivalent subunits (Sec61α, γ and β) contain 10, 1 and 1 transmembrane helices (Pohlschroder et al, 1997; van den Berg et al, 2004). The additional bacterial transmembrane domains of the SecE and SecG subunits, which are found only in E. coli and related bacteria, are not obligatory for translocation (Flower et al, 2000; Schatz et al, 1991). Sec61α and γ are reasonably well conserved with SecY and E, indicating that they function in a similar manner.
To understand this process properly we need to know the structure of the protein pore. Several low-resolution structures have been resolved by single-particle electron cryo-microscopy of the channel bound to a ribosome, either in the presence or absence of translocating polypeptide. Pictures from mammalian (Hanein et al, 1996; Menetret et al, 2000, 2005; Morgan et al, 2002), yeast (Beckmann et al, 1997, 2001) and bacterial systems (Meyer et al, 1999) show the channel bound as an oligomer to the ribosome. The most recent of these structures identifies significantly more detail (Mitra et al, 2005). Other structural and functional studies have implicated a dimer of SecYEG as the active species during protein translocation (Bessonneau et al, 2002; Breyton et al, 2002; Duong, 2003; Manting et al, 2000; Mitra et al, 2005; Tam et al, 2005; Tziatzios et al, 2004; Veenendaal et al, 2001).
To gain higher resolution images of the SecY complex, two structures have been resolved by crystallography. One is an 8 Å-resolution structure of the E. coli SecYEG, determined by electron microscopy of two-dimensional crystals of the protein within the membrane (Breyton et al, 2002; Collinson et al, 2001). The protein was visualized as a complex of two SecYEGs related to each other by a twofold symmetry axis close to the third and conserved transmembrane domain of SecE. This orientation has been referred to as a ‘back-to-back' dimeric formation (Fig 1B). The procedure for two-dimensional crystallization is similar to that used to reconstitute active translocation sites (Collinson et al, 2001), suggesting that this arrangement of the monomers is likely to be a form adopted by the native assembly.
A second structure of an archaeal homologue from Methanococcus jannaschii (SecYEβ) has been determined to atomic resolution by X-ray diffraction of three-dimensional crystals grown from detergent-solubilized protein (van den Berg et al, 2004). The structure was found to be monomeric, containing one copy of each of the three subunits. The transmembrane helices of SecY form a helical bundle in which helices 1–5 and 6–10 oppose one another, forming two halves of a ‘clam shell' (Fig 1C). SecE forms a ‘clamp' around SecY, with its highly tilted transmembrane helix connected to an amphipathic helix lying along the membrane interface.
Contrary to previous suggestions that the pore is formed at an oligomeric interface, the translocation channel is found in the centre of a single SecY protomer, between the two halves of the clam shell. The sites of most of the prl mutations can be mapped onto the lining of this putative channel, and residues in this region have been crosslinked to a translocating polypeptide (Cannon et al, 2005; van den Berg et al, 2004). The apparent constriction point in the channel comprises a ring of conserved hydrophobic residues, which might act to seal the pore preventing the leakage of water or ions.
A notable feature revealed by this structure is the reinsertion of a hydrophilic domain—found between transmembrane 1 and 2 of SecY—into the membrane forming an additional short helix. This helix sits in the periplasmic opening of the channel and seems to act as a ‘plug', effectively closing the channel to translocation substrates. During translocation, the plug has been proposed to move out of the channel and bind in a second pocket, close to the carboxyl terminus of SecE (van den Berg et al, 2004). This large movement of the plug might act as a switch, opening the channel as translocation partners and substrate are engaged. An interaction between the plug and SecE was previously suggested by genetic studies showing a synthetic phenotype when prl mutations in the plug region and SecE are combined (Flower et al, 1995). Residues of the plug helix can be crosslinked to a position on SecE, despite them being around 20 Å apart in the X-ray structure (Harris & Silhavy, 1999). This crosslinking is significantly enhanced in Sec complexes that are actively translocating substrate, suggesting that the movement of the plug is concomitant with channel opening (Tam et al, 2005).
During translocation the aperture of the channel must expand through the opening of the clam shell. Molecular probe accessibility and conductance measurements suggest that the pore might be up to 60 Å in diameter (Hamman et al, 1997; Wirth et al, 2003)—far too large to be formed by a single SecY complex. This larger estimate might have arisen as a result of the fluorescent probe ‘snorkelling' through its linker to a more spacious environment outside the membrane (Mitra et al, 2005). In any case, polypeptide substrates are transported in an unfolded state (Arkowitz et al, 1993; Matlack et al, 1998). Although there is no direct evidence with respect to the structure of the translocating chain, it is reasonable to assume that objects no larger than an α-helix need to pass through the central pore. Therefore, there seems to be no logical reason why such a large diameter would be required. A study of the molecular dynamics of the SecY complex using virtual balls to probe the channel suggested that the pore can expand to a diameter of around 16 Å (Tian & Andricioaei, 2006).
Incorporation of membrane proteins through the Sec complex requires that the transmembrane domains of the translocating protein are ejected laterally from the complex into the surrounding lipid, which could occur through the ‘lateral gate' at the opening of the clam shell (Fig 1C). Again, such movements have been observed during molecular dynamics simulations on the channel (Tian & Andricioaei, 2006).
The atomic model of the Sec complex can be overlaid onto the electron density map from the two-dimensional crystals, enabling the helices seen in the electron cryo-microscopy map to be assigned and an atomic model of the membrane-bound form to be built (Fig 1D; Bostina et al, 2005). A closer inspection of the two structures reveals subtle differences between them. In the membrane-bound dimer, the plug helix is displaced by 6 Å towards the periplasmic side of the membrane (Bostina et al, 2005). The dimeric association might prime the channel for translocation by slightly unplugging the pore. This idea has found biochemical support, as cysteine crosslinking between the plug and SecE was less efficient in a mutant that tends to be monomeric (Tam et al, 2005).
Both structures resolved by crystallography presented the Sec complex in its closed state, as no substrate or translocation partners were present. A new structure of the channel in the active conformation has recently been obtained by single-particle cryo-electron microscopy of purified E. coli SecYEG reconsituted with a ribosome-nascent chain complex (Mitra et al, 2005). The structure shows SecY complexes bound to the ribosome at two different sites: one bound to the polypeptide exit channel, as expected, and a second bound artefactually to the 5′ messenger RNA exit site. The complex bound to the polypeptide exit site seems to be active in translocation. At both sites, the amount of electron density observed was consistent with a dimer of SecYEG. The optimal fit of the electron density to the X-ray structure was achieved by using a ‘front-to-front' conformation, in apparent contradiction to the data from two-dimensional crystals (see Fig 1B). Regions of electron density unaccounted for by the fitted SecY complex might be due to the translocating polypeptide, the signal sequence and the SecY plug, which is apparently displaced from the pore in the translocating channel (Mitra & Frank, 2006). This is clearly an important advance necessary for the understanding of this reaction. However, the accuracy and consequences of this fit require further experimental support together with higher resolution such that each transmembrane α-helix can be individually and unambiguously assigned.
The nature of the oligomeric interface has been studied by disulphide crosslinking of the subunits through site-directed incorporation of single cysteine residues. Strong crosslinks have been observed between SecE subunits on adjacent Sec complexes (Kaufmann et al, 1999; Veenendaal et al, 2001). This is consistent with back-to-back dimerization, as implicated by the two-dimensional crystallography data (see Fig 1D). This crosslink reversibly inactivates the complex; however, crosslinking is enhanced in translocation-activated channels, suggesting that this interaction occurs in the active state of the complex (Kaufmann et al, 1999). It should, however, be noted that the crosslinking studies all refer to the post-translational mode of translocation. It cannot yet be formally excluded that the co-translational system studied by Mitra and colleagues (Mitra et al, 2005) might involve a different oligomeric conformation of the channel.
Conclusions
Our current picture of protein translocation and membrane protein integration by the Sec complex has emerged from an impressive array of complementary techniques, drawn from classical biochemistry, genetics and structural biology. This has enabled us to peer into the inner workings of this fascinating molecular machine at an atomic level and begin to piece together its mechanical operation. However, there is still much that we do not understand. On a structural level, it is still not clear how, and indeed why, dimers of the complex are formed. A recent low-resolution picture of the protein in its active translocating state, in the presence of a translocating polypeptide, is promising but raises yet more questions. These will undoubtedly be resolved by obtaining more structural detail and biochemical insight. Other aspects of the molecular mechanism, not covered in this review, still need to be addressed, such as how the translocon correctly identifies transmembrane helices to achieve the correct topology of membrane proteins. The nature and mechanism of the interaction of the Sec complex with its translocation partners, such as the ribosome, BiP and SecA, are other areas of debate. These questions are at the forefront of research by scientists across many disciplines, providing optimism for further advances in this field.

Ian Collinson & Alice Robson
Photograph by V. Gold
Acknowledgments
We thank F. Duong and K. Mitra for stimulating discussions and V. Gold for critical reading of the manuscript. We are also grateful to B. Mohsin and M. Bostina for help in preparation of Fig 1D. I.C. and A.R. are supported by the Biotechnology and Biological Sciences Research Council (grant number BB/C503538/1).
References
- Arkowitz RA, Joly JC, Wickner W (1993) Translocation can drive the unfolding of a preprotein domain. EMBO J 12: 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science 278: 2123–2126 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn C, Eswar N, Helmers J, Penczek P, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Bedouelle H, Bassford P, Fowler A, Zabin I, Beckwith J, Hofnung M (1980) Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature 285: 78–81 [DOI] [PubMed] [Google Scholar]
- Bessonneau P, Besson V, Collinson I, Duong F (2002) The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J 21: 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol 67: 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostina M, Mohsin B, Kühlbrandt W, Collinson I (2005) Atomic model of the E. coli membrane bound protein translocation complex SecYEG. J Mol Biol 352: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I (2002) Three-dimensional structure of the bacterial protein–translocation complex SecYEG. Nature 418: 662–665 [DOI] [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W (1990) The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62: 649–657 [DOI] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM Jr, Shibata Y, Rapoport TA (2005) Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol 169: 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport TA, Kühlbrandt W (2001) Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J 20: 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE (1993) The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell 73: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Crowley KS, Liao SR, Worrell VE, Reinhart GD, Johnson AE (1994) Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78: 461–471 [DOI] [PubMed] [Google Scholar]
- Derman AI, Puziss JW, Bassford PJ, Beckwith J (1993) A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J 12: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R (1987) A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol 105: 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville K, Price A, Eichler J, Economou A, Wickner W (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem 270: 20106–20111 [DOI] [PubMed] [Google Scholar]
- Duong F (2003) Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J 22: 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Wickner W (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78: 835–843 [DOI] [PubMed] [Google Scholar]
- Emr SD, Hanley-Way S, Silhavy T (1981) Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23: 79–88 [DOI] [PubMed] [Google Scholar]
- Flower A, Osborne R, Silhavy T (1995) The allele-specific synthetic lethality of prlA-prlG double mutants predicts interactive domains of SecY and SecE. EMBO J 14: 884–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A, Hines L, Pfennig P (2000) SecG is an auxiliary component of the protein export apparatus of Escherichia coli. Mol Gen Genet 263: 131–136 [DOI] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA (1993) Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75: 615–630 [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies K, Rapoport T (1992) A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71: 489–503 [DOI] [PubMed] [Google Scholar]
- Hamman B, Chen J, Johnson E, Johnson A (1997) The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell 89: 535–544 [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack K, Jungnickel B, Plath K, Kalies K, Miller K, Rapoport T, Akey C (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell 87: 721–732 [DOI] [PubMed] [Google Scholar]
- Harris CR, Silhavy TJ (1999) Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol 181: 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F, Lecker S, Schiebel E, Hendrick J, Wickner W (1990) The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63: 269–279 [DOI] [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J (2002) Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297: 2018–2026 [DOI] [PubMed] [Google Scholar]
- Kaufmann A, Manting EH, Veenendaal AK, Driessen AJ, van der Does C (1999) Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38: 9115–9125 [DOI] [PubMed] [Google Scholar]
- Lill R, Cunningham K, Brundage L, Ito K, Oliver D, Wickner W (1989) SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J 8: 961–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manting EH, van der Does C, Remigy H, Engel A, Driessen AJ (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J 19: 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K, Misselwitz B, Plath K, Rapoport T (1999) BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97: 553–564 [DOI] [PubMed] [Google Scholar]
- Matlack KE, Mothes W, Rapoport TA (1998) Protein translocation: tunnel vision. Cell 92: 381–390 [DOI] [PubMed] [Google Scholar]
- Menetret J, Neuhof A, Morgan D, Plath K, Radermacher M, Rapoport T, Akey C (2000) The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell 6: 1219–1232 [DOI] [PubMed] [Google Scholar]
- Menetret JF, Hegde RS, Heinrich SU, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW (2005) Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol 348: 445–457 [DOI] [PubMed] [Google Scholar]
- Meyer T, Menetret J, Breitling R, Miller K, Akey C, Rapoport T (1999) The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol 285: 1789–1800 [DOI] [PubMed] [Google Scholar]
- Milstein C, Brownlee GG, Harrison TM, Mathews MB (1972) A possible precursor of immunoglobulin light chains. Nat New Biol 239: 117–120 [DOI] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Matlack K, Rapoport T (1999) Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem 274: 20110–20115 [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J (2006) A model for co-translational translocation: ribosome-regulated nascent polypeptide translocation at the protein-conducting channel. FEBS Lett 580: 3353–3360 [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL 3rd, Ban N, Frank J (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DG, Menetret J-F, Neuhof A, Rapoport TA, Akey CW (2002) Structure of the mammalian ribosome-channel complex at 17 Å resolution. J Mol Biol 324: 871–886 [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport T (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81: 561–570 [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Prinz W, Hartmann E, Beckwith J (1997) Protein translocation in the three domains of life: variations on a theme. Cell 91: 563–566 [DOI] [PubMed] [Google Scholar]
- Pool MR (2005) Signal recognition particles in chloroplasts, bacteria, yeast and mammals. Mol Membr Biol 22: 3–15 [DOI] [PubMed] [Google Scholar]
- Rapoport T, Matlack K, Plath K, Misselwitz B, Staeck O (1999) Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol Chem 380: 1143–1150 [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Beckwith J (1990) Genetic analysis of protein export in Escherichia coli. Annu Rev Genet 24: 215–248 [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J (1991) One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J 10: 1749–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E, Driessen A, Hartl F, Wickner W (1991) Δμ H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64: 927–939 [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G (1991) A protein-conducting channel in the endoplasmic reticulum. Cell 65: 371–380 [DOI] [PubMed] [Google Scholar]
- Tam PC, Maillard AP, Chan KK, Duong F (2005) Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24: 3380–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P, Andricioaei I (2006) Size, motion, and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys J 90: 2718–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schägger H, Haase W, Duong F, Collinson I (2004) The bacterial protein-translocation complex: dimeric SecYEG associates with both one or two molecules of SecA. J Mol Biol 340: 513–524 [DOI] [PubMed] [Google Scholar]
- van den Berg L, Clemons WMJ, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- Veenendaal A, van der Does C, Driessen A (2001) Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J Biol Chem 276: 32559–32566 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Wirth A, Jung M, Bies C, Frien M, Tyedmers J, Zimmermann R, Wagner R (2003) The Sec61p complex is a dynamic precursor activated channel. Mol Cell 12: 261–268 [DOI] [PubMed] [Google Scholar]
- Xu Z, Knafels JD, Yoshino K (2000) Crystal structure of the bacterial protein export chaperone secB. Nat Struct Biol 7: 1172–1177 [DOI] [PubMed] [Google Scholar]


